SUMMARY
Sapoviruses (SaVs) are responsible for sporadic cases and outbreaks of acute gastroenteritis. Despite this, few studies in India have focused on the epidemiological investigation of SaV in cases of acute gastroenteritis. The aim of this study was to understand the molecular epidemiology, genetic diversity and clinical impact of SaV in diarrhoeic children from Pune, Western India. Between 2007 and 2011, a total of 985 faecal samples from diarrhoeic cases and non-diarrhoeic controls were collected and examined for the presence of SaV by nested RT–PCR. SaV was detected in 2·7% (21/778) of the cases and 1·9% (4/207) of the controls. We observed that the majority of SaV mono-infections caused severe gastroenteritis (67%) with clinical manifestations of diarrhoea (100%), vomiting (73%) and dehydration (80%). All known human SaV genogroups were detected in the study. At least eight genotypes were identified from cases and controls. Genogroups GIV and GV, along with genotypes GI.5, GII.4 and GII.6, were discovered for the first time in India. Two GII.4 study strains were found to be 98·5–99% identical, having a novel intra-genogroup recombinant (GII.1/GII.4) recently reported from the Philippines, suggesting probable evidence of recombination. The circulation pattern of SaV genotypes varied during the study period, with GII.1 being predominant in 2007 and 2009, GIV.1 in 2008, and GV.1 in 2011.
Key words: Acute gastroenteritis, genetic diversity, sapovirus, severe dehydration, Western India
INTRODUCTION
Acute gastroenteritis (AG) is one of the leading causes of morbidity and mortality worldwide, with ~70% of the cases attributed to viruses. Rotavirus (RV) is the foremost causative agent of viral gastroenteritis [1]. Sapovirus (SaV), a member of the family Caliciviridae, is known to cause diarrhoea in humans and animals. The family Caliciviridae is divided into five genera: Norovirus, Sapovirus, Vesivirus, Lagovirus and Nebovirus [2]. Of these, certain genogroups of norovirus (NoV) and SaV are known to infect humans. SaV infections are generally milder than NoV infections; however, severe cases and outbreaks of SaV have also been reported [3].
The non-enveloped SaV capsid encloses a single-stranded positive-sense RNA genome, flanked by Vpg at the 5′-end and a poly A tail at the 3′-end. It is ~7·1–7·7 kb in size. SaV strains are classified into five genogroups (GI–GV) on the basis of nucleotide sequence variation in the capsid region [4]. In these genogroups, GI, GII, GIV and GV are responsible for infection in humans, while GIII infects porcine hosts. In addition to these, nine new groups are proposed (GVI–GXIV), which cause infections in pigs, bats, minks and sea lions [5]. The number of open reading frames (ORFs) in the SaV genome varies between genogroups. However, irrespective of the genogroup, ORF1 codes for at least six of the non-structural proteins and the major structural capsid protein (VP1). The genome of GII and GIII strains consists of two ORFs [2]. A third ORF, the function of which is still unknown, has been predicted in GI, GIV and GV. The GI and GII strains are further classified into seven genotypes each (GI.1–GI.7 and GII.1–GII.7), while the GV strains include two genotypes (GV.1 and GV.2), and GIV strains have a single genotype (GIV.1) [4, 6]. In addition, inter- and intra-genogroup (inter- and intra-genotype) SaV recombinants have also been reported [7, 8].
Overall, SaVs have been detected in faecal samples of sporadic cases of AG at a frequency of 2·2–12·7%, worldwide [2]. They are also known to cause outbreaks of gastroenteritis, primarily in semi-closed settings such as day-care centres and schools [3, 9]. Several studies from Asia have documented the presence of SaV strains and genetic variations within both occurrences, namely sporadic cases and outbreaks of AG [6, 10, 11]. In India, a limited number of studies have been conducted to determine their prevalence in sporadic AG occurring in children. However, these studies have not addressed their clinical and epidemiological features and genetic diversity. To date, there is no report available on SaVs in sporadic cases and outbreaks of AG from Western India. Considering this gap in the literature, the present 5-year study was conducted to determine the epidemiology and genetic diversity of SaVs from cases in Pune, Maharashtra, Western India. The study also aims at understanding the clinical impact of SaV infections in children aged ⩽5 years.
