SUMMARY
During 2007–2010, 13 545 confirmed human verocytotoxin (VT)-producing Escherichia coli (VTEC) infections were reported in the European Union, including 777 haemolytic uraemic syndrome (HUS) cases. Clinical manifestations were reported for 53% of cases, 64% of which presented with diarrhoea alone and 10% with HUS. Isolates from 85% of cases were not fully serotyped and could not be classified on the basis of the Karmali seropathotype concept. There is no single or combination of phenotypic or genetic marker(s) that fully define ‘pathogenic’ VTEC. Isolates which contain the vtx2 (verocytotoxin 2) gene in combination with the eae (intimin-encoding) gene or aaiC (secreted protein of enteroaggregative E. coli) and aggR (plasmid-encoded regulator) genes have been associated with a higher risk of more severe illness. A molecular approach targeting genes encoding VT and other virulence determinants is thus proposed to allow an assessment of the potential severity of disease that may be associated with a given VTEC isolate.
Key words: Molecular classification, serogroup, seropathotype, VTEC, virulence factors
INTRODUCTION
Verocytotoxin-producing Escherichia coli (VTEC), also known as verotoxigenic E. coli, verocytotoxigenic E. coli, or Shiga toxin-producing E. coli (STEC), are characterized by the production of verocytotoxins (VTs). Disease symptoms associated with VTEC range from mild to severe bloody diarrhoea with haemorrhagic colitis (HC), haemolytic uraemic syndrome (HUS) and thrombocytopenia, although asymptomatic carriage has also been reported. Age is a risk factor with the most severe forms of disease occurring in children.
Enterohaemorrhagic E. coli (EHEC) were originally defined as a subset of VTEC that were associated with HC, and later as specific O groups (namely O157, O26, O145, O111, O121, O103, O28). In addition to the verocytotoxin production (vtx)-encoding genes, EHEC usually carry the attaching and effacing eae gene located in the locus of the enterocyte effacement (LEE) pathogenicity island (PAI) [1]. EHEC strains are typically isolated from cases of severe disease (i.e. HC and HUS), but are poorly defined because there is no commonly accepted definition of this pathotype.
The most prevalent VTEC serotype associated with human illness worldwide has been O157:H7, although an increasing number of infections due to non-O157 serotypes have been recorded in recent years. For example, about half of all confirmed cases of VTEC in the European Union (EU) in 2011 were reported as non-O157 [2]. The most common non-O157 VTEC serotypes from confirmed cases of human infection in the EU from 2007 to 2010 were O103:H2, O26:H11, O117:H7, O91:H–, O145:H–, O63:H6, O128:H2, O111:H–, and O146:H21, although infections with at least 200 other serotypes were notified [3]. Many other non-O157 VTEC serotypes have been identified in food-producing animals, but their capacity to cause disease in humans is unknown.
A further class of pathogenic E. coli referred to as ‘enteroaggregative’ E. coli (EAEC) are characterized by their ability to aggregatively adhere to tissue culture cells in a distinct ‘stacked and brick-like’ manner, which is mediated by aggregative adherence fimbriae (AAF). Such strains usually produce an enteroaggregative heat-stable toxin (EAST1), which is encoded by plasmid-borne astA genes [4]. In 2011 an E. coli O104:H4 outbreak resulted in 4321 confirmed cases of infection and 852 cases of HUS, with 54 deaths reported in 14 EU countries, the USA and Canada in 2011 [5–7]. Whole genome sequence analysis revealed the outbreak strain to be a unique combination of virulence factors typical of EAEC plus a VT2-encoding gene acquired by transformation [5, 8]. Thus the outbreak strain had a combination of classical VTEC and EAEC virulence markers, but lacked eae genes previously thought to be essential for the expression of high virulence capability in EHEC [8].
There is no single or combination of genetic marker(s) that defines the potential of a VTEC strain to cause human disease, as various bacterial and host-related factors contribute to the virulence of VTEC. Strains that produce Vtx2 are more likely to cause HUS than those that produce Vtx1 alone [9]. Furthermore the presence of the eae gene, or of genetic determinants associated with the capability of adhering to the intestinal mucosa with a stacked brick pattern, have both been found in VTEC strains causing HUS [9, 10].
Several strategies have been proposed to allow an assessment of the potential of different types of VTEC to cause disease. Karmali et al. [11] proposed a VTEC seropathotype classification based on serotype, reported frequency and severity of disease in humans and association with outbreaks. This classification system utilizes a gradient ranging from seropathotype A – high risk of causing severe symptoms of infection – to seropathotypes D and E – minimal risk. This seropathotype approach has been of considerable value in defining pathogenic VTEC serotypes of potential importance in cases of human infection [12–14] but was seriously challenged by the major outbreak of E. coli O104:H4 in 2011 [5].
