SUMMARY
This descriptive longitudinal study was conducted to investigate the faecal shedding of Shiga toxin-producing E. coli (STEC) in finishing swine and to characterize the swine STEC isolates that were recovered. Three cohorts of finishing swine (n = 50/cohort; total 150 pigs) were included in the longitudinal study. Individual faecal samples were collected every 2 weeks (8 collections/pig) from the beginning (pig age 10 weeks) to the end (pig age 24 weeks) of the finishing period. STEC isolates were recovered in at least one sample from 65·3% (98/150) of the pigs, and the frequency distribution of first-time STEC detection during the finishing period resembled a point-source outbreak curve. Nineteen O:H serotypes were identified among the STEC isolates. Most STEC isolates (n = 148) belonged to serotype O59:H21 and carried the stx2e gene. One O49:H21 STEC isolate carried the stx2e and eae genes. High prevalence rates of STEC during the finishing period were observed, and STEC isolates in various non-O157 serogroups were recovered. These data enhance understanding of swine STEC epidemiology, and future research is needed to confirm whether or not swine STEC are of public health concern.
Key words: Epidemiology, longitudinal, serotype, Shiga-like toxin-producing E. coli, swine
INTRODUCTION
Shiga toxin-producing Escherichia coli (STEC) represent a subset of E. coli that produce a cytotoxin known as Shiga toxin (Stx) encoded by the stx gene [1]. Stx has two types (Stx1, Stx2), and each Stx type has multiple variants (Stx1c, Stx1d, Stx2c, Stx2d, Stx2e, Stx2f, Stx2 g) [2]. STEC infection is associated with outbreaks and sporadic cases of diarrhoea and severe clinical diseases in humans, including haemorrhagic colitis (HC) and the potentially fatal haemolytic uraemic syndrome (HUS) [2]. Therefore, STEC are a critical public health concern, and they cause more than 170 000 cases of human illness yearly in the USA [3]. The infections are often acquired by consuming contaminated food (meat, dairy products, produce, and other foods) and water [2]. While STEC serotype O157:H7 is viewed as the serotype associated with most outbreaks and severe diseases, non-O157 STEC-associated outbreaks have been increasingly documented [4]. Notably, more than 50% of human STEC infections have been attributed to non-O157 STEC [3], but our current knowledge of non-O157 STEC is very limited.
Although cattle are considered the primary STEC reservoirs for human infections, food products from other animal species, including pork products, have been implicated as vehicles in STEC transmission [5–11]. A recent STEC outbreak was associated with consuming cuts of pork from a whole roasted pig [11]. Even though investigators were unable to trace back the original sources of STEC in these pork products, there were possible sources of contamination, including via cross-contamination from foodstuffs from other animal species or other ingredients, during swine processing, or the organisms were from swine [8–11]. Further research is warranted to elucidate whether swine are a source of STEC contamination.
Unlike cattle, swine may have clinical disease associated with STEC infection [12]. Oedema disease, often occurring in post-weaning or young finishing-age pigs, is caused by STEC strains carrying the stx2e gene [12]. Cross-sectional epidemiological studies have been performed to estimate the prevalence of STEC in clinically healthy swine in multiple regions of the world. STEC prevalence in these studies ranged from 0% [13] to 68·3% [14]. In the USA, earlier studies focused on the detection of STEC O157:H7, and the prevalence estimates were low, ranging from 0% [15] to 1·9% [16]. In one more recent survey, STEC isolates were recovered from 28·5% of the faecal samples collected, and all of the isolates belonged to non-O157 serotypes [17]. The role that swine play in the epidemiology of STEC, specifically non-O157 STEC, needs further investigation.
Previous studies clearly indicate that pigs can shed STEC, particularly non-O157 STEC serotypes. Yet, these studies are limited as they employed cross-sectional study designs relying on point estimates of STEC prevalence, and furthermore, only recently have efforts been made to identify and characterize non-O157 serotypes. Little is known about faecal shedding of STEC in clinically healthy swine over time. This longitudinal study was conducted to fill this gap by investigating prevalence of STEC in swine over the finishing period, and characterizing the recovered STEC isolates.
METHODS
Study design
This longitudinal study was conducted on two finishing sites (sites A and B) within one all-in, all-out multi-site production system in the Midwestern USA. This means that the pigs originated from the same sow herd, but were reared separately from birth to marketing. These two sites were selected based on convenience and the producers' willingness to participate in the study. Faecal samples were collected from three cohorts of finishing pigs (one cohort on site A and two cohorts on site B) from age 10 weeks until age 24 weeks (approximately market age). For cohort 1, the farm visits began in May 2011 and ended in August 2011. For cohort 2, the farm visits began in July 2011 and ended in October 2011. For cohort 3, the farm visits began in November 2011 and ended in February 2012.
