SUMMARY
There has been a rapid rise in the prevalence of cases of monophasic Salmonella Typhimurium (mST) in both humans and farm animals, and it has been found in pigs, cattle and poultry. It is therefore vital to have a good understanding of how to efficiently detect infected farms. The objective of this project was to determine sample type sensitivity in the detection of Salmonella to detect infected groups of animals on both pig (breeder, grower and finisher sites) and cattle (beef and dairy) farms, using data collected from a study investigating farms that were positive for mST, and to explore any variation between different age groups and management practices. A Bayesian approach in the absence of a gold standard was adopted to analyse the individual and pooled faecal sample data collected from each epidemiological group on each of the farms. The sensitivity of pooled sampling depended on the prevalence of infection in the group being sampled, with a higher prevalence leading to higher sensitivity. Pooled sampling was found to be more efficient at detecting positive groups of animals than individual sampling, with the probability of a random sample from a group of animals with 5% prevalence testing positive being equal to 15·5% for immature pigs (3·6% for an individual faecal sample, taking into account the sensitivity and infection prevalence), 7·1% for adult pigs (1·2% for individual sampling), 30% for outdoor cattle (2% for individual sampling) and 34% for indoor cattle (1% for individual sampling). The mean prevalence of each epidemiological group was higher in outdoor farms than indoor for both pigs and cattle (mean within-farm prevalence of 29·4% and 38·7% for outdoor pigs and cattle, respectively, compared to 19·8% and 22·1% for indoor pigs and cattle)
Key words: Bayesian modelling, monophasic Salmonella, pooled sampling, Salmonella, surveillance
INTRODUCTION
Salmonellosis is one of the major causes of gastroenteritis in the UK and worldwide, and while cases of Salmonella Typhimurium and S. Enteritidis have been decreasing, there has been a rise, over the last 20 years, of monophasic strains of S. Typhimurium (mST), antigenic formula 1,4,[5],12:i:- [1, 2]. Along with the increase in human cases, there has also been a marked increase in mST from pigs in recent years, and mST has also been recovered from cattle and poultry [3, 4].
The rapid emergence of various mST strains to become among the most common serovars in pigs and humans in multiple countries shows that they have the potential to spread rapidly, compared to other serovars and strains of S. Typhimurium that were initially more common [3, 5]. This rapid emergence is reflected in the close genetic relationship of strains of related phage types, e.g. the most recent DT193 ampicillin, streptomycin, sulfonamide, and tetracycline ASSuT-resistant strains [3]. The mechanism for this rapid spread is unclear, but international trade in pigs and pig meat is likely [6]. It is also thought that mST strains may have the ability to partially evade the hosts’ immune response compared to other S. Typhimurium strains [2], thus promoting extensive spread within a farm, environmental contamination and onward dissemination by the usual epidemiological routes before the herd immunity effect becomes established.
In order to isolate Salmonella from a farm environment a selection of different samples can be collected: individual faecal samples, pooled faecal samples, dust and environmental samples. Initial work investigated the sensitivity in detection of Salmonella by collecting individual faecal samples from pigs, which were then analysed individually or pooled in differing quantities, in the laboratory [7, 8]. Pooling faecal samples achieved almost 100% sensitivity for pools with >50% prevalence of positive faecal samples [7] and fewer pooled samples were required in order to detect the same level of prevalence compared to individual rectal swabs, in breeding pigs [8]. For Salmonella detection in cattle, a previous study [9] reported that pooling up to 20 faecal samples provided a reliable method to detect Salmonella; however, a later study [10] found no difference between pooled and individual sampling in the detection of verocytotoxin-producing Escherichia coli O157 in cattle. Later work [11, 12] has demonstrated differences in the sensitivity of sample types in poultry, with pooled faecal samples reported to be more sensitive in detecting Salmonella than individual faecal sampling. The collection of pooled faecal samples is often more practical and cost-effective than collecting individual faecal samples from every animal present in an epidemiological group, and is less invasive than if samples are collected from the rectum, rather than from the floor. Previous studies considered each animal group as a whole, and no distinction was made between animals of different ages or the different management practices on the farms.
The objective of this project was to determine sample type sensitivity for the detection of Salmonella in groups of animals on both pig (breeder, grower and finisher sites) and cattle (beef and dairy) farms, using data collected from a study investigating farms that were positive for mST, and to explore any variation between different age groups and management practices. The positive samples collected therefore predominantly contained mST and the aim of the work was to identify effective sampling procedures for different farm types and housing situations.
METHODS
Farms positive for mST were identified using the Animal Health and Veterinary Laboratories Agency (AHVLA) surveillance database, contacted via their private veterinary surgeon and visited repeatedly over a 3-year period.
