SUMMARY
This study is a part of the surveillance study on childhood diarrhoea in the Andaman and Nicobar Islands; here we report the drug resistance pattern of recent isolates of Shigella spp. (2006–2011) obtained as part of that study and compare it with that of Shigella isolates obtained earlier during 2000–2005. During 2006–2011, stool samples from paediatric diarrhoea patients were collected and processed for isolation and identification of Shigella spp. Susceptibility to 22 antimicrobial drugs was tested and minimum inhibitory concentrations were determined for third-generation cephalosporins, quinolones, amoxicillin-clavulanic acid combinations and gentamicin. A wide spectrum of antibiotic resistance was observed in the Shigella strains obtained during 2006–2011. The proportions of resistant strains showed an increase from 2000–2005 to 2006–2011 in 20/22 antibiotics tested. The number of drug resistance patterns increased from 13 in 2000–2005 to 43 in 2006–2011. Resistance to newer generation fluoroquinolones, third-generation cephalosporins and augmentin, which was not observed during 2000–2005, appeared during 2006–2011. The frequency of resistance in Shigella isolates has increased substantially between 2000–2006 and 2006–2011, with a wide spectrum of resistance. At present, the option for antimicrobial therapy in shigellosis in Andaman is limited to a small number of drugs.
Key words: Antibiotic resistance, diarrhoea, paediatrics, resistance to drugs
INTRODUCTION
Shigella is a major cause of dysentery throughout the world and is responsible for 5–10% of diarrhoeal illness in many areas [1]. It has been estimated that 91 million individuals worldwide contract shigellosis each year of which 1·1 million die [2]. About, 410 000 (40%) of these deaths occur in Asian children [3]. Antibiotic therapy reduces the duration and severity of shigellosis and is therefore recommended for the treatment of moderate to severe dysentery [4]. Appropriate antibiotic treatment of shigellosis depends on identifying resistance patterns [5]. Rapid emergence of resistance warrants the need for continuous monitoring of sensitivity patterns [6, 7] and the choice of antibiotic should be governed by periodically updated local antibiotic sensitivity patterns of Shigella isolates [4].
Hospital-based bacteriological surveillance has identified shigellosis as endemic and a major cause of acute childhood diarrhoea in the Andaman and Nicobar Islands [8, 9], a centrally administered territory of India with a population of about 380 000. The distribution of Shigella spp. and serotypes has changed over time [8, 10] as have their drug resistance patterns. The major objective of this study was to analyse the trends in antimicrobial resistance in Shigella isolated from these remote islands of India over a period of 12 years (2000–2011), and to describe the distribution of Shigella serotypes and their resistant phenotypes.
PATIENTS AND SAMPLES
A total of 162 Shigella isolates obtained from 1886 paediatric diarrhoea cases (0–14 years) during 2000–2011 were included in the study. The study was approved by the Regional Medical Research Centre Ethics Clearance Committee. All the Shigella strains were identified, confirmed following standard techniques and serotyped using commercially available antisera (Denka Seiken Co. Ltd, Japan) [11].
Antimicrobial susceptibility testing
Antibiotic susceptibility testing was performed following the disk diffusion method [12]; 22 commercially available disks (HiMedia Laboratories, India) of ampicillin (AMP), amoxicillin-clavulanic acid/augmentin (AMC), azithromycin (AZM), carbenicillin (CAR), imipenem (IMP), amikacin (AMK), chloramphenicol (CHL), co-trimoxazole (CoT), nalidixic acid (NAL), norfloxacin (NOR), ciprofloxacin (CIP), ofloxacin (OFX), gatifloxacin (GAT), gentamicin (GEN), nitrofurantoin (NIT), tetracycline (TET), cephalothin (CEF), cefuroxime (CXM), cefixime (CFM), ceftriaxone (CRO), cefotaxime (CTX) and ceftazidime (CAZ). Escherichia coli ATCC 25 922 and Staphylococcus aureus ATCC 25 923 were included in each test as control strains. Extended spectrum β-lactamase (ESBL) production was detected using the double-disk synergy method with ceftazidime-clavulanic acid (CAC, 30/10 μg) and ceftriaxone-clavulanic acid (CAC, 30/10 μg) [13]. The minimum inhibitory concentration (MIC) for cephalosporins (ceftriaxone, ceftazidime, cefotaxime), quinolones (nalidixic acid, ciprofloxacin, norfloxacin), augmentin and gentamicin were determined by E-test (AB Biodisk, Sweden).
