SUMMARY
Leptospirosis is the most widespread zoonosis in humans worldwide. In the United States, widespread detection of antibodies to leptospirosis have been identified in feral swine (Sus scrofa) with the highest detection of serovars, Bratislava, Icterohaemorrhagiae, and Pomona. Over the past few years, feral swine populations have expanded their geographical range and distribution in the United States with reports in at least 39 of 50 states. Since feral swine serve as reservoirs for serovars that can infect humans, it is important to understand the risk of transmission. In order to learn more about the probability that feral swine shed infectious leptospires, we collected kidneys and paired serum when possible from 677 feral swine in 124 counties of 29 states. These counties had previously been identified as antibody positive for Leptospira interrogans serovars Bratislava, Canicola, Grippotyphosa, Hardjo, Icterohaemorrhagiae or Pomona. Although exposure to these same six serovars of leptospirosis continued to be high (53% overall) in the counties we sampled, we detected leptospiral DNA in only 3·4% of feral swine kidneys tested. Based on these results, it appears that although feral swine can serve as a source of infection to humans, especially in those who are more likely to encounter them directly such as wildlife biologists, veterinarians, and hunters, the risk may be relatively low. However, further studies to examine the relationship between leptospiral shedding in the urine and kidneys in addition to culturing the organism are recommended in order to better understand the risk associated with feral swine.
Key words: Feral swine, kidney, Leptospira, leptospirosis, Sus scrofa
INTRODUCTION
Leptospirosis is a widespread zoonosis that is transmitted to humans and other animals via direct contact with the urine of infected animals, or indirectly with contaminated water or soil [1, 2]. Although human cases of leptospirosis are more commonly reported in the tropics, cases are still reported in non-tropical countries such as the United States usually through occupational or recreational activities [3]. Worldwide, human cases are likely underreported because symptoms are similar to many other diseases and can include fever with sudden onset, severe headache, chills, cough, acute abdominal pain, severe myalgia, rash, nausea, diarrhoea, and vomiting, or even death [4].
There are about 300 serovars of Leptospira interrogans, a pathogenic species of bacteria that causes leptospirosis, with certain serovars more adapted to particular hosts [5]. For example, serovar Bratislava is associated with horses (Equus caballus) and pigs (Sus scrofa), Canicola in dogs (Canis lupus familiaris), Hardjo in cattle (Bos taurus), Grippotyphosa in raccoons (Procyon lotor), Icterohaemorrhagiae in rats (Rattus spp.), and Pomona in pigs [1, 3]. Almost any mammal can serve as a carrier, but rats are considered the major carriers for most zoonotic leptospires [2]. Pigs have been identified as reservoir hosts for the disease in humans [6], which is significant since pigs have similar anatomy, genetics and physiology to humans [7], and could serve as a source of infection to those with the potential for occupational exposure. The most common serovars infecting domestic swine in the United States are Bratislava, Grippotyphosa, and Pomona, with occasional reports of Canicola and Icterohaemorrhagiae [8]. Infection in pigs rarely causes death and is generally characterized by abortion, weakness, inability to stand, muscle spasms, and muscle rigidity [4]. In the United States, feral swine (Sus scrofa) are defined as escaped domestic swine, Eurasian wild boars or hybrids of the two, and while current populations are estimated to exceed 6 million nationwide, geographical distribution and population size have increased rapidly over the last few decades through natural range expansion and accidental or intentional release [9]. Since numerous viral and bacterial pathogens have been documented in feral swine including leptospirosis [9, 10], there is concern that humans with a greater likelihood of exposure such as wildlife biologists, veterinarians, hunters and outdoor adventure racers may be at a higher risk of becoming infected. However, with the recent range expansion, feral swine have increasingly been reported in urban environments such as golf courses, city parks, residents’ backyards, and cemeteries further increasing the risk of exposure.