Epidemiological groups of animals were identified on each farm, and were groups of animals that were housed together or kept in the same pen/field, and which were related to stage of production, e.g. farrowing sow, weaners, calves, adults, etc. These were classified as adults (pig: boars, dry sows, farrowing sows, service area; cattle: bulls, calving pen, cows and calves, dry cows, milkers, sick cows) and immature (pig: finishers, growers, in-pig gilts, maiden gilts, sick pen, weaners; cattle: heifers, stores and steers, weaned calves, unweaned calves, youngstock). Animals were also categorized by management type: indoor pig, outdoor pig, indoor cattle and outdoor cattle. A total of 16 positive pig farms (seven indoor, nine outdoor) and 16 positive cattle farms (nine indoor, seven outdoor) were sampled on up to four sampling occasions, leading to a total number of epidemiological groups of 69, 112, 38 and 30 for indoor pigs, outdoor pigs, indoor cattle and outdoor cattle, respectively (Table 1).
Table 1.
Summary of the number of groups, and the number of pooled and individual samples taken from each species and housing status
| Farm no. | Species, housing status | Number of positive groups (immature) | Number of animals per group (range) | Mean samples per group (range) | |
|---|---|---|---|---|---|
| Individual | Pooled | ||||
| 1 | Pig indoor | 12 (10) | 345 (80–600) | 30 (15–60) | 21 (5–40) |
| 2 | Pig indoor | 5 (4) | 255·6 (18–600) | 67·6 (16–119) | 30·4 (4–45) |
| 3 | Pig indoor | 3 (3) | 106·7 (60–200) | 27·3 (12–40) | 4·3 (3–6) |
| 4 | Pig indoor | 4 (4) | 25 (15–30) | 24·8 (15–30) | 5 (5–5) |
| 5 | Pig indoor | 28 (17) | 157·5 (20–500) | 46·9 (10–62) | 22·2 (6–54) |
| 6 | Pig indoor | 16 (13) | 202·5 (20–600) | 38 (14–60) | 18·7 (2–43) |
| 7 | Pig indoor | 1 (1) | 300 (300–300) | 60 (60–60) | 13 (13–13) |
| 8 | Pig outdoor | 12 (12) | 50 (50–50) | 13·3 (10–30) | 1 (1–1) |
| 9 | Pig outdoor | 4 (4) | 100 (100–100) | 8 (8–8) | 2 (2–2) |
| 10 | Pig outdoor | 4 (4) | 100 (100–100) | 12 (12–12) | 2 (2–2) |
| 11 | Pig outdoor | 2 (2) | 500 (100–900) | 45 (30–60) | 10 (2–18) |
| 12 | Pig outdoor | 3 (3) | 433·3 (200–800) | 60 (60–60) | 8·7 (4–16) |
| 13 | Pig outdoor | 4 (2) | 107·8 (11–300) | 30·8 (11–60) | 7·8 (4–10) |
| 14 | Pig outdoor | 23 (8) | 65·7 (10–340) | 23·1 (6–60) | 8·5 (1–64) |
| 15 | Pig outdoor | 29 (16) | 44·9 (6–360) | 18·7 (6–60) | 4·1 (1–28) |
| 16 | Pig outdoor | 31 (0) | 68·1 (10–240) | 29·5 (6–102) | 5·1 (1–24) |
| 17 | Cattle indoor | 3 (1) | 82·3 (47–120) | 55·7 (47–60) | 17·7 (15–20) |
| 18 | Cattle indoor | 4 (2) | 5·5 (5–6) | 5·5 (5–6) | 2·5 (2–3) |
| 19 | Cattle indoor | 5 (2) | 24·6 (3–80) | 20·6 (3–60) | 6 (2–13) |
| 20 | Cattle indoor | 2 (1) | 90 (10–170) | 35 (10–60) | 9·5 (8–11) |
| 21 | Cattle indoor | 15 (4) | 60·9 (7–250) | 32·8 (7–120) | 7·2 (3–17) |
| 22 | Cattle indoor | 2 (0) | 55 (50–60) | 60 (60–60) | 15 (14–16) |
| 23 | Cattle indoor | 1 (0) | 120 (120–120) | 120 (120–120) | 20 (20–20) |
| 24 | Cattle indoor | 4 (1) | 63 (27–150) | 52 (28–84) | 12·5 (10–16) |
| 25 | Cattle indoor | 2 (1) | 92 (4–180) | 31 (1–61) | 6·5 (4–9) |
| 26 | Cattle outdoor | 76 (1) | 72·5 (25–120) | 42·5 (25–60) | 3·5 (1–6) |
| 27 | Cattle outdoor | 38 (0) | 60 (60–60) | 60 (60–60) | 6 (6–6) |
| 28 | Cattle outdoor | 532 (2) | 62·9 (14–130) | 48·7 (14–79) | 5·6 (3–12) |
| 29 | Cattle outdoor | 114 (1) | 88 (38–170) | 60 (60–60) | 8·7 (8–9) |
| 30 | Cattle outdoor | 228 (1) | 41·5 (7–120) | 31·8 (7–60) | 4·2 (2–14) |
| 31 | Cattle outdoor | 38 (1) | 43 (43–43) | 43 (43–43) | 13 (13–13) |
| 32 | Cattle outdoor | 114 (1) | 73·3 (60–80) | 44 (12–60) | 8·3 (8–9) |
Sample collection and analysis
Samples were collected in parallel for each epidemiological group present. The number of samples collected depended on the number of animals in each group, which varied between groups, with as many individual faeceal samples as animals in the group, up to a maximum of 60. Individual faecal samples (2 g) were incubated in 18 ml buffered peptone water (BPW); pooled floor faeces, collected using a moist 29 × 31 cm gauze swab (Robinson Healthcare, UK), were transported and incubated in 225 ml BPW. Samples were analysed for the presence of Salmonella using the ISO6579: Annex D method [13], using Rambach agar as the plating medium. Positive samples were serotyped according to the White–Kauffmann–Le Minor scheme [14] and phage-typed according to current versions of the Health Protection Agency phage-typing schemes (see [15, 16] for further identification).