Statistical analysis
The study period was divided into two equal periods of 6 years each, 2000–2005 and 2006–2011. The antibiotic sensitivity (ABST) pattern of these 74 Shigella strains obtained during 2000–2005 were compared and statistically analysed with the ABST patterns of the 88 Shigella strains obtained during 2006–2011. The proportions of isolated Shigella strains resistant to each of the antibacterial drugs were calculated. Proportions of all Shigella strains resistant to each drug in 2000–2005 and in 2006–2011 were compared and statistical significance was tested by χ2 test using Epi Info v. 7 software (www.cdc.gov/epiinfo/). A P value of <0·05 was considered statistically significant.
RESULTS
Serotype distribution
The age distribution of 1886 diarrhoea patients who were positive for Shigella ranged from 6 months to 12 years (mean age 3·5 years) with 66% being male. Of the 162 Shigella isolates, 88 (54·3%) were S. flexneri, 55 (33·9%) S. sonnei, 11 (9%) S. dysenteriae, and eight (2%) S. boydii (Table 1). Of S. flexneri isolated, subserotype 2a (35·2%) accounted for the majority of cases followed by 4a (3·7%), 3a (3·1%), 1a (2·5%) and X variant (2·5%) (Table 1). In S. dysenteriae, subserotype 2 (3·7%) was the prevalent subserotype followed by subserotype 1 (1·8%). S. sonnei was observed to be competing with the S. flexneri counterpart as the predominant serotype isolated in the years 2003, 2005 and 2009.
Table 1.
The serotype distribution of Shigella isolates in the Andaman Islands during 2000–2011
| Species | 2000 (N = 12) | 2001 (N = 7) | 2002 (N = 14) | 2003 (N = 13) | 2004 (N = 18) | 2005 (N = 10) | 2006 (N = 3) | 2007 (N = 3) | 2008 (N = 27) | 2009 (N = 17) | 2010 (N = 32) | 2011 (N = 6) | Overall (N = 162) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. flexneri | |||||||||||||
| 2a | 4 (33·3) | — | 1 (7·1) | 1 (7·7) | 6 (33·3) | 1 (10·0) | 2 (66·7) | 3 (100·0) | 17 (62·9) | 6 (35·3) | 13 (41·0) | 3 (50·0) | 57 (35·2) |
| 2b | — | — | 2 (14·3) | — | 1 (5·5) | — | — | — | — | — | — | — | 3 (1·8) |
| 4a | — | 1 (14·3) | — | — | 1 (5·5) | — | — | — | 3 (11·1) | — | 1 (3·1) | — | 6 (3·7) |
| 3b | — | — | — | — | — | — | — | — | 2 (7·4) | — | — | — | 2 (1·2) |
| 3a | — | 1 (14·3) | 1 (7·1) | — | 1 (5·5) | 1 (10·0) | — | — | — | — | 1 (3·1) | — | 5 (3·1) |
| 1a | — | — | 3 (21·4) | — | — | — | — | — | — | — | 1 (3·1) | — | 4 (2·5) |
| 1b | — | — | 3 (21·4) | — | — | — | — | — | — | — | — | — | 3 (1·8) |
| 6 | — | — | — | — | 1 (5·5) | — | — | — | — | 1 (5·9) | — | — | 2 (1·2) |
| X | — | — | 1 (7·1) | 1 (7·7) | — | — | — | — | — | — | 2 (6·3) | — | 4 (2·5) |
| Y | — | — | — | — | 2 (11·1) | — | — | — | — | — | — | — | 2 (1·2) |
| S. sonnei | 1 (8·3) | 5 (7·1) | 2 (14·3) | 10 (76·9) | 6 (33·3) | 8 (80·0) | — | — | 2 (7·4) | 8 (47·1) | 11 (34·4) | 2 (33·3) | 55 (33·9) |
| S. dysenteriae | — | ||||||||||||
| 1 | — | — | — | 1 (7·7) | — | — | 1 (33·3) | — | — | 1 (5·9) | — | — | 3 (1·8) |
| 2 | 1 (8·3) | — | — | — | — | — | — | — | 3 (11·1) | — | 2 (6·3) | — | 6 (3·7) |
| 3 | — | — | — | — | — | — | — | — | — | 1 (5·9) | — | — | 1 (0·6) |
| 5 | — | — | 1 (7·1) | — | — | — | — | — | — | — | — | — | 1 (0·6) |
| S. boydii 1 | 6 (50·0) | — | — | — | — | — | — | — | — | — | 1 (3·1) | 1 (27·7) | 8 (4·9) |
Values given are n (%).