Site A was a wean-to-finish facility, in which weaned pigs (pig aged 3–4 weeks) were placed into the barns and raised to market age. Site B was a finishing facility, in which pigs, after being housed in a nursery facility (from age 3–10 weeks), were moved into the barns and raised until market age. After a batch of pigs of the same age was placed into the barn, no new pigs were introduced into the barn. These two sites had similar building designs. Each site had two separate buildings, and two separate barns were within each building. Thus, there were four barns on each site. Each barn had 12 pens and was capable of housing a total of 1000 pigs (total site inventory of 4000 pigs). In each barn, there were eight ‘large’ and four ‘small’ pens. About 100–125 pigs were placed in each of eight large pens, and 50 pigs were housed in two of the four small pens. The remaining small pens were used for housing sick pigs or pigs deemed to be at high risk for disease. Pen dividers allowed pig-to-pig contact between pens.
Sample collection
A total of 150 individually identified finishing pigs (n = 50/cohort; three cohorts) were included in this study. For each cohort, the same proportional sampling scheme was followed. When pigs were aged 10 weeks, 50 pigs in a single barn were randomly selected: six pigs per pen in six large pens, five pigs per pen in two large pens, and two pigs per pen in two of the four small pens (hospital and at-risk pens were empty at the time of placement). The pigs were selected based on random numbers generated by Microsoft Office Excel 2007 (Microsoft, USA).
From age 10 weeks, faecal samples were collected from the selected pigs every 2 weeks for a total of eight collections (16 weeks). A total number of 1200 faecal samples (50 pigs/cohort; eight collections/cohort; three cohorts) were planned for collection. At each collection period, health conditions (e.g. diarrhoea, lameness, clinically healthy) of the selected pigs were observed and recorded by research personnel, and health records (e.g. use of medication, signs of clinical symptoms, numbers of pigs that died, potential cause of death) documented by the producers were also recorded.
Individual faecal samples were collected directly from the finishing pigs by gloved hands, and new gloves were used for each pig. Faecal samples were placed into sterile VWR® microbiology/urinalysis specimen containers (VWR International, USA). The specimen containers were placed into a cooler under ambient temperature for transportation to the laboratory at Michigan State University in East Lansing, MI. Faecal samples were stored at 2·7°C for up to 48 h prior to shipping for culture depending on the availability of the shipping service. The faecal samples where then shipped overnight on ice packs to the laboratory located at the Eastern Regional Research Centre of the U.S. Department of Agriculture in Wyndmoor, PA. All faecal samples were processed 24–72 h after collection at the farm.
STEC detection and isolation in swine faecal samples
The sample enrichment method was modified from Grant et al. [18]. In summary, a 5-g portion of each swine faecal sample was added to 95 ml tryptic soy broth (TSB) at pH 3·0 in a filter Stomacher bag. The bag was subjected to pummelling in a Stomacher for 30 s, and then incubated at room temperature for 10–15 min. One hundred millilitres of TYTP (TSB + 12·0 g/l yeast extract, 12·5 g/l Trizma base, and 1·0 g/l sodium pyruvate, with a final pH of 8·7) were then added, and samples were incubated without rotation for 15 h at 41°C.
DNA was then extracted using the PrepSEQ Rapid Spin Sample Preparation kit (Life Technologies Corporation, USA) according to the manufacturer's instructions. A multiplex PCR assay was performed using primers Stx1/2-F and mod-Stx1/2-R and probes Stx1-P [FAM] and Stx2-P [FAM], targeting stx1, stx2 and all variants except stx2f and Eae-F and mod-Eae-R primers and EaeP [MAXN] probe, targeting the eae gene [19], which encodes for the outer membrane protein intimin, which is important for attachment to the intestinal epithelial cells. The PCR assay was performed using 2 μl template DNA and the TaqMan® Environmental Master Mix 2·0 (Life Technologies Corporation) as described by Wasilenko and co-workers [19]. The PCR assay was performed in an Applied Biosystems 7500 thermal cycler (Applied Biosystems, USA) using a protocol consisting of 95 °C for 10 min, and then 40 cycles of 95°C for 15 s and 60°C for 60 s.
Enrichment samples that were positive for the stx gene were then plated onto CHROMagar STEC (DRG International Inc., USA). Although CHROMagar STEC, which contains tellurite, prevents growth of tellurite-sensitive STEC strains, CHROMagar STEC has been suggested to allow growth of 75–86·4% of STEC of various serotypes [20–22]. From each plate, three presumptive positive colonies were picked and confirmed as STEC using the stx1/2-eae multiplex PCR assay described above. The presence of stx gene types (stx1, stx2) and stx2e variant carried by the confirmed STEC isolates was determined by PCR assays described in previous publications [17, 23].