Statistical methods
The Bayesian method of estimating the sensitivity of pooled and individual samples was adopted [17, 18], as used in a previous study [8]. In short, this method uses data on the number of positive pooled and individual samples to estimate the sensitivity of pooled sampling to detect a Salmonella-infected group of animals, relative to the within-group prevalence, and the sensitivity of individual samples. The specificity of the culture method was assumed to be 100% in line with previous studies [7, 8, 11, 12].
The data for herd i and pen/group j are the number of individual test-positive animals, yij out of nij tested and the number of positive pooled faecal samples xij out of mij tested. We assumed that the data {yij} and {xij} follow binomial distributions:
where ηincl, ηpool are the individual faecal sample and pooled faecal sample sensitivity, respectively, and πij is the prevalence of infection in herd i and group j.
The sensitivity of pooled faecal sampling to detect a Salmonella-infected group of animals was assumed to follow the same relationship vs. the within-group prevalence as that found in a previous study [7], i.e. that the sensitivity of a pooled faecal sample of weight w consisting of a proportion πof positive faecal samples is given by:
| (1) |
where C and ρ are parameters to be estimated, and determine the estimated sensitivity of the pooled sampling. C represents the concentration of Salmonella clusters in faeces and ρ is a parameter that relates the probability of successful culture to the concentration of Salmonella clusters in the sample. Equation (1) describes the dilution effect of mixing positive and negative samples, since it was found in a previous study [7] that the sensitivity of pooled sampling reduces as the proportion of positive samples in the pool reduces. This means that the value of ηpool will vary between groups according to πij. The weight of a pooled faecal sample was assumed to be 25 g.
The model was fitted to the observed sampling data using WinBUGS 3·1 (http://www.mrc-bsu.cam.ac.uk/software/bugs/the-bugs-project-winbugs/). Convergence was assessed by inspection of history plots for each parameter and running the model with several sets of starting values and testing convergence using Gelman–Rubin statistics, as implemented in WinBUGS 3·1. Final model estimates were produced using 10 000 iterations as a burn-in followed by 5000 iterations of the model.
As a Bayesian method, priors are required to be specified for each of the parameters to be estimated. In this case relatively uninformative priors were used to reflect the possibility that the sensitivity of detection of monophasic Salmonella in pigs may differ from that previously estimated for Salmonella [8], and that there are no previous studies estimating the sensitivity of pooled samples for Salmonella in cattle. The list of priors is given in Table 2.
Table 2.
Summary of the priors in the Bayesian model used to estimate the sensitivity of pooled and individual faecal sampling for detection of Salmonella
| Parameter | Description | Prior | Source |
|---|---|---|---|
| πij | Salmonella prevalence on farm i and epidemiological group j. | Beta(1,1) | Uninformative |
| ηincl | Sensitivity of individual faecal samples | Beta(1,1) | Uninformative |
| C | Salmonella concentration in pig faeces | Normal(5,0·05) | Mean similar to Arnold et al. [7], but greater uncertainty |
| Ρ | Parameter determining how test sensitivity varies with C | Normal(0·5,0·12) | Mean similar to Arnold et al. [7], but greater uncertainty |
Hypothesis testing
The sensitivity of individual and pooled sampling was compared for each species between adult and immature animals, and between indoor and outdoor animals. Similarly, within-farm prevalence was compared between indoor and outdoor animals for each species, and the mean pen/group prevalence was compared between immature and adult animals for each species.
The hypothesis tests related to comparisons of test sensitivity were performed by use of Deviance Information Criterion (DIC) [19], which is a Bayesian analogue of Akaike's Information Criterion. To assist in the interpretation of DIC, a DIC weight (wDIC) was calculated for each model being compared, which gives an estimate of the probability that each model is the best model for the data at hand, and is calculated according to
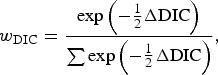 |
where ΔDIC was the difference between the model in question and the minimum value of DIC for the models being compared, and the denominator was the sum of the differences over all the models being compared. The best-fitting model out of those compared will be that with the highest DIC weight, and a value close to 1 indicates strong evidence that it is the best model. Using this approach, the following model comparisons were made:
Sample sensitivities vary between age group and housing status for both pigs and cattle (different estimates for C and ρ by age, and housing status for both pigs and cattle).
Sample sensitivities are the same for adults and young. To test this, models were fitted assuming common estimates for C and ρ for cattle and pigs by age group.
Sample sensitivities are the same for different housing status. To test this, models were fitted assuming common estimates for C and ρ for cattle/pigs for both indoor/outdoor (but retaining the model from step 2 with the lowest DIC regarding sensitivities with respect to age group by species).