The primary objectives of this paper are (a) to interrogate centrally collected EU data to determine which serogroups have been most commonly reported in confirmed VTEC cases in the EU over the 4-year period 2007–2010; (b) assess the distribution of vtx and eae genes in isolates of VTEC from cases of human infection in the EU over this period with a view to investigating the relationship between vtx type, the presence or absence of the eae gene and the severity of disease; (c) to evaluate the seropathotype concept in light of recent developments with VTEC infections in the EU, as exemplified by the increasing frequency of infections with non-O157 VTEC strains and the 2011 outbreak of VTEC O104:H4; and (d), to propose a new classification scheme based on the molecular virulence markers in VTEC.
METHODS
Case-based data from The European Surveillance System (TESSy data) were provided by the European Centre for Disease Prevention and Control (ECDC) from cases of human infection with VTEC reported in the EU from 2007 to 2010. Only laboratory-confirmed VTEC cases were selected as defined by the following criteria (1) isolation of an E. coli strain that produces VT or harbours vtx1 or vtx2 gene(s); (2) isolation of non-sorbitol-fermenting (NSF) E. coli O157; (3) direct detection of vtx1 or vtx2 gene(s) nucleic acid; (4) direct detection of free VT in faeces. These criteria may or may not fulfil the clinical criteria for VTEC diarrhoea (i.e. any person with at least the following two: diarrhoea or abdominal pain; and for HUS: any person with acute renal failure and at least one of the following two: microangiopatic haemolytic anaemia or thrombocytopenia) [3]. The information in the dataset included: the age of the patient, the clinical outcome (alive/dead), hospitalization (yes/no), HUS (yes/no), and clinical manifestation (bloody diarrhoea, diarrhoea, asymptomatic). Additional data included the characteristics of the VTEC isolate (O and H antigen, the presence or absence of eae, vtx1 and vtx2 genes and VT production). Data for VTEC cases occurring in 2011 was based on the information available in the EU Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2011 [2].
RESULTS
VTEC serogroups associated with confirmed reported cases in the EU together with the distribution of vtx and eae genes
The reported confirmed VTEC cases in humans in the EU from 2007 to 2010 by serogroup are listed in Table 1, together with the virulence genes (eae, vtx1, vtx2) found in these VTEC serogroups. In this period, 13 545 confirmed VTEC infections were reported to ECDC of which 9489 (77·0%) had the ‘O’ antigen reported. The most commonly reported serogroup was O157 (70·2% of cases with ‘O’ antigen determined) followed by O26 (8·2%) and O103 (3·9%).
Table 1.
The serogroups and vtx-eae gene characteristics of reported confirmed VTEC serogroups from cases of human infection from 2007–2010 (based on The European Surveillance System data as provided by the European Centre for Disease Prevention and Control)
| Serogroup | Total† | Subtotal‡ | Virulence* characteristics | |||||
|---|---|---|---|---|---|---|---|---|
| eae, vtx1 | eae, vtx2 | eae, vtx1, vtx2 | vtx1 | vtx2 | vtx1, vtx2 | |||
| O157 | 6658 | 5412 | 31 | 3833 | 1530 | 1 | 13 | 4 |
| O26 | 780 | 309 | 205 | 63 | 30 | 9 | 2 | 0 |
| O103 | 370 | 176 | 168 | 4 | 2 | 1 | 1 | 0 |
| O145 | 220 | 108 | 25 | 81 | 1 | 0 | 1 | 0 |
| O91 | 202 | 69 | 0 | 1 | 1 | 35 | 5 | 27 |
| O111 | 134 | 67 | 37 | 13 | 13 | 1 | 2 | 1 |
| O117 | 76 | 64 | 2 | 0 | 1 | 60 | 1 | 0 |
| O146 | 102 | 62 | 0 | 1 | 1 | 3 | 30 | 27 |
| O128 | 108 | 61 | 1 | 7 | 5 | 7 | 19 | 22 |
| O121 | 56 | 49 | 1 | 45 | 1 | 1 | 1 | 0 |
| O113 | 61 | 35 | 0 | 5 | 0 | 2 | 18 | 10 |
| O63 | 55 | 29 | 0 | 25 | 4 | 0 | 0 | 0 |
| O174 | 32 | 23 | 0 | 0 | 0 | 1 | 12 | 10 |
| O156 | 34 | 21 | 6 | 0 | 0 | 13 | 0 | 2 |
| O76 | 34 | 21 | 2 | 0 | 0 | 15 | 1 | 3 |
| O5 | 25 | 18 | 9 | 0 | 1 | 4 | 1 | 3 |
| O55 | 31 | 14 | 3 | 4 | 0 | 7 | 0 | 0 |
| O8 | 20 | 11 | 0 | 2 | 1 | 0 | 7 | 1 |
| O153 | 16 | 11 | 0 | 7 | 0 | 1 | 2 | 1 |
| O84 | 13 | 10 | 8 | 0 | 0 | 0 | 2 | 0 |
| O181 | 12 | 10 | 0 | 0 | 0 | 4 | 0 | 6 |
| Other§ | 450 | 231 | 26 | 51 | 10 | 42 | 80 | 22 |
| NT¶ | 4035 | 467 | 88 | 112 | 41 | 88 | 89 | 49 |
| Total | 13 524 | 7278 | 612 (8·4%) | 4254 (58·5%) | 1642 (22·6%) | 295 (4·1%) | 287 (3·9%) | 188 (2·6%) |
eae, Intimin-coding gene; vtx1, verocytotoxin 1 gene; vtx2, verocytotoxin 2 gene.