Serotyping of the STEC isolates
At least one confirmed STEC isolate was selected from every positive sample (154 STEC isolate-positive samples/1040 collected samples) for O:H serotype characterization. For O serogrouping, the confirmed STEC isolates were submitted to the E. coli Reference Centre at the Pennsylvania State University in University Park, PA. Antisera were used against serogroups O1 to O181, except for O31, O47, O72, O94, and O122, which are not designated. All STEC isolates with O serogroup information were submitted for H-typing at the French Agency for Food, Environmental and Occupational Health and Safety (Maisons-Alfort, France). Selected flagellar antigen genes (H2, H7, H8, H11, H19, H21, H28, H16, H25, H4, H32) were determined as part of a high-throughput real-time PCR system modified from previously published methods [24]. The STEC isolates which were not H typable by the real-time PCR system were submitted to the E. coli Reference Centre (University Park, PA) for H-typing by PCR restriction fragment length polymorphism of the flagellar antigen gene.
Sample size calculation
The sample size was estimated by using the population survey formula in the program Epi Info v. 7 (CDC, USA). An earlier study by Fratamico et al. [17] reported a 28·5% individual animal prevalence of STEC in finishing swine in the USA We chose an estimated prevalence of 30·0% with a 10·0% confidence limit and a population size of 1000, and the confidence was set as 90·0%. This resulted in a target sample size of 50 for each cohort to estimate prevalence at each sampling period.
Variables and statistical methods
All data were recorded and managed in Microsoft Office Excel 2007 (Microsoft). A pig was considered STEC positive when at least one confirmed STEC isolate was recovered from the faecal sample. The outcome of interest was the proportion (prevalence rate) of pigs positive for STEC at each farm visit. After data collection was completed, data validation was performed by comparing the records of farm visits and STEC isolation results from the laboratory to the database. Descriptive statistics of the proportion of pigs positive for STEC at each farm visit was performed in Microsoft Office Excel 2007 (Microsoft).
For each pig positive for STEC, the duration of STEC shedding was defined as the time interval (days) between the first and last sampling date of positive STEC isolation, which was before three consecutive STEC negative samples. An additional 14 days was counted for each positive sampling date to account for sampling intervals. Right-censored data included pigs that died or were shipped to market before the observation of three consecutive STEC-negative samples. Survival analysis using the Kaplan–Meier method was performed to analyse the duration of STEC shedding in finishing swine by Stata v. 13 (StataCorp LP, USA). The equality of survivor curves by cohort, gender, and STEC serotype was examined by the Peto-Peto-Prentice test. P values <0·05 were considered significant.
RESULTS
Finishing pigs and sample collection
A total of 150 pigs (50 pigs in each cohort, three cohorts in total) were included in the longitudinal sampling. Ten (6·7%) out of the 150 pigs died during the study period. Diarrhoea was observed in at least 12·0% (6/50) of the pigs in cohort 1, 12·0% (6/50) of the pigs in cohort 2, and 10·0% (5/50) of the pigs in cohort 3, and samples were collected in these pigs with diarrhoea. There were seven instead of eight farm visits for cohort 1 due to marketing of the pigs prior to the final collection, and eight farm visits occurred for cohort 2 and for cohort 3. Eight farm visits were completed for 43·3% (65/150) of the pigs, and seven farm visits were completed for 36·7% (55/150) of the pigs. At the end of sample collection, a total of 1040 faecal samples were collected. The main reasons why samples may not have been collected at farm visits were that the pigs were too ill, the pigs were dead, or the pigs were shipped for marketing before sample collection (n = 14 in cohort 1, n = 0 in cohort 2, n = 9 in cohort 3). The sample collection time and demographic information of the finishing pigs are summarized in Table 1.
Table 1.
