SUMMARY
This study was undertaken to survey the presence of Salmonella in 200 chicken samples collected from Mansoura, Egypt. Salmonella was detected in 16% (8/50), 28% (14/50), 32% (16/50) and 60% (30/50) of whole chicken carcasses, drumsticks, livers and gizzards, respectively, with an overall prevalence of 34% (68/200) among all samples. One hundred and sixty-six isolates were identified biochemically as Salmonella, and confirmed genetically by PCR, based on the presence of invA and stn genes. The spvC gene, however, was detected in only 25·3% (42/166) of the isolates. Isolates were serotyped as Salmonella Enteritidis (37·3%), S. Typhimurium (30·1%), S. Kentucky (10·8%), S. Muenster (8·4%), S. Virchow (4·8%), S. Anatum (4·8%), S. Haifa (1·2%), and four were non-typable. Antimicrobial susceptibility tests of the Salmonella isolates revealed that 100% were resistant to each of erythromycin, penicillin, and amoxicillin, while 98·8%, 96·4%, 95·2%, and 91·6% were resistant to nalidixic acid, sulphamethoxazole, oxytetracycline, and ampicillin, respectively. Multidrug resistance was evident for 92·8% of the isolates. The high contamination level of chicken meat with multidrug-resistant Salmonella can constitute a problem for public health.
Key words: Antimicrobial resistance, chickens, PCR, Salmonella serovars, virulence genes
INTRODUCTION
Salmonella is one of the most important causative agents of foodborne infection in both developed and developing countries [1]. In the USA, it has been estimated that about 87% of all Salmonella-confirmed cases are foodborne, with 10% due to person-to-person infection, and 3% to pets [2]. Poultry and poultry products are among the main food sources most often incriminated in outbreaks of human salmonellosis [3, 4] and infection frequently occurs as a result of cross-contamination from equipment, utensils and workers' hands with subsequent handling of raw carcasses and products, in addition to the consumption of undercooked poultry meat [5].
In recent years, there has been an increase in the incidence of human salmonellosis that is more difficult to treat due to the appearance of multidrug-resistant strains, especially S. Typhimurium, which have been isolated from various foods of animal origin worldwide [6]. The high prevalence of antimicrobial-resistant bacteria throughout the food industry is probably due to widespread overuse of common antimicrobials as therapeutics, prophylactics or growth promoters in food animals [7].
Although all the serovars of S. enterica are considered potentially pathogenic, there are considerable differences in their virulence to humans and this has been attributed to the absence or presence of plasmids carrying virulence-associated genes [8]. The invasion A (invA) gene is unique to all Salmonella serovars and is an internationally recognized marker for the rapid detection of Salmonella genus [9]. The invA gene is required for invasion of the organism into host cells [10], while the enterotoxin (stn) gene encodes a protein which mediates severe diarrhoea has also been utilized as a PCR target for the detection of Salmonella strains [11]. The Salmonella plasmid virulence gene, spvC, is believed to increase the growth rate of salmonellae in host cells and affect their interaction with the host immune system [12].
Due to the increased incidence of resistant or multi-resistant Salmonella isolates worldwide against many commonly used antimicrobials, this study set out to determine the prevalence, serotypes, presence of virulence genes, and antimicrobial resistance of Salmonella isolates from chicken carcasses and giblets from retail outlets in Mansoura city, Egypt.
MATERIALS AND METHODS
Sample collection and bacteriological analysis
In total, 200 chicken samples (50 each of whole chicken carcasses, drumsticks, gizzards and livers) were randomly collected from 25 retail shops and supermarkets, of different sanitation levels, distributed in Mansoura city, Egypt on 10 occasions during the period June–November 2012. Each of the 25 shops was visited twice for sampling. On each sampling occasion, five shops were visited, and four samples (whole chicken carcass, drumstick, gizzard and liver) were taken from each shop. Each sample was packaged individually into a sterile impermeable polyethylene bag, labelled and transferred within 1 h in an icebox at ~4°C to the food hygiene laboratory for bacteriological analyses. The test portion for analysis was 25 g outer skin of whole chicken carcasses, skin plus muscle of drumsticks, and tissue from gizzards or livers excised aseptically with a sterile scalpel for each individual sample.
