Abstract
Alterations in two highly conserved N-linked glycosylation sites within the gp120 envelope glycoprotein of human immunodeficiency virus type I (HIV-1) implicated in the phenotype of a noncytopathic HIV-1 variant were introduced independently and in combination into a cytopathic, infectious HIV-1 clone by site-specific mutagenesis. Neither mutation affected the synthesis of HIV-1 envelope glycoproteins. However, one of the mutations restricted the ability of HIV-1 envelope to localize on the cell membrane and thus markedly impaired virus assembly. The HIV-1 assembly defect could be overcome in trans if site-specific mutants were packaged in HeLa cells constitutively producing wild-type HIV-1 envelope glycoprotein. In addition to inefficient virus assembly, this mutation impaired the ability of the virus to infect CD4+ T cells, but did not affect CD4-independent infection of muscle cells. These results suggest additional functions of posttranslational modification in virus replication (i.e., envelope glycoprotein transport). Given that such modifications can restrict CD4-mediated uptake without affecting CD4-independent uptake, variations in posttranslational env processing between different HIV-1 genotypes may affect virus tropism in vivo.
INTRODUCTION
The envelope glycoprotein of human immunodeficiency virus type 1 (HIV-1) plays a central role in cell-specific tropism and HIV-1-mediated cytopathogenesis. The selective tropism of HIV-1 for CD4-bearing cells1,2 is characterized by specific recognition of the CD4 receptor by virus determinants within the envelope glycoprotein gp120.3 Syncytium-dependent cytopathogenesis of HIV-1 and virus-cell fusion during HIV-1 infection are also dependent upon gp120-CD4 interaction.4,5
The envelope protein of HIV undergoes extensive posttranslational modification. The gp120 envelope glycoprotein of the HIV-1 IIIb isolate6 contains 24 N-linked glycosylation sites, all of which are utilized.7 The very high degree of conservation between certain glycosylation sites in the HIV-1 envelope glycoprotein indicates an important function for these posttranslational modifications in virus replication. For example, glycosylation of gp120 is required for infectivity of HIV-1.8–10 Since the HIV-1 envelope-CD4 interaction is central to the cytopathogenic process,4,5,11 it is possible that alterations in posttranslational processing would affect HIV-1-mediated cytopathogenicity. Indeed, posttranslational modifications of feline leukemia virus12 distinguish nonpathogenic variants from those that induce fatal immunodeficiency disease.12
We previously reported the cloning and characterization of an HIV-1 variant diminished in the ability to induce cytolysis. The determinants for this phenotype map to the envelope glycoprotein,13 specifically to two highly conserved N-linked glycosylation sites within gp120 which were absent from the noncytopathic variant. We proposed that these posttranslational differences interfere with processing of gp120 and may restrict the transport of envelope glycoprotein to the surface of infected cells.13 Mutations in these highly conserved N-linked glycosylation sites were introduced individually and in combination into a cytopathic, infectious clone of HIV-1. Our results demonstrate that the loss of a single, highly conserved N-glycosylation site restricts HIV-1 envelope glycoprotein localization on the cell surface and impedes virus assembly. This impaired virus assembly was alleviated by the loss of the second N-linked glycosylation site, which itself had no effect on virus assembly. In addition to the effect on envelope localization, the former alteration reduced the ability of mutant virions to infect CD4+ T cells but not CD4− muscle cells. These results indicate that posttranslational modifications within HIV-1 envelope glycoprotein are important in envelope processing and virus assembly. Additionally, since these modifications can restrict viral infectivity for T cells without affecting the ability of virions to infect muscle cells, variations in posttranslational env modification may influence HIV-1 tropism.
MATERIALS AND METHODS
Preparation of oligonucleotides
Oligodeoxyribonucleotides were synthesized on an Applied Biosystems synthesizer (Model 380B) using phosphoramidite chemistry. Oligonucleotides were purified on 16% polyacrylamide gels and then desalted using a Sep-Pak C18 column (Waters Associates) as described elsewhere.14
Oligonucleotide-directed mutagenesis
Mutants were generated in an infectious molecular clone of HIV-1, pHXB2gpt.15 A2.7-kb Sal I-BamHI fragment containing the entire gp120 envelope (env) coding region was subcloned into M13, mpl8. Site-directed mutations were introduced by the method of Kunkel,16 using the muta-gene M13 in vitro mutagenesis kit (Bio-Rad). Mutagenic oligonucleotides were GCTTGACAAGTTGTggtACCTCAGTCATTAC for the HXB2 M7 mutation and CTGTCAATTTCAtcGAtAATGCTAAAACC for the HXB2 M8 mutation (nucleotide changes that were introduced are identified by lower case letters). The M7/8 double mutation was derived by annealing both M7 and M8 mutagenic primers to uracil-containing single stranded DNA. These changes correspond to an asparagine197 to glycine mutation in M7 and a threonine276 to isoleucine mutation in M8. To facilitate identification of mutants, the M7 mutation introduced a KpnI site while the M8 mutation introduced a ClaI site at positions 6816 and 7056 of the HXB2 sequence, respectively.6 Site-directed mutations were confirmed by DNA sequencing using synthetic oligonucleotide primers and the dideoxy chain termination procedure.17 Following mutagenesis, the mutated 2.7 kb Sal I-BamHI env fragment was excised from the replicative form of M13 and inserted in the SalI and BamHI sites of pHXB2gpt. Location of the M7 and M8 mutations in the HIV-1 envelope glycoprotein coding region and the resulting altered amino acids, are shown on Figure 1.
FIG. 1.
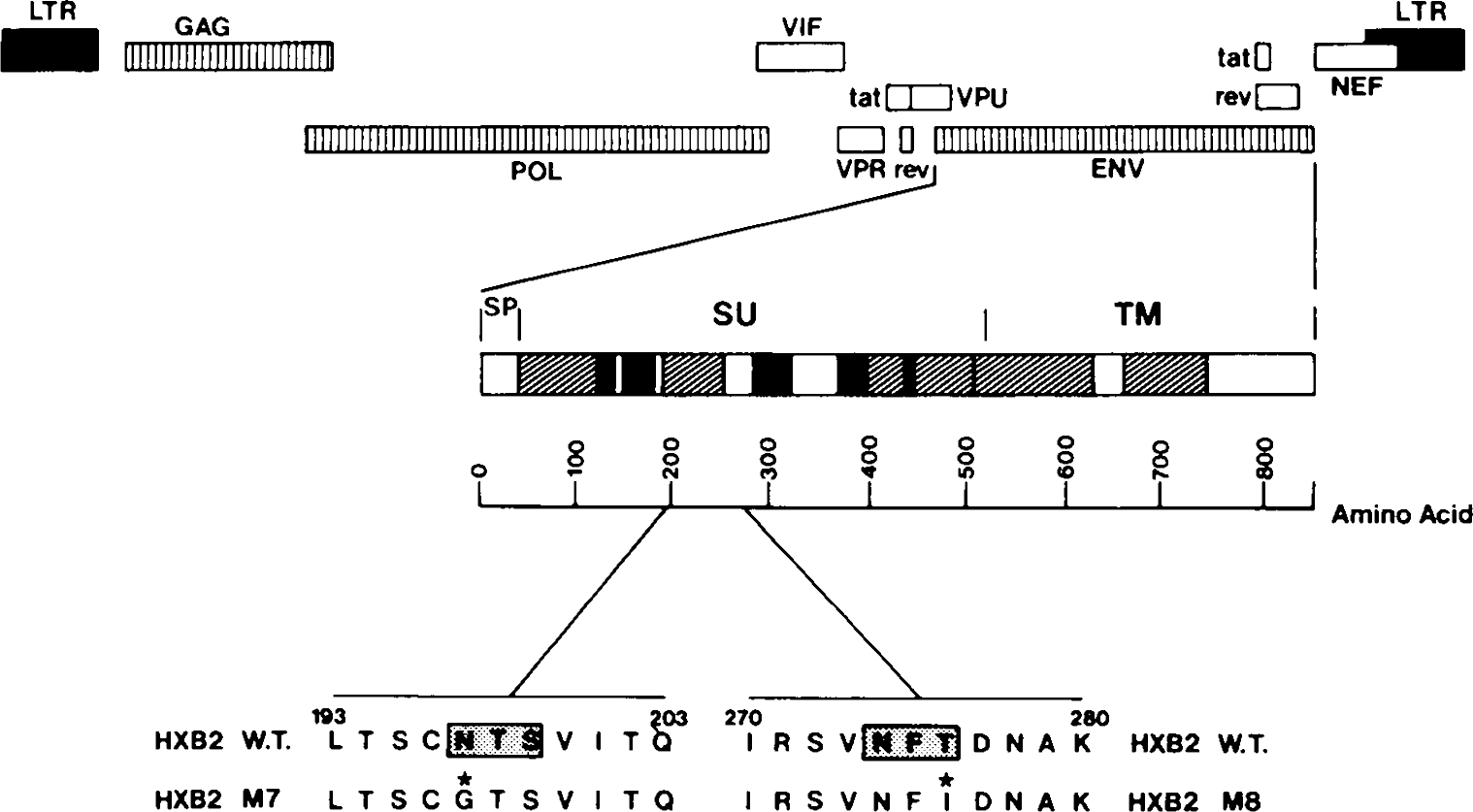
Designation of HIV-1 HXB2 env mutants. The major coding regions of the HIV-1 genome are indicated together with hyperconserved (unshaded), conserved (hatched), and hypervariable (solid) regions of the HIV-1 envelope glycoprotein. Major regions of the HIV-1 envelope include the signal peptide (SP), external (SU), and transmembrane (TM) domains. Locations of the N-glycosylation site sequences (asparagine-X-serine/threonine) which are mutated in the M7 and M8 mutants are numbered according to the sequence of Ratner.6 The shaded box represents the tripeptide N-glycosylation sequence and the asterisk identifies the amino acid mutated.
