SUMMARY
Salmonella enterica commonly colonizes the intestinal tract of cattle and is a leading cause of foodborne illness. A previously described investigation into the prevalence of S. enterica on a dairy farm revealed an 8-year-long asymptomatic S. enterica epidemic caused by serotypes Cerro and Kentucky in the lactating herd. To investigate the source of the S. Kentucky strains, the genomes of two S. Kentucky isolates were sequenced; one collected prior to the epidemic (2004) and one collected during the epidemic (2010). Comparative genomic analysis demonstrated significant polymorphisms between the two strains. PCR primers targeting unique and strain-specific regions were developed, and screening of the archived isolates identified the index case of the asymptomatic S. Kentucky epidemic as a heifer that was raised off-site and transported onto the study farm in 2005. Analysis of isolates collected from all heifers brought onto the farm demonstrated frequent re-introduction of clones of the epidemic strain suggesting transmission of pathogens between farms might occur repeatedly.
Key words: Food safety, genomics, Salmonella
Salmonella spp. are a major cause of morbidity and mortality in humans and domesticated animals worldwide. The aetiological agents of most salmonellosis cases in humans and other mammals are members of the Salmonella enterica species, and the intensity or level of illness is often serotype-specific [1, 2]. All S. enterica serotypes are considered potentially pathogenic to humans, but not all serotypes cause symptomatic infections in other mammals. Non-human hosts may subclinically harbour these organisms and therefore pose an under-appreciated public health risk to humans.
Salmonella enterica subsp. enterica serovar Kentucky is an occasional pathogen of humans and was the 44th most isolated serotype from diagnosed salmonellosis cases in humans from 1999 to 2009 in the United States [3]. Currently, human infections with highly drug-resistant S. Kentucky ST198 strains have spread globally suggesting it is a serotype with clear global health significance [4, 5]. S. Kentucky is often subclinically shed from dairy cattle and previous studies demonstrated the incidence of infection in cows on S. Kentucky-positive farms ranging between 1·8% and 97% [6–9]. Thus, its frequent isolation from dairy farms and ability to cause illness in humans suggests that monitoring this serotype, among others, may limit the public health risk of zoonotic salmonellae for humans who consume dairy and beef products and those who work in the dairy industry or have contact with animals at agricultural fairs.
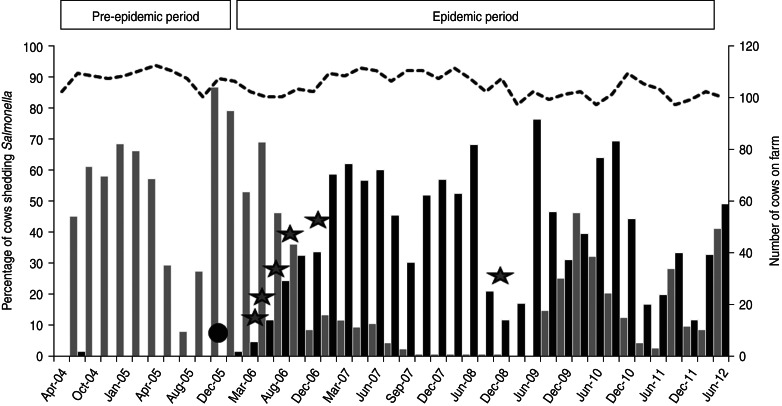
As part of an on-going longitudinal investigation (2004–2012) into the presence of human foodborne pathogens on dairy farms in the northeast United States, a commercial dairy farm in Pennsylvania was investigated for the presence of S. enterica [9]. A long-term S. enterica epidemic in the herd was observed and the two primary serotypes were identified as Cerro and Kentucky [9]. Further work demonstrated that S. Kentucky was prevalent on other dairy farms surrounding the study farm demonstrating a regional presence of this serotype [10]. From April 2004 to December 2005, the majority of recovered S. enterica isolates from the study farm were serotyped as Cerro. Two cows were shedding S. Kentucky and four were shedding S. Typhimurium for only one sample collection date each, as reported previously [9]. During this period, S. Kentucky was isolated 16 times (twice from mature cows, 11 times from composite manure samples, once from flies, once from a tractor tyre swab, and once from standing water near the tractor tyre) (Fig. 1). S. Typhimurium became extinct on the farm in June 2004 and, from January 2006 onwards, S. Kentucky was isolated with increasing frequency [9]. A shift from Cerro to Kentucky dominance was gradually observed, followed by co-existence of the two serotypes on the farm (Fig. 1) [9]. Cows shedding either serotype did not display symptoms of an infection. Sporadic isolations of S. Oranienburg (two cows), S. Enteriditis (one cow), S. Muenster (one cow) and S. Montevideo (one cow) were observed during the study period [9].