Includes all reported confirmed VTEC cases in humans.
Includes the reported confirmed VTEC cases in humans for which the vtx-eae gene characteristics are reported.
Includes other serogroups than already listed. This group includes 83 serogroups.
NT, Untyped/untypable and cases where ‘O’ antigen was reported as unknown.
The presence and type of vtx and eae genes were reported for isolates from 7278 (54%) of the 13 545 cases. About 60% of all isolates were eae- and vtx2-positive. Of the O157 isolates, 71% had the eae-vtx2 combination and 28% were eae-vtx1-vtx2 positive. About 76%, 97% and 75% of isolates from cases due to O23, O103 and O111, respectively, carried vtx1 (alone or in combination with vtx2) and eae while 11% of isolates did not carry the eae gene (770 of 7278) including all but two reported cases caused by serogroup O91 and the majority (>95%) of reported cases caused by O117 and O146 serogroups were also eae negative.
In 2011, 5301 confirmed VTEC cases of human infection were reported to ECDC of which 4506 had the ‘O’ antigen reported. The most commonly reported serogroup was O157 (48·5% of cases with ‘O’ antigen determined) followed by O104 (23·6%) and O26 (6·4%). One hundred and sixteen out of the 118 isolates of serogroup O104 were eae negative and vtx2 positive [2].
Relationships between vtx type, the presence and absence of the eae gene, and severity of disease
The clinical outcome of the 13 545 confirmed VTEC infections in humans from 2007 to 2010 was reported for only a fraction of cases as follows: (1) information on hospitalization was reported for 1315 cases (10%) only, of which 522 (or 39·7%) were hospitalized. This low reporting of hospitalizations can partly be explained because this parameter was only collected in 2009 and 2010; (2) the HUS status was reported for 7920 cases (59%) , of which 777 (9·8%) developed HUS; (3) the clinical manifestation (expressed as bloody diarrhoea, diarrhoea or asymptomatic) was reported for 53% of cases.
The percentage of cases, hospitalizations and HUS associated with specific eae-vtx gene combinations is shown in Table 2. Most hospitalized cases (86·3%) and HUS cases (89·2%), for which information was reported on virulence factors were either eae-vtx2 positive or eae-vtx1-vtx2 positive.
Table 2.
Virulence characteristics of reported confirmed VTEC cases in 2007–2010 including all cases, hospitalized cases only and haemolytic uraemic syndrome (HUS) cases only (based on The European Surveillance System data as provided by the European Centre for Disease Prevention and Control)
| Cases | Total† | Subtotal‡ | Virulence* gene detection | |||||
|---|---|---|---|---|---|---|---|---|
| eae, vtx1 | eae, vtx2 | eae, vtx1, vtx2 | vtx1 | vtx2 | vtx1, vtx2 | |||
| Hospitalized§¶ | 522 | 313 | 22 (7·0%) | 185 (59·1%) | 85 (27·2%) | 4 (1·3%) | 10 (3·2%) | 7 (2·2%) |
| HUS§ | 777 | 371 | 10 (2·7%) | 294 (79·2%) | 37 (10·0%) | 2 (0·5%) | 24 (6·5%) | 4 (1·1%) |
| All§ | 13 545 | 7278 | 612 (8·4%) | 4254 (58·5%) | 1642 (22·6%) | 295 (4·1%) | 287 (3·9%) | 188 (2·6%) |
eae, Intimin-coding gene; vtx1, verocytotoxin 1 gene; vtx2, verocytotoxin 2 gene.
Includes all reported confirmed VTEC cases in humans.
Includes the reported confirmed VTEC cases in humans for which the vtx-eae gene characteristics are reported.
The percentage (within parentheses) is calculated using the corresponding total number of cases (either 7218; 313 or 371) as denominator.
Data on hospitalizations have only been collected for the last two years (2009 and 2010).
Relationship between serogroup, age of the patient and occurrence of HUS
During 2007–2010, the age of the patients was provided for 584 HUS cases where the serogroup of the isolate had been reported. Overall, 64·2% of HUS cases were reported in children up to 4 years of age and 26·0% in the 5–14 years age group. Within these groups, VTEC O157 was identified in 65·7% of cases, followed by VTEC O26 (18·6%).