Each test portion was transferred into a sterile homogenizer flask containing 225 ml of sterile buffered peptone water (Oxoid, UK) and homogenized for 1 min in a stomacher (Seward Medical, UK). The homogenate was incubated at 37°C for 24 h then 0·1 and 1 ml volumes were aseptically added to 10 ml each of Rappaport Vassilliadis (RV) broth (Oxoid) and Muller–Kauffmann tetrathionate/novobiocin (MKTTn) broth (Oxoid), and incubated at 42°C for 24 h and 37°C for 24 h, respectively. These broths were subcultured on xylose-lysine-desoxycholate (XLD) agar (Oxoid) and Brilliant Green agar (BGA) with sulfadiazine (Neogen Corp., USA) which were incubated at 37°C for 24 h, and at 35°C for 24 h, respectively. Up to five typical (pink colonies with or without black centres) or suspected colonies of presumptive Salmonella were subcultured onto nutrient agar slopes and incubated at 37°C for 24 h for further biochemical and serological identification.
Identification of presumptive Salmonella isolates was performed according to standard methods [13] and carbohydrate fermentation profile using the API Rapid 20E system (bioMérieux, France) according to the manufacturer's instructions. Biochemically confirmed Salmonella isolates were serotyped by slide agglutination with O and H polyvalent antisera (Wellcome Diagnostic, UK).
Molecular analysis
Genomic DNA was prepared by a method described previously [14]. Salmonella Typhimurium (RIMD 1 985 009) and Escherichia coli K12DH5α were used as positive and negative control strains, respectively, for the presence and absence of invA, stn and spvC genes. The primers for PCR amplification of invA (244 bp) were as described previously [15]. For PCR amplification of stn, two oligonucleotide primers (forward: 5′-CTTAATCGCGCCGCCATGCTGTT-3′; reverse: 5′-CATGAACTGGCGCAGGTGAT-3′) were constructed to produce an amplified band size of 480 bp. For amplification of spvC, two primers (forward: 5′-AACGGTTCCTCACGTAAAGCCTGT-3′; reverse: 5′- ACCAAATGCGGAAGATGCCGGTAT-3′) produced an amplified band size of 580 bp. PCR was performed in a 15-μl volume comprising 1 μl Salmonella DNA template, 1·6 μl each of forward and reverse primers (3 pmol each), 3 μl dNTPs (2 mm), 7·5 μl of 2 × PCR buffer for KOD FX, and 0·3 μl KOD FX DNA polymerase (Toyobo Co. Ltd, Japan). After an initial denaturation at 94°C for 2 min, 35 cycles (98°C for 10 s, 58°C for 30 s, 68°C for 30 s) were performed followed by a final extension at 68°C for 7 min. Amplified genes were verified by DNA sequencing with the BigDye Terminator v. 3.1 Cycle Sequencing kit (Applied BioSystems, USA) according to the manufacturer's instructions on an ABI Prism 3100 automated sequencer (Applied Biosystems). Nucleotide sequence data were analysed with GENETYXMAC software, v. 12 (GENETYX Corp., Japan). Homology searches of the obtained sequences against the already published genes in the GenBank were performed using Standard Nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
Antimicrobial susceptibility tests
The antimicrobial susceptibility of Salmonella isolates was determined by an agar disk diffusion standard method [16] on Mueller–Hinton agar (Oxoid). Antibiotic discs were obtained from Difco (USA) and bioMérieux at the following drug concentrations: erythromycin (15 μg), nalidixic acid (30 μg), penicillin (10 IU), amoxicillin (30 μg), oxytetracycline (30 μg), sulphamethoxazole (25 μg), ampicillin (10 μg), streptomycin (10 μg), neomycin (30 μg), chloramphenicol (30 μg), norfloxacin (10 μg), ciprofloxacin (5 μg), kanamycin (30 μg), and gentamicin (10 μg). Isolates were classified as susceptible, intermediate or resistant according to National Committee for Clinical Laboratory Standards criteria [16], with intermediate susceptibility counted as resistant. E. coli ATCC 25 922 was used as a reference strain for antibiotic disc control. The multiple antibiotic resistance (MAR) index for each resistance pattern was calculated from the number of resistances to antimicrobials/total number of antimicrobials tested.