Cell culture and virus rescue
Mononuclear cells were isolated from peripheral blood of HIV-1 seronegative individuals by Ficoll-hypaque gradient centrifugation. Monocytes and macrophages were depleted by adherence to plastic. Nonadherent T cells were suspended at a density of 2 × 105 cells/ml in RPMI-1640 supplemented with 15% heat-inactivated fetal calf serum (FCS). Lymphocytes were activated by an overnight incubation in RPMI culture medium containing phytohemagglutinin (PHA) at 2 μg/ml. MT-4 cells (an HTLV-I-transformed CD4+ cell line),18 and Mo-T cells (an HTLV-II-transformed CD4+ cell line)19 were maintained in RPMI-1640 medium containing 10% FCS. The human cervical carcinoma-derived epithelial cell line, HeLa was maintained in Dulbecco’s minimal essential medium (DMEM) containing 5% FCS. The HeLa T4+ cell line which constitutively expresses the CD4 receptor20 was maintained in DMEM supplemented with 1 μg/ml G418 (Geneticin, Gibco). The HeLa-tat-III and HeLa-env-III cell lines which express HIV-1 tat and env proteins, respectively21 were maintained in DMEM supplemented with 250 ng/ml xanthine, 25 ng/ml mycophenolic acid, 10 ng/ml thymidine, 60 ng/ml hypoxanthine, and 10% dialyzed FCS. The human medullablastoma-derived cell line, Te67122 now shown to be derived from a rhabdomyosarcoma,23 was maintained in DMEM supplemented with 10% FCS.
To obtain viral stocks from wild-type and in vitro-mutated DNA clones, HeLa cells were seeded at approximately 30–50% confluence and transfected by the calcium phosphate–DNA coprecipitation method (10 μg of DNA/106 cells). Two days after transfection, cell culture supernatant was removed and virus production measured by an HIV-1 gag p24-specific ELISA (Coulter Immunology). Cell associated p24 was determined in 5 × 104 cells and extracellular p24 quantitated in 200 μl of culture supernatant.
Reverse transcriptase assay
Mg2+-dependent reverse transcriptase activity was measured in 50 μl of 50 mM Tris hydrochloride (pH 7.5)-5 mM dithiothreitol, 100 mM KCl, 10 mM MgCl2, 10 mM [3H]dTTP, 0.1% Triton X-100 containing 2 μg of poly (A), 0.4 μg of oligo-(dt)12–18, and an appropriate amount of virions. The solution was incubated at 37°C for 1 h, and the polymerized [3H]dTTP was precipitated with 10% trichloroacetic acid (TCA) and collected on a membrane filter (Millipore Corp.). The filter was washed with 10% trichloroacetic acid and ethanol, air dried and radioactivity quantitated in a liquid scintillation counter.
Immunofluorescence analysis
Infected cells were washed in phosphate-buffered saline, spotted on glass slides, dried, and fixed in acetone at −20°C for 15 minutes. Fixed cells were reacted with HIV-1-positive serum from an asymptomatic HIV-1-seropositive hemophiliac (antibody titer, 1:5160). After a 30-minute incubation at 37°C, cells were reacted with fluorescein isothiocyanate-conjugated goat anti-human immunoglobulin G, and virus antigen-positive cells were counted under an epifluorescence microscope (Nikon Inc.).
Syncytium assay
HeLa T4+ cells were plated in 25 cm3 flasks at 25–30% confluence and transfected with 10 μg of each of the wild-type and mutant proviral clones by calcium phospate/DNA coprecipitation. At 48 h after transfection, cultures were overlayed with methylene blue and the number of syncytia containing three or more nucleii were determined in 25 random fields (300 cells/field) at 100× magnification under a Nikon labphot photomicroscope. HeLa T4+ cells transfected with an HIV-1 gag/pol expression vector (p220 gag/pol), which does not express HIV-1 env proteins, served as a negative control.
Polymerase chain reaction analysis
Infected cells were washed with ice cold PBS, centrifuged 3 minutes at 14,000 g, and the cell pellet resuspended in Tris HCl (10 mM, pH 8.0)-EDTA (10 mM). Virus was inactivated by heating at 65°C for 15 minutes. Sodium dodecyl sulfate (SDS) was added to a final concentration of 0.1% and incubation continued for 30 minutes at 65°C. Cell lysates were incubated for 30 minutes at 37°C in the presence of 1 μg/ml proteinase K followed by two extractions with a mixture of phenol/chloroform/isoamyl alcohol (25:24:1). Cell extracts were finally ethanol precipitated in the presence of 10 μg of yeast tRNA and the DNA resuspended in Tris-HCl EDTA (10 mM:1 mM).
Detection of HIV-1 DNA by polymerase chain reaction (PCR) was performed essentially as described elsewhere.24 Briefly, DNA from an equivalent of 5 × 104 cells was subjected to 30 rounds of amplification in a 50 μl reaction mixture containing 100 picomoles of plus and minus strand primers complementary to the HIV-1 pol gene24 or to the α-tubulin gene25 (nucleotides 489–507, plus primer, nucleotides 756–777, minus primer), 200 μM of each deoxynucleotide, 50 mM KCl, 10 mM Tris HCl, (pH 8.3), 2.5 mM MgCl2, and two units Thermus aquaticus (Taq) enzyme.26 Each cycle comprised a one minute denaturation step (95°C), a 2 minute annealing step (61°C), and a 3 minute extension step (72°C). Aliquots (20 μl) of the PCR reaction were resolved on 0.5% agarose/2% Nu-Sieve agarose-Tris Borate gels containing 0.5 μg/ml ethidium bromide. Following electrophoresis, the gels were denatured, neutralized and transferred to Nytran nylon membranes (Schleicher and Schuell). The membranes were prehybridized in a solution containing 5X SSPE (1 X SSPE is 0.18 M NaCl, 10 mM sodium phosphate pH 7.7, 1 mM EDTA, 0.1% SDS and 5X Denhardt’s solution). Hybridization to γ-ATP-labeled primer probe proceeded overnight in the same solution, and was followed by two washes at 5°C above the Tm in a solution containing 5X SSPE and 0.1% SDS. Blots were autoradiographed overnight at −70°C. Details of PCR conditions and primers used are available from M. Stevenson upon request.