In 2011, a total of 1006 confirmed cases developed HUS [2, 8]. Of these, 318 were reported to be due to the E. coli O104:H4 outbreak strain. The VTEC serogroup was not reported for isolates from the majority of the 411 German HUS cases where the E. coli O104:H4 outbreak originated [4]. These are likely to have been caused by the outbreak strain, as 845 German HUS cases associated with the outbreak were reported elsewhere [8]. By age group (provided for 577 HUS cases with known serogroup), 28·1% of the HUS cases were reported in children up to 4 years of age, followed by 20·8% in the 25–44 years age group. E. coli O157 was the most commonly reported VTEC serogroup in the 0–14 years age group while O104 was the predominant serotype in confirmed cases in the remaining age groups (Fig. 1).
Fig. 1.
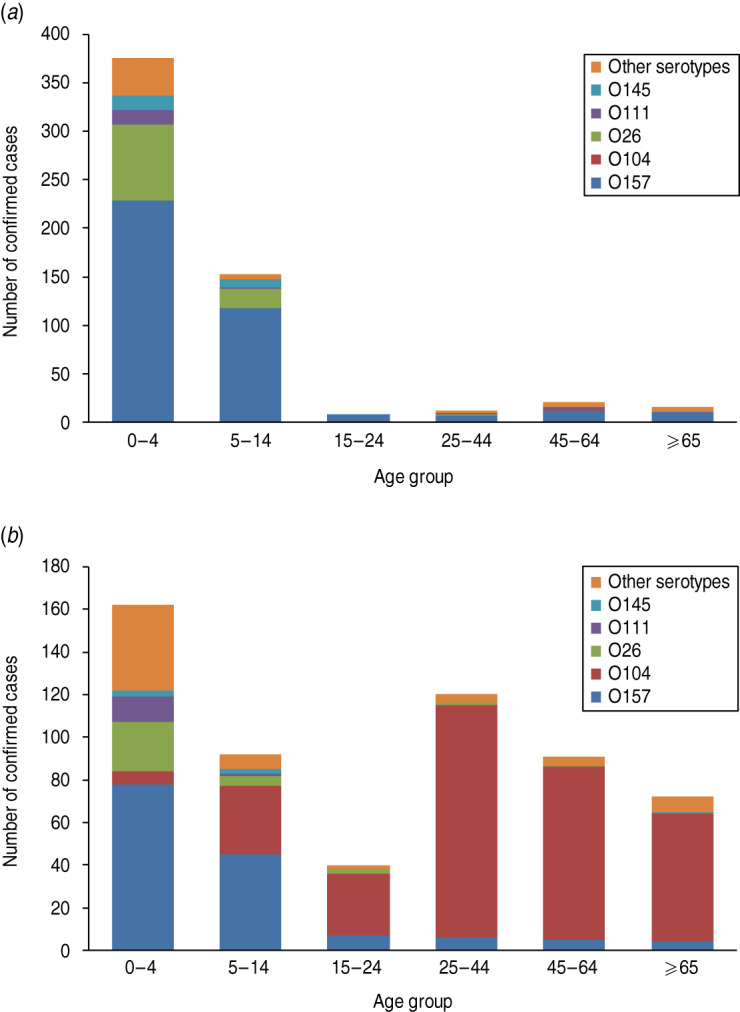
Haemolytic uraemic syndrome (HUS) by age and serogroup in reporting EU Member States. (a) 2007–2010 (n = 584) (based on TESSy data as provided by ECDC); (b) 2011 (n = 577) [2].
Evaluation of the seropathotype model
For the purposes of this paper the seropathotype model of Karmali et al. [11] has been evaluated using data on VTEC infections reported to ECDC over the period 2007–2010. Two approaches have been used, the original approach of Karmali et al. [11] (approach 1), and a modification of this approach (approach 2) based on the health outcome of reported and confirmed human VTEC cases.
Original Karmali seropathotype approach (approach 1)
The 13 545 confirmed VTEC infections reported to ECDC in the period 2007–2010 were assigned to seropathotypes in accordance to Karmali et al. [11]. The proportions of cases in the different categories are shown in Table 3. Of the 13 545 isolates, 11 488 (85%) were not fully serotyped and therefore could not be classified by seropathotype. Of the cases where isolates were fully serotyped, 1047 (51%) belonged to seropathotype A, 323 (16%) to seropathotype B, 24 (1%) to seropathotype C, 104 (5%) to seropathotype D and 14 (0·7%) to seropathotype E. The serotypes of 545 isolates (27%) were not considered by Karmali et al. [11] and therefore could not be included in approach 1. The majority of cases were associated with seropathotypes A and B and included 71% of the deaths, 76% of the hospitalizations, 86% of the HUS cases and 88% of cases with bloody diarrhoea.
Table 3.