RESULTS
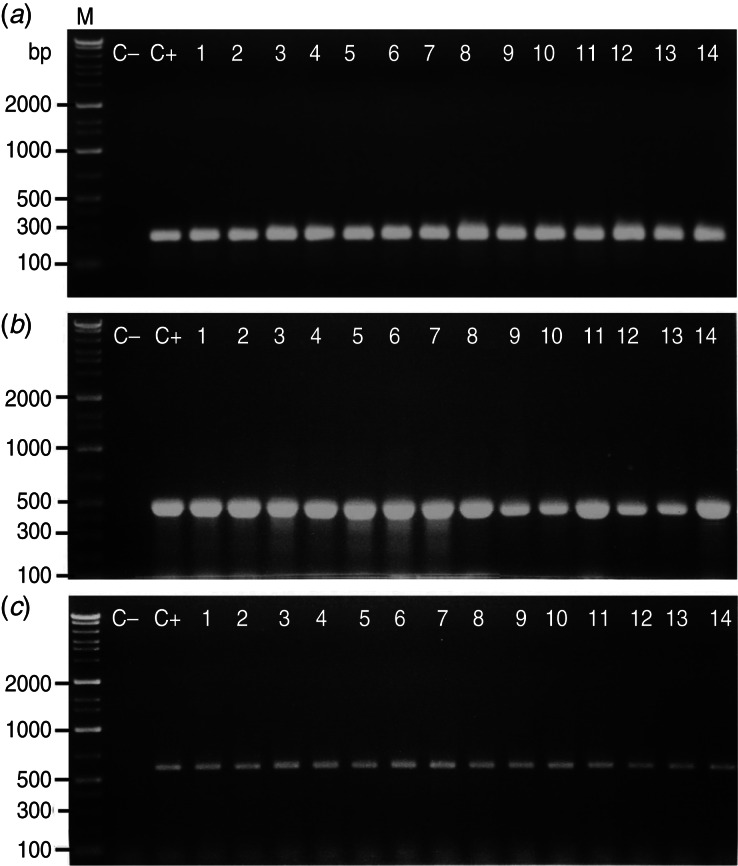
Salmonella spp. were detected in 34% (68/200) of all chicken samples, distributed as 16% (8/50), 28% (14/50), 32% (16/50) and 60% (30/50) among whole chicken carcasses, drumsticks, livers, and gizzards, respectively. By PCR, all 166 isolates were positive for the invA and stn genes; while 25·3% (42/166) carried the spvC gene (Fig. 1) and were mainly S. Typhimurium and S. Enteritidis. The specific primers used in the present study exhibited good amplification efficiency for detection of the target genes and produced sufficient DNA for sequence analysis to confirm the identity of the amplified genes.
Fig. 1.
Representative agarose gel electrophoresis for PCR products showing the proper molecular size of (a) 244 bp for the amplified invA gene, (b) 480 bp for the amplified enterotoxin (stn) gene, and (c) 580 bp for the amplified Salmonella plasmid virulence (spvC) gene in Salmonela isolates recovered from chicken meat and giblets. Chromosomal DNA from the Salmonella isolates (n = 166) was used as a template for PCR amplification using specific primer sets for invA, stn and spvC genes. Three microliters of the PCR product were separated by electrophoresis on 1·2% agarose gel and visualized under UV light. M, DNA marker (gene ladder 100 bp) used as a reference for fragment size; lane C–, E. coli K12 DH5α used as negative control strain; lane C+, Salmonella Typhimurium (RIMD 1985009) used as positive control strain; lanes 1–14, representative positive strains.