METHODS
Sample collection
A 5-year retrospective study was conducted on faecal samples (n = 778) collected from children (⩽5 years) hospitalized with AG in Pune, Maharashtra, Western India, between January 2007 and December 2011. Faecal samples collected from non-diarrhoeic controls (n = 207) during the study period were also included. A sporadic case of AG was defined as the passage of ⩾3 episodes of loose (watery) stools per day, with or without associated symptoms like vomiting, fever or abdominal pain. Informed consent permitting the collection and utilization of faecal samples was obtained from the parents or legal guardians of the children. Detailed case-reporting forms (CRFs) covering demographic and clinical information were completed for each registered case. The clinical severity of cases was determined from CRFs using the 20-point Vesikari scoring system [12]. This study was approved by the Institutional Human Ethical Committee of the National Institute of Virology, Pune. All faecal samples were analysed for the presence of enteric viruses such as RV, NoV, adenovirus (AdV), astrovirus (AstV) and enterovirus (EV). Detection of group A rotavirus was carried out by ELISA (Dako Cytomation, UK), while NoV, AdV, AstV and EV were detected by conventional polymerase chain reaction (PCR) using specific primers targeting the RdRp (126 bp), Hexon (300 bp), ORF1a (289 bp) and 5′-UTR region (537/400 bp), respectively.
Sample processing and nucleic acid extraction
Clarified 30% (w/v or v/v) faecal suspensions were prepared in 0·01 m PBS (pH 7·4). Viral RNA extraction was carried out using QIAamp viral RNA mini kit (Qiagen, Germany) according to the manufacturer's protocol.
SaV detection in clinical samples
Reverse transcription (RT)–PCR was performed in a single 25 μl reaction mixture containing 5 μl of extracted RNA, 5 μl of 5× RT–PCR buffer, and 1 μl RT–PCR enzyme mix (Qiagen One-Step RT–PCR kit, Germany), using a nested set of primers described previously [13]. RT was performed at 47 °C for 30 min, according to the manufacturer's protocol. The thermal cycling conditions used for the first-round PCR were as follows: initial denaturation at 94 °C for 10 min, followed by 35 cycles at 94 °C for 1 min, 47 °C for 1 min, and 72 °C for 1 min, followed by final extension for 10 min at 72 °C. Nested PCR was carried out under identical conditions by changing the annealing temperature to 42 °C. The PCR products were resolved on 2% agarose gel, and positive amplicons (~420 bp) were visualized under a UV transilluminator using a gel documentation system.
Nucleotide sequencing and phylogenetic analysis
All positive amplicons were subjected to cycle sequencing using BigDye Terminator v. 3.1 Cycle Sequencing kit (Applied Biosystems, USA), and were further sequenced using the ABI 3130XL Genetic Analyzer (Applied Biosystems). Alignment of study sequences, along with prototype strains and reference strains deposited in GenBank and phylogenetic analysis, were performed using MEGA 6 software [14]. Nucleotide sequences identified in the study have been submitted to GenBank under accession numbers KU317432–KU317451.
Statistical analysis
The positivity between groups was compared using Fisher's exact test, while the difference in median values between mono- and mixed infection groups was compared using the Mann–Whitney U test. A P value of ⩽0·05 was considered as significant.
RESULTS
Detection of SaV in faecal samples from AG cases and controls
During the 5-year study period, SaV was detected at a frequency of 2·7% (21/778) in children hospitalized with AG, and 1·9% (4/207) in asymptomatic controls. Although the frequency of detection in diarrhoeic cases was more than that in non-diarrhoeic controls, this finding was not statistically significant (P = 0·54, Fisher's exact test). Co-infection with other enteric viruses was observed in 28·6% (6/21) of the positive cases and 25·0% (1/4) of the controls. Dual infection with RV (33·3%), AdV (33·3%) and AstV (16·7%) was observed in 83·3% cases (5/6), while triple infection with RV and EV was observed in 16·7% cases (1/6). In the SaV-positive control sample, dual infection with EV was observed.
Age distribution of SaV-positive cases
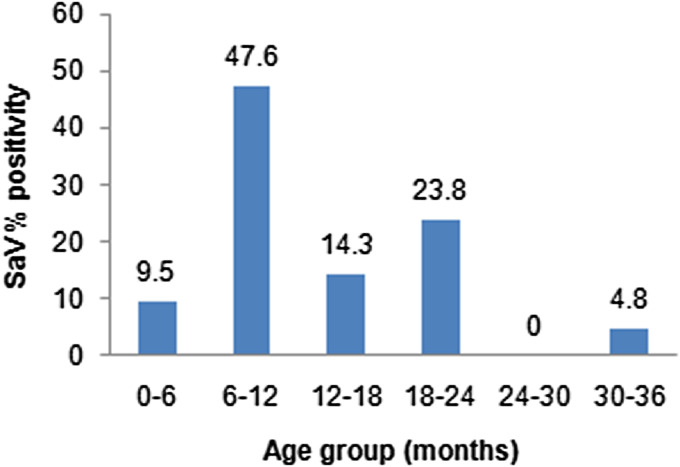
SaV was the sole infection in children aged ⩽3 years, in gastroenteritis cases as well as controls. In SaV-positive AG, 12/21 cases (57·1%) belonged to children aged <12 months, while 8/21 (30·1%) and 1/21 (4·8%) cases were observed in children aged between 12–24 and 24–36 months, respectively (Fig. 1).