Resistant trends
A wide spectrum of antibiotic resistance was observed in the Shigella strains isolated during 2006–2011 (Table 2). All the Shigella strains isolated during this period were resistant to ampicillin. Resistance to tetracycline and co-trimoxazole was observed in 81 (92%) and 70 (79%) isolates, respectively. During the study period, out of 88 Shigella strains, 85 (97%) were nalidixic acid resistant, 11 of which were sensitive to the fluoroquinolones. Norfloxacin resistance was observed in 72 (82%) Shigella strains, 67 (76%) were ciprofloxacin resistant and 63 (72%) were ofloxacin resistant, respectively (Table 2). The MICs of the isolates ranged from 64 to >256 μg/ml for nalidixic acid, 4 to >256 μg/ml for ciprofloxacin and 16 to >256 μg/ml for norfloxacin. Fifteen (17%) stains of Shigella spp. were resistant to all four third-generation cephalosporins (cefixime, ceftriaxone, cefotaxime, ceftazidime) tested. The MICs of the resistant isolates ranged from 32 to >256 μg/ml for ceftriaxone, 4 to >256 μg/ml for cefotaxime, 4 to >256 μg/ml for ceftazidime and 4 to >256 μg/ml for amoxicillin-clavulanic acid, which are all above the breakpoint for reduced susceptibility according to Clinical and Laboratory Standards Institute (CLSI) guidelines [11]. All of the cephalosporin-resistant Shigella strains were confirmed as producing ESBLs by combination disk test. Of aminoglycosides, gentamicin resistance was observed in 19 (22%) (MIC 0·5 to >256 μg/ml) and amikacin resistance in 34 (39%) strains (Table 2). Azithromycin resistance was observed in 43 (49%) Shigella strains isolated during 2006–2011.
Table 2.
Proportions of Shigella isolates from the Andaman Islands resistant to various antibacterial drugs during 2000–2005 and 2006–2011 and statistical significance of the difference (χ2 test)
| Antimicrobial agents | Resistance | P value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 2006–2011 | 2000–2005 | ||||||||||
| S. flexneri (N = 55) | S. sonnei (N = 23) | S. dysenteriae (N = 8) | S. boydii (N = 2) | All Shigella isolates (N = 88) | S. flexneri (N = 33) | S. sonnei (N = 32) | S. dysenteriae (N = 3) | S. boydii (N = 6) | All Shigella isolates (N = 74) | ||
| Ampicillin | 100·0 (55) | 100·0 (23) | 100·0 (8) | 100·0 (2) | 100·0 (88) | 51·5 (17) | 37·5 (12) | 66·7 (2) | 50·0 (3) | 45·9 (34) | <0·05 |