Transmission of leptospirosis by feral swine is of particular concern since the disease can infect humans, dogs, domestic pigs and other livestock, and widespread exposure has been documented in feral swine [11–13]. Although serological prevalence of leptospirosis in feral swine has been examined in the United States and other countries [11, 13–17], we are unaware of any national scale efforts in the United States to quantify leptospiral shedding in the species. Since pathogenic leptospires have a predilection for the kidneys of infected swine [4], we tested kidneys and paired serum of feral swine collected in counties previously identified as antibody positive [11]. Our objectives were to assess the likelihood that feral swine are actively infected and thus at risk of transmitting the pathogen in their urine, and to examine the association between shedding and serological prevalence to better understand the risk that feral swine pose to humans, dogs, and livestock.
METHODS
Sample collection
The U.S. Department of Agriculture's Wildlife Services removes feral swine for wildlife damage management purposes and routinely collects sera from a subset for pathogen surveillance. Since removal of feral swine is dependent upon access to the landowner's property, samples are collected opportunistically. Sera are tested for various pathogens at the time of collection and additional sera are archived at −80 °C for future testing. Prior to this study, archived sera were tested to identify counties with antibody-positive feral swine (target counties). A county was considered positive if sera tested positive by microagglutination for serovars Bratislava, Canicola, Grippotyphosa, Hardjo, Icterohaemorrhagiae, or Pomona [11]. In this study, a kidney and serum sample were collected from the same feral swine when possible in target counties. From April 2012 to September 2014, 677 kidneys and serum from 642 of the same feral swine were collected in 124 counties in 29 states.
Blood was collected immediately after death via cardiac puncture or orbital bleed, the blood was allowed to clot, and then centrifuged. Sera were aliquoted into 2 ml cryogenic vials, labelled with a barcode unique to the feral swine, and stored at −80 °C until testing. The kidney was removed post-mortem by extracting it through a 6- to 8-inch incision at the mid paralumbar region. A single kidney was then placed into a Ziploc® (S. C. Johnson & Sons Inc., USA) or Whirlpak® (Nasco, USA) bag, labelled with a barcode unique to the feral swine, and stored at −80 °C until shipping.
Feral swine were classified as juvenile (<2 months), sub-adult (2 months to 1 year), or adult (>1 year) by examining the erupted teeth on the lower jaw [18]. In addition, sex, collection site, GPS coordinates, county, and state were recorded on a standardized datasheet.
Microagglutination test (MAT)
Serum was tested at Colorado State University Veterinary Diagnostic Laboratory (CSUVDL) with MAT as described previously [11, 19]. Briefly, a panel to detect antibodies to six serovars of Leptospira interrogans (Bratislava, Canicola, Grippotyphosa, Hardjo, Icterohaemorrhagiae, Pomona) was conducted; all reference serovars were obtained from the National Veterinary Services Laboratories in Ames, Iowa. A titre of ⩾1:100 was considered positive. MAT results were reported as the endpoint dilution of serum where 50% agglutination of cells was observed with twofold dilutions, starting at 1:100. Titres >1:6400 were not measured to their endpoint since titres ⩾1:800 were considered evidence of recent or current infection [13, 20]. The serovar in which agglutination was detected at the highest dilution was considered the infective serogroup, which for some samples included multiple serovars.
Nucleic acid isolation and real-time PCR (qPCR)
Half-centimetre square slices of tissue were dissected from the renal medulla of each kidney and homogenized using a ceramic bead-beater tube for 60 s, centrifuged at 5000 g, and the supernatant was removed. DNA was extracted using the QIAamp DNA mini-kit (Qiagen, USA), according to the manufacturer's specifications. Finally, the DNA was eluted in 100 µl water.
The Genesig® standard kit for quantification of leptospirosis genomes (Primerdesign Ltd, UK) which targets the outer membrane lipl32 was used to for detection of Leptospira by qPCR. Samples with cycle threshold (Ct) values <35 were treated as putative positives. Samples with Ct values >35 were considered negative and no further testing was performed.
Sequencing
Kidneys that tested positive by qPCR were subjected to additional conventional PCR, in order to produce a band for sequencing. Extracted DNA was subjected to conventional PCR [21] targeting the 23S rDNA region of L. interrogans followed by agarose gel electrophoresis on all PCR products. Bands produced by positive samples were cut out of the gel and processed for sequencing using the QIAquick PCR Purification kit (Qiagen) according to the manufacturer's specifications. DNA was sequenced by a commercial sequencing company. Standard nucleotide BLAST was used to confirm that sequences matched Leptospira spp. Samples were considered positive if both PCR tests and sequencing results were positive.