Seven different serovars were distinguished among the Salmonella isolates. S. Enteritidis (37·3%) was the most frequent, followed by S. Typhimurium (30·1%), S. Kentucky (10·8%), S. Muenster (8·4%), S. Virchow (4·8%), S. Anatum (4·8%), and S. Haifa (1·2%). Four (2·4%) isolates could not be serotyped.
The distribution of Salmonella serovars among the chicken samples is shown in Table 1. S. Enteritidis and S. Typhimurium were recovered from all chicken parts, but S. Kentucky was not found in whole chicken carcass samples. Similarly, S. Muenster and the non-typable isolates were present in only liver and gizzard samples. S. Virchow was present in only drumstick and gizzard samples, and S. Anatum and S. Haifa were present in gizzard samples alone.
Table 1.
Distribution of Salmonella serovars (n = 166) among chicken samples
| Serotypes | S. Enteritidis | S. Typhimurium | S. Kentucky | S. Muenster | S. Virchow | S. Anatum | S. Haifa | Untyped |
|---|---|---|---|---|---|---|---|---|
| Whole carcasses | 6 | 4 | — | — | — | — | — | — |
| Drumsticks | 8 | 8 | 6 | — | 4 | — | — | — |
| Livers | 12 | 6 | 4 | 2 | — | — | — | 2 |
| Gizzards | 36 | 32 | 8 | 12 | 4 | 8 | 2 | 2 |
| Total | 62 | 50 | 18 | 14 | 8 | 8 | 2 | 4 |
All Salmonella isolates were resistant to erythromycin, penicillin, and amoxicillin (Table 2), while high resistance rates (>90%) were found for nalidixic acid (98·8%), sulphamethoxazole (96·4%), oxytetracycline (95·2%), and ampicillin (91·6%). The lowest rates were recorded for ciprofloxacin (63·9%), kanamycin (41·0%), and gentamicin (21·7%). The analysis of resistance profiles and MAR indexes of isolates by serotype is shown in Table 3. The great majority (92·8%, 154/166) of isolates showed resistance to ⩾3 of the 14 antimicrobials tested. Two-thirds of isolates (63·2%, 105/166) had a MAR index above the average (0·582) and within these isolates 13 resistance profiles encompassing eight to all 14 antimicrobials were identified. Among S. Enteriditis, 50/62 (80·6%), and all of 50 S. Typhimurium isolates were resistant to ⩾9 antimicrobials.
Table 2.
Percentages of antimicrobial susceptibility of Salmonella species isolated from chicken carcasses and products (n = 166 isolates)
| Antimicrobial agent | Sensitive | Intermediate | Resistant |
|---|---|---|---|
| No. (%) | No. (%) | No. (%) | |
| Erythromycin (E) | — | — | 166 (100) |
| Nalidixic acid (NA) | 2 (1·2) | 6 (3·6) | 158 (95·2) |
| Penicillin (P) | — | 12 (7·2) | 154 (92·8) |
| Amoxicillin (AMX) | — | 14 (8·4) | 152 (91·6) |
| Oxytetracycline (T) | 8 (4·8) | 10 (6·0) | 148 (89·2) |
| Sulphamethoxazole (SXT) | 6 (3·6) | 22 (13·2) | 138 (83·1) |
| Ampicillin (AM) | 14 (8·4) | 28 (16·9) | 124 (74·7) |
| Streptomycin (S) | 30 (18·1) | 24 (14·4) | 112 (67·5) |
| Neomycin (N) | 26 (15·7) | 38 (22·9) | 102 (61·4) |
| Chloramphenicol (C) | 36 (21·7) | 52 (31·3) | 78 (47·0) |
| Norfloxacin (NOR) | 54 (32·5) | 62 (37·4) | 50 (30·1) |
| Ciprofloxacin (CP) | 60 (36·1) | 74 (44·6) | 32 (19·3) |
| Kanamycin (K) | 98 (59·0) | 52 (31·3) | 16 (9·6) |
| Gentamycin (G) | 130 (78·3) | 34 (20·5) | 2 (1·2) |
Table 3.