Fig. 1.
Age distribution of sapovirus (SaV) in acute gastroenteritis cases.
Seasonal distribution of SaV-positive cases
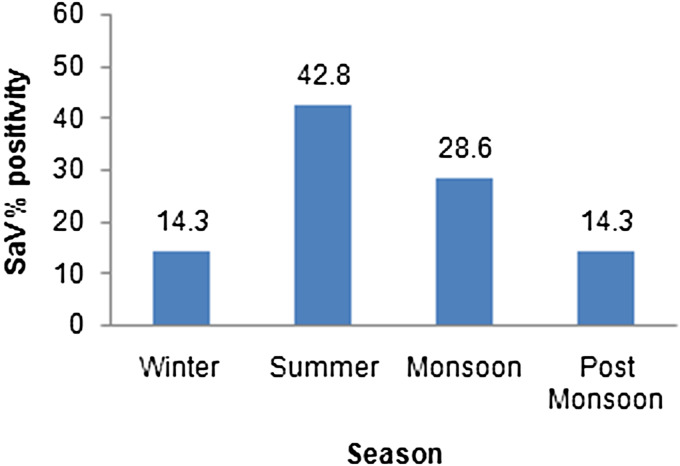
In AG cases, SaV infections were reported throughout the year, with peak infections recorded in summer: 9/21 (42·8%) cases occurred in summer, 6/21 (28·6%) during monsoon, 3/21 (14·3%) each in post-monsoon and winter seasons, respectively (Fig. 2). In controls, SaV was detected only during monsoon (50%) and winter (50%).
Fig. 2.
Seasonal distribution of sapovirus (SaV) in acute gastroenteritis.
Clinical features of SaV infection in AG cases
Clinical features of SaV mono-infections (15/21) and mixed infections (6/21) are presented in Table 1. While diarrhoea was primarily observed in all positive cases, occurrence of fever and vomiting varied between the two groups. Children with mono-infections experienced vomiting more frequently than those with mixed infections. In contrast, fever was observed more frequently in mixed infections. However, these differences were not statistically significant (P = 0·29 and P = 0·55, respectively, using Fisher's exact test). Dehydration was assessed on the basis of presence or absence of clinical features such as lethargy, restless/irritable behaviour, sunken eyes, skin pinch, and feeding habits of the child. Some degree of dehydration was observed in both groups (80% in SaV mono-infections and 83% in mixed infections). A high number of children with mono-infection had severe dehydration (60%). The median duration and frequency of diarrhoea in children with mono-infections were slightly lower than that observed in children with mixed infections, whereas the duration and frequency of vomiting was higher for mono-infections. The median duration of hospitalization for mono-infection cases was significantly longer than mixed infection cases (P = 0·039, Mann–Whitney U test). Overall, the severity assessment of positive cases revealed that 67% of SaV mono-infections resulted in severe gastroenteritis.
Table 1.
Comparison of clinical characteristics between sapovirus (SaV) mono- and mixed infections in children with acute gastroenteritis, 2007–2011
| SaV mono-infection | SaV mixed infection | |
|---|---|---|
| (N = 15) | (N = 6) | |
| Clinical characteristics | n (%) | n (%) |
| Fever | 8 (53) | 6 (100) |
| Diarrhoea | 15 (100) | 6 (100) |
| Median (min–max) | ||
| Duration of diarrhoea (days) | 2 (1–5)a1 | 2·5 (1–6)b1 |
| Frequency of diarrhoea (episodes/24 h) | 6 (4–10)a2 | 6·5 (4–8)b2 |
| Vomiting | 11 (73) | 3 (50) |
| Median (min–max) | ||
| Duration of vomiting (days) | 2 (0–4)a3 | 0·5 (0–3)b3 |
| Frequency of vomiting (episodes/24 h) | 3 (0–6)a4 | 1 (0–3)b4 |
| Degree of dehydration | ||
| Severe (>5%) | 9 (60) | 2 (33) |
| Some (⩽5%) | 3 (20) | 3 (50) |
| None (0%) | 3 (20) | 1 (17) |
| Vesikari score | ||
| Mild (0–5) | − | − |
| Moderate (6–10) | 4 (26) | 3 (50) |
| Severe (11–15) | 10 (67) | 1 (16·7) |
| Very severe (16–20) | 1 (7) | 2 (33·3) |
| Duration of hospitalization, median (min–max) | 5 (2–7)a5 | 2 (1–5)b5 |
a1 vs. b1, a2 vs. b2, a3 vs. b3, a4 vs. b4: P > 0·05 (n.s.), a5 vs. b5: P = 0·039 (S).
n.s., Non-significant; S, significant.