| Nalidixic acid | 96·4 (53) | 100·0 (23) | 87·5 (7) | 100·0 (2) | 96·6 (85) | 45·5 (15) | 100·0 (32) | 33·3 (1) | 0 | 64·9 (48) | 0.000002 |
| Tetracycline | 94·5 (52) | 82·6 (19) | 100·0 (8) | 100·0 (2) | 92·0 (81) | 51·5 (17) | 68·8 (22) | 66·7 (2) | 0 | 55·4 (41) | 0.000001 |
| Norfloxacin | 87·3 (48) | 73·9 (17) | 75·0 (6) | 50·0 (1) | 81·8 (72) | 0 | 6·3 (2) | 33·3 (1) | 0 | 4·1 (3) | <0·05 |
| Co-trimoxazole | 74·5 (41) | 91·3 (21) | 75·0 (6) | 100·0 (2) | 79·5 (70) | 81·8 (27) | 100·0 (32) | 66·7 (2) | 16·7 (1) | 83·7 (62) | 0·625018 |
| Ciprofloxacin | 76·4 (42) | 73·9 (17) | 87·5 (7) | 50·0 (1) | 76·1 (67) | 0 | 6·3 (2) | 33·3 (1) | 0 | 4·1 (3) | <0·05 |
| Ofloxacin | 74·5 (41) | 65·2 (15) | 75·0 (6) | 50·0 (1) | 71·6 (63) | 0 | 0 | 0 | 0 | 0·0 (0) | <0·05 |
| Carbenicillin | 74·5 (41) | 21·7 (5) | 75·0 (6) | 50·0 (1) | 60·2 (53) | 0 | 0 | 0 | 0 | 0·0 (0) | <0·05 |
| Azithromycin | 29·1 (16) | 78·3 (18) | 87·5 (7) | 100·0 (2) | 48·8 (43) | 0 | 0 | 0 | 0 | 0·0 (0) | <0·05 |
| Amikacin | 29·1 (16) | 52·2 (12) | 62·5 (5) | 50·0 (1) | 38·6 (34) | 0 | 0 | 0 | 0 | 0·0 (0) | <0·05 |
| Gatifloxacin | 29·1 (16) | 47·8 (11) | 75·0 (6) | 50·0 (1) | 38·6 (34) | 0 | 0 | 0 | 0 | 0·0 (0) | <0·05 |
| Cephalothin (cephalexin) | 36·4 (20) | 34·8 (8) | 62·5 (5) | 50·0 (1) | 38·6 (34) | 0 | 0 | 0 | 0 | 0·0 (0) | <0·05 |
| Chloramphenicol | 41·8 (23) | 4·3 (1) | 37·5 (3) | 00·0 (0) | 30·6 (27) | 36·4 (12) | 03·1 (1) | 33·3 (1) | 0 | 18·9 (14) | 0·125047 |
| Cefuroxime | 25·5 (14) | 8·7 (2) | 50·0 (4) | 50·0 (1) | 23·8 (21) | 0 | 0 | 0 | 0 | 0·0 (0) | 0·000002 |
| Gentamicin | 21·8 (12) | 13·0 (3) | 25·0 (2) | 100·0 (2) | 21·5 (19) | 0 | 0 | 0 | 0 | 0·0 (0) | 0·000062 |
| Cefixime | 16·4 (9) | 8·7 (2) | 37·5 (3) | 50·0 (1) | 17·0 (15) | 0 | 0 | 0 | 0 | 0·0 (0) | 0·000548 |
| Ceftriaxone | 16·4 (9) | 8·7 (2) | 37·5 (3) | 50·0 (1) | 17·0 (15) | 0 | 0 | 0 | 0 | 0·0 (0) | 0·000548 |
| Cefotaxime | 16·4 (9) | 8·7 (2) | 37·5 (3) | 50·0 (1) | 17·0 (15) | 0 | 0 | 0 | 0 | 0·0 (0) | 0·000548 |
| Ceftazidime | 16·4 (9) | 8·7 (2) | 37·5 (3) | 50·0 (1) | 17·0 (15) | 0 | 0 | 0 | 0 | 0·0 (0) | 0·000548 |
| Amoxicillin-clavulanic acid (augmentin) | 16·4 (9) | 8·7 (2) | 37·5 (3) | 50·0 (1) | 17·0 (15) | 0 | 0 | 0 | 0 | 0·0 (0) | 0·000548 |
| Nitrofurantoin | 7·3 (4) | 4·3 (1) | 37·5 (3) | 50·0 (1) | 10·2 (9) | 0 | 6·3 (2) | 0 | 0 | 2·7 (2) | 0·113457 |
| Imipenem | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 (0·0) | |
Values given are % (n).