Antibiotic resistant profiles and multiple resistance index (MAR) of Salmonella isolates from chicken carcasses and products (n = 166 isolates)
| Salmonella serovars | Antimicrobial resistance profile | No. of isolates | MAR index |
|---|---|---|---|
| S. Enteritidis | E, NA, P, AMX, T, SXT, AM, S, N, C, NOR, CP, K | 4 | 0·928 |
| S. Enteritidis | E, NA, P, AMX, T, SXT, AM, S, N, C, NOR, CP | 8 | 0·857 |
| S. Enteritidis | E, NA, P, AMX, T, SXT, AM, S, N, C, NOR | 8 | 0·785 |
| S. Enteritidis | E, NA, P, AMX, T, SXT, AM, S, N, C | 20 | 0·714 |
| S. Enteritidis | E, NA, P, AMX, T, SXT, AM, S, N | 10 | 0·642 |
| S. Enteritidis | E, NA, P, AMX, T, SXT, AM, S | 4 | 0·571 |
| S. Enteritidis | E, NA, P, AMX, T, SXT | 2 | 0·428 |
| S. Enteritidis | E, NA, P, AMX, T | 4 | 0·357 |
| S. Enteritidis | E, NA, P, AMX | 2 | 0·285 |
| S. Typhimurium | E, NA, P, AMX, T, SXT, AM, S, N, C, NOR, CP, K, G | 2 | 1 |
| S. Typhimurium | E, NA, P, AMX, T, SXT, AM, S, N, C, NOR, CP, K | 10 | 0·928 |
| S. Typhimurium | E, NA, P, AMX, T, SXT, AM, S, N, C, NOR, CP | 8 | 0·857 |
| S. Typhimurium | E, NA, P, AMX, T, SXT, AM, S, N, C, NOR | 10 | 0·785 |
| S. Typhimurium | E, NA, P, AMX, T, SXT, AM, S, N, C | 8 | 0·714 |
| S. Typhimurium | E, NA, P, AMX, T, SXT, AM, S, N | 12 | 0·642 |
| S. Kentucky | E, NA, P, AMX, T, SXT, AM, S | 2 | 0·571 |
| S. Kentucky | E, NA, P, AMX,T, SXT, AM | 4 | 0·500 |
| S. Kentucky | E, NA, P, AMX, T, SXT | 4 | 0·428 |
| S. Kentucky | E, NA, P, AMX, T | 2 | 0·357 |
| S. Kentucky | E, NA, P, AMX | 2 | 0·285 |
| S. Kentucky | E, NA, P | 2 | 0·214 |
| S. Kentucky | E | 2 | 0·071 |
| S. Muenster | E, NA, P, AMX,T, SXT, AM | 6 | 0·500 |
| S. Muenster | E, NA, P, AMX,T, SXT | 2 | 0·428 |
| S. Muenster | E, NA | 2 | 0·142 |
| S. Muenster | E | 4 | 0·071 |
| S. Virchow | E, NA, P, AMX, T, SXT, AM, S, N | 2 | 0·642 |
| S. Virchow | E, NA, P, AMX, T, SXT, AM, S | 4 | 0·571 |
| S. Virchow | E, NA, P, AMX,T, SXT, AM | 2 | 0·500 |
| S. Anatum | E, NA, P, AMX,T, SXT | 6 | 0·428 |
| S. Anatum | E, NA, P, AMX,T | 2 | 0·357 |
| S. Haifa | E | 2 | 0·071 |
| Salmonella spp. | E, NA, P, AMX, T | 2 | 0·357 |
| Salmonella spp. | E, NA | 2 | 0·142 |
| Average = 0·582 | |||
E (erythromycin, 15 μg); NA (nalidixic acid, 30 μg); P (penicillin, 10 IU); AMX (amoxicillin, 30 μg); T (oxytetracycline, 30 μg); SXT (sulphamethoxazole, 25 μg); AM (ampicillin, 10 μg); S (streptomycin,10 μg); N (neomycin,10 μg); C (chloramphenicol, 30 μg); NOR (norfloxacin, 10 μg); CP (ciprofloxacin, 5 μg); K (kanamycin, 30 μg); G (gentamicin, 10 μg).