Statistical analysis
Descriptive statistics and exact binomial 95% confidence intervals (CIs) were generated using Microsoft Excel (Microsoft Corp., USA). We investigated the effects of sex and age class on the antibody status of individual feral swine. Generalized linear mixed models were fit using maximum likelihood in the R statistical platform using packages MuMIn (version 1.15.6) [22] and lme4 (version 1.1–10) [23]. Model selection was performed using Akaike's Information Criterion (AIC) and the model with the lowest AIC value was viewed as the most parsimonious. The generalized linear mixed model with a binomial distribution examined the effect of sex and age on leptospirosis antibody status (positive/negative). County was included as a random effect to account for correlation associated with samples collected close together in space. This analysis was not extended to PCR-positive feral swine because of the low number of positives.
RESULTS
Sample composition
The samples were collected from adults (81·4%), sub-adults (14·0%), and juveniles (4·6%). The models with the most support for antibody-positive feral swine included both age and sex. Sex provided 56% of model weights, while age class provided 28% of model weights (Supplementary Table S1); however, parameter estimates indicated that neither sex nor age were significantly associated with serological results (Supplementary Table S2). Samples were collected from about the same number of males (49·8%) and females (50·2%) (Table 1).
Table 1.
Antibody prevalence and antigen prevalence of feral swine serum and kidneys tested for exposure to Leptospira interrogans serovars Bratislava, Canicola, Grippotyphosa, Hardjo, Icterohemorrhagiae, and Pomona by age class and gender
| Variable | Kidneys | Bratislava | Canicola | Grippotyphosa | Hardjo | Icterohaemorrhagiae | Pomona | All serovars |
|---|---|---|---|---|---|---|---|---|
| Age class (no. of kidneys, serum) | ||||||||
| Adult (551, 522) | 2·5 (1·5–4·2) | 33·0 (29·1–37·1) | 10·3 (8·0–13·3) | 9·2 (7·0–12·0) | 4·6 (3·1–6·8) | 28·4 (24·7–32·4) | 15·3 (12·5–18·7) | 56·7 (52·4–60·9) |
| Sub-adult (95, 93) | 3·2 (1·1–8·9) | 19·4 (12·6–28·5) | 6·5 (3·0–13·4) | 5·4 (2·3–12·0) | 3·2 (1·1–9·1) | 14·0 (8·4–22·5) | 4·3 (1·7–10·5) | 33·3 (24·6–43·4) |
| Juvenile (31, 27) | 19·4 (9·2–36·3) | 25·9 (13·2–44·7) | 7·4 (2·1–23·4) | 25·9 (13·2–44·7) | 18·5 (8·2–36·7) | 14·8 (5·9–32·5) | 11·1 (3·9–28·1) | 48·2 (30·7–66·0) |
| Gender (no. of kidneys, serum) | ||||||||
| Male (337, 318) | 3·0 (1·6–5·4) | 27·4 (22·8–32·5) | 8·2 (5·6–11·7) | 9·4 (6·7–13·2) | 5·4 (3·4–8·4) | 21·7 (17·5–26·6) | 15·1 (11·6–19·4) | 50·0 (44·5–55·5) |
| Female (340, 324) | 3·8 (2·3–6·4) | 34·0 (29·0–39·3) | 11·1 (8·1–15·0) | 9·3 (6·6–12·9) | 4·6 (2·8–7·5) | 29·6 (24·9–34·8) | 12·0 (8·9–16·0) | 55·9 (50·4–61·2) |
Values given are % positive (95% confidence interval).