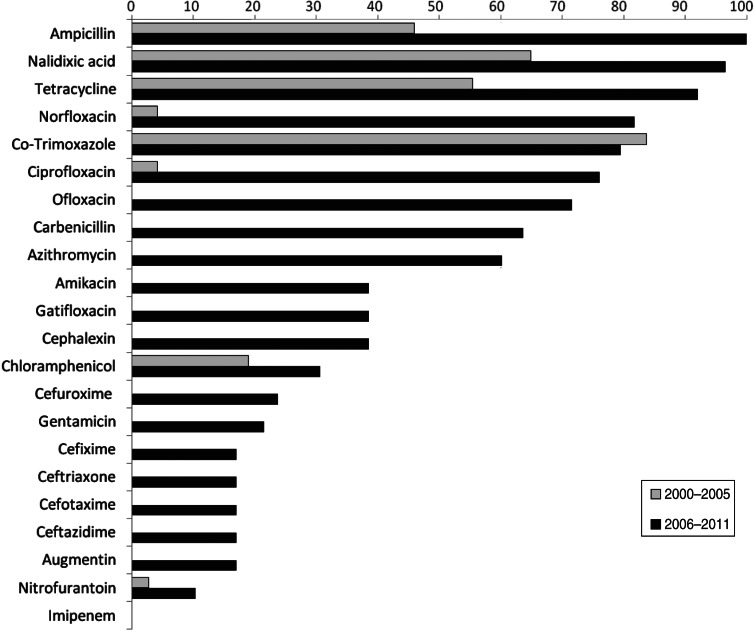
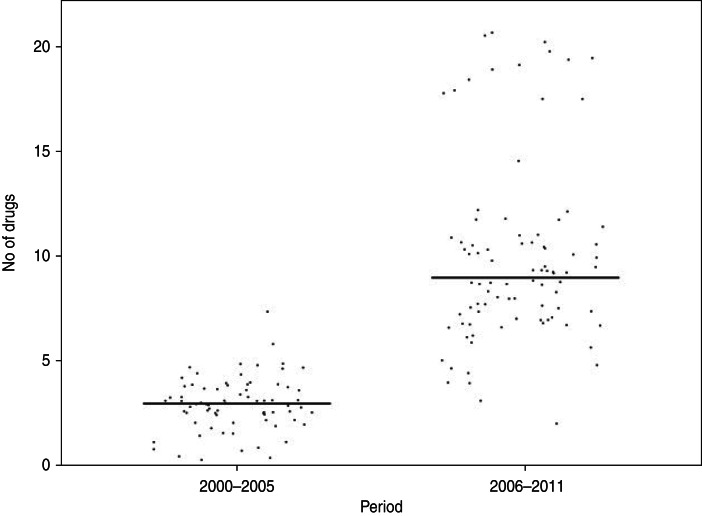
Of the Shigella isolates obtained during 2000–2005, nalidixic acid, tetracycline and co-trimoxazole were the only three drugs against which more than 50% of the strains showed resistance during 2006–2011 there were eight drugs, including ampicillin, norfloxacin, ciprofloxacin, ofloxacin, and carbenicillin, to which more than 50% of the strains were resistant (Table 2). The proportions of resistant strains showed an increase from 2000–2005 to 2006–2011 in 20/22 antibiotics tested, the only exception being co-trimoxazole and imipenem (Table 2, Fig. 1). All the strains were susceptible to imipenem. A decrease (83·7% to 79·5%) in resistance to co-trimoxazole was observed (Fig. 1). The increase in the proportions of resistant strains between 2000–2005 and 2006–2011 was statistically significant at the 5% level in 18/21 drugs against which resistance was observed (Table 2). The most marked increases in the prevalence of resistance were against nalidixic acid (64·9–96·6%), ampicillin (45·9–100%), norfloxacin (4·1–81·8%), ciprofloxacin (4·1–76·1%), tetracycline (55·4–92%), ofloxacin (0–71·6%) and carbenicillin (0–60·2%) (Table 2). In 2000–2005, the number of drug resistance patterns observed was only 13 whereas during 2006–2011 a total of 43 patterns were observed, indicating a widening of the spectrum of drug resistance during the intervening period. In 2000–2005, only 14 (19%) of the isolated strains were resistant to more than three drugs and none was resistant to more than five drugs whereas during 2006–2011, 80/88 isolated strains (91%) were resistant to more than five drugs. The median number of drugs shigellae were resistant to was three in 2000–2005 and 10 during 2006–2011 (Fig. 2). Two different clusters of drug resistance were observed in the Shigella strains of 2006–2011. In addition to the major cluster of 10, another cluster was also observed (n = 20).