Envelope ELISA
Goat anti HIV-1 gp120SF2 polyclonal antiserum (titer > 100,000) was diluted 1:10 in PBS. Antibodies were adsorbed to a 96-well plate (Dynatech) for 1 h at 37°C then for 12 h at room temperature. Wells were rinsed four times in PBS containing 0.1% Triton X-100 (PBS-Triton). Next 5 × 104 HIV-1 HXB2 or mutant infected cells suspended in 200 μl RPMI-1640, containing 10% normal goat serum and 0.1% Triton X-100 were added to each well and incubated for 12–16 hours at room temperature. The wells were rinsed with PBS–Triton and a 200 μl solution of PBS-Triton containing a 1:100 dilution of pooled HIV-1 positive human serum (titer 1:5120) and 10% normal goat serum was added. After 2 h at 37°C, the wells were rinsed in PBS-Triton and incubated for 1 h at 37°C with 200 μl PBS-Triton containing 1 mg/ml biotinylated goat anti-human IgG (BRL) and 10% normal goat serum. The wells were washed and developed using streptavidin-conjugated horseradish peroxidase (Coulter Immunology). Plates were analyzed in a Vmax kinetic microplate reader (Molecular Devices) at a wavelength of 450 nm. Linearity of the ELISA measurements was verified by comparison with values obtained from a dilution series of known amounts of purified recombinant HIVSF2 gp120 or HIV III B gp120.
RESULTS
We previously identified two N-linked glycosylation site alterations implicated in the phenotype of a noncytopathic HIV-1 isolate (HIV mfD).13 Both mutations mapped to the second highly conserved domain of gp120,27 and were located at envelope amino acid positions 196 and amino acid 273 of a noncytopathic HIV-1 clone, HIV mfD13 (envelope amino acid positions 197 and 276 of HXB26). To determine the role of these conserved N-glycosylation sites in HIV-1 replication, mutations were introduced either singly or in combination into a cytopathic, infectious clone of HIV-1 (pHXB2gpt).15 In the HXB2 M7 mutation, the N-linked glycosylation site (asparagine-threonine-serine) was altered by substitution of the asparagine residue with a glycine at amino acid position 197 (Fig. 1). Alteration of the N-glycosylation site sequence in mutant HXB2 M8 was by substitution of a threonine residue at amino acid 276 with isoleucine (Fig. 1).
Characterization of HIV envelope N-glycosylation mutants
To obtain virus stocks from mutated and wild-type virus clones, HeLa cells were seeded at 30–50% confluence and transfected with proviral DNA by calcium phosphate DNA coprecipitation. At 48 h after trnsfection, viral antigen production was measured with a gag p24 ELISA and equal amounts of virus (based on p24 measurements) were used to inoculate MT4 cells. It should be emphasized that virus inoculums were based on the level of gag p24 rather than tissue culture infectious dose units13 due to differences in infectivity of the env mutants. Using the experimental conditions described here, defective virions (i.e., core particles lacking env protein) did not contribute significantly to the supernatant gag p24 measurements following virus rescue. Thus, cell associated and extracellular gag p24 levels (see below) reflected those of the envelope glycoproteins (Table 1). Additionally, we could not detect significant levels of gag p24 release from a gag p24 expression vector following transient transfection (see below). Replication kinetics of wild-type and env-mutant viruses, was monitored by increase in antigen positive cells with time (immunofluorescence staining) (Fig. 2), HIV-1 gag p24 production (Fig. 2) and reverse transcriptase activity in culture supernatant (results not shown). An increase in antigen-positive cells, HIV-1 p24, or reverse transcriptase activity, provided evidence of a spreading HIV-1 infection in the MT4 cell line. Transfected HeLa cells also were cocultivated with MT4 cells in the presence of polybrene 24 h after transfection and the cultures were monitored by immunofluorescence and p24 ELISA (Fig. 2). The replication kinetics of the HXB2 M8 mutant were indistinghishable from wild-type virus. Cultures infected or cocultivated with either wild-type or M8 mutant virions, displayed high levels of virus replication as early as 10 days postinfection. These cultures were not monitored beyond 20 days postinfection due to the extensive cytopathic effect (CPE) after this time. Conversely, HXB2 mutants M7 and M7/8 were unable to establish a spreading viral infection in MT4 cells, even after 40 days in culture (Fig. 2). Thus, we were unable to address cytopathic properties of the M7 and M7/8 mutants. Similar results were obtained using cell-free virions and cocultivation with either primary phytohemagglutainin (PHA) activated peripheral blood lymphocytes (PBL) or HTLV-II transformed Mo-T cells (not shown). The capacity of wild-type and mutant HIV clones to induce syncytia was determined 48 h after transfection of HeLa T4+ cells. Only the HXB2 M7 clone was incapable of syncytium induction in these cells while the other clones displayed syncytium-inducing properties (Table 1). By comparison, a CD4 binding mutant (HIV Mf M1, M. Stevenson, unpublished data) was unable to induce syncytia in the HeLa T4 system (Table 1) due to mutagenesis of a critical HIV-1 env-CD4 binding epitope.3
Table 1.
Biological Properties of HIV-1 env Mutants
| Clone |
Infectivity
a
|
Replication inb CD4+ cells | Syncytiumc induction |
envd
Production (ng/106 cells) |
||
|---|---|---|---|---|---|---|
| MT4 | Te671 | Cell associated | Extra cellular | |||
|
| ||||||
| HXB2 M7 | + | + | – | +/– | 13.0 ± 1.8 | 2.7 ± 0.6 |
| HXB2 M7/8 | + | + | – | ++ | 11.4 ± 1.6 | 9.0 ± 1.1 |
| HXB2 M8 | +++ | + | + | +++ | 10.5 ± 1.2 | 8.3 ± 0.7 |
| HXB2 Wild-Type | +++ | + | + | +++ | 12.0 ± 1.3 | 10.1 ± 0.8 |
| HIV Mf Mle | + | + | – | – | ND | ND |
Determined by PCR 24 h after infection with HIV-1 (200pg p24/105 cells).
Determined as ability to set up spreading infection in MT4 Mo-T or PBL cultures.
Determined 48 h after transfection of HeLa T4+ cells.
Determined by ELISA. Values represent mean (n = 3–5) ± SD.
HIV-Mf Ml mutant (M. Stevenson, unpublished data) contains an Alanine → Histidine mutation [amino acid 433 of HXB2 env6] in an HIV-1 env epitope critical for CD4 binding.3
ND, Not done.
FIG. 2.