RESULTS
The summary of the number of positive samples by species, age group, indoor/outdoor and whether pooled or individual is given in Table 3, and the summary of the number of epidemiological groups detected is given in Table 4. These results support the use of pooled sampling as an efficient sampling approach to detect infected groups of animals, with a much higher observed proportion of pooled samples positive compared to individual samples (Table 3). There were also differences in the proportion of samples positive between groups, especially between cattle and pigs for individual sampling.
Table 3.
Summary of the total number of faecal samples positive for Salmonella according to whether individual /pooled and the species sampled
| Species, indoor/outdoor | Number of samples positive/number tested | |||
|---|---|---|---|---|
| Individual | Pooled | |||
| Immature | Adult | Immature | Adult | |
| Pig indoor | 437/2040 (0·21) | 47/819 (0·06) | 462/901 (0·51) | 128/469 (0·27) |
| Pig outdoor | 472/1833 (0·26) | 51/786 (0·06) | 144/288 (0·50) | 54/289 (0·19) |
| Cattle indoor | 12/152 (0·08) | 74/1012 (0·07) | 43/67 (0·64) | 117/217 (0·54) |
| Cattle outdoor | 17/295 (0·06) | 116/424 (0·27) | 15/40 (0·38) | 29/51 (0·57) |
Table 4.
Summary of the number of groups tested and the number of groups positive for Salmonella, according to species, whether adult or immature, and whether pooled or individual faecal samples
| Species, indoor/outdoor | Number of groups positive/number tested | |||
|---|---|---|---|---|
| Individual | Pooled | |||
| Immature | Adult | Immature | Adult | |
| Pig indoor | 42/52 (0·81) | 11/17 (0·65) | 51/52 (0·98) | 17/17 (1) |
| Pig outdoor | 64/79 (0·81) | 26/33 (0·79) | 68/79 (0·86) | 23/33 (0·7) |
| Cattle indoor | 6/12 (0·5) | 15/26 (0·58) | 12/12 (1) | 26/26 (1) |
| Cattle outdoor | 5/7 (0·71) | 15/23 (0·65) | 6/7 (0·86) | 23/23 (1) |
The proportion of groups detected by pooled sampling was higher than for individual sampling for almost all species/indoor or outdoor/immature or adult combinations, except for adult outdoor pigs, despite there being far fewer pooled samples collected (Table 4).
Estimation of pooled and individual sampling sensitivity
Use of DIC indicated that the best-fitting model (model 4, Table 5) had common estimates of sensitivity between young and adult cattle, i.e. there was no evidence of a difference in the sensitivity of sampling between young and adult cattle. Models that assumed common sensitivity between the sensitivity of sampling indoor and outdoor cattle had a very low DIC weight, showing that there was strong evidence of a difference between the sensitivity of sampling between indoor and outdoor cattle. Therefore, the estimates of sensitivity for sampling of cattle were derived with data from all age groups amalgamated, i.e. including immature, adult and mixed groups together. For individual sampling, when all farms were included in the analysis, the individual sampling for outdoor cattle was estimated to be higher than that of indoor cattle, in contrast to the pooled sampling (where sampling indoor cattle had a higher sensitivity than outdoor cattle). However, this result was heavily influenced by one dairy farm which had a very high proportion of individual samples positive (a total of 83/120 individual samples positive and 15/17 pooled samples positive). The impact of this one farm on the estimates of sensitivity was investigated by repeating the analysis with this farm excluded, and it was found that for the remaining farms the estimate of the sensitivity of individual sampling for outdoor cattle was then lower than that of indoor cattle, i.e. consistent with the pooled sampling estimates.
Table 5.
Summary of results of model comparisons by use of Deviance Information Criterion (DIC) and the associated estimate that each model is the best for the estimation of sensitivity of pooled and individual sampling for cattle and pigs
| Model | Model parameters determining sensitivities for pooled and individual sampling | DIC | P (best model) | |
|---|---|---|---|---|
| Cattle | Pigs | |||
| 1 | Differ by housing and age | Differ by housing and age | 1804·3 | 0·01 |
| Test differences by age group | ||||
| 2 | Common by age, differ between indoor/outdoor | Differ by age and by indoor/outdoor | 1799·7 | 0·13 |
| 3 | Common by age, differ between indoor/outdoor | Common by age, differ between indoor/outdoor | 1813·4 | 0·0001 |
| Test differences by housing status | ||||
| 4 | Differ between indoor/outdoor (common by age) | Common between indoor/outdoor (differ by age) | 1795·9 | 0·86 |
| 5 | Common between indoor/outdoor (common by age) | Common between indoor/outdoor (differ by age) | 1809·4 | 0·001 |
There was strong evidence of difference between the sensitivity of sampling between immature and adult pigs (model 2 vs. model 3, Table 5, showing a very low DIC weight for a model assuming common sensitivity between immature and adult pigs). The sensitivity of individual sampling for immature pigs was much higher than that of adult pigs (Table 6).
Table 6.