Sample time periods, demographic information and numbers of faecal samples of finishing pigs
| Cohort 1 | Cohort 2 | Cohort 3 | |
|---|---|---|---|
| Production site | A | B | B |
| Sample collection period | 31 May 2011 (visit 1) | 18 July 2011 (visit 1) | 21 November 2011 (visit 1) |
| 22 August 2011 (visit 7) | 22 October 2011 (visit 8) | 18 February 2012 (visit 8) | |
| Number of animals in each cohort | 50 | 50 | 50 |
| Number of animals died during study period | 2 | 6 | 2 |
| Sex | |||
| Male | 23 | 17 | 24 |
| Female | 27 | 33 | 26 |
| Number of faecal samples collected | 320 | 357 | 363 |
Distribution of STEC-positive pigs over the finishing period
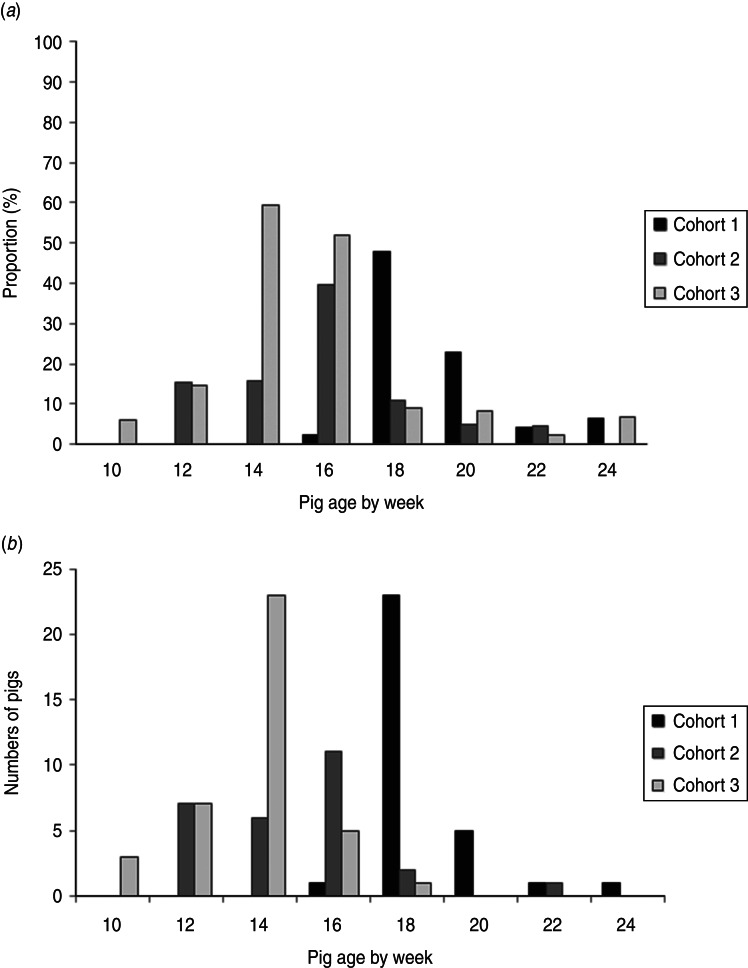
STEC isolates were recovered from at least one sample from 65·3% (98/150) of the pigs. Specifically, STEC isolates were recovered in a least one sample from 62·0% (31/50) of the pigs in cohort 1, 54·0% (27/50) of the pigs in cohort 2, and 80·0% (40/50) of the pigs in cohort 3. Figure 1a illustrates the proportions (prevalence rates) of pigs with STEC isolates by pig age (in weeks) over the finishing period, and Figure 1b displays numbers of pigs at the age of first-time STEC detection. The highest prevalence rates of STEC-positive pigs were detected when pigs were aged 18 weeks in cohort 1 (47·9%), 16 weeks in cohort 2 (39·5%), and 14 weeks in cohort 3 (59·2%). The numbers of pigs at the age of first-time STEC detection peaked at the same age as the proportions of STEC-positive pigs in every cohort. At the end of finishing period, the prevalence rates of STEC-positive pigs were lower: 6·3% in cohort 1, 0% in cohort 2, and 6·7% in cohort 3. STEC isolates were recovered in one faecal sample in 36·7% (55/150), two samples in 20·7% (31/150), three samples in 7·3% (11/150), and four samples in 0·7% (1/150) of the pigs. No STEC isolates were ever recovered from samples of the pigs that had diarrhoea.
Fig. 1.
(a) Proportion of pigs from which Shiga toxin-producing E. coli (STEC) were isolated by pig age over the finishing period. (b) Frequency distribution of pigs at the age of first-time STEC isolation.
Characterization of the STEC isolates
Of the 1040 swine faecal samples, a total of 285 STEC isolates were recovered from 83·2% (154/185) stx gene-positive samples. The presence of virulence genes (stx1, stx2, stx2e, eae) were determined by PCR in all the 285 STEC isolates. Most of the STEC isolates (97·9%, 279/285) carried the stx2e gene. Four STEC isolates carried the stx1 gene. Two STEC isolates carried the stx2 gene and were negative for stx2e. The eae gene was detected in only one of the 285 STEC isolates.
For O:H serotype characterization, at least one isolate was selected from every STEC-positive pig at each farm visit. A total of 200 STEC isolates were submitted for O:H serotype determination. Of the 200 STEC isolates, ten different O serogroups were identified while some isolates (n = 29) were O serogroup non-typable. Nine H types were identified in the 200 STEC isolates, and together, 19 different O:H serotypes were identified. A majority (73·6%, 148/201) of the STEC isolates were categorized as serotype O59:H21. The results of O:H serotype and virulence gene (stx1, stx2, stx2e, eae) characterization are summarized in Table 2.
Table 2.