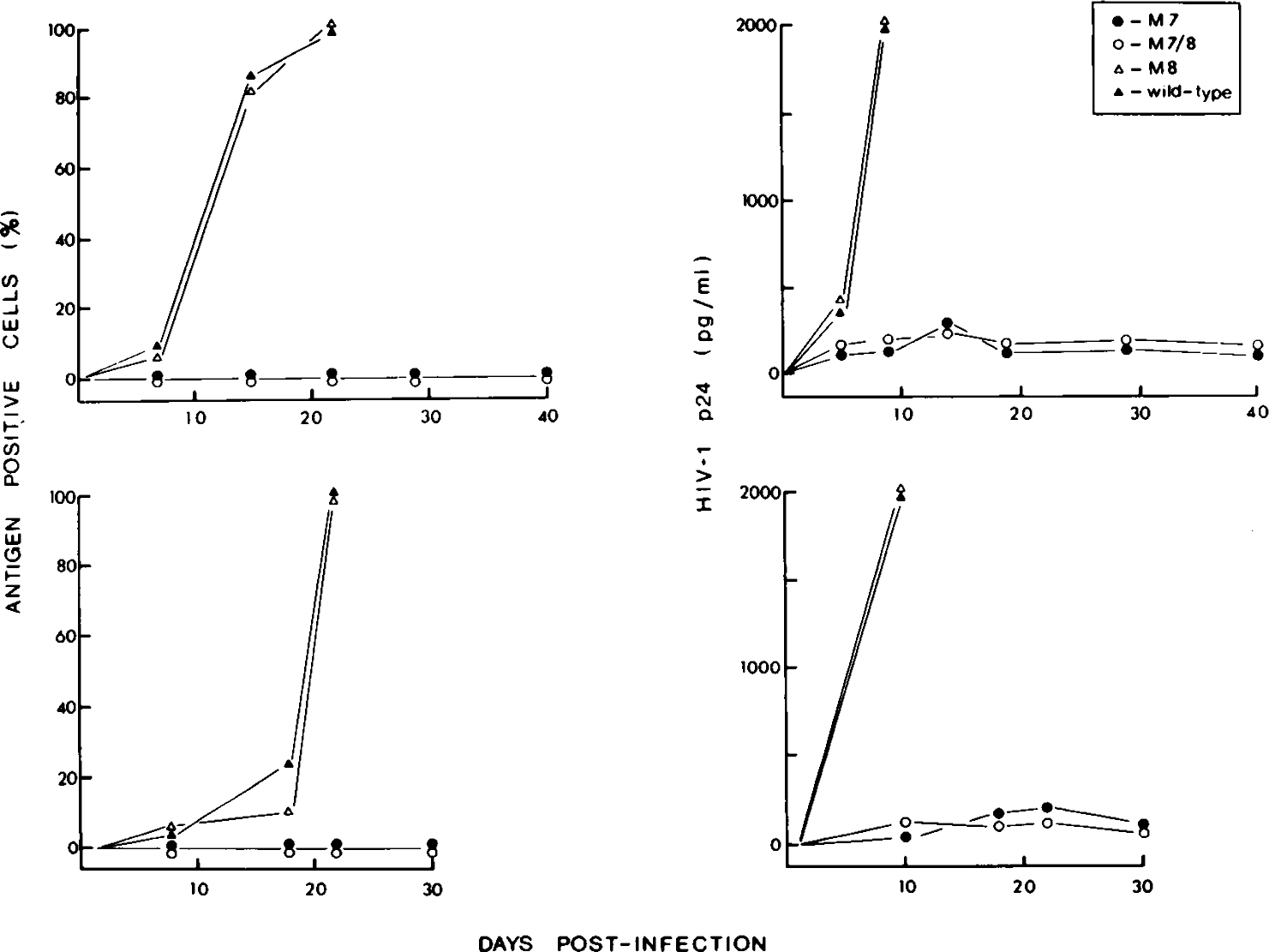
Replicative capacity of wild-type and env mutant HXB2 proviruses. Cloned HXB2 wild-type and env mutant virus stocks were obtained 48 h after transfection of HeLa cultures, filtered (0.8 μm), and used to directly infect MT4 cells (200 pg/5 × 105 cells) (upper panels). Alternatively, 24 h after transfection, HeLa cell cultures were cocultivated with MT4 cells in the presence of polybrene (lower panels). HIV-1 replication was monitored over a 40-day period (direct infection), or 30-day period (cocultivation) by immunofluorescence analysis (left-hand panels) and by HIV-1 gag p24-specific ELISA (right-hand panels). M8 and wild-type HXB2 virus-infected cultures were not monitored beyond 20 days postinfection due to the extensive cytopathic effect (CPE) in these cultures.
The HXB2 M7 env mutation affects CD4 dependent but not CD4 independent HIV-1 infection
Given the inability of viruses bearing the M7 N-glycosylation mutation to establish a spreading infection in CD4+ cells, we determined whether this mutation blocked virus-CD4 interaction. The polymerase chain reaction (PCR) method was adapted to evaluate the infectivity of wild-type and mutant proviruses for CD4+ and CD4− cells. Virus was rescued from the transfected HeLa cultures and equal amounts of virus (based on p24 measurements) were used to infect CD4+ MT4 cells (which are exquisitely sensitive to HIV-1 infection28) and the CD4− rhabdomyosarcoma-derived cell line Te671 (which is infectible by HIV-1 despite the absence of detectible CD429–31). At 24 hours after infection, total cellular DNA was extracted and subjected to 30 rounds of PCR as detailed in materials and methods. Under the conditions of the PCR procedure used here, the amount of PCR product is representative of the initial target copy number within the infected cells and subsaturating PCR conditions have been used to quantitate efficiency of HIV-1 infection in activated and quiescent T-cells.24 Extensive washing of cultures after transfection ensured that there was no background signal due to carryover of plasmid DNA from the transfections prior to virus rescue. For example, CD4− HeLa cells exposed to virus supernatants prepared in this manner, were free of viral DNA by PCR (not shown). By PCR analysis, it was evident that the infectivity of the HXB2 M7 and M7/8 mutants was reduced when compared to wild-type HXB2 or M8 mutant (Fig. 3). By comparison, the infectivity of HXB2 M7 and M7/8 virions was indistinguishable from wild-type or M8 mutants in CD4− Te671 cells (Fig. 3). We also compared the infectivity of wild-type HXB2 and N-glycosylation mutants with a CD4-binding mutant (HIV Mf M1) (Fig. 3). In this mutant, an alanine residue (residue 433 of HXB2 env6) in a highly conserved region of gp120 shown to be critical for interaction with CD43 was replaced with a histidine residue. The HIV Mf M1 mutant was also deficient in the ability to infect CD4+ cells while infection of CD4− Te671 cells was unaffected (Fig. 3). Uninfected cultures (Fig. 3) or cultures infected with heat-inactivated virus (56°C, 30 min) (not shown) did not give a signal after PCR analysis.
FIG. 3.
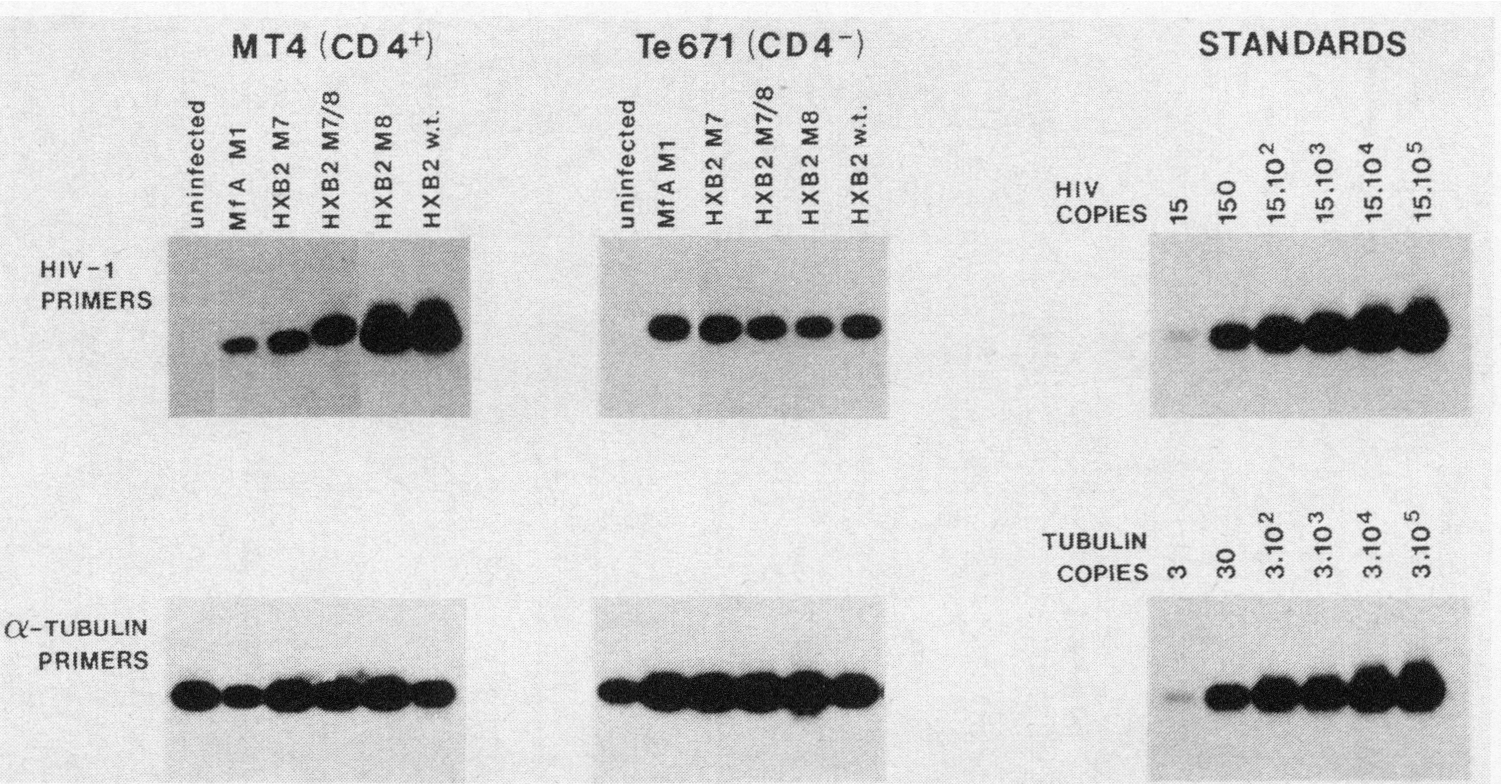
Infectivity of HXB2 wild-type and env mutant proviruses in CD4+ MT4 cells and CD4− Te671 cells. Cloned virus stocks were rescued 24 h after transfection of HeLa cell cultures and used to infect MT4 cells (CD4+) and Te671 cells (CD4−). After an additional 24 h, total cellular DNA was isolated following proteinase K/SDS treatment and the presence of HIV-1 DNA in the equivalent of 5 × 104 cells was determined by PCR using HIV-1 pol-specific primers24 while amplification with α-tubulin-specific primers ensured the presence of equivalent amounts of DNA in each sample. The MfA M1 mutant (M. Stevenson, unpublished data) contains an alanine to histidine substitution at residue 433 in a critical CD4-binding epitope within HIV-1 gp120.3 For comparison, a series of log dilutions of DNA prepared from a chronically infected CD4+ cell line24 was amplified using identical conditions. DNA from uninfected MT4 and Te671 cultures provided a negative control.