Only 12 (7·2%) of the 166 isolates exhibited resistance to just one or two antimicrobials and these comprised the less frequently occurring serotypes, S. Haifa, S. Muenster and S. Kentucky.
DISCUSSION
Our results of the prevalence of Salmonella in whole chickens and drumsticks are in agreement with the prevalence rates of Salmonella in poultry meats recorded in different countries which range from 19·2% in fresh and frozen chicken carcasses in South Africa [17], 22% in Louisiana (USA) retail stores [18], and 27% in retail market broiler chicken carcasses in Colombia [19]. Higher Salmonella contamination rates have been reported from several studies ranging from 34% in Turkey [5] to 66% in Thailand [20]. By contrast, only 0·6% of 168 samples of meat parts of broiler chickens tested in an earlier survey in Turkey [21] were contaminated with Salmonella, while all of the 127 poultry carcasses tested in Brazil were negative for these organisms [22]. Similarly for chicken giblets (gizzards, liver, heart) reported rates vary depending on the survey country, notably 86% in Thailand [20], 34·5% and 41% of livers and gizzards, respectively, in Ethiopia [23] and 3% in Argentina [24].
The wide variation in Salmonella prevalence in chicken meat from different studies could be attributed to geographical differences, sampling techniques, bacteriological methods as well as slaughter hygiene and cross-contamination of products at different stages of chicken dressing and preparation. The observed greater contamination of gizzard and liver samples over other samples may reflect greater manipulation of these organs in addition to contamination from the crop and intestinal contents during evisceration. As a result of the control programme of Salmonella in chickens reared for meat production in the UK, the number of S. Enteritidis- and S. Typhimurium-infected breeding chicken flocks is currently very low owing to the introduction of strict control measures among which include management, cleaning and disinfection, hen vaccination, pest control, biosecurity, monitoring, and the potential use of other aids in the control of Salmonella [25]. Wider application of such programmes may therefore be beneficial in reducing contamination rates in some countries.
The invA gene has been widely used for the detection of Salmonella spp. in food samples and its presence is highly associated with other virulence genes such as the stn gene which contributes to the pathogenicity process, primarily diarrhoea [26]. In S. Typhimurium and S. Enteriditis the virulence plasmid is known to increase the growth rate of the microorganisms at sites beyond the intestine and aid colonization of deeper tissue. Our finding of spv genes among these serovars is therefore consistent with early reports of their association with highly invasive serovars.
Gizzard samples proved to be the most contaminated (30/50) of tissue samples and yielded each of the seven serovars detected from all samples. The absence of certain serovars such as S. Virchow, S. Anatum, and S. Haifa from some of the other sample types no doubt reflects their low overall frequency and perhaps less cross-contamination during preparation.
The predominance of S. Enteriditis and S. Typhimurium in this survey echoes the results of several other surveys of foodborne salmonellosis in the literature, albeit with differences in counties in the rates of these serovars [27]. However, low rates of S. Enteritidis (5·9%) were notable from Turkey [4], and 1·3% (1/73) from Brazil [22]; similarly low frequencies of S. Typhimurium have been reported from other surveys [17, 28]. S. Kentucky accounted for 10·8% of all our isolates which contrasts markedly with 59·5% and 41% in studies from the USA and Ireland, respectively [29, 30].
As expected, resistance to erythromycin, nalidixic acid and penicillin was almost universal and high resistance rates were evident for most of the antimicrobials tested which is consistent with the literature [5, 28, 31]. In the context of agents that would be considered for the treatment of diarrhoeal salmonellosis, it was surprising to find relatively poor levels of clear susceptibility to chloramphenicol (22%) norfloxacin (32·5%), and ciprofloxacin (36%). Although 78% of isolates were susceptible to gentamicin, this drug would generally be used parenterally and only for extraintestinal infections. The relatively lower rates of resistance to norfloxacin, ciprofloxacin, kanamycin and gentamicin could be attributed to their limited use in animal production. Our findings corroborate the widely held view that poultry is a major source of multidrug-resistant Salmonella, and underlines the value of antibiotic susceptibility surveys for selecting appropriate treatment options for salmonellosis caused by strains of poultry origin. The data also serve to highlight the need for implementation of antimicrobial stewardship programmes in developing countries, including Egypt, to optimize their use for treatment, and reduce the spread and development of antimicrobial-resistant strains.