Distribution of Shiga toxin-producing E. coli isolates by O:H serotype, Shiga toxin gene subtypes, and eae
| Serotype | Total number of isolates | Number of isolates positive for | |||
|---|---|---|---|---|---|
| stx2e | Other stx2 variants | stx1 variants | eae | ||
| O15:H45 | 1 | 1 | 0 | 0 | 0 |
| O15:H+* | 1 | 0 | 1 | 0 | 0 |
| O20:H21 | 1 | 1 | 0 | 0 | 0 |
| O49:H21 | 1 | 1 | 0 | 0 | 1 |
| O59:H19 | 3 | 3 | 0 | 0 | 0 |
| O59:H21 | 148 | 148 | 0 | 0 | 0 |
| O59:H21/H4† | 3 | 3 | 0 | 0 | 0 |
| O59:H19/H21‡ | 2 | 2 | 0 | 0 | 0 |
| O59/O54§:H21 | 2 | 2 | 0 | 0 | 0 |
| O89:H19 | 1 | 1 | 0 | 0 | 0 |
| O98:H12 | 4 | 0 | 0 | 4 | 0 |
| O98:H19 | 1 | 1 | 0 | 0 | 0 |
| O115:H19 | 1 | 1 | 0 | 0 | 0 |
| O119:H21 | 1 | 1 | 0 | 0 | 0 |
| O167:H21 | 1 | 1 | 0 | 0 | 0 |
| ONT¶:H19 | 19 | 19 | 0 | 0 | 0 |
| ONT¶:H4 | 7 | 7 | 0 | 0 | 0 |
| ONT¶:H21 | 2 | 2 | 0 | 0 | 0 |
| ONT¶:H27 | 1 | 0 | 1 | 0 | 0 |
| Total | 200 | 194 | 2 | 4 | 1 |
H+, flagellar gene fliC present, there were two bands for PCR.
These isolates were positive for H21 and H4 genes by part of the high-throughput real-time PCR platform.
These isolates were positive for H19 and H21 genes by part of the high-throughput real-time PCR platform.
These isolates were positive with both O59 and O54 antisera.
ONT, O antigen non-typable.
Estimation of duration of STEC shedding in finishing swine
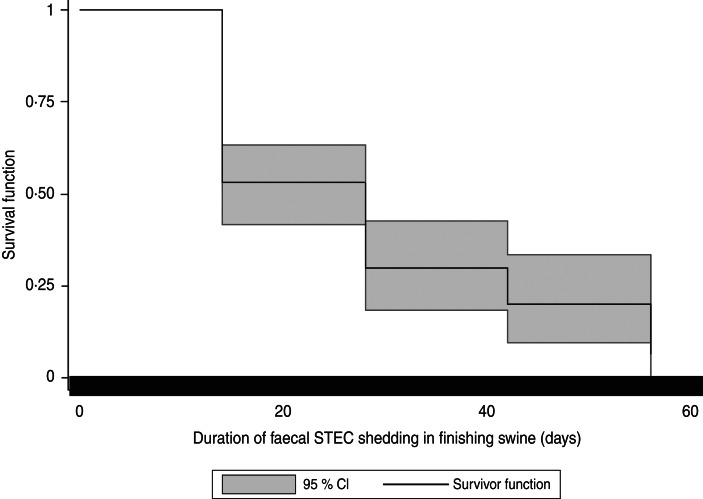
The pigs (n = 19) with samples positive for STEC isolates belonging to more than one serotype were excluded from the survival analysis. Therefore, data from 79 pigs with samples positive for STEC isolates for one serotype were included in the survival analysis. In these 79 pigs, data from 34·2% (27/79) of the pigs were right-censored. In the 27 right-censored pigs, 7·4% (2/27) of the pigs died during the study period, and 40·7% of them (11/27) were shipped to market before the study ended. The event (three consecutive STEC negative samples) was not observed in the remaining 14 of the 27 censored pigs before the study ended. In the 79 pigs included in the survival analysis, the median of the duration of STEC faecal shedding was 28 days. Kaplan–Meier analysis suggested that the cumulative probability of finishing swine shedding STEC for 28 days was 30·0%. The shortest shedding duration was 14 days at a probability of 53·2%. The longest shedding duration was 56 days at a probability of 6·7%. Figure 2 illustrates the Kaplan–Meier survival curve of duration of faecal STEC shedding in finishing swine. No significant differences were observed in survival curves by cohorts (P = 0·20), gender (P = 0·96), and STEC serotypes (P = 0·84).
Fig. 2.
Kaplan–Meier survival curves for duration of faecal Shiga toxin-producing E. coli (STEC) shedding in finishing swine.
DISCUSSION
This study represents the first longitudinal study of STEC shedding in commercial swine. Our results indicated that finishing swine shed STEC at relatively high prevalence rates during the finishing period. Previous studies have reported a wide range of STEC prevalence in swine, but most of them were cross-sectional study designs, challenging direct comparison to this study [13–15, 17]. In addition to study designs, comparison is limited because of the type of sample collected, sample collection method, and STEC isolation protocols, which vary widely across these previous reports. Although only one production company participated in this study, this company sufficiently represents the majority of conventional swine production systems in the USA (all-in, all-out multi-site production, etc.)