The M7 mutation also affects envelope transport and virus assembly
Transfection of HeLa cells with HXB2 M7 proviral mutants consistently gave low levels of extracellular virus production as evidenced by gag p24 (Fig. 4) and envelope glycoprotein (Table 1) measurements. Thus it was possible that, in addition to affecting CD4 interaction, the M7 mutation may alter envelope processing and viral assembly. To investigate this, we first determined whether the M7 mutation affected the synthesis of the mutant envelope glycoprotein. HeLa cells were transfected by calcium phosphate/DNA coprecipitation and HIV-1 envelope production was determined by ELISA as detailed in Materials and Methods. Production of envelope glycoproteins by wild-type and mutant proviruses, as evidenced by a quantitative gp120 ELISA assay, was similar (Table 1); however, extracellular env levels (Table 1) reflected those obtained from analysis of gag p24. Thus, despite similar levels of cell associated gag p24 and env glycoprotein in cultures transfected with wild-type and mutant proviral clones, extracellular levels of gag p24 and env glycoprotein were significantly lower in M7 transfected cultures. To more accurately determine whether there was indeed a defect in viral assembly, wild-type and env mutant proviral HIV-1 clones were cotransfected into HeLa cells, HeLa T4+ cells (which constitutively express the CD4 receptor20) and HeLa-tat-III and HeLa-env-III cells (which constitutively express HIV-1 tat and env proteins, respectively21). These cell lines also were transfected with an episome-based HIV-1 gag/pol expression vector p220 gag/pol (M. Stevenson, unpublished data). This vector contains the complete HIV-1 gag/pol open reading frame under control of the highly constitutive cytomegalovirus (CMV) immediate early promoter,32 in an Epstein-Barr virus-based episomal replicon.33 This vector was used to determine the degree of HIV-1 gag p24 shed from transfected HeLa cells in the absence of HIV-1 env production, and thus provide an indication of env independent HIV-1 core assembly and release. At 48 hours after transfection with the various plasmid constructs, cells and culture supernatants were assayed for gag p24. In all cases, extracellular gag p24 levels exceeded cell associated gag p24 in cells transfected with M7/8, M8 and wild-type proviruses (Fig. 4). This pattern was reproducible between multiple transfections (n = 3–5) performed on different days. In contrast, the majority of gag p24 in HeLa cells and HeLa T4 cells transfected with the HXB2 M7 mutant was cell associated (Fig. 4). A marginal increase in extracellular p24 was observed when the M7 mutant was transfected into HeLa cells constitutively expressing the HIV-1 tat-gene product (Fig. 4). However, the block to release of HIV-1 from transfected cells could be relieved when the M7 mutant was packaged in HeLa-env-III cells, which constitutively express wild-type envelope glycoprotein (Fig. 4). The differences in virus production from the various HIV-1 clones were not due to large differences in transfection efficiency since all clones produced abundant levels of cell-associated gag p24 and envelope glycoproteins (Table 1) (Fig. 4). It is important to note that the extracellular HIV-1 gag p24 production was a true indication of virus release and not simply an indication of envelope-independent HIV-1 core particle shedding from transfected cells.34,35 Thus, cell-associated and extracellular gag p24 levels in transfected HeLa cells (Fig. 4) mirrored those of the envelope glycoproteins (Table 1). The p220 gag/pol vector which expresses HIV-1 core proteins in the absence of HIV-1 envelope glycoprotein demonstrated that, in our system, envelope independent assembly and release of HIV-1 core particles contributed only approximately 10% of the total extracellular p24 levels. The relative efficiency of envelope independent core particle release in the HeLa cultures is in agreement with results obtained in COS cells,36 in which envelope independent release of core particles represented approximately 15% of the total cell-associated gag p24 production. Of interest was the ability of the M8 env mutation to overcome the block to virus assembly induced by the M7 env mutation (Fig. 4). Indeed, despite similar levels of cell-associated gag p24 in the various cell lines, extracellular levels of gag p24 from the M7/8 env clone were higher than in cultures transfected with the wild-type clone. Thus, the presence of the second mutation had a compensatory effect upon the defect introduced by the M7 glycosylation mutation.
FIG. 4.
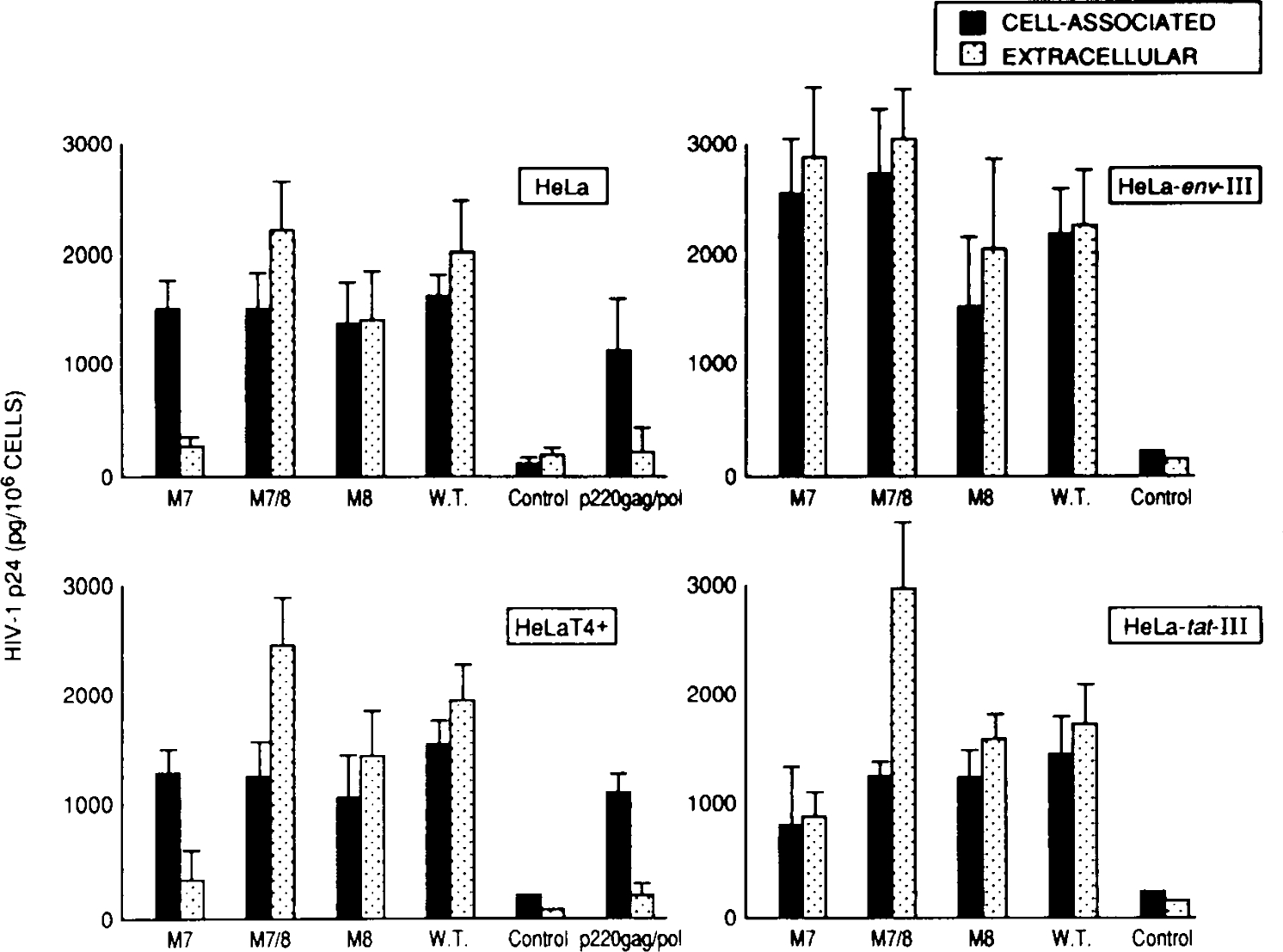
Assembly characteristics of wild-type and env HXB2 mutant clones. HeLa cell cultures seeded at 30–50% confluence were transfected with 10 μg of cloned proviral DNA. At 48 hours after transfection, HIV-1 gag p24 antigen production was determined from 200 μl of transfected culture supernatant (extracellular) and from 5 × 104 cells (cell associated). HeLa cell cultures transfected include the parental, cervical carcinoma-derived epithelial cell line (HeLa); HeLa T4+, which constitutively expresses CD4 antigen;20 and HeLa-env-III and HeLa-tat-III, which constitutively express HIV-1 env and tat proteins, respectively.21 The p220 gag/pol episomal gag/pol expression vector (M. Stevenson, unpublished data) contains the HIV-1 gag/pol open reading frame under control of the cytomegalovirus, immediate early promoter in an Epstein-Barr virus-derived episomal replicon.33 This vector which produces gag/pol polyproteins, in the absence of envelope glycoproteins, gives an indication of HIV-1 envelope-independent core assembly and release. Untransfected HeLa-cell clones served as a negative control in the HIV gag p24 ELISA.