Fig. 1.
Percentage of cows shedding Salmonella enterica on the study farm. Black bars indicate percentage of cows shedding S. Kentucky and grey bars indicate percentage of cows shedding S. Cerro (primary y-axis). Dotted black line shows the number of cows on the farm (secondary y-axis). The black circle indicates initial introduction of the epidemic strain onto the farm. Grey stars indicate introductions of heifers shedding the epidemic strain.
Identification of the source, or index case of S. enterica epidemics and contamination is integral to preventing larger outbreaks as well as identifying agricultural management practices that promote transmission or contamination. The aim of this study was to identify the source of the S. Kentucky on this dairy farm using comparative genomics followed by PCR identification of epidemic-associated genotypes.
Monitoring of the herd and environmental samples (feed, source and trough water, composite manure, environmental waters and streambed sediments in the surrounding area, and flies) was conducted 4–6 times per year [11, 12]. The herd size ranged between 98 and 112 lactating cows. Calves were monitored post-weaning prior to leaving the home herd for a heifer-rearing facility, and prior to their re-introduction as pre-fresh heifers to their home herd. Processing of samples and identification of S. enterica were as described previously [11, 12]. For each sample at least six randomly chosen presumptive Salmonella colonies were selected for confirmation. Serotyping was determined on select isolates [13]. Farm management records were collected during the course of the study.
To identify potential unique genomic markers of the outbreak strain we compared the genome sequence of a ‘pre-epidemic strain’ to that of an ‘epidemic strain’. The pre-epidemic strain (strain 0253) was isolated from a randomly selected cow in 2004 and the epidemic strain (strain 5349) was isolated from a different randomly selected cow in 2010. Genomic DNA from each strain was isolated from overnight cultures using a DNeasy blood and tissue kit (Qiagen, USA). The genomes were sequenced using the Genome Sequencer FLX+ 454 Life Sciences (Roche, USA) and the Sequencing Reagents XL+ kit according to the manufacturer's instructions. De novo assemblies were performed using Roche Newbler software v. 2.6 (Roche). The assembled genomes were annotated using rapid annotations based on subsystem technology (RAST) [14]. To identify significant regions of non-homogeneity between the two S. Kentucky isolates, the genome of S. Kentucky 5349 was aligned with the S. Kentucky 0253 genome using BLASTP.
PCR primers were developed to target unique regions of the epidemic strain. To amplify DNA from an indel region of nupG, primers nupG-F 5′-ctcactaccctgggctcgta-3′ and nupG-R 5′-tcaggaagaacggaatggtc-3′ were used. To amplify the DNA from an indel region of the putative transport protein primers PTP-F 5′-ccgattctgcagtggttttt-3′ and PTP-R 5′-acaataagatttgcggcaatg-3′ were used. Annealing temperature for NupG was 55°C and the annealing time was 60 s. Annealing temperature for PTP was 53·3°C and the annealing time was 60 s. PCR products were separated by size in a 1% agarose gel and visualized under UV light. Amplicon sizes were estimated by comparison to PCR ladders of known sizes. To determine the source of the epidemic strain all S. Kentucky isolates from the beginning of sampling in September 2004 onwards were screened until an isolate with both epidemic-specific indels was found.