Fig. 1.
Prevalence of antibiotic resistance in Shigella strains from the Andaman Islands obtained during 2000–2005 and 2006–2011.
Fig. 2.
Distribution of Shigella strains isolated from the Andaman Islands in 2000–2005 and 2006–2011 by the number of antimicrobials the strain is resistant to.
DISCUSSION
In the present study, S. flexneri in general and 2a serotype in particular were observed to be the most frequent and predominant serotype isolated in the islands, with S. sonnei occasionally competing. S. sonnei and S. flexneri have been observed to be the predominant Shigella spp. in developed and developing countries, respectively [14, 15]. However, several reports of the predominance of S. sonnei have been reported from less developed and developing countries [15–19].
Changing patterns of antimicrobial susceptibilities and acquisition of resistance by enteric pathogens to increasing number of antibacterial drugs is becoming a serious problem, particularly in developing countries where shigellosis is a common occurrence [13, 20].
The present study demonstrates the increasing prevalence and spectrum of antimicrobial resistance of Shigella isolates during the last decade in these islands.
Resistance to ampicillin used to treat shigellosis in young children, was observed in the 1990s in the islands [8, 9]. This has been increasing over the years and at present all the isolates are resistant to this drug. Prevalence of resistance to tetracycline showed a tenfold increase from 9% to 94% during this decade. Resistance to nalidixic acid was first reported in these islands in the late 1990s [21] and by 2006–2011 almost all the isolated strains had become resistant to it. Subsequently other quinolones such as norfloxacin, ciprofloxacin and ofloxacin became the primary choice for antibacterial therapy; however, at present more than 80% of the strains are resistant to these. Although fluoroquinolones are recommended as the drugs of choice for shigellosis by the World Health Organization [22], emergence of fluoroquinolone resistance in Shigella spp. has now been documented in many countries [23–25]. The present study shows that Shigella strains in the islands are rapidly acquiring resistance to alternate drugs such as third-generation cephalosporins which are being used commonly at present. Some of the drugs such as nitrofurantoin and aminoglycosides were included although these are not recommended for treatment of shigellosis, resistance to these could be used as a phenotypic characteristic to study the evolution of the pathogen over a period of time.
Widespread selective pressure and efficient dissemination channels for multidrug-resistant organisms are major factors that might have contributed to the rapid emergence and spread of resistant organisms [26]. The genetic transfer of drug resistance genes may not be of immediate concern for the treating shigellosis patients, but will pose a potential problem in the future.
Antibiotic use is a main driver of selection pressure that contributes to resistance. Most antibiotics are used unnecessarily, in commercially driven agriculture, and by physicians uncertain of a diagnosis or treating largely self-limiting bacterial or viral infections. Multiple genetic factors (plasmid and chromosomal-borne resistant determinants and efflux pump mediated) together with the increasing and indiscriminate use of antibiotics created the opportunity for the emergence of highly resistant clinical isolates associated with multidrug resistance [27].