Health outcome of reported confirmed* human VTEC cases during 2007–2010 as categorized by the seropathotype concept of Karmali et al. [11]. Based on The European Surveillance System data as provided by the European Centre for Disease Prevention and Control
| Sero-pathotype† | Total | Death | Hospitalization | HUS | Clinical manifestation | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Yes (%) | No | Yes (%) | No | Yes (%) | No | Bloody diarrhoea | Diarrhoea | Asymptomatic | ||
| A | 1047 [50·9] | 5 (1·23) [71·4] | 400 | 46 (59·0) [55·4] | 32 | 96 (16·3) [72·7] | 493 | 286 (52·1) [79·0] | 236 (43·0) [26·3] | 27 (4·9) [35·1] |
| B | 323 [15·7] | 0 (0) [0] | 283 | 17 (37·0) [20·5] | 29 | 18 (6·2) [13·6] | 273 | 31 (11·7) [8·6] | 222 (84·1) [24·7] | 11 (4·2) [14·3] |
| C | 24 [1·2] | 0 (0) [0] | 16 | 2 (100) [2·4] | 0 | 1 (5·6) [0·8] | 17 | 0 (0) [0] | 15 (93·8) [1·7] | 1 (6·3) [1·3] |
| D | 104 [5·1] | 0 (0) [0] | 80 | 3 (75·0) [3·6] | 1 | 0 (0) [0] | 88 | 5 (6·3) [1·4] | 70 (88·6) [7·8] | 4 (5·1) [5·2] |
| E | 14 [0·7] | 0 (0) [0] | 12 | 1 (100) [1·2] | 0 | 0 (0) [0] | 10 | 1 (9·1) [0·3] | 9 (81·8) [1·0] | 1 (9·1) [1·3] |
| NLK | 545 [26·5] | 2 (0·48) [28·6] | 414 | 14 (36·8) [16·9] | 24 | 17 (3·8) [12·9] | 430 | 39 (9·3) [10·8] | 346 (82·8) [38·5] | 33 (7·9) [42·9] |
| NFT | 11 488 | 11 (0·21) | 5296 | 439 (38·3) | 707 | 645 (10·0) | 5 832 | 1820 (31·5) | 3689 (63·8) | 275 (4·8) |
| Total | 13 545 | 18 (0·28) | 6501 | 522 (39·7) | 793 | 777 (9·8) | 7 143 | 2182 (30·6) | 4587 (64·4) | 352 (4·9) |
Confirmed cases are laboratory confirmed and may or may not fulfil the clinical criteria as described in the case definition. For the majority of these confirmed VTEC cases, the clinical outcome was not reported: the case fatality was not reported for 52% of these cases, hospitalization was not reported for 90% and haemolytic uraemic syndrome (HUS) status was unknown for 41%. The clinical manifestation (expressed as bloody diarrhoea, diarrhoea or asymptomatic) was not reported for 47% of the cases. Percentages of cases are given within parentheses based on rows ( ) and within square brackets based on columns [ ].
Seropathotype A includes O157:H7, O157:NM; seropathotype B includes O26:H11, O103:H2, O111:NM, O121:H19, O145:NM; seropathotype C includes includes O91:H21, O104:H21, O113:H21, O5:NM, O121:NM, O165:H25; seropathotype D includes O7:H4, O69:H11, O103:H25, O113:H4, O117:H7, O119:H25, O132:NM, O146:H21, O171:H2, O172:NM, O174:H8, Orough:H2; seropathotype E includes O6:H34, O8:H19, O39:H49, O46:H38, O76:H7, O84:NM, O88:H25, O98:H25, O113:NM, O136:H12, O136:NM, O153:H31, O156:NM, O163:NM; NLK, serotypes that were fully serotyped but were not listed by Karmali et al. [11]; NFT, strains that were not fully serotyped.
From the total cases assigned to seropathotype A and for which reported outcome was available, five (1·2%) were reported as fatal. Similarly two cases (0·5%) from the group fully serotyped but not listed by Karmali et al. [11] [subsequently listed in this communication as ‘Not Listed by Karmali’ (NLK)] had a reported fatal outcome. The fatal cases in this latter group were associated with strains typed as O105:H18 and O17:H41. Hospitalization rates for the different seropathotypes were: seropathotype A, 59%; B, 37%; C, 100% (only two cases identified); D, 75% (only three cases); E, 100% (only one case); and NLK, 37%. Similarly HUS and bloody diarrhoea was reported from: seropathotype A (16% and 52%, respectively); seropathotype B (6% and 12%); seropathotype C (6%, and 0%); seropathotype D (0% and 1·4%), seropathotype E (0% and 0·3%); and NLK (4% and 11%).
Of the 777 HUS cases, isolates from 645 cases (83%) were not fully serotyped and therefore could not be classified. Of the remaining 132 cases, 115 (87%) belonged to seropathotypes A–C and 17 (13%) were classified as NLK. The latter included serotypes O145:H28, 091:H10, O111:H8, O128:H2, O121:H2, O76:H19, O174:H21, O174:H2, O1:H42, O86:H27, O80:H2, O123:H2, O105:H18, and O7:H6.
‘Modified’ Karmali seropathotype approach (approach 2)
In order to be able to account for serotypes not listed by Karmali et al. [11], a modified approach (approach 2), based on the health outcome of confirmed VTEC cases reported in the EU has been developed. This second approach has been applied to the VTEC data reported to ECDC during 2007–2010.