The estimated sensitivity of individual faecal sampling for detection of Salmonella in pigs and cattle according to age and whether indoor/outdoor
| Species | Housing status | Age | Bayesian estimates | |
|---|---|---|---|---|
| Median | 95% CrI | |||
| Pig | Indoor/outdoor | Immature | 0·72 | 0·65–0·8 |
| Pig | Indoor/outdoor | Adult | 0·24 | 0·18–0·3 |
| Cattle* | Outdoor | Immature/adult | 0·58 | 0·46–0·72 |
| Cattle | Indoor | Immature/adult | 0·40 | 0·32–0·5 |
| Cattle† | Outdoor | Immature/adult | 0·20 | 0·15–0·28 |
CrI, Credibility interval.
Includes one farm with an extremely high prevalence of positive samples.
Excludes one farm with an extremely high prevalence of positive samples.
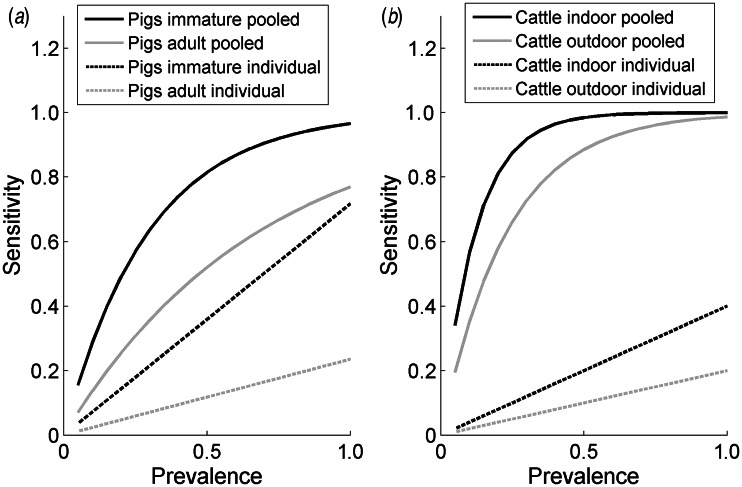
The estimates of the sensitivity of pooled and individual sampling confirm the efficiency of pooled sampling as a method of detection of Salmonella in groups of animals for both species (Fig. 1), as there is a much greater likelihood per sample of detecting Salmonella in a pooled sample compared to an individual sample (the likelihood of a positive individual sample in Fig. 1 is assumed to be the product of the within-group prevalence and the individual sample sensitivity). For example, the probability of a random sample from a group of animals with 5% prevalence testing positive was equal to 15·5% for immature pigs (3·6% for an individual faecal sample, taking into account the sensitivity and infection prevalence), 7·1% for adult pigs (1·2% for individual sampling), 30% for outdoor cattle (2% for individual sampling) and 34% for indoor cattle (1% for individual sampling). The greater per sample probability of detecting Salmonella in a pooled sample compared to an individual sample is estimated to be particularly high for cattle, as the sensitivity of individual sampling for cattle is relatively low.
Fig. 1.
The estimated per sample sensitivity of individual and pooled sampling to detect a given within-group prevalence of Salmonella in (a) pigs and (b) cattle according to age (pigs) and housing status (indoor/outdoor, cattle).
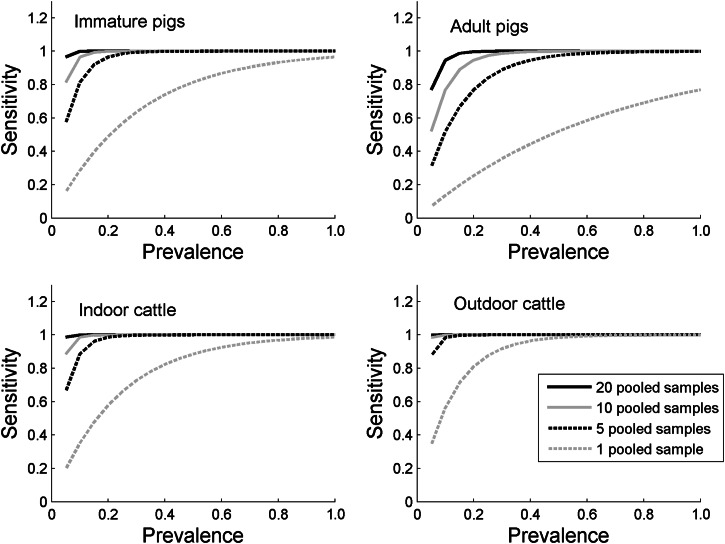
Figure 2 shows the impact of increasing the number of pooled samples on the probability of detecting Salmonella in a group of animals. As the number of pooled samples is increased, the probability of detecting low-prevalence Salmonella increases, and 10 pooled samples has a >80% chance of detecting a group of animals with a 5% prevalence in both indoor and outdoor cattle, and immature pigs, although only a 52% probability of detecting Salmonella in adult pigs. A much greater number of individual samples would be required for equivalent likelihood of detection; for example, assuming a large herd, to detect a 5% prevalence in a group of indoor cattle with an 80% chance would require 80 individual samples, with 44 individual samples being required for immature pigs.