Nucleoside permease subunit G (nupG), a housekeeping gene conserved among Gram-negative bacteria, encodes an inner membrane porin that is involved in transport of nucleosides. Mutants of this gene in S. Typhi have been demonstrated to be viable and can grow on minimal media [15]. The BLASTP analysis of strains 5349 and 0253 demonstrated this gene to encode a tandem 159 bp duplication of the sequence between nucleotides 158 and 318 resulting in an amino acid (aa) that is 53 aa longer in the epidemic strain (strain 5349) than that of the pre-epidemic strain (strain 0253). Another open reading frame (ORF) annotated as a putative transport protein exhibited a 93 bp deletion in the epidemic strain when aligned with the pre-epidemic strain.
From the beginning of the study period until the index case was detected, 1212 cow faecal, 152 composite manure, 97 water (trough and source), 90 feed (mixed and components), and 452 other (flies, waste lagoon, tyres, etc.) samples were collected. Of these, only 16 samples were positive for S. Kentucky. The first isolate with the markers of the epidemic strain was recovered from the faeces of a heifer that had been moved from an off-site independent heifer-rearing facility to the study farm in November 2005 (Fig. 1). The sample was collected prior to this animal entering the study farm indicating that the strain colonized the intestinal tract of the heifer prior to contact with other animals and the environment of the study farm. All 16 S. Kentucky isolates recovered prior to this event, like the sequenced pre-epidemic strain, did not encode the markers of the epidemic strain suggesting that, although the pre-epidemic strain was persistent on the farm, it did not become established in the bovine population. Farm records also indicate that, at this time, the heifer-rearing facility from which the epidemic strain originated was newly contracted thereby exposing heifers from the study farm to a novel population of heifers from multiple farms. Further, PCR targeting the epidemic-strain markers of Salmonella isolates collected from eight separate heifers brought onto the farm between January 2006 and November 2008 resulted in positive amplicons for six heifers, demonstrating this strain was carried onto the farm multiple times after the initial introduction (Fig. 1). It is important to note that more heifers were brought onto the farm during this time-frame, but only eight were sampled. During this period, no calves leaving the study farm were positive for S. Kentucky, indicating that epidemic-strain infections originated at the heifer-rearing facility and subsequently were transported onto the study farm.
Heifer raisers specialize in rearing female calves away from their birth farm in a facility that is often utilized by several farmers within a region. The heifers from the study farm were moved to the heifer-rearing facility after weaning (~6–7 months) and were generally returned to their home farm within 16–18 months, shortly before giving birth to their first calf. Previous studies demonstrated that off-site rearing of heifers is significantly associated with the introduction of Salmonella serotypes to a herd that were not previously identified in that herd [16–18]. This indicates a potential biosecurity issue as specialized heifer-rearing operations are becoming more common. Currently about 1 in 10 dairy farms in the United States utilizes heifer-rearing operations [19].
Monitoring transported cattle for carriage of human infectious agents, such as S. enterica, prior to their integration in a herd may help reduce the risk of pathogen transmission between herds. This may identify cows that carry agents harmful to other members of the herd or the human population that work with the herd or consume the meat and dairy products produced by the herd. Although, such monitoring remains economically infeasible for most dairy operations, decreasing costs of rapid molecular monitoring may allow for more rapid detection of pathogens in the future.
Results of this study demonstrate genomically that livestock movement, commingling with livestock from other farms, or utilizing facilities used by several other farms may result in transmission of pathogens to novel geographical locations and potentially increase the risk of pathogen exposure to larger human and animal populations. Results of this study also demonstrate a need to further evaluate the genomic variability within Salmonella enterica subsp. enterica serovars to better elucidate those elements and/or polymorphisms in the genome that result in emergence of specific strains within a susceptible host population.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the technical support provided by Jakeitha Sonnier (USDA, ARS) and Charles Wang (FDA).
Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Paulin SM, et al. Analysis of Salmonella enterica serotype-host specificity in calves: avirulence of S. enterica serotype Gallinarum correlates with bacterial dissemination from mesenteric lymph nodes and persistence in vivo. Infection and Immunity 2002; 70: 6788–6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stevens MP, Humphrey TJ, Maskell DJ. Molecular insights into farm animal and zoonotic Salmonella infections. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences 2009; 364: 2709–2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.CDC. (http://www.cdc.gov/ncezid/dfwed/edeb/reports.html#Salmonella), 2010.
- 4.Le Hello S, et al. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. Journal of Infectious Diseases 2011; 204: 675–684. [DOI] [PubMed] [Google Scholar]
- 5.Le Hello S, et al. Highly drug-resistant Salmonella enterica serotype Kentucky ST198-X1: a microbiological study. Lancet Infectious Diseases 2013; 13: 672–679. [DOI] [PubMed] [Google Scholar]
- 6.Wells SJ, et al. Fecal shedding of Salmonella spp. by dairy cows on farm and at cull cow markets. Journal of Food Protection 2001; 64: 3–11. [DOI] [PubMed] [Google Scholar]
- 7.Edrington TS, et al. Variation in the faecal shedding of Salmonella and E. coli O157:H7 in lactating dairy cattle and examination of Salmonella genotypes using pulsed-field gel electrophoresis. Letters in Applied Microbiology 2004; 38: 366–372. [DOI] [PubMed] [Google Scholar]
- 8.Blau DM, et al. Salmonella in dairy operations in the United States: prevalence and antimicrobial drug susceptibility. Journal of Food Protection 2005; 68: 696–702. [DOI] [PubMed] [Google Scholar]
- 9.Van Kessel JS, et al. Dynamics of Salmonella serotype shifts in an endemically infected dairy herd. Foodborne Pathogens and Disease 2012; 9: 319–324. [DOI] [PubMed] [Google Scholar]
- 10.Van Kessel JS, et al. Regional distribution of two dairy-associated Salmonella enterica serotypes. Foodborne Pathogens and Disease 2013; 10: 448–452. [DOI] [PubMed] [Google Scholar]
- 11.Van Kessel JS, et al. Longitudinal study of a clonal, subclinical outbreak of Salmonella enterica subsp. enterica serovar Cerro in a U.S. dairy herd. Foodborne Pathogens and Disease 2007; 4: 449–461. [DOI] [PubMed] [Google Scholar]
- 12.Van Kessel JS, et al. Environmental sampling to predict fecal prevalence of Salmonella in an intensively monitored dairy herd. Journal of Food Protection 2008; 71: 1967–1973. [DOI] [PubMed] [Google Scholar]
- 13.Herrera-León S, et al. Blind comparison of traditional serotyping with three multiplex PCRs for the identification of Salmonella serotypes. Research in Microbiology 2007; 158: 122–127. [DOI] [PubMed] [Google Scholar]
- 14.Aziz RK, et al. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008; 9: 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bucarey SA, et al. The Salmonella enterica Serovar Typhi tsx gene, encoding a nucleoside-specific porin, is essential for prototrophic growth in the absence of nucleosides. Infection and Immunity 2005; 73: 6210–6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hegde NV, et al. Dissemination of Salmonella enterica subsp. enterica serovar Typhimurium var. Copenhagen clonal types through a contract heifer-raising operation. Journal of Clinical Microbiology 2005; 43: 4208–4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vanselow B, et al. Salmonella and on-farm risk factors in healthy slaughter-age cattle and sheep in eastern Australia. Australian Veterinary Journal 2007; 85: 498–502. [DOI] [PubMed] [Google Scholar]
- 18.Adhikari B, et al. The role of animal movement, including off-farm rearing of heifers, in the interherd transmission of multidrug-resistant Salmonella. Journal of Dairy Science 2009; 92: 4229–4238. [DOI] [PubMed] [Google Scholar]
- 19.United States Department of Agriculture. Animal and Plant Health Inspection Service. Off-Site heifer raising on U.S. dairy operations, 2007. (http://www.aphis.usda.gov/animal_health/nahms/dairy/index.shtml). 2007.