Leptospiral detection
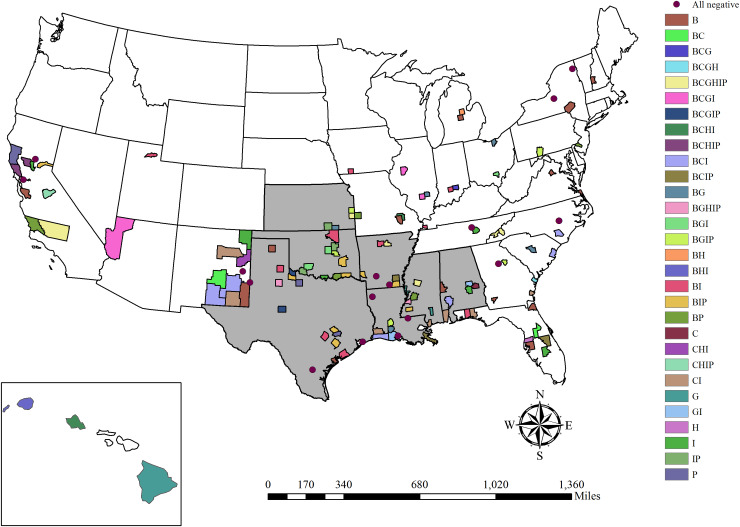
Twenty-three (3·4%, 95% CI 2·3–5·1) out of 677 kidneys collected in seven states were identified as positive (Table 1, Fig. 1). Corresponding serum from the feral swine with positive kidneys was antibody positive for all animals from which there was serum available to test (20/23), and all of these tested positive for at least one serovar ⩾800 except for two (Table 2). The apparent prevalence based on kidneys was higher in Mississippi than any other state (26·4%, 95% CI 16·4–39·6) (Supplementary Table S3).
Fig. 1.
Locations where kidney and serum samples were collected from feral swine. All samples were tested for exposure to Leptospira interrogans serovars Bratislava (B), Canicola (C), Grippotyphosa (G), Hardjo (H), Icterohaemorrhagiae (I), and Pomona (P). Counties are shaded by antibody positive serovar (titre ⩾100) and states are shaded if any positive kidneys were identified.
Table 2.
Location where kidney and serum samples from feral swine were collected that tested positive for Leptospira interrogans by PCR with corresponding antibody prevalence estimates by L. interrogans serovar with 95% confidence intervals (CI). Corresponding serum was positive for at least one serovar from all feral swine except three collected in Holmes County, Mississippi for which there was no serum to test
| Serovars (95% CI) | |||||||
|---|---|---|---|---|---|---|---|
| State/county (no. of kidneys, serum) | % pos. (kidneys) (95% CI) | Bratislava | Canicola | Grippotyphosa | Hardjo | Icterohaemorrahagiae | Pomona |
| Alabama (31, 31) | 3·2 (0·6–16·2) | 16·1 (7·1–32·6) | 12·9 (5·1–28·9) | 6·5 (1·8–20·7) | 0 (0–11·0) | 38·7 (23·7–56·2) | 0 (0–11·0) |
| Clarke (10, 10) | 10·0 (1·8–40·4) | 30·0 (10·8–60·3) | 20·0 (5·7–51·0) | 0 (0–27·8) | 0 (0–27·8) | 60·0 (31·3–83·2) | 0 (0–27·8) |
| Arkansas (25, 25) | 8·0 (2·2–25·0) | 48·0 (30·0–66·5) | 8·0 (2·2–25·0) | 4·0 (.7–19·5) | 4·0 (0·7–19·5) | 20·0 (8·9–39·1) | 44·0 (26·7–62·9) |
| Ashley (5, 5) | 20·0 (3·6–62·5) | 40·0 (11·8–76·9) | 0 (0–43·5) | 0 (0–43·5) | 0 (0–43·5) | 20·0 (3·6–62·5) | 100 (56·6–100) |
| Drew (5, 5) | 20·0 (3·6–62·5) | 60·0 (23·1–88·2) | 20·0 (3·6–62·5) | 0 (0–43·5) | 0 (0–43·5) | 20·0 (3·6–62·5) | 80·0 (37·6–96·4) |
| Kansas (28, 28) | 7·1 (2·0–22·7) | 39·3 (23·6–57·6) | 3·6 (.6–17·7) | 10·7 (3·7–27·2) | 7·1 (2·0–22·7) | 28·6 (15·3–47·1) | 14·3 (5·7–31·5) |