Variations in STEC prevalence rates over time were observed in this study, and this highlights the importance of longitudinal sampling to determine the STEC shedding status on farm. A shape similar to a point-source outbreak curve was observed when plotting the numbers of pigs at the age of first-time STEC detection, and the outbreak-like curves were observed from the results of all three of the cohorts. This may suggest that the pigs were exposed to a single source of STEC infection in the beginning of the finishing period. The STEC prevalence rates by pig age were different in these three cohorts. The highest prevalence rates of STEC-positive pigs occurred at ages 18, 16, and 14 weeks in these three cohorts. At the cohort level, no significant change of barn environment and farm management protocols occurred during the study period in these three cohorts. At the individual pig level, no significant factors (e.g. sex and health condition) were associated with the STEC-positive pigs. Common sources of STEC infection may be from the finishing barn environment, the finishing diet, or other factors. To further determine whether the STEC strains were from a common source of infection, it will be essential for future studies to examine the genetic relatedness of the swine STEC strains by molecular genotyping methods. Moreover, expanding the scope of observations to more finishing sites will be crucial to understanding the potential cohort-level or individual risk factors associated with the dynamics of STEC prevalence on swine farms.
The duration of detectable shedding in this study was similar to that described in experimentally challenged swine. Regardless of the inoculation dose, STEC O157:H7 strains were recovered in faecal samples 14–16 days after inoculation [25]. However, when inoculated with a higher dose at 1010 c.f.u/strain per animal, STEC O157:H7 strains were able to be recovered in faecal samples 58–60 days after inoculation [25]. Together, these data suggest that some pigs in this study might have been exposed to a higher dose of STEC during the finishing period, and this resulted in a longer duration of STEC shedding. Moreover, re-infection of STEC from other pigs in the same barn may occur. Re-infection of STEC from pigs in neighbouring barns may also occur because of potential pig-to-pig contact through the pen dividers in the barns in this study. Additionally, the differences of shedding duration may be attributed to differential colonization ability of different STEC strains and the host itself. One study reported no difference in duration of STEC shedding comparing pigs housed on crates and on cement floors [26]. However, what the potential factors are and how these factors contribute to the duration of STEC shedding in pigs remains largely unknown. A better understanding of the factors associated with the duration of shedding will be helpful for strategizing future plans for controlling STEC shedding on farms.
The STEC isolates recovered in this study belonged to a number of different serotypes. However, none belonged to O serogroups O138, O139, O141, and O147, which are more frequently associated with oedema disease in swine [12]. The serotypes were different from those reported in the swine STEC isolates from the NAHMS 2000 study [17]. Different STEC isolation methods may contribute to the observations of different serotypes. For example, CHROMagar STEC, which was used in this study, contains tellurite, and some STEC strains do not grow on this medium [20–22]. Nevertheless, CHROMagar STEC was suggested to be useful for recovery of STEC isolates from faecal samples, and it allowed the growth of 75–86·5% of STEC strains belonging to a wide variety of serotypes that were examined in previous studies [20–22].
The majority of the STEC isolated in the current study were serotype O59:H21. This particular serotype has not yet been reported in human cases but was reported in STEC isolates from food (beef, pork, others) [27]. Some serotypes and O serogroups of the swine STEC isolated in this study have been reported in human cases. For instance, STEC O59:H19 [28], O20 [29], O49, O98, and O119 [4] have been associated with human cases. Results of this study show that swine carry a variety of different STEC serotypes, and this was observed with sampling only three groups of pigs in one swine production company.
Most swine STEC isolates in this study carried the stx2e gene. Interestingly, Stx2e-producing E. coli strains have not only been recovered in pigs, but also in humans with HUS [30] and uncomplicated diarrhoea [31–34]. Although no source of infection was ascertained in these human cases, the association between Stx2e-producing E. coli and human illness requires further examination. The eae gene was detected in only one STEC isolate (O49:H21, carrying the stx2e gene) in this study. The presence of the eae gene in swine STEC was not consistent with some previous studies. Some studies reported that the eae gene was not detected in swine STEC [35], while others have reported detection of eae in STEC O157:H7 strains recovered from swine colon faecal samples and carcass swabs at slaughter houses [36]. Although it is well-known that intimin is essential in STEC attachment in humans [37], it has been suggested that intimin is not required for STEC O157:H7 colonization in pigs [38]. This may partially explain the fact that only one out of 285 STEC isolates in this study was eae-positive. However, outbreaks and cases associated with eae-negative STEC strains were reported [39]. Many novel adhesins have been reported in eae-negative STEC, such as the STEC autoagglutinating adhesin encoded by the saa gene [40]. To better assess the potential risks posed by swine STEC, there is a need to more extensively examine the virulence gene profiles in swine STEC isolates.
Swine have not been viewed as an important STEC reservoir. However, the findings of this study provide insights into the epidemiology of faecal STEC shedding in finishing swine. The swine STEC isolates recovered in this study belonged to various non-O157 STEC serotypes, and E. coli O157:H7 was not isolated. There is increasing awareness of the public health burden associated with non-O157 STEC infections [3]. In addition, the recent emergence of highly virulent non-O157 STEC possessing unusual virulence gene combinations stress the need to further understand these pathogens [39]. From this study, STEC isolates of various non-O157 serotypes were recovered and relatively high prevalence rates of STEC isolation were observed during the swine finishing period. Future study is warranted to confirm whether or not swine are an important source of human STEC infections, specifically non-O157 STEC.