Fig. 2.
The estimated sensitivity of 1, 5, 10 and 20 pooled samples to detect Salmonella according to the prevalence of infection in the pen/group of animals, and according to species, age and housing status.
Within-group and within-farm prevalence
The mean within-farm prevalence (estimated for each farm from the mean of the prevalence of infection in each group/pen in that farm) was higher in outdoor farms than indoor for both pigs and cattle (mean within-farm prevalence of 29·4% and 38·7% for outdoor pigs and cattle, respectively, compared to 19·8% and 22·1% for indoor pigs and cattle). However, there was large between-farm variability in the within-farm prevalence, with some low-prevalence outdoor farms and high-prevalence indoor farms (see online Supplementary Figs S1–S3).
There was little difference in the mean pen/group prevalence between immature and adult animals for pigs (indoor pigs: 20·7% adult, 25·1% immature; outdoor pigs: 30·9% adult, 32·0% immature), or for indoor cattle (29·0% adult, 34·0% immature). The within-group prevalence was similarly variable as for pigs (Supplementary Figs S1, S4, S5). For outdoor cattle, there was greater mean group prevalence for adult cattle (45·4%) compared to immature (11·7%), although the difference was largely due to one dairy farm which had a particularly high prevalence in adults in the herd.
DISCUSSION
The results show a clear benefit of pooled sampling compared to individual sampling, in line with previous studies looking at pooled sampling in pigs [7, 8], as there was a much greater proportion of pooled samples positive compared to individual samples, and a far greater proportion of infected groups detected for pooled sampling, despite fewer pooled samples being taken compared to individual samples.
Immature pigs are often grouped in larger numbers than adult pigs. This may have an impact on the pooled sampling as pools from the immature pigs will include faeces from a larger selection of pigs than the pooled samples collected from the adult pigs. Younger pigs are also likely to shed higher numbers of Salmonella organisms because of more recent exposure to infection and their more limited immune response [20].
The increased sensitivity of samples collected from indoor cattle compared to those collected from outdoor cattle is likely to be due to the easily accessible pooled fresh faecal material in communal farm areas on indoor farms [21]. Cattle kept on grass range over a large area and therefore locating fresh faecal samples can be more difficult. Pooled faecal samples may be collected from around water/feed troughs so may contain soil as well as faecal material; however, Salmonella can survive in soil for significant periods [22] and these pooled samples resulted in a higher sensitivity than individual faeces collected from the same grazing herds.
The lack of any evidence for a difference between detection results from the immature and adult cattle may be due to younger animals being kept in smaller groups and on straw, making sampling more difficult than for adult cattle groups despite a greater likelihood of infection and high-level shedding in younger animals [23]. An earlier study [9] reported that pooling of individually collected cattle faeces produced reliable results when there were ≤20 animals per group, but in naturally pooled faeces from a large number of animals the chance of inclusion of material from a high-level shedder in the pool is increased [24]. In the present study the cattle, especially milking herds and beef cattle, were often kept in larger groups (maximum 250, mean 59), and pooled samples were consistently more sensitive than individual samples. This difference is likely to be in part due to the collection of naturally pooled faecal samples using a hand swab, which has been shown to be more sensitive than pooling faecal picks [25] rather than the artificial pooling of collected individual samples.
Unlike previous studies of the sensitivity of pooled sampling in pigs, this study was able to compare the sensitivity of sampling types between immature and adult pigs in the same study. The results indicate an important difference between them, with a much higher sensitivity of faecal sampling in young pigs, suggesting that fewer pooled samples would be needed to detect Salmonella in young pigs compared to adult pigs. This is evident in Figure 2, where 10 pooled samples would have a >80% chance of detecting a 5% prevalence in immature pigs compared to only a 50% chance in adult pigs. This suggests that the sensitivity of sampling carried out in the EU baseline survey of breeding pigs [25, 26] is also likely to have been low in many cases.
The Bayesian median estimate of the sensitivity of pooled sampling for adult pigs (both pooled and individual samples), at around 70%, was lower in the present study compared to previous studies [7, 26], where at 100% prevalence a pooled sample was estimated to have ~90% sensitivity. However the credible interval for the estimate of pooled sample sensitivity at 100% prevalence was relatively wide, with an upper 97·5 percentile of 85%, i.e. close to the Bayesian median from previous studies, and so the results from the present study are consistent with findings from previous studies based on naturally infected pigs, with respect to pooled sampling of adult pigs. The sensitivity of individual samples was also lower in the present study compared to that found in an earlier study [8], where the individual sampling was estimated to have a median estimate of 79%. This may be partly due to differences in the amount of faeces cultured for each individual sample between the two studies (2 g vs. 10 g) [27], and the fact that in the former study the pooled faecal samples were created artificially by combining carefully measured aliquots of positive and negative faeces from highly positive farms. However, the naturally pooled faeces in the current study would have combined material from more than 30 pigs in most cases so the amount of material per contributing pig would have been very low, and variable. This suggests that sample weight was not the most critical factor in this case, but may have been more important for pooled faeces as the chance of inclusion of material from individual positive animals in the pool would be reduced [28]. There may, however, be negative aspects of culturing large volumes of faeces, as this may increase the competitive environment and lead to overgrowth of target organisms by competitors. This is thought to be the greatest limitation to detection and is farm or epidemiological group dependent [29]. The sensitivity of pooled sampling may also be adversely affected by a very low within-herd prevalence of low Salmonella counts, but in this situation a large number of individual samples would need to be tested for efficient detection, which is likely to be impractical in most cases [30].