| Bourbon (14, 14) | 7·1 (1·3–31·5) | 42·9 (21·4–67·4) | 7·1 (1·3–31·5) | 7·1 (1·3–31·5) | 14·3 (4·0–39·9) | 28·6 (11·7–54·7) | 14·3 (4·0–39·9) |
| Linn (1, 1) | 100 (20·7–100) | 100 (20·7–100) | 0 (0–79·4) | 100 (20·7–100) | 0 (0–79·4) | 100 (20·7–100) | 100 (20·7–100) |
| Louisiana (46, 41) | 2·2 (0·4–11·3) | 12·2 (5·3–25·5) | 9·8 (3·9–22·6) | 12·2 (5·3–25·5) | 0 (0–8·6) | 26·8 (15·7–41·9) | 4·9 (1·4–16·1) |
| Acadia (7, 5) | 14·3 (2·6–51·3) | 20·0 (3·6–62·5) | 0 (0–43·5) | 20·0 (3·6–62·5) | 0 (0–43·5) | 0 (0–43·5) | 0 (0–43·5) |
| Mississippi (53, 47) | 26·4 (16·4–39·6) | 46·8 (33·3–60·8) | 6·4 (2·2–17·2) | 27·7 (16·9–41·8) | 17·0 (8·9–30·1) | 21·3 (12·0–34·9) | 42·3 (29·5–56·7) |
| Claiborne (11, 11) | 45·5 (21·3–72·0) | 81·8 (52·3–94·9) | 0 (0–25·9) | 9·1 (1·6–37·7) | 9·1 (1·6–37·7) | 36·4 (15·2–64·6) | 72·7 (43·4–90·3) |
| Hinds (4, 4) | 25·0 (4·6–69·9) | 50·0 (15·0–85·0) | 0 (0–49·0) | 0 (0–49·0) | 0 (0–49·0) | 0 (0–49·0) | 50·0 (15·0–85·0) |
| Holmes (25, 19) | 32·0 (17·2–51·6) | 52·6 (31·7–72·7) | 10·5 (2·9–31·4) | 52·6 (31·7–72·7) | 36·8 (19·2–59·0) | 21·1 (8·5–43·3) | 47·4 (27·3–68·3) |
| Oklahoma (61, 53) | 1·6 (0·3–8·7) | 28·3 (18·0–41·6) | 0 (0–6·8) | 7·6 (3·0–17·9) | 0 (0–6·8) | 30·2 (19·5–43·5) | 17·0 (9·2–29·2) |
| Okfuskee (5, 5) | 20·0 (3·6–62·5) | 40·0 (11·8–76·9) | 0 (0–43·5) | 40·0 (11·8–76·9) | 0 (0–43·5) | 60·0 (23·1–88·2) | 40·0 (11·8–76·9) |
| Texas (60, 58) | 3·3 (0·9–11·4) | 40·0 (28·1–52·5) | 5·2 (1·8–14·1) | 6·9 (2·7–16·4) | 1·7 (0·3–9·1) | 20·7 (12·3–32·8) | 34·5 (23·6–47·3) |
| Dickens (15, 15) | 13·3 (3·7–37·9) | 40·0 (19·8–64·3) | 0 (0–20·4) | 6·7 (1·2–29·8) | 6·7 (1·2–29·8) | 20·0 (7·1–45·2) | 60·0 (35·8–80·2) |
Sera
Of 642 serum samples, 53·0% (95% CI 49·1–56·8) were antibody positive for at least one serovar. Of the antibody-positive samples (n = 160), 47·1% were positive for multiple servoars. Bratislava was the most commonly identified serovar (30·7%), followed by Icterohaemorrhagiae (25·7%), Pomona (13·6%), Canicola (9·7%), Grippotyphosa (9·4%), and Hardjo (5·0%, Table 3). The most common co-infections were with Bratislava and Icterohaemorrhagiae (n = 34), Pomona and Bratislava (n = 22), and Icterohaemorrhagiae and Canicola (n = 15). Based on antibody titre, feral swine were most commonly identified as actively infected with serovars Bratislava and Pomona (Table 3).