ACKNOWLEDGEMENTS
We acknowledge the producers who participated in this research, MSU students: Dr Alda Pires, Alexandra Buckley, Allan Mergener, Tasha Likavec, Joel Sparks for assistance with sample collection and shipment. We thank Dr Chitrita DebRoy and the E. coli Reference Centre at The Pennsylvania State University, Dr Patrick Fach and Dr Sabine Delannoy at the French Agency for Food, Environmental and Occupational Health and Safety for assistance with isolate characterization.
This work was supported by the National Pork Board (project no. 12-069).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.O'Brien AD, LaVeck GD. Purification and characterization of a Shigella dysenteriae 1-like toxin produced by Escherichia coli. Infection and Immunity 1983; 40: 675–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gyles CL. Shiga toxin-producing Escherichia coli: an overview. Journal of Animal Science 2007; 85: E45–62. [DOI] [PubMed] [Google Scholar]
- 3.Scallan E, et al. Foodborne illness acquired in the United States – major pathogens. Emerging Infectious Diseases 2011; 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.CDC. 2012. National Shiga toxin-producing Escherichia coli (STEC) surveillance annual summary, 2009 (http://www.cdc.gov/ncezid/dfwed/pdfs/national-stec-surv-summ-2009-508c.pdf). Accessed 22 October 2013.
- 5.CDC. Escherichia coli O157:H7 outbreak linked to commercially distributed dry-cured salami- Washington and California, 1994. Morbidity and Mortality Weekly Report 1995; 44: 157–160. [PubMed] [Google Scholar]
- 6.CDC. Community outbreak of hemolytic uremic syndrome attributable to Escherichia coli O111:NM-South Australia, 1995. Morbidity and Mortality Weekly Report 1995; 44: 550–558. [PubMed] [Google Scholar]
- 7.Paton AW, et al. Molecular microbiological investigation of an outbreak of hemolytic-uremic syndrome caused by dry fermented sausage contaminated with Shiga-like toxin-producing Escherichia coli. Journal of Clinical Microbiology 1996; 34: 1622–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams RC, et al. Illness outbreak associated with Escherichia coli O157:H7 in Genoa salami. Canadian Medical Association Journal 2000; 162: 1409–1413. [PMC free article] [PubMed] [Google Scholar]
- 9.MacDonald DM, et al. Escherichia coli O157:H7 outbreak linked to salami, British Columbia, Canada, 1999. Epidemiology and Infection 2004; 132: 283–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conedera G, et al. A family outbreak of Escherichia coli O157 haemorrhagic colitis caused by pork meat salami. Epidemiology and Infection 2007; 135: 311–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trotz-Williams LA, et al. Pork implicated in a Shiga toxin-producing Escherichia coli O157:H7 outbreak in Ontario, Canada. Canadian Journal of Public Health 2012; 103: e322–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fairbrother JM, Gyles CL. Colibacillosis. In: Zimmerman JJ, Karriker LA, Ramirez A, Schwartz KJ, Stevenson GW, eds. Diseases of Swine, 10th edn. West Sussex, UK: John Wiley and Sons Inc., 2012, pp. 723–747. [Google Scholar]
- 13.Pinaka O, et al. Shiga toxin-producing Escherichia coli in Central Greece: prevalence and virulence genes of O157:H7 and non-O157 in animal faeces, vegetables, and humans. European Journal of Clinical Microbiology and Infectious Diseases 2013; 32: 1401–1408. [DOI] [PubMed] [Google Scholar]
- 14.Borie C, et al. Prevalence and characterization of enterohaemorrhagic Escherichia coli isoated from healthy cattle and pigs slaughtered in Santiago, Chile. Archivos de Medicina Veterinaria 1997; 29: 205–212. [Google Scholar]
- 15.Bush E, U.S. swine herd appears free of Escherichia coli O157:H7. Food Safety Digest 1997; January–Februrary, 4. [Google Scholar]
- 16.Feder I, et al. Isolation of Escherichia coli O157:H7 from intact colon faecal samples of swine. Emerging Infectious Diseases 2003; 9: 380–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fratamico PM, et al. Prevalence and characterization of Shiga toxin-producing Escherichia coli in swine faeces recovered in the National Animal Health Monitoring System's Swine 2000 study. Applied and Environmental Microbiology 2004; 70: 7173–7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant MA, Mogler MA, Harris DL. Comparison of enrichment procedures for shiga toxin-producing Escherichia coli in wastes from commercial swine farms. Journal of Food Protection 2009; 72: 1982–1986. [DOI] [PubMed] [Google Scholar]
- 19.Wasilenko JL, et al. Influence of primer sequences and DNA extraction method on detection of non-O157 Shiga toxin-producing Escherichia coli in ground beef by real-time PCR targeting the eae, stx, and serogroup-specific genes. Journal of Food Protection 2012; 75: 1939–1950. [DOI] [PubMed] [Google Scholar]