The M7 env mutation restricts HIV-1 envelope localization on the cell surface
The HeLa packaging experiments suggested a virus assembly defect due to the presence of the M7 env mutation. This defect could be corrected in trans if complemented with wild-type envelope glycoprotein and indicated that the defect in assembly was env-dependent. The M7 mutation, however, did not affect envelope synthesis (Table 1), although the HXB2 M7 clone displayed weak syncytium-inducing properties (Table 1). Thus it was possible that the M7 mutation restricted envelope localization on the cell surface which is necessary both for virus formation and for syncytium induction. To determine cell surface env expression, HeLa cells expressing mutated and wild-type envelope glycoprotein were subjected to the cytotoxic effects of a Pseudomonas exotoxin-soluble CD437 fusion protein. Pseudomonas exotoxin-soluble CD4 fusion protein (CD4-Pe) is cytotoxic for cells expressing HIV-1 envelope glycoprotein37 and thus would be expected to provide a measure of the level of envelope glycoprotein on the cell surface available for CD4-Pe attack. HeLa cells were transfected with the various mutated and wild-type proviral clones; then 12 h after transfection, cultures were challenged with 300 ng/ml of CD4-Pe. After an additional 24 h, the amount of HIV-1 DNA in challenged and unchallenged control cultures was measured by PCR (Fig. 5). The relative amount of HIV-1 DNA in cells transfected with wild-type proviruses or proviruses bearing M7/8 and M8 mutations was dramatically reduced following treatment with CD4-Pe (Fig. 5) suggesting that these clones directed expression of HIV-1 envelope glycoprotein to the cell surface. In contrast, cells transfected with the HXB2 M7 clone were unaffected by challenge with CD4-Pe and the level of HIV-1 DNA in these cultures remained unchanged following CD4-Pe challenge (Fig. 5). Thus, despite synthesis of envelope glycoprotein by HXB2 clones containing the M7 mutation (Table 1), there was a restriction upon env localization on the cell surface. The insensitivity of the M7 env mutation to CD4-Pe was not due to inability of M7 env glycoprotein to bind CD4 since the M7/8 mutation, which also displayed reduced infectivity for CD4-bearing cells, was as sensitive to CD4-Pe challenge as wild-type HXB2 env glycoprotein (Fig. 5). However, the possible influence of CD4 affinity of M7 on sensitivity to CD4-Pe and compensatory effects of the M8 mutation cannot be excluded at present.
FIG. 5.
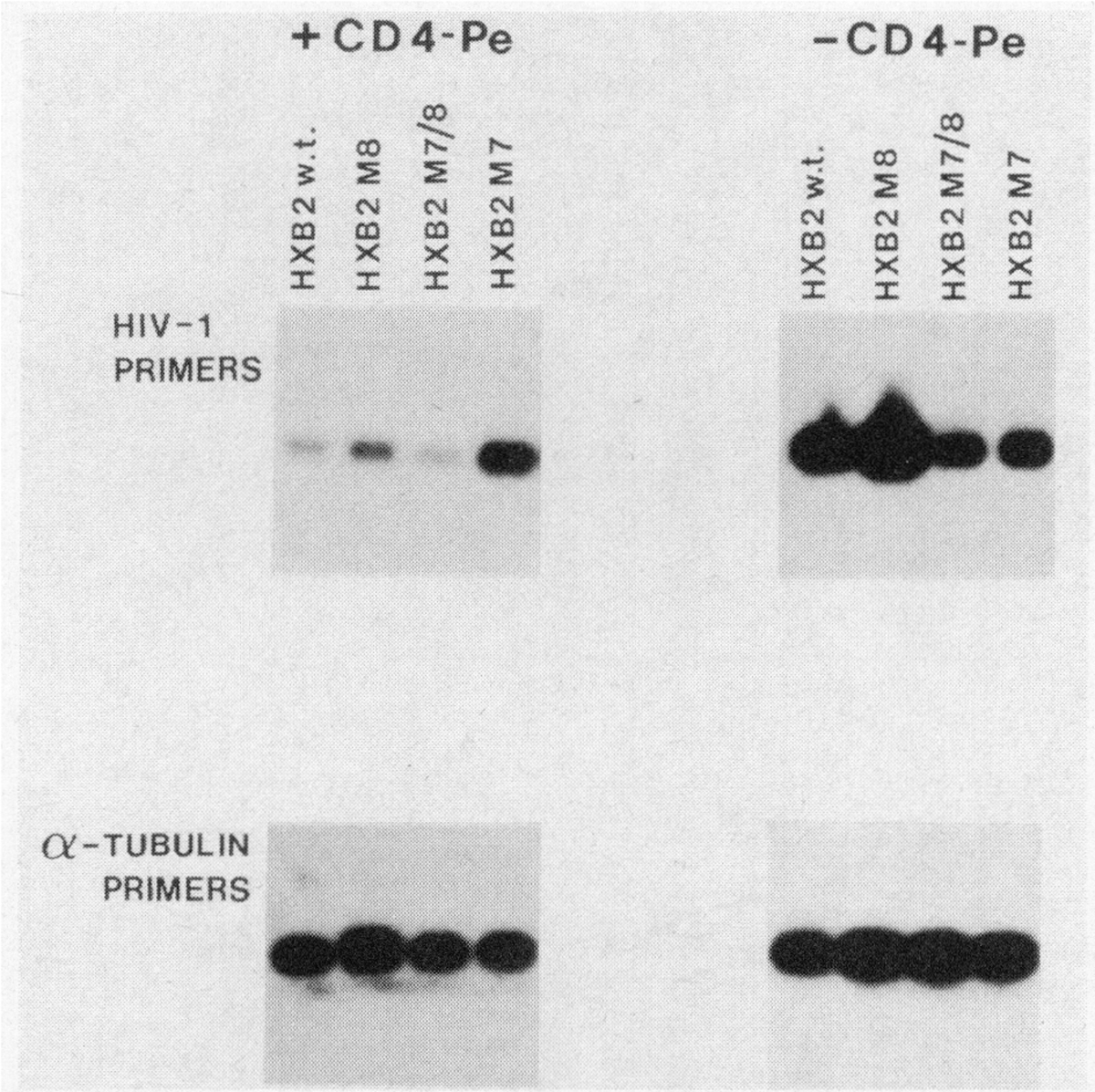
Plasma membrane localization of HIV env glycoprotein is restricted by the M7 env mutation. HeLa cells were transfected with the various mutated and wild-type proviral DNA clones and 12 hours after transfection, HeLa cultures were challenged with 300 ng/ml of a Pseudomonas exotoxin-soluble CD4 fusion protein (CD4-Pe).37 After an additional 24 h, proviral HIV-1 DNA in the transfected cultures was assayed by semiquantitative PCR using HIV-1 pol-specific primers. The relative amount of proviral DNA with and without CD4-Pe challenge is shown, while PCR analysis with α-tubulin-specific primers confirmed that similar amounts of DNA were analyzed in each sample.