Nucleotide sequence and phylogenetic analysis
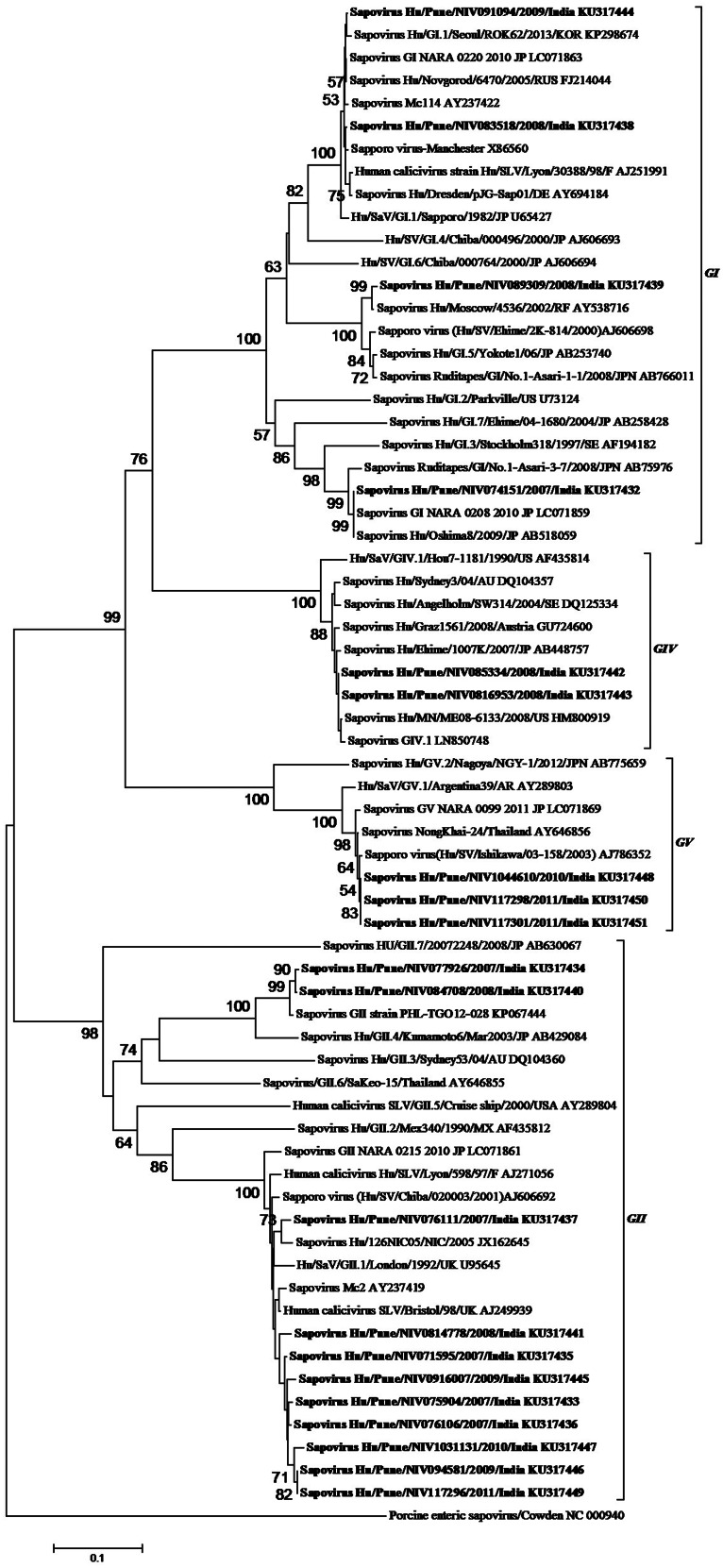
Nucleotide sequence analysis of the SaV capsid region (~420 bp) showed a high degree of variation in study strains. All four human SaV genogroups were detected, and at least eight different genotypes could be identified from AG cases: GI.1 (n = 2), GI.3 (n = 1), GI.5 (n = 1), GII.1 (n = 9), GII.4 (n = 2), GIV.1 (n = 3), GV.1 (n = 3), and in controls: GI.1 (n = 1), GI.2 (n = 1), GII.1 (n = 1) and GII.6 (n = 1). Phylogenetic analysis of the SaV strains from AG cases indicated that these strains were distributed in seven genoclusters within GI, II, IV and V (Fig. 3). Out of these 21 strains, 11 (52·3%) clustered with GII9 GII.1, and two GII.4 strains forming a separate cluster with a recombinant strain TG012-28, four (19%) with GI in three clusters (two GI.1, one GI.3, one GI.5), three (14·3%) with GV, and three (14·3%) with GIV. One GIV strain could not be incorporated in the tree due to sub-optimal amplification (~280 bp).