The estimates of probability that a truly infected animal gives a positive test result for Salmonella from the sampling of individual faeces are generally very low (Table 6). This is a reflection of the intermittent shedding of Salmonella by healthy carrier animals [31], the non-uniform and clustered distribution of the organism within a faecal dropping, meaning that the portion of the dropping that is sampled may not contain any Salmonella [7, 27] and the detection sensitivity of the test used, which is always imperfect [32, 33] Test sensitivity varies according to the numbers of Salmonella present (typically low for most healthy carriers) in the sample [34] and the level of competing bacteria, particularly other Enterobacteriaceae [29].
There has been a previous study comparing the sensitivity of culture for faecal sampling with that of ELISA and PCR [33]. This found a sensitivity of >90% for faecal culture for both pigs and cattle. However, the study was based on artificially contaminated samples, so it is difficult to assess its relevance to sampling faeces from infected pigs.
The statistical model to estimate within-group prevalence was based on assuming a binomial distribution for the number of observed positive samples. This is appropriate when the group size is much larger than the number of samples being taken. For the case of sampling individual faeces, this means that the number of individual faeces in the pen, house or field being sampled needs to be much greater than the number of faeces sampled. The number of individual faeces was usually greater than the number of animals, except where new bedding had been placed or where the faeces were rapidly trodden into the bedding, and there could be up to ten times as many individual faeces identifiable as there were animals. Therefore, while there may be a few cases where hypergeometric sampling would provide a better description of the data, we believe that binomial sampling is a good approximation in most pens.
The farms in the present study were known Salmonella-positive farms, so the mean within-herd prevalence estimates are likely higher than those for infected pig and cattle farms in the population. This also means that there is likely to be a higher proportion of positive samples than would be found on random positive farms. However, the group-level prevalence estimates were highly variable, ranging from 1% to 98%.
In conclusion, while differences in the detection of Salmonella were observed between different age groups of pigs and different management types for cattle, pooling of faecal samples is a more sensitive and effective method to determine the Salmonella status of both pig and cattle herds than the analysis of individual faeces.
ACKNOWLEDGEMENTS
This study was supported by Department for Environment, Farming and Rural Affairs (Defra) project grant OZ0342.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268814002453.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Mossong J, et al. Outbreaks of monophasic Salmonella enterica serovar 4,[5],12:i:- in Luxembourg, 2006. Eurosurveillance 2007; 12: E11–E12. [DOI] [PubMed] [Google Scholar]
- 2.Switt AIM, et al. Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i. Foodborne Pathogens and Disease 2009; 6: 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.European Food Safety Authority (EFSA). Scientific Opinion on monitoring and assessment of the public health risk of ‘Salmonella Typhimurium-like’ strains. EFSA Journal 2010; 8: 1826. [Google Scholar]
- 4.Hauser E, et al. Pork Contaminated with Salmonella enterica Serovar 4,5,12:i:-, an emerging health risk for humans. Applied Environmental Microbiology 2010; 76: 4601–4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Centre for Disease Control (CDC). Salmonella surveillance: annual summary, 2006. US Department of Health and Human Services Centers for Disease Control 2008. (http://www.cdc.gov/ncidod/dbmd/phlisdata/Salmonella.htm).
- 6.Health Protection Agency. National increase in Salmonella Typhimurium PT U323. Health Protection Weekly Report 2012; 6: Issue 40 (http://www.hpa.org.uk/hpr/archives/2012/news4012.htm#stu323). [Google Scholar]
- 7.Arnold ME, et al. A modelling approach to estimate the sensitivity of pooled faecal samples for isolation of salmonella in pigs. Journal of the Royal Society Interface 2005; 2: 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnold ME, Cook AJC. Estimation of sample sizes for pooled faecal sampling for the detection of Salmonella in pigs. Epidemiology and Infection 2009; 137: 1734–1741. [DOI] [PubMed] [Google Scholar]
- 9.Kivela SL, et al. Pooled faecal samples compared with individual samples for detection of Salmonella in cattle. Bovine Practitioner 1999; 33: 74–75. [Google Scholar]
- 10.Arnold ME, et al. Investigation into the effectiveness of pooled fecal samples for detection of verocytotoxin-producing Escherichia coli O157 in cattle. Journal of Veterinary Diagnostic Investigation 2008; 20: 21–27. [DOI] [PubMed] [Google Scholar]
- 11.Arnold ME, et al. A comparison of pooled and individual bird sampling for detection of salmonella in commercial egg laying flocks. Preventive Veterinary Medicine 2011; 99: 176–184. [DOI] [PubMed] [Google Scholar]
- 12.Arnold ME, Carrique-Mas JJ, Davies RH. Estimation of the sensitivity of environmental sampling for detection of Salmonella Enteriditis in commercial egg-laying flocks relative to the within-flock prevalence. Epidemiology and Infection 2010; 138: 330–339. [DOI] [PubMed] [Google Scholar]
- 13.Anon. Detection of Salmonella spp. in animal faeces and in samples of the primary production stage. Draft amendment Annex D 2005; ISO 6579:2002/DAM.ISO 2005.