DISCUSSION
In this report we describe HIV-1 envelope modifications which both restrict CD4-dependent virus infectivity and virus assembly. The inefficient virus assembly caused by the presence of the M7 env mutation was due to inefficient localization of envelope glycoprotein on the cell surface. The role of posttranslational modification in envelope processing and virus replication has been extensively studied and agents which inhibit the glycosylation process affect virus infectivity and syncytium formation.8,10,38 Others have reported that the oligosaccharides on gp120 are not important for receptor binding.39 Studies with vesicular stomatitis virus suggest that carbohydrate does not promote intracellular transport and plasma membrane localization of envelope glycoprotein directly, but rather influences polypeptide folding or oligomerization which are critical for transport.40 As demonstrated in the present report, the ability to correct defects in virus assembly by the presence of a second N-glycosylation mutation (as in HXB2 B7/8) would suggest that envelope conformation is more important for envelope transport than is carbohydrate content. Indeed, envelope mutations which do not affect envelope glycosylation have also been shown to affect envelope localization on the plasma membrane.39
The infectivity of HXB2 virus bearing the M7 env mutation was reduced in CD4+ cells. This reduced infectivity, although not completely abrogating infectivity for CD4+ cells, prevented viruses bearing this mutation from setting up a spreading infection in CD4+ cells. Willey et al.27 have described a region within the second conserved domain of gp120 that is critical for infectivity. Of the several N-linked glycosylation mutants studied in that work, one mutant lacking the glycosylation site at envelope amino acid position 276 replicated in CD4+ cells with wild-type kinetics.27 In agreement with that result, removal of the same glycosylation consensus sequence in mutant HIV-1 HXB2 M8 also did not affect replication of the virus in CD4+ cells. In this report, M7 and M7/8 env mutants which could not establish a spreading infection in CD4+ cells, still retained the ability to bind CD4 and were able to infect CD4-bearing cells as evidenced by the presence of double-stranded proviral DNA within M7-infected cells. This, however, did not lead to a productive viral infection. Thus it is possible that if env-CD4 affinity is reduced, the effects of this reduced affinity would be amplified after the multiple rounds of reinfection required to set up a spreading viral infection. In support of this, we have constructed a number of amino acid substitution and deletion mutations within a critical CD4-binding epitope of gp120 described by Lasky et al.3 The removal or alteration of critical amino acids in this CD4-binding epitope does not completely prevent ability of HIV-1 to infect CD4 cells, but does restrict the ability of mutants to set up a productive infection (Stevenson et al., manuscript in preparation). It is of interest that the M7 env mutation, although resulting in a replication-defective virus when introduced into the prototype HXB2 clone, did not affect replication of wild-type viruses (HIV mfC and D)13 bearing this mutation although they exhibited a noncytopathic phenotype. Thus, wild-type HIV mfC and D viruses possess sequences which compensate for the loss of the N-glycosylation site caused by the M7 env mutation. This adds an added degree of complexity to studies aimed at attributing certain biological properties of the virus (e.g., tropism, cytopathogenicity) to specific genetic determinants of the viral genome.
Env mutations affecting CD4-dependent virus infectivity described here apparently had no effect upon CD4-independent infection of muscle cells. A CD4-independent mechanism of HIV-1 uptake has been implicated in a variety of cell types including neural cells and muscle cells.29–31,41,42 Should HIV-1 infection of brain and muscle cells indeed proceed via a CD4-independent route, it is possible that a discrete region of gp120 not involved in CD4 binding recognizes the second receptor. Thus, posttranslational mutations which reduce CD4-dependent uptake of HIV may not affect CD4-independent HIV infection. It is possible that HIV-1 env sequence diversity leads to differences in posttranslational modification, thus influencing the ability of the virus to utilize CD4-dependent or independent pathways for infection and influencing HIV-1 tropism in vivo.
ACKNOWLEDGMENTS
We thank Lee Ratner for the gift of the pHXB2 gpt infectious HIV-1 clone, Gary Tarpley for CD4-Pe, Stephen Pyle and Larry Arthur for HIV-1 gp120IIIB, Bill Sugden for the p220 episomal expression vector, Rodney McComb for the Te 671 cell line, Jonathan Goldsmith and Steve Suvalsky for clinical materials and antisera, Angela Mann for preparation of all PCR-related reagents, Trevor Stanwick for technical assistance, and Karen Hansen for manuscript preparation. The MT4, HeLa T4, HeLa tat III, and HeLa env III cell lines and antiserum to HIV-1 gp120SF2 and recombinant HIVSF2 gp120 were obtained through the AIDS Research and Reference Reagent Program Division of AIDS, NIAID, National Institutes of Health. MS was supported by Grants A125582 and A130386 from the National Institutes of Health, Grant 962-7-RGR from the American Foundation for AIDS Research, and the Nebraska Research Initiative. SH was supported by NIH postdoctoral training Grant T32 CA09476.