Table 3.
Number of feral swine serum samples testing positive for six serovars of Leptospira interrogans by titre and apparent antibody prevalence of each serovar
| Serovar (n = 642) | Titre | |||||||
|---|---|---|---|---|---|---|---|---|
| 100 | 200 | 400 | 800 | 1600 | 3200 | ⩾6400 | % positive (95% CI) | |
| Bratislava | 51 | 57 | 51 | 22 | 12 | 0 | 4 | 30·7 (27·2–34·4) |
| Canicola | 29 | 21 | 8 | 2 | 2 | 0 | 0 | 9·7 (7·6–12·2) |
| Grippotyphosa | 24 | 13 | 8 | 4 | 3 | 4 | 4 | 9·4 (7·3–11·9) |
| Hardjo | 11 | 15 | 3 | 1 | 2 | 0 | 0 | 5·0 (3·6–7·0) |
| Icterohaemorrhagiae | 92 | 58 | 10 | 2 | 2 | 0 | 1 | 25·7 (22·5–29·2) |
| Pomona | 18 | 19 | 12 | 15 | 9 | 7 | 7 | 13·6 (11·1–16·4) |
CI, confidence interval.
DISCUSSION
Leptospiral infection appears to be infrequent in feral swine in the United States since only 3·4% of feral swine kidneys tested positive even though they were collected in counties that previously had been identified as antibody positive. The low apparent prevalence of leptospires is surprising since wild boars sampled during similar studies in Japan and Germany identified apparent prevalences of 15% and 10%, respectively [16, 24]. One possible explanation for the apparent discrepancy between serology (53% antibody positive) and detection of pathogenic leptospires is that feral swine may shed the organism intermittently after being exposed, similar to that documented in domestic pigs [10], and the organism may exist at a level below PCR detection between shedding events thus further reducing the likelihood of detection. Another reason for the disparate results may be that feral swine maintain antibodies long after exposure, or, repeated exposures to the bacterial spirochaete while wallowing in swampy habitats may shorten the period of bacterial shedding. Yet another possibility is that the leptospires were not present in the specific portion of the kidney that was tested.
Many of the feral swine had low-level antibody titres (100) to serovar Icterohaemorrhagiae, and although seropositive according to our cut-off value of ⩾100, it is unlikely they were shedding pathogenic Leptospira. Detection of serovar Icterohaemorrhagiae is not surprising either since this serovar is common in rats [3], and it is likely that feral swine encountered rats directly or indirectly through contaminated urine, and subsequently developed antibodies. Serovar Icterohaemorrhagiae is often identified as the infecting serovar in human cases of leptospirosis [25–27], but since we only identified five feral swine with titres ⩾800 (Table 3), it appears that the risk of feral swine transmitting this serovar to humans or other animals is low. Although infection with serovars Pomona and Bratislava is not as common in humans as other serovars, cases have been reported [27–29], and titres against these two serovars were highest in the feral swine we tested (Table 3). Based on the high titres indicating active infection, and the high frequency of seropositive animals for Bratislava and Pomona serovars, humans that come into contact with feral swine are most likely to be exposed to these serovars. It is not surprising that these serovars were commonly identified in the feral swine we tested because they have been reported previously in both domestic swine [6, 8] and feral swine [15, 16]. Serovars Pomona and Bratislava have also been commonly identified in feral swine not only in the United States [11–13, 30], but also in countries around the world such as Spain [14], Italy [31], Australia [15, 32], Japan [24], and Germany [16]. In addition, transmission of serovar Pomona by infected pigs to calves has been documented, [6] suggesting that feral swine may also transmit the pathogen to cattle, thus further impacting livestock.