Fig. 3.
Phylogenetic tree based on partial nucleotide sequence analysis of the capsid region of sapovirus (SaV) (~420 bp). The tree was generated using neighbour-joining method with 1000 bootstrap replicates, in MEGA 6. Scale bar indicates nucleotide substitutions per site. SaV strains detected in the study are indicated by bold font.
GI and GII strains displayed more diversity than GIV and GV strains, which appeared to be almost identical (Table 2). Nucleotide sequence identity for study strains between the same genotype was >95%, while genotypes of the same genogroup varied between 20% and 30%. GII.1 strains showed the highest level of variation (96·1–99·3%) among all the genotypes detected in the study. GI.1, GII.1, GIV.1 and GV.1 strains most closely resembled their prototype strains, with nucleotide identity >95%, while GI.3, GI.5 and GII.4 strains showed greater divergence to their respective prototypes. The SaV genotypes and their variants identified in this study were most closely related to strains from Japan, the Philippines, Russia, Thailand, UK and the United States.
Table 2.
Nucleotide sequence identities of sapovirus (SaV) strains detected in acute gastroenteritis cases
| Nucleotide identity (%) | |||||
|---|---|---|---|---|---|
| SaV genotype (n) | Prototype strain | Within study strains | With prototype strain | Maximum identity with reference strain | |
| GI.1 (2) | Sapporo/1982/JP | 99·3 | 99·3 | 99·5–99·8 | FJ214044 (Russia) |
| GI.3 (1) | Stockholm318/97/SE | – | 90·5 | 100 | LC071859 (Japan), AB518059 (Thailand) |
| GI.5 (1) | Yokote1/06/JP | – | 96·5 | 99·3 | AY538716 (Russia) |
| GII.1 (9) | London/1992/UK | 96·1–99·3 | 95·5–96·3 | 97·8–98·5 |
AJ249939 (UK) AY237419 (Thailand) |
| GII.4 (2) | Kumamoto6/JP | 99·5 | 91·6 | 98·5–99 | KP067444 (Philippines) |
| GIV.1 (2) | Hou7-1181/US/1990 | 100 | 95·4 | 99·7 | HM800919 (USA) |
| GV.1 (2) | Arg39/Argentina/1995 | 100 | 96·4 | 99·5 | AJ786352 (Japan), AY646856 (Thailand) |
The circulation pattern of SaV strains varied throughout the study period and showed a changing trend (Table 3). While GII strains were detected throughout the study period, the occurrence of GI, IV and V strains varied greatly. Although genotype GII.1 was consistently detected, it was observed to be predominant only in 2007 and 2009. GIV.1 emerged in 2008 and was the predominant genotype that year whereas GV.1 emerged in 2010 and became the predominant genotype in 2011. In 2010, the frequency of detection of GII.1 was similar to that of GV.1 (Table 3).
Table 3.
Year-wise distribution of sapovirus genotypes in acute gastroenteritis cases and their changing trend, 2007–2011
| Year | Genotypes in circulation |
|---|---|
| 2007 (n = 6) | GII.1 (4), GI.3 (1), GII.4 (1) |
| 2008 (n = 6) | GIV.1 (2), GII.1 (1), GII.4 (1), GI.1 (1), GI.5 (1) |
| 2009 (n = 4) | GII.1 (2), GI.1 (1) |
| 2010 (n = 2) | GII.1 (1), GV.1 (1) |
| 2011 (n = 4) | GV.1 (2), GII.1 (1), GIV.1 (1) |
Predominant strains are indicated by bold font.