In conclusion, we have demonstrated that a high proportion of chicken carcasses and giblets sold in Mansoura, Egypt were contaminated with Salmonella, predominantly S. Typhimurium and S. Enteritidis, the great majority of which were multidrug resistant. Hence, chicken meat and their products constitute a significant problem for public health and this calls for better antimicrobial stewardship to reduce the unnecessary use of antimicrobials in the food industry.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Cardinale E, et al. Epidemiological analysis of Salmonella enterica spp. enterica serovars Hadar, Brancaster and Enteritidis from humans and broiler chickens in Senegal using pulsed-field gel electrophoresis and antibiotic susceptibility. Journal of Applied Microbiology 2005; 99: 968–977. [DOI] [PubMed] [Google Scholar]
- 2.Buzby JC, Roberts T. The economics of enteric infections: human foodborne disease costs. Gastroenterology 2009; 136: 1851–1862. [DOI] [PubMed] [Google Scholar]
- 3.Capita R, et al. Occurrence of salmonellae in retail chicken carcases and their products in Spain. International Journal of Food Microbiology 2003; 81: 169–173. [DOI] [PubMed] [Google Scholar]
- 4.Tauxe R. Emerging foodborne diseases: an evolving public health challenge. Emerging Infectious Diseases 1997; 3: 425–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yildirim Y, et al. Incidence and antibiotic resistance of Salmonella spp. on raw chicken carcasses. Food Research International 2011; 44: 725–728. [Google Scholar]
- 6.Threlfall EJ. Antimicrobial drug resistance in Salmonella: problems and perspectives in food and water-borne infection. FEMS Microbiological Reviews 2002; 26: 141–148. [DOI] [PubMed] [Google Scholar]
- 7.Capita R, Alonso-Calleja C. Antibiotic-resistant bacteria: a challenge for the food industry. Critical Reviews in Food Science and Nutrition 2013; 53: 11–48. [DOI] [PubMed] [Google Scholar]
- 8.Porwollik S, et al. Characterization of Salmonella enterica subspecies I genovars by use of microarrays. Journal of Bacteriology 2004; 186: 5883–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rahn K, et al. Amplification of an invA gene sequence of Salmonella Typhimurium by polymerase chain reaction as a specific method of detection of Salmonella. Molecular and Cellular Probes 1992; 6: 271–279. [DOI] [PubMed] [Google Scholar]
- 10.Torpdahl M, et al. Genotypic characterization of Salmonella by multilocus sequence typing, pulsed-field gel electrophoresis and amplified fragment length polymorphism. Journal of Microbiological Methods 2005; 63: 173–184. [DOI] [PubMed] [Google Scholar]
- 11.Prager R, et al. Salmonella enterotoxin (stn) gene is prevalent among strains of Salmonella enterica but not among Salmonella bongori and other Enterobacteriaceae. FEMS Immunology & Medical Microbiology 1995; 12: 47–50. [DOI] [PubMed] [Google Scholar]
- 12.Gulig PA, et al. Molecular analysis of spv virulence genes of the Salmonella virulence plasmids. Molecular Microbiology 1993; 7: 823–830. [DOI] [PubMed] [Google Scholar]
- 13.International Organization for Standardization. Microbiology of food and animal feeding stuffs – horizontal method for the detection of Salmonella spp., 4th edn. ISO 6579:2002. The International Organization for Standardization, Geneva, Switzerland 2002 (http://www.aait.org.cn/web/images/upload/2013/07/11/201307111148308281.pdf). Accessed 14 June 2014. [Google Scholar]
- 14.Choo E, et al. Prevalence and genetic diversity of Bacillus cereus in dried red pepper in Korea. Journal of Food Protection 2007; 70: 917–922. [DOI] [PubMed] [Google Scholar]