- 20.Hirvonen JJ, Siitonen A, Kaukoranta S-S. Usability and performance of CHROMagar STEC medium in detection of Shiga toxin-producing Escherichia coli strains. Journal of Clinical Microbiology 2012; 50: 3586–3590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wylie JL, et al. Evaluation of a new chromogenic agar medium for detection of Shiga toxin-producing Escherichia coli (STEC) and relative prevalences of O157 and non-O157 STEC in Manitoba, Canada. Journal of Clinical Microbiology 2013; 51: 466–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gouali M, et al. Evaluation of CHROMagar STEC and STEC O104 chromogenic agar media for detection of Shiga toxin-producing Escherichia coli in stool sepcimens. Journal of Clinical Microbiology 2013; 51: 894–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blanco M, et al. Genes coding for enterotoxins and verotoxins in porcine Escherichia coli strains belonging to different O:K:H serotypes: relationship with toxic phenotypes. Journal of Clinical Microbiology 1997; 35: 2958–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bugarel M, et al. Virulence gene profiling of enterohemorrhagic (EHEC) and enteropathogenic (EPEC) Escherichia coli strains: a basis for molecular risk assessment of typical and atypical EPEC strains. BMC Microbiology 2011; 11: 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Booher SL, Cornick NA, Moon HW. Persistence of Escherichia coli O157:H7 in experimentally infected swine. Veterinary Microbiology 2002; 89: 69–81. [DOI] [PubMed] [Google Scholar]
- 26.Cornick NA, Helgerson AF. Transmission and infectious dose of Escherichia coli O157:H7 in swine. Applied and Environmental Microbiology 2004; 70: 5331–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beutin L, et al. Identification of human-pathogenic strains of Shiga toxin-producing Escherichia coli from food by a combination of serotyping and molecular typing of Shiga toxin genes. Applied and Environmental Microbiology 2007; 73: 4769–4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galli L, et al. Virulence profile comparison between LEE-negative Shiga toxin-producing Escherichia coli (STEC) strains isolated from cattle and humans. Veterinary Microbiology 2010; 143: 307–313. [DOI] [PubMed] [Google Scholar]
- 29.Kappeli U, et al. Human infections with non-O157 Shiga toxin-producing Escherichia coli, Switzerland, 2000–2009. Emerging Infectious Diseases 2011; 17: 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas A, et al. Isolation of Vero cytotoxin-producing Escherichia coli serotypes O9ab:H- and O101:H-carrying VT2 variant gene sequences from a patient with haemolytic uraemic syndrome. European Journal of Clinical Microbiology and Infectious Diseases 1994; 13: 1074–1076. [DOI] [PubMed] [Google Scholar]
- 31.Pierard D, et al. Diarrhoea associated with Escherichia coli producing porcine oedema disease verotoxin. Lancet 1991; 338: 762. [DOI] [PubMed] [Google Scholar]
- 32.Muniesa M, et al. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infection and Immunity 2000; 68: 4850–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Friedrich AW, et al. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. Journal of Infectious Diseases 2002; 185: 74–84. [DOI] [PubMed] [Google Scholar]
- 34.Beutin L, et al. Characterization of Shiga toxin-producing Escherichia coli strains isolated from human patients in Germany over a 3-year period. Journal of Clinical Microbiology 2004; 42: 1099–1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fratamico PM, et al. Characterization of Shiga toxin-producing Escherichia coli strains isolated from swine faeces. Foodborne Pathogen and Disease 2008; 5: 827–838. [DOI] [PubMed] [Google Scholar]
- 36.Lenahan M, et al. Molecular characterization of Irish E. coli O157:H7 isolates of human, bovine, ovine and porcine origin. Journal of Applied Microbiology 2009; 107: 1340–1349. [DOI] [PubMed] [Google Scholar]
- 37.Frankel G, Phillips AD. Attaching effacing Escherichia coli and paradigms of Tir-triggered actin polymerization: getting off the pedestal. Cellular Microbiology 2008; 10: 549–556. [DOI] [PubMed] [Google Scholar]
- 38.Jordan DM, Booher SL, Moon HW. Escherichia coli O157:H7 does not require intimin to persist in pigs. Infection and Immunity 2005; 73: 1865–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bielaszewska M, et al. Characterisation of the Escherichia coli strain associated with an outbreak of haemolytic uraemic syndrome in Germany, 2011: a microbiological study. The Lancet Infectious Diseases 2011; 11: 671–676. [DOI] [PubMed] [Google Scholar]
- 40.Paton AW, et al. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infection and Immunity 2001; 69: 6999–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]