Using approach 2, VTEC serotypes in seropathotype groups A–C were combined with those associated with HUS in the 2007-2010 data and designated ‘HUS-associated serotypes’ (HAS). In Table 4 the clinical outcome of the confirmed VTEC cases during 2007–2010 is shown and categorized by this approach.
Table 4.
Health outcome of reported confirmed* human VTEC cases during 2007–2010 as categorized based on the reported haemolytic uraemic syndrome (HUS) cases of human VTEC in the EU in 2007–2010 [grouped as HAS (A/B/C)]. Based on The European Surveillance System data as provided by the European Centre for Disease Prevention and Control
| Sero-pathotype† | Total | Death | Hospitalization | HUS | Clinical manifestation | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Yes (%) | No | Yes (%) | No | Yes (%) | No | Bloody diarrhoea | Diarrhoea | Asymptomatic | ||
| A/B/C (HAS) | 1340 [65·1] | 6 (0·94) [85·7] | 631 | 59 (49·2) [71·1] | 32 | 132 (15·6) [100·0] | 717 | 311 (40·6) [85·9] | 413 (53·9) [46·0] | 43 (5·6) [55·8] |
| D | 717 [34·9] | 1 (0·17) [14·3] | 574 | 24 (49·0) [28·9] | 0 | 0 (0) [0] | 594 | 51 (9·0) [14·1] | 485 (85·1) [54·0] | 34 (6·0) [44·2] |
| NFT | 11 488 | 11 (0·21) | 5296 | 439 (38·3) | 707 | 645 (10·0) | 5 832 | 1820 (31·5) | 3689 (63·8) | 275 (4·8) |
| Total | 13 545 | 18 (0·28) | 6501 | 522 (39·7) | 793 | 777 (9·8) | 7 143 | 2182 (30·6) | 4587 (64·4) | 352 (4·9) |
Confirmed cases are laboratory confirmed and may or may not fulfil the clinical criteria as described in the case definition. For the majority of these confirmed VTEC cases, the clinical outcome was not reported: the case fatality was not reported for 52% of these cases, hospitalization was not reported for 90% and HUS status was unknown for 41%. The clinical manifestation (expressed as bloody diarrhoea, diarrhoea or asymptomatic) was not reported for 47% of the cases. Percentages of cases are given within parentheses based on rows ( ) and within square brackets based on columns [ ].
HAS, HUS-associated serotypes. Includes the serotypes that have been associated with reported confirmed HUS cases of human VTEC in EU in 2007–2010: O157:H7, O157:H–, O121:H19, O26:H11, O174:H2, O111:H–, O145:H–, O145:H28, O1:H42, O128:H2, O111:H8, O104:H21, O174:H21, O7:H6, O76:H19, O80:H2, O86:H27, O121:H2, O123:H2, O105:H18, O91:H10; seropathotype D includes the serotypes that have been fully serotyped but have not been associated with the reported confirmed HUS cases of human VTEC in EU in 2007–2010; NFT, strains that were not fully serotyped.
Based on approach 2, 1340 (86% of cases where isolates were fully serotyped) cases have been classified as seropathotype group HAS. As the majority of cases with the most severe forms of the disease were associated with Karmali seropathotypes A/B/C, the HAS group now includes the serotypes causing 86% of the deaths, 71% of the hospitalizations, 100% of the HUS cases (due to the concept design) and 86% of the cases with bloody diarrhoea. Using this modified approach, when new information becomes available serotypes can be re-classified and the lists updated.
Molecular approach to VTEC strain classification
There are insufficient data to perform a quantitative risk assessment relating the presence of a particular combination of virulence genes and/or serogroup to any of the disease outcomes. Achieving a balance between specificity and sensitivity of virulence prediction is therefore difficult. The modified Karmali approach (approach 2) does not resolve the underlying problem with isolates that have not been fully serotyped. Additionally, and particularly when considering non-human isolates, a classification based solely on the presence of vtx genes is inadequate since there are numerous vtx-containing isolates belonging to serotypes that have not been associated with severe disease in humans. To overcome these problems an approach based on the presence of genes encoding specific virulence characteristics determinants in addition to the presence of vtx genes is proposed.
The additional virulence-associated genes suggested for this classification are eae (encoding intimin), aaiC (secreted protein of EAEC) and aggR (plasmid-encoded regulator gene). These genes are the best markers of the only two adherence factors that have been commonly associated with clinical isolates of VTEC.
This approach (approach 3) delivers a new scheme that describes the categorization of VTEC according to potential risk for public health. These risks have been categorized as group I (high risk for diarrhoea and HUS), group II (high risk for diarrhoea and unknown risk for HUS) through to group III (unknown risk) (see Table 5).
Table 5.