The study confirmed that older antimicrobials, such as ampicillin, co-trimoxazole, tetracycline and nalidixic acid, no longer have a place in the treatment of shigellosis. The emergence of resistance to several new drugs such as flouroquinolones, third-generation cephalosoprins, and macrolides in shigellae is a cause of great concern not only at the local level but at regional and national levels also. Spread of antibiotic resistance has been recognized by the WHO as an extremely serious problem as it greatly complicates the treatment of infectious diseases. Therefore there is an urgent need to fight the spread of antibiotic resistance. The need for new antibiotics to address the emerging resistance microorganisms cannot be overstated. To complement the strategy of introducing new drugs onto the market, concerted action must be taken now to conserve existing drugs. Strategies to realign economic incentives, fill information gaps, and minimize interruptions in the supply chain will require no less innovation [28]. Proof that antibiotic prescribing contributes to resistance in the individual patient and not just at the societal level is the kind of evidence that needs to be communicated to clinicians [29, 30]. In the present scenario of the rapid emergence of wide spectrum of drug resistance, developing a vaccine against the most prevalent strain may be a suitable alternative. Therefore, any shift in the trends of seroprevalence and antimicrobial resistance patterns of shigellae strains needs to be monitored very carefully in any geographical region.
CONCLUSION
The wide distribution of multidrug-resistant Shigella isolates and the emergence of ESBL-producing and fluoroquinolone-resistant isolates clearly emphasizes the need for strict control of antimicrobial prescriptions, improved sanitation and vaccine development, rather than new antibiotics, as a long-term solution to the problem that disproportionately affects developing countries. A network of laboratories for real-time monitoring of antibiotic resistance in enteric pathogens and timely dissemination of such information to clinicians for modification of treatment strategy is a pressing need. For far too long antimicrobial resistance has been an unrecognized and neglected problem. The time to act is now.
ACKNOWLEDGEMENTS
The authors are grateful to Professor S. C. Sehgal and Dr P. Vijayachari, former and present Directors of RMRC (ICMR), Port Blair, respectively, and Dr Asit Ranjan Ghosh for initiation and execution of diarrhoeal disease surveillance in the islands. The authors are also grateful to the Indian Council of Medical Research for providing a financial grant for the study (Project No. Tribal-26/2006/ECD-II).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Ahmed AM, et al. Genetic characterization of multidrug resistance in Shigella sp. from Japan. Journal of Medical Microbiology 2006; 55: 1685–1691. [DOI] [PubMed] [Google Scholar]
- 2.Kotloff KL, et al. Global burden of Shigella infections: implications for vaccine development and implementation of control strategies. Bulletin of the World Health Organization 1999; 77: 651–666. [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Shigellosis: disease burden, epidemiology and case management. Weekly Epidemiological Record 2005; 80: 94–99. [PubMed] [Google Scholar]
- 4.Christopher PR, et al. Antibiotic therapy for Shigella dysentery. Cochrane Database of Systematic Reviews 2010. Issue 1, Art. No.: CD006784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashkenazi S, et al. Growing antimicrobial resistance of Shigella isolates. Journal of Antimicrobial Chemotherapy 2003; 51: 427–429. [DOI] [PubMed] [Google Scholar]
- 6.Bennish ML, Salam MA. Rethinking options for the treatment of shigellosis. Journal of Antimicrobial Chemotherapy 1992; 30: 243–247. [DOI] [PubMed] [Google Scholar]
- 7.Zafar A, et al. Frequency of isolation of various subtypes and antimicrobial resistance of Shigella from urban slums of Karachi, Pakistan. International Journal of Infectious Disease 2009; 13: 668–672. [DOI] [PubMed] [Google Scholar]
- 8.Ghosh AR, Sehgal SC. Existing status of shigellosis in Andaman & Nicobar Islands. Indian Journal of Medical Research 1996; 103: 134–137. [PubMed] [Google Scholar]
- 9.Ghosh AR, Sehgal SC. Shigella infections among children in Andaman – an archipelago of tropical islands in Bay of Bengal. Epidemiology and Infection 1998; 121: 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roy S, et al. Distribution of Shigella enterotoxin genes and secreted autotransporter toxin gene among diverse species and serotypes of Shigella isolated from Andaman Islands, India. Tropical Medicine and International Health 2006; 11: 1694–1698. [DOI] [PubMed] [Google Scholar]