REFERENCES
- 1.Dalgeish AG, Beverley PCL, Clapham PR, Crawford DH, Greaves MF, and Weiss RA: The CD4 (T-4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature (London) 1987;312:763–766. [DOI] [PubMed] [Google Scholar]
- 2.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, and Montaigner L: T-lymphyocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature (London) 1987;312:767–768. [DOI] [PubMed] [Google Scholar]
- 3.Lasky LA, Nakamura GM, Smith DH, Fennie C, Shimasaki C, Patzer E, Berman P, Gregory T, and Capon DJ: Delineation of a region of the human immunodeficiency virus type 1 gp120 glycoprotein critical for interaction with CD4 receptor. Cell 1987;50:975–985. [DOI] [PubMed] [Google Scholar]
- 4.Lifson JD, Feinberg MB, Rayes GR, Rabin L, Banapour B, Chakrabarti S, Moss B, Wong-Staal F, Steimer KS, and Engleman E: Induction of CD4-dependent cell fusion by the HTLV-III/LAV envelope glycoprotein. Nature (London) 1986;323:725–729. [DOI] [PubMed] [Google Scholar]
- 5.Sodroski J, Goh WC, Rosen C, Campbell K, and Haseltine W: Role of the HTLV-III/LAV envelope in syncytium formation and cytopathicity. Nature (London) 1986;322:470–474. [DOI] [PubMed] [Google Scholar]
- 6.Ratner L, Haseltine W, Patarca R, Livak KJ, Starcich P, Josephs SJ, Doran ER, Rafalski JA, WHitehorn EA, Baumeister K, Ivanoff L, Petteway SR Jr., Pearson ML, Lautenberger JA, Papas TS, Ghrayeb J, Chang NT, Gallo RC, and Wong-Staal F: Complete nucleotide sequence of the AIDS virus, HTLV-III. Nature (London) 1985;313:277–284. [DOI] [PubMed] [Google Scholar]
- 7.Leonard CK, Spellman MW, Riddle L, Harris RJ, Thomas JN, and Gregory TJ: Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type I recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem 1990;265:10373–10382. [PubMed] [Google Scholar]
- 8.Gruters RA, Neefjes JJ, Tersmette M, de Goede RAY, Tulp A, Huisman HG, Miedema F, and Ploegh HL: Interference with HIV-induced syncytium formation and viral infectivity by inhibitors of trimming glucosidase. Nature (London) 1987;330:74–77. [DOI] [PubMed] [Google Scholar]
- 9.Matthews TJ, Weinhold KJ, Lyerly HK, Langlois AJ, Wigzell H, and Bolognesi DP: Interaction between the human T-cell lymphotropic virus type IIIB envelope glycoprotein gp120 and a surface antigen CD4: Role of carbohydrate in binding and cell fusion. Proc Natl Acad Sci (USA) 1987;84:5424–5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walker B, Kowalski M, Goh WC, Kozarsky K, Krieger M, Rosen C, Rohrschneider L, Haseltine WA, and Sodroski J: Inhibition of human immunodeficiency virus syncytium formation and virus replication by castanospermine. Proc Natl Acad Sci (USA) 1987;84:81202–8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevenson M, Meier C, Mann AM, Chapman N, and Wasiak A: Envelope glycoprotein of HIV induces interference and cytolysis resistance in CD4+ cells: mechanism for persistence in AIDS. Cell 1988;53:483–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poss ML, Mullins JI, and Hoover EA: Post-translational modifications distinguish the envelope glycoprotein of the immunodeficiency disease-inducing feline leukemia virus retrovirus. J Virol 1989;63:189–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stevenson M, Haggerty S, Lamonica C, Mann AM, Meier C, and Wasiak A: Cloning and characterization of human immunodeficiency virus type 1 variants diminished in the ability to induce syncytium-independent cytolysis. J Virol 1990;64:3792–3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanchez-Pescador R and Urdea MA: Use of purified synthetic deoxynucleotide primers for rapid dideoxynucleotide chain termination sequencing. DNA 1984;3:339–343. [DOI] [PubMed] [Google Scholar]
- 15.Fisher AG, Feinberg MB, Josephs SF, Harper ME, Marselle LM, Reyes G, Gonda MA, Aldovini A, Debouk C, Gallo RC, and Wong-Staal F: The trans-activator gene of HTLV-III is essential for virus replication. Nature 1986;320:367–371. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel TA: Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci (US) 1985;82:488–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanger F, Nicklen S, and Coulson AR: DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci (USA) 1977;74:5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miyoshi I, Taguchi H, Kubonishi S, Yoshimoto S, Ohrauki Y, Shirashi Y, and Akagi T: Type-C virus-producing cell lines derived from adult T-cell leukemia. Jpn J Cancer Res 1982;28:219–228. [Google Scholar]
- 19.Chen ISY, Quan SG, and Golde DW: Human T-cell leukemia virus type II transforms normal human lymphocytes. Proc Natl Acad Sci (USA) 1983;80:7006–7009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maddon PJ, Dalgleish AG, McDougal JS, Clapham PR, Weiss RA, and Axel R: The T4 gene encodes the AIDS virus receptor and is expressed in the immune system and the brain. Cell 1986;47:333–348. [DOI] [PubMed] [Google Scholar]
- 21.Terwilliger A, Proulx JP, Sodroski J, and Haseltine WA: Cell lines that express stably env gene products from three strains on HIV-1. J AIDS 1988;1:317–323. [PubMed] [Google Scholar]
- 22.He X, Skapek SX, Wikstrand CJ, Friedman HS, Trojanowski JQ, Kemshead JT, Cokaham HB, Bigner SH, and Bigner DD: Phenotypic analysis of four human medulloblastoma cell lines and transplantable xenografts. J Neuro Pathol Exp Neurol 1989;48:48–68. [DOI] [PubMed] [Google Scholar]
- 23.Stratton MR, Reeves BR, and Cooper CS: Misidentified cell (left). Nature 1989;337:311–312. [DOI] [PubMed] [Google Scholar]
- 24.Stevenson M, Stanwick TL, Dempsey MP, and Lamonica CA: HIV-1 replication is controlled at the level of T-cell activation and proviral integration. EMBO J 1990;9:1551–N1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cowan NJ, Dobner PR, Fuchs EV, and Cleveland DW: Expression of human α-tubulin genes: interspecies conservation of 3′ untranslated regions. Mol Cell Biol 1983;3:1738–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis KB, and Erlich HA: Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science 1988;239:487–491. [DOI] [PubMed] [Google Scholar]
- 27.Willey RL, Rutledge RA, Dias S, Folkes T, Theodore T, Buckler CE, and Martin MA: Identification of conserved and divergent domains within envelope gene of the acquired immunodeficiency syndrome retrovirus. Proc. Natl Acad Sci (USA) 1986;83:5038–5042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harada S, Koyanagi Y, and Yamamoto N: Infection of HTLV-III/LAV in HTLV-I-carrying cells MT-2 and MT-4 and application in a plaque assay. Science 1985;229:563–566. [DOI] [PubMed] [Google Scholar]
- 29.Clapham PR, Weber JN, Whitby D, McIntosh K, Dalgleish AG, Maddon PJ, Deen KC, Sweet RW, and Weiss RA: Soluble CD4 blocks the infectivity of diverse strains of HIV and SIV for T-cells and monocytes but not for brain and muscle cells. Nature (London) 1989;337:368–370. [DOI] [PubMed] [Google Scholar]
- 30.Harouse JM, Kunsch C, Hartie HT, Laughlin MA, Hoxie JA, Wigdahl B, and Gonzalez-Scarano F: CD4-independent infection of human neural cells by human immunodeficiency virus type 1. J Virol 1989;63:2527–2533, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weber J, Clapham P, McKeating J, Stratton M, Robey E, and Weiss R: Infection of brain cells by diverse human immunodeficiency virus isolates: Role of CD4 as receptor. J Gen Virol 1989;70:2653–2660. [DOI] [PubMed] [Google Scholar]
- 32.Boshart M, Weber F, Jahn G, Dorsch-Häsler K, Fleckenstein B, and Schaffner W: A very strong enhancer is located up-stream of an immediate early gene of human cytomegalovirus. Cell 1985;41:521–530. [DOI] [PubMed] [Google Scholar]
- 33.Yates JL, Warren N, and Sugden B: Stable replication of plasmids derived from Epstein-Barr virus in various mammalian cells. Nature 1985;313:812–815. [DOI] [PubMed] [Google Scholar]
- 34.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, and De Wilde MP: Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell 1989;59:103–112. [DOI] [PubMed] [Google Scholar]
- 35.Karacostas V, Nagashima K, Gonda MA, and Moss B: Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc Natl Acad Sci (USA) 1989;86:8963–8967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trono D, Feinberg MB, and Baltimore D: HIV-1 gag mutants can dominantly interfere with the replication of the wild-type virus. Cell 1989;59:113–120. [DOI] [PubMed] [Google Scholar]
- 37.Chaudhary VK, Mizukami T, Fuerst TR, FitzGerald DJ, Moss B, Pastan I, and Berger EA: Selective killing of HIV-infected cells by recombinant human CD4-Pseudomonas exotoxin hybrid protein. Nature 1988;335:369–372. [DOI] [PubMed] [Google Scholar]
- 38.Dewar RL, Vasudevachari MB, Natarajan V, and Salzman NP: Biosynthesis and processing of human immunodeficiency virus type 1 envelope glycoproteins: Effects of monensin on glycosylation and transport. J Virol 1989;63:2452–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cordonnier A, Riviére Y, Montagnier L, and Emerman M: Effects of mutations in hyper-conserved regions of the extra-cellular glycoprotein of human immunodeficiency virus type 1 on receptor binding. J Virol 1989;63:4464–4468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pitta AM, Rose JK, and Machamer CE: A single-amino-acid substitution eliminates the stringent carbohydrate requirement for intracellular transport of a viral glycoprotein. J Virol 1989;63:3801–3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kunsch C, Hartie HT, and Wigdahl B: Infection of human fetal dorsal route ganglion glial cells with human immunodeficiency virus type 1 involves an entry mechanism independent of the CD4 T4A epitope. J Virol 1989;63:5054–5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li XL, Moudgil T, Vinters HV, and Ho DD: CD4-independent productive infection of a neuronal cell line by human immunodeficiency virus type 1. J Virol 1990;64:1383–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]