Although other studies have identified a higher seropositivity in adults [16], we did not detect an association between exposure and age (Supplementary Table S2). There were also no strong associations between sexes in seropositivity (Supplementary Table S2) which is similar to other studies [16, 24]. The apparent prevalence of leptospires detected in Mississippi was higher than in any other state. Although the warm and generally humid climate in Mississippi is ideal for leptospiral survival, it is surprising that the same trend was not observed in other states throughout the southeastern United States with a similar climate. However, this is not the first report of high apparent prevalence of leptospirosis in a wildlife species in Misssissippi. An antibody apparent prevalence of 77% (n = 50) was reported in cottontail (Sylvilagus floridanus) and swamp (S. aquaticus) rabbits sampled in the Mississippi Delta, and focal nephritis (possible indicator of past or present leptospiral infection) was detected in 92% of kidneys [33]. Since this region of the state has a swampy habitat it may be more likely to support leptospiral survival and be a locally important area of high risk. Perhaps the reason that we detected such a high apparent prevalence of leptospiral shedding was that our targeted sampling coincided with focal areas of high prevalence. Additional sampling in the southeast is recommended to determine if this pattern is consistent or repeatable.
Based on our apparent leptospiral prevalence data, using a serological titre of ⩾800 to identify active infection may not be accurate since, based on this criteria, the prevalence of actively infected animals would have been 12·3% (n = 79, Table 3), which is much higher than what we actually detected. However, since kidneys were not considered leptospiral positive unless both PCR assays and sequencing were positive, our conservative testing protocol may have underestimated the true prevalence.
Since we identified leptospires in the kidneys by PCR and not by culture, we are unsure which serovar was most commonly being shed in the urine (via the kidney). We recommend additional studies to culture the organism to allow comparison between serovar excretion in the urine and exposure indicated by the serological results. This could also further elucidate the risk to humans associated with leptospiral shedding in feral swine. Since leptospirosis is an emerging disease issue and is considered the most important zoonosis worldwide, a better understanding of the pathogen distribution in feral swine is essential. As this reservoir continues to expand its geographical range and distribution, the potential for dispersing pathogenic leptospires that may infect humans is further increased. Studies in other countries have suggested that surveillance should be conducted not only in feral swine but also in humans in the same area to better assess the risk and directionality of transmission [16], and we recommend this approach in the United States as well to ensure the health of those exposed through occupational or recreational exposure.
ACKNOWLEDGEMENTS
We thank all of the wildlife biologists and technicians who participated by collecting the samples included in this paper and spending many hours trapping feral swine and preparing samples for testing. We also thank Christina Weller and Sean McCluskey who provided technical support for this project. Mention of trade names or commercial products in this work is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture.
Supplementary material
For supplementary material accompanying this paper visit http://doi.org/10.1017/S0950268816002247.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Adler B, de la Peña Moctezuma A. Leptospira and leptospirosis. Veterinary Microbiology 2010; 140: 287–296. [DOI] [PubMed] [Google Scholar]
- 2.Evangelista KV, Coburn J. Leptospira as an emerging pathogen: a review of its biology, pathogenesis and host immune responses. Future Microbiology 2010; 5: 1413–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bharti AR, et al. Leptospirosis: a zoonotic disease of global importance. Lancet Infectious Diseases 2003; 3: 757–771. [DOI] [PubMed] [Google Scholar]
- 4.Campagnolo ER, et al. Analysis of the 1998 outbreak of leptospirosis in Missouri in humans exposed to infected swine. Journal of the American Veterinary Medical Association 2000; 216: 676–682. [DOI] [PubMed] [Google Scholar]