DISCUSSION
Worldwide, SaV is detected at a frequency ranging between 0·9% and 12·7% by conventional RT–PCR [4, 15]. Higher prevalence (17–18%) has been observed in recent studies using the real-time PCR approach [16, 17]. Previous reports available from southern (Vellore) and northern (New Delhi) regions of India, along with Japan, have reported SaV infections in 5·1–12·7% of cases hospitalized with AG [11, 18, 19]. In the present study, SaV was detected in 2·7% of children with AG and in 1·9% of asymptomatic children. Our data is similar to other studies from eastern (Kolkata) and northern (Lucknow) India, wherein the frequency of detection of SaV in children with AG was reported between 0·9% and 3·2% [15, 20]. SaV prevalence in asymptomatic controls was reported to be lower than in AG cases. In a study from Denmark, SaV infections were observed in 3·2% of the diarrhoeal cases and in 1·2% of the non-diarrhoeic controls [21]. In an earlier study from Vellore, India, SaV prevalence was reported to be higher in AG cases than in controls (5·1% vs. 3·5%) [19]. Similarly to our finding, the difference was not statistically significant (P = 0·4). SaVs are often reported to cause mixed infections with other enteric pathogens. High rates of co-infections were reported in earlier studies from India (61%) and Burkina Faso (56%) [17, 19]. The lower rate of co-infection (28·6%) in our study was comparable to similar studies from France (26·2%) and the Philippines (34%) [8, 22]. The reason for these differences could be explained by the fact that only a few other enteric pathogens were examined in our study. In the present study, SaV infections were observed only in children aged <3 years. The occurrence of infections was observed to decrease with increasing age (1 > 2 > 3 years), with 95·2% of the infections observed in children aged <2 years. Comparable data on age susceptibility to SaV infections was previously reported from the United States and Nicaragua, where ~75% and 93% of infections were observed by age 2 years, respectively [17, 23]. Globally, SaV infections have mainly been reported in winter months [10, 23–25]. However, in our study, the infections were observed throughout the year, with peak activity during the summer months. Although human calicivirus infections are detected more frequently during winter, sporadic SaV cases and outbreaks have been reported in summer months as well. Our data on seasonality is in agreement with studies from Finland and Japan, where highest SaV prevalence was reported during the warmer months [26, 27].
Although SaVs are reported to cause milder infections than RVs and NoVs, they are frequently known to cause hospitalization [28]. In our study, the most common clinical manifestations in children with SaV mono-infections were diarrhoea (100%), dehydration (80%) and vomiting (73%). In similar studies from Bangladesh and Burkina Faso, cases with SaV infection experienced diarrhoea more frequently than vomiting [10, 16]. Variable levels of dehydration in SaV-positive gastroenteritis cases, have been reported from countries like Bangladesh (88%) and Nicaragua (40%) [10, 17]. However, the status of dehydration in mono- and mixed infections was not clearly differentiated in these studies. In the present study, dehydration was observed in 80% and 83% of mono- and mixed infection cases, respectively. Data from Burkina Faso showed similar rates of dehydration in mono- (75%) and mixed (77%) infection cases [16]. However, while 60% of cases with mono-infection were observed to have severe dehydration in our study, only 10% of the mono-infection cases were reported to have severe dehydration in Burkina Faso. Greater severity of dehydration in our study could be explained by the fact that 73% of the SaV mono-infections were accompanied by vomiting, while only 45% from Burkina Faso were reported to experience vomiting. Although the median duration and frequency of diarrhoea and vomiting in our study did not vary significantly between the two groups, median duration of hospitalization was significantly longer for mono-infection cases than mixed infection cases. Our study showed that SaVs could cause severe infections in sporadic AG cases, resulting in long periods of hospitalization. Currently, similar detailed epidemiological studies on age distribution, seasonality and clinical severity of SaV infections have not been reported from India.
SaVs display a wide range of genetic diversity, which varies with geographical location. To date, human SaVs are classified into four genogroups and 17 genotypes, based on >20·1% variation in their complete capsid sequences [4, 6]. Of these, several genotypes of GI and GII were predominantly detected in AG cases worldwide, while GIV and GV genotypes are detected at lower frequencies [24, 29, 30]. Earlier reports available from Asian countries have documented the predominance of genotypes GI.1, GI.2 and GII.1 in AG cases [10, 31, 32]. Limited data with respect to genetic diversity of SaVs in AG cases are available from India. SaV GII.1 was predominantly detected in studies from Kolkata (east) and Vellore (south), while GI.1 was predominant in New Delhi (north) [15, 18, 19]. A total of six genotypes (GI.1–3 and GII.1–3) were reported to circulate in Vellore between 2001 and 2004. A recent study from Lucknow (north India) reported the presence of SaV in AG cases; however, the genetic variation among the strains was not addressed [20]. Our study reports the presence of at least eight genotypes of SaV from AG cases and asymptomatic controls. Genotypes GI.5 and GII.6, along with GIV.1 and GV.1, were reported for the first time in India. The analysis of genetic diversity within the genogroups for GIV and GV strains, was limited by their low frequency of detection in the study. The wide genetic diversity of the SaV strains detected in our study could be due to detection targeting of the RdRp-capsid junction with our assay, compared to assays targeting only the RdRp region in other studies from India. Two of the GII.4 study strains formed a separate cluster with a recombinant strain from the Philippines. As described earlier, this GII.1/GII.4 recombinant strain shared nucleotide and amino-acid identity of 94% and 99% with GII.1 in the non-structural region, and 92% and 99% similarity with GII.4 in the capsid region [8]. While the two GII.4 strains showed 9·5% divergence from the GII.4 prototype, they shared a 98·5–99% sequence identity with the Philippines strain. It is likely that these two GII.4 strains are recombinant SaV strains as suggested in the Philippines study. However, sequencing of the complete RdRp and capsid regions is mandatory to confirm this. In our study, the annual distribution of SaV genotypes varied greatly in Pune, Western India, with GII.1 being predominant in 2007 and 2009, GIV.1 in 2008, and GV.1 in 2011. Similar genotypic variation and shift was previously observed in Chiang Mai province, Thailand and the Kumamoto prefecture in Japan [33–35]. The GV.1 study strains shared an amino-acid identity of 99% with other GV.1 strains that were reported from an outbreak that occurred in Japan, indicating that SaV strains detected from sporadic cases could also be responsible for outbreaks of gastroenteritis. In view of the present data on genetic diversity and the changing trend of SaV strains identified in Pune, a constant surveillance of SaVs from other parts of India is mandatory.