Proposed* molecular approach for the categorisation of VTEC (vtx present)
| Group | Genes† | Serogroups | Potential risk‡ | HUS/HC§ |
|---|---|---|---|---|
| Diarrhoea | ||||
| I | eae-positive or (aaiC and aggR)-positive | O157, O26, O103, O145, O111, O104 | High | High |
| II | eae-positive or (aaiC and aggR)-positive | Any other | High | Unknown |
| III | eae-negative and (aaiC plus aggR)-negative | Any other | Unknown | Unknown |
As yet this proposed molecular approach must be regarded as provisional. This is because screening VTEC for the presence of eae, aaiC and aggR is not routinely undertaken by all laboratories reporting data to The European Surveillance System.
Additional to the presence of vtx genes. eae, Intimin-coding gene; aaiC, chromosomally encoded gene encoding secreted protein of EAEC; aggR, plasmid-encoded regulator gene.
Needs epidemiological studies for confirmation.
HUS, Haemolytic uraemic syndrome; HC, haemorrhagic colitis.
The listed serogroups under group I reflect the five serogroups (O157, O26, O103, O145, O111), which, with the addition of serogroup O104 are generally recognized as those most frequently associated with clinical cases. The proposal for the inclusion of aaiC and aggR encoding genes is due to the 2011 E. coli O104:H4 outbreak (see above), which may be an exceptional event. Future surveillance will provide data that can be used to review the inclusion of aaiC and aggR and may identify new virulence markers that should be used in this molecular approach.
VTEC strains categorized into group I should be regarded as representing the higher risk. For VTEC that would fall under group II there remains uncertainty as to whether or not they are able to cause HUS. For VTEC falling into group III, there is uncertainty whether or not they are able to cause disease and we are unable to make a scientific judgement based on the present knowledge of virulence characteristics.
DISCUSSION
Microbiological and epidemiological data on cases of human infection with VTEC and reported in the EU from 2007–2010 would appear to broadly fit the Karmali seropathotype model in terms of incidence and severity. The major drawback of this classification system is the limited number (n = 39) of serotypes included, while more than 200 different serotypes have been reported to the ECDC in the period under examination. Thus, 27% of the confirmed VTEC infections that were reported to ECDC in this period could not be classified using the Karmali approach.
In the above assessment, incomplete serotype identification has resulted in the exclusion of 85% of reported VTEC cases. Such exclusions could give rise to a biased outcome. For example, about half of the isolates with missing ‘H’ antigen information are from cases of O157 infection (5610 cases). Assuming that these would have been typed as O157:H7 or O157:H–, many of these cases would have been assigned to seropathotype group A, thereby expanding this group to 6657 (87%) of cases, 78% of fatal cases, 91% of hospitalizations, 91% of HUS cases and 95% of cases with bloody diarrhoea.
There are also important exceptions reported in the scientific literature. Before the 2011 outbreak, E. coli O104:H4 had a low incidence in human disease and was not associated with outbreaks, and therefore would be classified as belonging to seropathotype D. In the 2011 outbreak, 3816 human cases likely to be due to VTEC O104:H4 were reported (including 54 deaths) and of those, 845 cases developed HUS [2]. In addition, of the six foodborne VTEC outbreaks where full serotyping data was reported during the period 2007–2011 [2, 15–17], only E. coli O157:H7 could be assigned to a seropathotype group (group A), as the other serogroups were not included in the list of Karmali et al. [11].
With the modified Karmali seropathotype approach (approach 2) 86% of fully serotyped VTEC cases were classified as seropathotype group HAS. The majority of infections with the most severe forms of the disease were associated with Karmali seropathotypes A/B/C. Under this scheme, the HAS group now includes the serotypes causing 86% of the deaths, 71% of the hospitalizations, 100% of the HUS cases (due to the concept design) and 86% of the cases with bloody diarrhoea.
Using approach 2, for cases where isolation of VTEC has been successful and full serotyping has been undertaken, all serotypes associated with severe disease are automatically categorized in the HAS group. This approach is limited by the lack of routine serotyping and may be superseded by increased application of data of the frequency of different virulence genes.
The molecular approach (approach 3), based on the presence of vtx in combination with eae or aaiC and aggR genes has the advantage of overcoming problems associated with the lack of flagella ‘H’ antigen typing information. Nevertheless, because screening VTEC for the presence of aaiC and aggR is not routinely undertaken by all laboratories reporting data to TESSy, this approach must be regarded as provisional. Similarly, information on the presence of eae-vtx is not always available. For example, during 2007–2010 such data were available for only 371/777 (47·7%) reported HUS cases. Additionally the whole virulence gene asset of putative pathogenic VTEC is not completely known. Furthermore VTEC eae, aaiC and aggR negative strains have been isolated from patients with diarrhoea and HUS. Further research is required to gain a more complete understanding of the different combinations of virulence factors capable of mediating disease in humans.