- 14.Grimont PAD, Weill F.-X. Antigenic formulae of the Salmonella serovars. WHO Collaborating Centre for Reference and Research on Salmonella; 2007, Paris, France. [Google Scholar]
- 15.Anderson ES, et al. Bacteriophagetyping designations of Salmonella Typhimurium. Journal of Hygiene 1977; 78: 297–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bale JA, et al. Kauffmann-White scheme – 2007. Salmonella identification – serotypes and antigenic formulae. London: Health Protection Agency, 2007. [Google Scholar]
- 17.Branscum AJ, Gardner IA, Johnson WO. Bayesian modeling of animal- and herd-level prevalences. Preventive Veterinary Medicine 2004; 66: 101–112. [DOI] [PubMed] [Google Scholar]
- 18.Branscum AJ, Johnson WO, Gardner IA. Sample size calculations for disease freedom and prevalence estimation surveys. Statistics in Medicine 2006; 25: 2658–2674. [DOI] [PubMed] [Google Scholar]
- 19.Spiegelhalter DJ, et al. Bayesian measures of model complexity and fit. Journal of the Royal Statistical Society, Series B 2002; 64: 583–640. [Google Scholar]
- 20.Pires A, Funk JA, Bolin CA. Longitudinal study of Salmonella shedding in naturally infected finishing pigs. Epidemiology and Infection 2013; 141: 1928–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lombard JE, et al. Comparison of individual, pooled, and composite fecal sampling methods for detection of Salmonella on US dairy operations. Journal of Food Protection 2012; 75: 1562–1571. [DOI] [PubMed] [Google Scholar]
- 22.Semenov AV, van Overbeek L, van Bruggen AH. Percolation and survival of Escherichia coli O157: H7 and Salmonella enterica serovar Typhimurium in soil amended with contaminated dairy manure or slurry. Applied Environmental Microbiology 2009; 75: 3206–3215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen LR, Kudahl AB, Østergaard S. Age-structured dynamic, stochastic and mechanistic simulation model of Salmonella Dublin infection within dairy herds. Preventive Veterinary Medicine 2012; 105: 59–74. [DOI] [PubMed] [Google Scholar]
- 24.Fegan N, et al. Quantification and prevalence of Salmonella in beef cattle presenting at slaughter. Journal of Applied Microbiology 2004; 97: 892–898. [DOI] [PubMed] [Google Scholar]
- 25.EFSA. Analysis of the baseline survey on Salmonella in holdings with breeding pigs in the EU, in 2008. Part B: Factors associated with Salmonella pen positivity. EFSA Journal 2011; 9: 2329. [Google Scholar]
- 26.EFSA. Analysis of the baseline survey on the prevalence of Salmonella in holdings with breeding pigs in the EU, 2008, Part A: Salmonella prevalence estimates. EFSA Journal 2011; 7: 1377. [Google Scholar]
- 27.Funk JA, Davies PR, Nichols MA. The effect of fecal sample weight on detection of Salmonella enterica in swine feces. Journal of Veterinary Diagnostic Investigation 2000; 12: 412–418. [DOI] [PubMed] [Google Scholar]
- 28.Cannon RM, Nicholls TJ. Relationship between sample weight, homogeneity, and sensitivity of fecal culture for Salmonella enterica. Journal of Veterinary Diagnostic Investigation 2002; 14: 60–62. [DOI] [PubMed] [Google Scholar]
- 29.Baggesen DL, et al. Growth inhibitory factors in bovine faeces impairs detection of Salmonella Dublin by conventional culture procedure. Journal of Applied Microbiology 2007; 103: 650–656. [DOI] [PubMed] [Google Scholar]
- 30.Singer RS, et al. Use of pooled samples for the detection of Salmonella in feces by polymerase chain reaction. Journal of Veterinary Diagnostic Investigation 2006; 18: 319–325. [DOI] [PubMed] [Google Scholar]
- 31.Belœil PA et al. Risk factors for Salmonella enterica subsp. enterica shedding by market-age pigs in French farrow-to-finish herds. Preventive Veterinary Medicine, 2004; 63: 103–120. [DOI] [PubMed] [Google Scholar]
- 32.Davies PR et al. comparison of methods for isolating Salmonella bacteria from faeces of naturally infected pigs. Journal of Applied Microbiology 2000; 89: 169–177. [DOI] [PubMed] [Google Scholar]
- 33.Ericksson E, Aspan A. Comparison of culture, ELISA and PCR technology for Salmonella detection in cattle, pigs and poulty. BMC Veterinary Research 2007; 3:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gopinath S et al. Shedding light on Salmonella carriers. Trends in Microbiology 2012; 20: 320–327. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S0950268814002453.
click here to view supplementary material