- 15.Chiu CH, Ou JT. Rapid identification of Salmonella serovars in feces by specific detection of virulence genes, invA and spvC, by an enrichment broth culture multiplex PCR combination assay. Journal of Clinical Microbiology 1996; 34: 2619–2622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Supplement M100-S11. Villanova, PA, USA: 2001. [Google Scholar]
- 17.van Nierop W, et al. Contamination of chicken carcasses in Gauteng, South Africa, by Salmonella, Listeria monocytogenes and Campylobacter. International Journal of Food Microbiology 2005; 99: 1–6. [DOI] [PubMed] [Google Scholar]
- 18.Lestari SI, et al. Prevalence and antimicrobial resistance of Salmonella serovars in conventional and organic chickens from Louisiana retail stores. Journal of Food Protection 2009; 72: 1165–1172. [DOI] [PubMed] [Google Scholar]
- 19.Donado-Godoy P, et al. Prevalence of Salmonella on retail broiler chicken meat carcasses in Colombia. Journal of Food Protection 2012; 75: 1134–1138. [DOI] [PubMed] [Google Scholar]
- 20.Jaowapa J, et al. Occurrence of salmonellae in raw broilers and their products in Thailand. Journal of Food Protection 1994; 57: 808–810. [DOI] [PubMed] [Google Scholar]
- 21.Cetinkaya F, et al. Shigella and Salmonella contamination in various foodstuffs in Turkey. Food Control 2008; 19: 1059–1063. [Google Scholar]
- 22.de Freitas CG, et al. PCR multiplex for detection of Salmonella Enteritidis, Typhi and Typhimurium and occurrence in poultry meat. International Journal of Food Microbiology 2010; 139: 15–22. [DOI] [PubMed] [Google Scholar]
- 23.Molla B, Mesfin A. A survey of Salmonella contamination in chicken carcass and giblets in central Ethiopia. Revue de Medecine Veterinaire 2003; 154: 267–270. [Google Scholar]
- 24.Favier GI, et al. Prevalence, antimicrobial susceptibility and molecular characterization by PCR and pulsed field gel electrophoresis (PFGE) of Salmonella spp. isolated from foods of animal origin in San Luis, Argentina. Food Control 2013; 29: 49–54. [Google Scholar]
- 25.Defra. UK National Control Programme for Salmonella in chickens (Gallus gallus) reared for meat (broilers) (https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/183080/salmonella-broilers.pdf). Accessed 14 June 2014.
- 26.Chopra AK, et al. Cloning and expression of the Salmonella enterotoxin gene. Journal of Bacteriology 1987; 169: 5095–5100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foley SL, et al. Salmonella challenges: prevalence in swine and poultry and potential pathogenicity of such isolates. Journal of Animal Science 2008; 86: E149–E162. [DOI] [PubMed] [Google Scholar]
- 28.Dogru AK, et al. Serotype identification and antimicrobial resistance profiles of Salmonella spp. isolated from chicken carcasses. Tropical Animal Health and Production 2010; 42: 893–897. [DOI] [PubMed] [Google Scholar]
- 29.Parveen S, et al. Prevalence and antimicrobial resistance of Salmonella recovered from processed poultry. Journal of Food Protection 2007; 70: 2466–2472. [DOI] [PubMed] [Google Scholar]
- 30.Whyte P, et al. The prevalence and PCR detection of Salmonella contamination in raw poultry. Veterinary Microbiology 2002; 89: 53–60. [DOI] [PubMed] [Google Scholar]
- 31.Álvarez-Fernández E, et al. Prevalence and antimicrobial resistance of Salmonella serotypes isolated from poultry in Spain: comparison between 1993 and 2006. International Journal of Food Microbiology 2012; 153: 281–287. [DOI] [PubMed] [Google Scholar]