- 11.World Health Organization. Manual for Laboratory Investigation of Acute Enteric Infections, 1987; CDD/83.3. Geneva, Switzerland: World Health Organization. [Google Scholar]
- 12.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; Twenty Second Informational Supplement, M100–S22, 2012; Wayne, PA, USA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 13.Bhattacharya D, et al. Rapid emergence of third generation cephalosporin resistance in Shigella sp. isolated in Andaman and Nicobar Islands, India. Microbial Drug Resistance 2011; 17: 329–32. [DOI] [PubMed] [Google Scholar]
- 14.Greenhill C. Infectious disease: Genome sequencing reveals how Shigella sonnei spread around the world. Nature Reviews Gastroenterology and Hepatology 2012; 9: 487. [DOI] [PubMed] [Google Scholar]
- 15.Dutta S, et al. Shifting serotypes, plasmid profile analysis and antimicrobial resistance pattern of Shigella strains isolated from Kolkata, India during 1995–2000. Epidemiology and Infection 2002; 129: 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Khan AI, et al. Shigella serotypes among hospitalized patients in urban Bangladesh and their antimicrobial resistance. Epidemiology and Infection 2004; 132: 773–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vasilev V, et al. Variability of Shigella flexneri serotypes in Israel during a period of two years: 2000 and 2001. Epidemiology and Infection 2004; 132: 51–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Farshad S, et al. Characterization of Shigella strains in Iran by plasmid profile analysis and PCR amplification of ipa genes. Journal of Clinical Microbiology 2006; 44: 2879–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Von Seidlein L, et al. A multicentre study of Shigella diarrhoea in six Asian countries: disease burden, clinical manifestations and microbiology. PLoS Medicine 2006; 3: e353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Niyogi SK, et al. Changing patterns of serotypes and antimicrobial susceptibilities of Shigella species isolated from children in Calcutta, India. Japanese Journal of Infectious Disease 2001; 54: 121–122. [PubMed] [Google Scholar]
- 21.Ghosh AR, et al. Emergence of nalidixic acid-resistant Shigella sonnei in acute-diarrhoea patients on Andaman & Nicobar Islands, India. Antimicrobial Agents and Chemotherapy 2003; 47: 1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization. Guidelines for the control of shigellosis, including epidemics due to Shigella dysenteriae type 1. Geneva: WHO, 2005. (http://whqlibdoc.who.int/publications/2005/9241592330.pdf). [Google Scholar]
- 23.Naheed A, et al. Fluoroquinolone resistant Shigella dysenteriae type 1 in northeastern Bangladesh. Lancet Infectious Disease 2004; 4: 607–608. [DOI] [PubMed] [Google Scholar]
- 24.Hirose K, et al. Antimicrobial susceptibility of Shigella sonnei isolates in Japan and molecular analysis of S. sonnei isolates with reduced susceptibility to fluoroquinolones. Antimicrobial Agents and Chemotherapy 2005; 49: 1203–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy S, et al. Emergence of fluoroquinolone resistance in Shigella isolated from Andaman & Nicobar Islands, India. Indian Journal of Medical Research 2010; 131: 720–722. [PubMed] [Google Scholar]
- 26.Okeke IN, et al. Antimicrobial resistance in developing countries. Part I: recent trends and current status. Lancet Infectious Disease 2005; 5: 481–493. [DOI] [PubMed] [Google Scholar]
- 27.Bhattacharya D, et al. Shigellosis in Bay of Bengal Islands, India: clinical and seasonal patterns, surveillance of antibiotic susceptibility patterns, and molecular characterization of multidrug-resistant Shigella strains isolated during a 6-year period from 2006 to 2011. European Journal of Clinical Microbiology and Infectious Diseases 2013; 33: 157–170. [DOI] [PubMed] [Google Scholar]
- 28.Yadav P. Countering drug resistance in the developing world: an 11 assessment across the value chain and recommendations for policy interventions. Working Paper 183. Center for Global Development, 2009. (www.cgdev.org/content/publications/detail/1422842). [Google Scholar]
- 29.Costelloe C, et al. Effect of antibiotic 12 prescribing in primary care on antimicrobial resistance in individual patients: systematic review and meta-analysis. British Medical Journal 2010; 340: 2096. [DOI] [PubMed] [Google Scholar]
- 30.So AD, et al. Tackling antibiotic resistance: concerted action is needed to provide new technologies and conserve existing drugs. British Medical Journal 2010; 340: c2071.20483948 [Google Scholar]