Although the rate of SaV detection from Western India was slightly lower than that previously reported from other parts of India, the number of SaV mono-infection cases with severe gastroenteritis having longer periods of hospitalization, was higher in this study. The genotypic data from the study region reveals circulation of multiple genotypes, with the presence of genotypes GIV.1 and GV.1 being documented for the first time in India. Our study further emphasizes the need for continuous surveillance of SaVs from other parts of India for better management of AG, which is one of the major public health concerns in developing countries like India.
ACKNOWLEDGEMENTS
Authors are grateful to Dr D. T. Mourya, Director, National Institute of Virology, Pune for his constant support during this study.
Financial support provided to the first author by the University Grants Commission (UGC) New Delhi, India is gratefully acknowledged. The authors are grateful to the Indian Council of Medical Research (ICMR), New Delhi, for funding the project.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Chow CM, Leung AKC, Hon HL. Acute gastroenteritis: from guidelines to real life. Clinical and Experimental Gastroenterology 2010; 3: 97–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oka T, et al. Comprehensive review of human sapoviruses. Clinical Microbiology Reviews 2015; 28: 32–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Svraka S, et al. Epidemiology and genotype analysis of emerging sapovirus-associated infections across Europe. Journal of Clinical Microbiology 2010; 48: 2191–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oka T, et al. Human sapovirus classification based on complete capsid nucleotide sequences. Archives of Virology 2012; 157: 349–352. [DOI] [PubMed] [Google Scholar]
- 5.Scheuer KA, et al. Prevalence of porcine noroviruses, molecular characterization of emerging porcine sapoviruses from finisher swine in United States, and unified classification scheme for sapoviruses. Journal of Clinical Microbiology 2013; 51: 2344–2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shibata S, et al. Complete genome sequence of a novel GV.2 sapovirus strain, NGY-1, detected from a suspected food borne gastroenteritis outbreak. Genome Announcements 2015; 3: 2–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansman GS, et al. Intergenogroup recombination in sapoviruses. Emerging Infectious Diseases 2005; 11: 1916–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, et al. Molecular detection and characterization of sapovirus in hospitalized children with acute gastroenteritis in Philippines. Journal of Clinical Virology 2015; 68: 83–88. [DOI] [PubMed] [Google Scholar]
- 9.Hansman GS, et al. Outbreak of gastroenteritis due to sapovirus. Journal of Clinical Microbiology 2007; 45: 1347–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dey SK, et al. Prevalence of sapovirus infection among infants and children with acute gastroenteritis in Dhaka city, Bangladesh during 2004–2005. Journal of Medical Virology 2007; 79: 633–638. [DOI] [PubMed] [Google Scholar]
- 11.Harada S, et al. Surveillance of pathogens in outpatients with gastroenteritis and characterization of sapovirus strains between 2002–2007 in Kumamoto prefecture, Japan. Journal of Medical Virology 2009; 81: 1117–1127. [DOI] [PubMed] [Google Scholar]
- 12.Ruuska T, Vesikari T. Rotavirus disease in Finnish children: use of numerical scores for clinical severity of diarrhoeal episodes. Scandinavian Journal of Infectious Diseases 1990; 22: 259–267. [DOI] [PubMed] [Google Scholar]
- 13.Okada M, et al. The detection of human sapoviruses with universal and genogroup-specific primers. Archives of Virology 2006; 151: 2503–2509. [DOI] [PubMed] [Google Scholar]
- 14.Tamura K, et al. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nayak MK, et al. A new variant of norovirus GII.4/2007 and inter-genotype recombinant strains of NVGII causing watery diarrhoea among children in Kolkata, India. Journal of Clinical Virology 2009; 45: 223–229. [DOI] [PubMed] [Google Scholar]
- 16.Matussek A, et al. Molecular characterization and genetic susceptibility of sapovirus in children with diarrhoea in Burkina Faso. Infection Genetics and Evolution 2015; 32: 396–400. [DOI] [PubMed] [Google Scholar]