The model based on the detection of virulence-associated genes will require periodic revision in light of new epidemiological and microbiological information. If adopted, the performance of this proposed approach needs to be verified with well-characterized isolates from cases of human infection and from food-producing animals and foods. Routine surveillance that includes molecular testing for known/new virulence genes together with accurate reporting of clinical presentation will help to further classify VTEC strains according to risk.
ACKNOWLEDGEMENTS
The authors thank the members of the Biological Hazards Panel that adopted the EFSA Opinion on ‘VTEC-seropathotype and scientific criteria regarding pathogenicity assessment’ [EFSA Journal 2013; 11(4): 3138]. This paper is largely based on the mentioned Opinion prepared under the auspices of EFSA. W.M. and E.L. are employed by the European Food Safety Authority (EFSA). The present article is published under the sole responsibility of the authors and may not be considered as an EFSA scientific output. The positions and opinions presented in this article are those of the authors alone and are not intended to represent the views or scientific works of EFSA. The authors also thank the European Centre for Disease Prevention and Control (ECDC) for providing data extracted from The European Surveillance System – TESSy. The views and opinions of the authors expressed herein do not necessarily state or reflect those of the ECDC. The accuracy of the authors' statistical analysis and the findings they report are not the responsibility of ECDC. ECDC is not responsible for conclusions or opinions drawn from the data provided. ECDC is not responsible for the correctness of the data and for data management, data merging and data collation after provision of the data. ECDC shall not be held liable for improper or incorrect use of the data.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Clements A, et al. Infection strategies of enteric pathogenic Escherichia coli. Gut Microbes 2012; 3: 71–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.EFSA and ECDC (European Food Safety Authority, European Centre for Disease Prevention and Control). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2011. EFSA Journal 2013; 11: 3129, 250 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EFSA Panel on Biological Hazards (BIOHAZ). Scientific Opinion on VTEC-seropathotype and scientific criteria regarding pathogenicity assessment. EFSA Journal 2013; 11: 3138, 106 pp. [Google Scholar]
- 4.Nataro JP, Kaper JB. Diarrheagenic Escherichia coli. Clinical Microbiology Reviews 1998; 11: 142–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank C, et al. Large and ongoing outbreak of haemolytic uraemic syndrome, Germany, May 2011. Eurosurveillance 2011, 16: 2–4. [PubMed] [Google Scholar]
- 6.Buchholz U, et al. German outbreak of Escherichia coli O104:H4 associated with sprouts. New England Journal of Medicine 2011; 365: 1763–1770. [DOI] [PubMed] [Google Scholar]
- 7.Karch H, et al. The enemy within us: lessons from the 2011 European Escherichia coli O104:H4 outbreak. EMBO Molecular Medicine 2012; 4: 841–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank C, et al. Epidemic profile of Shiga-toxin-producing Escherichia coli O104:H4 outbreak in Germany – preliminary report. New England Journal of Medicine 2011; 365: 1771–1780. [DOI] [PubMed] [Google Scholar]
- 9.Friedrich AW, et al. Escherichia coli harboring Shiga toxin 2 gene variants: frequency and association with clinical symptoms. Journal of Infectious Diseases 2002; 185: 74–84. [DOI] [PubMed] [Google Scholar]
- 10.Bosilevac JM, Koohmaraie M. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Applied and Environmental Microbiology 2011; 77: 2103–2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karmali MA, et al. Association of genomic O island 122 of Escherichia coli EDL 933 with verocytotoxin-producing Escherichia coli seropathotypes that are linked to epidemic and/or serious disease. Journal of Clinical Microbiology 2003; 41: 4930–4940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Caprioli A, et al. Non-O157 Shiga toxin-producing Escherichia coli infections in Europe. Emerging Infectious Diseases 1997; 3: 578–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.EFSA (European Food Safety Authority). Scientific Opinion of the Panel on Biological Hazards (BIOHAZ) on monitoring of verotoxigenic Escherichia coli (VTEC) and identification of human pathogenic VTEC types. EFSA Journal 2007; 579, 1–61. [Google Scholar]
- 14.Coombes BK, Gilmour MW, Goodman CD. The evolution of virulence in non-O157 Shiga toxin-producing Escherichia coli. Frontiers in Microbiology 2011; 2: 1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.EFSA and ECDC (European Food Safety Authority, European Centre for Disease Prevention and Control). The Community Summary Report on Trends and sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in the European Union in 2008. EFSA Journal 2010; 8: 1496, 363 pp. [Google Scholar]
- 16.EFSA and ECDC (European Food Safety Authority, European Centre for Disease Prevention and Control). The European Union Summary Report on Trends and sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2009. EFSA Journal 2011; 9: 2090, 378 pp. [Google Scholar]
- 17.EFSA and ECDC (European Food Safety Authority, European Centre for Disease Prevention and Control). The European Union Summary Report on Trends and Sources of Zoonoses, Zoonotic Agents and Food-borne Outbreaks in 2010. EFSA Journal 2012; 10: 2597, 442 pp. [DOI] [PMC free article] [PubMed] [Google Scholar]