- 17.Bucardo F, et al. Predominance of norovirus and sapovirus in Nicaragua after implementation of universal rotavirus vaccination. PLoS ONE 2014; 9: e98201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monica B, et al. Human caliciviruses in symptomatic and asymptomatic infections in children in Vellore, south India. Journal of Medical Virology 2007; 79: 544–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rachakonda G, et al. Genetic diversity of noroviruses and sapoviruses in children with acute sporadic gastroenteritis in New Delhi, India. Journal of Clinical Virology 2008; 43: 42–48. [DOI] [PubMed] [Google Scholar]
- 20.Jain A, et al. Aetiology of childhood viral gastroenteritis in Lucknow, north India. Indian Journal of Medical Research 2015; 141: 469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olesen B, et al. Aetiology of diarrhoea in young children in Denmark: a case-control study. Journal of Clinical Microbiology 2005; 43: 3636–3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lekana-Douki SE, et al. Molecular epidemiology of enteric viruses and genotyping of rotavirus A, adenovirus and astrovirus among children under 5 years old in Gabon. International Journal of Infectious Diseases 2015; 34: 90–95. [DOI] [PubMed] [Google Scholar]
- 23.Chhabra P, et al. Aetiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. Journal of Infectious Diseases 2013; 208: 790–800. [DOI] [PubMed] [Google Scholar]
- 24.Johnsen CK, et al. Genetic diversity of sapovirus infections in Danish children 2005–2007. Journal of Clinical Virology 2009; 46: 265–269. [DOI] [PubMed] [Google Scholar]
- 25.Phan TG, et al. Human astrovirus, norovirus (GI, GII) and sapovirus infections in Pakistani children with diarrhoea. Journal of Medical Virology 2004; 73: 256–261. [DOI] [PubMed] [Google Scholar]
- 26.Pang XL, et al. Human caliciviruses in acute gastroenteritis of young children in the community. Journal of Infectious Diseases 2000; 181 (Suppl. 2S): 288–294. [DOI] [PubMed] [Google Scholar]
- 27.Phan TG, et al. Emergence of rare sapovirus genotype among infants and children with acute gastroenteritis in Japan. European Journal of Clinical Microbiology and Infectious Diseases 2007; 26: 21–27. [DOI] [PubMed] [Google Scholar]
- 28.Zintz C, et al. Prevalence and genetic characterization of caliciviruses among children hospitalized for acute gastroenteritis in the United States. Infection Genetics and Evolution 2005; 5: 281–290. [DOI] [PubMed] [Google Scholar]
- 29.Gallimore CI, et al. Characterization of sapoviruses collected in the United Kingdom from 1989 to 2004. Journal of Medical Virology 2006; 78: 673–682. [DOI] [PubMed] [Google Scholar]
- 30.Medici MC, et al. Molecular detection and epidemiology of astrovirus, bocavirus and sapovirus in Italian children admitted to hospital with acute gastroenteritis, 2008–2009. Journal of Medical Virology 2012; 84: 643–650. [DOI] [PubMed] [Google Scholar]
- 31.Ren Z, et al. Etiological study of enteric viruses and the genetic diversity of norovirus, sapovirus, adenovirus and astrovirus in children with diarrhoea in Chongqing, China. BMC Infectious Diseases 2013; 13: 412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang G, et al. Genetic diversity of sapoviruses in non-hospitalized adults with sporadic cases of acute gastroenteritis in Shanghai, China. Journal of Clinical Virology 2014; 59: 250–254. [DOI] [PubMed] [Google Scholar]
- 33.Khamrin P, et al. Genetic diversity of noroviruses and sapoviruses in children hospitalized with acute gastroenteritis in Chiang Mai, Thailand. Journal of Medical Virology 2007; 79: 1921–1926. [DOI] [PubMed] [Google Scholar]
- 34.Khamrin P, et al. Emergence of new norovirus variants and genetic heterogeneity of noroviruses and sapoviruses in children admitted to hospital with diarrhoea in Thailand. Journal of Medical Virology 2010; 82: 289–296. [DOI] [PubMed] [Google Scholar]
- 35.Harada S, et al. A confirmation of sapovirus re-infection gastroenteritis cases with different genogroups and genetic shifts in the evolving sapovirus genotypes, 2002–2011. Archives of Virology 2012; 157: 1999–2003. [DOI] [PubMed] [Google Scholar]