- 5.Ahmed N, et al. Multilocus sequence typing method for identification and genotypic classification of pathogenic Leptospira species. Annals of Clinical Microbiology and Antimicrobials 2006; 5: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnstein T, Baker JA. Leptospirosis in swine caused by Leptospira pomona. Journal of Infectious Diseases 1954; 94: 53–64. [DOI] [PubMed] [Google Scholar]
- 7.Meurens F, et al. The pig: a model for human infectious diseases. Trends in Microbiology 2012; 20: 50–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bolin CA, Cassells JA. Isolation of Leptospira interrogans serovars bratislava and hardjo from swine at slaughter. Journal of Veterinary Diagnostic Investigation 1992; 4: 87–89. [DOI] [PubMed] [Google Scholar]
- 9.Bevins SN, et al. Consequences associated with the recent range expansion of nonnative feral swine. Bioscience 2014; 64: 291–299. [Google Scholar]
- 10.Meng X, Lindsay D, Sriranganathan N. Wild boars as sources for infectious diseases in livestock and humans. Philosophical Transactions of the Royal Society of London, Series B: Biological Sciences 2009; 364: 2697–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pedersen K, et al. Widespread detection of antibodies to Leptospira in feral swine in the United States. Epidemiology and Infection 2015; 143: 2131–2136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saliki JT, Rodgers SJ, Eskew G. Serosurvey of selected viral and bacterial diseases in wild swine from Oklahoma. Journal of Wildlife Diseases 1998; 34: 834–838. [DOI] [PubMed] [Google Scholar]
- 13.Chatfield J, et al. Serosurvey of leptospirosis in feral hogs (Sus scrofa) in Florida. Journal of Zoo and Wildlife Medicine 2013; 44: 404–407. [DOI] [PubMed] [Google Scholar]
- 14.Vicente J, et al. Antibodies to selected viral and bacterial pathogens in European wild boars from southcentral Spain. Journal of Wildlife Diseases 2002; 38: 649–652. [DOI] [PubMed] [Google Scholar]
- 15.Mason R, et al. Leptospira interrogans antibodies in feral pigs from New South Wales. Journal of Wildlife Diseases 1998; 34: 738–743. [DOI] [PubMed] [Google Scholar]
- 16.Jansen A, et al. Leptospirosis in urban wild boars, Berlin, Germany. Emerging Infectious Diseases 2007; 13: 739–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vengust G, et al. Leptospira antibodies in wild boars (Sus scrofa) in Slovenia. European Journal of Wildlife Research 2008; 54: 749–752. [Google Scholar]
- 18.Matschke GH. Aging European wild hogs by dentition. Journal of Wildlife Management 1967; 31: 109–113. [Google Scholar]
- 19.OIE. Leptospirosis. In: Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. Paris, France: Office International des Epizooties, 2008, pp. 251–264. [Google Scholar]
- 20.U.S. Department of Health & Human Services. Leptospirosis (Leptospira interrogans) 2013 case definition (https://wwwn.cdc.gov/nndss/conditions/leptospirosis/case-definition/2013/). Accessed 5 July 2016.
- 21.Woo T, et al. Identification of pathogenic Leptospira genospecies by continuous monitoring of fluorogenic hybridization probes during rapid-cycle PCR. Journal of Clinical Microbiology 1997; 35: 3140–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton K. MuMIn: Multi-Model Inference, 2016 (http://CRAN.R-project.org/package=MuMIn).
- 23.Bates D, et al. Fitting linear mixed-effects models using lme4. arXiv preprint arXiv:14065823 2014.
- 24.Koizumi N, et al. Prevalence of Leptospira spp. in the kidneys of wild boars and deer in Japan. Journal of Veterinary Medical Science 2009; 71: 797–799. [DOI] [PubMed] [Google Scholar]
- 25.Ciceroni L, et al. Epidemiological trend of human leptospirosis in Italy between 1994 and 1996. European Journal of Epidemiology 2000; 16: 79–86. [DOI] [PubMed] [Google Scholar]
- 26.Adesiyun AA, et al. Human leptospirosis in the Caribbean, 1997–2005: characteristics and serotyping of clinical samples from 14 countries. Revista Panamericana Salud Publica 2011; 29: 350–357. [PubMed] [Google Scholar]
- 27.Christova I, Tasseva E, Manev H. Human leptospirosis in Bulgaria, 1989–2001: epidemiological, clinical, and serological features. Scandinavian Journal of Infectious Diseases 2003; 35: 869–872. [DOI] [PubMed] [Google Scholar]
- 28.Desmarchelier P. Human leptospirosis in New South Wales, 1975–1985. Medical Journal of Australia 1987; 146: 353–357. [DOI] [PubMed] [Google Scholar]
- 29.Whitney EAS, et al. Prevalence of and risk factors for serum antibodies against Leptospira serovars in US veterinarians. Journal of the American Veterinary Medical Association 2009; 234: 938–944. [DOI] [PubMed] [Google Scholar]
- 30.Corn J, et al. Survey of selected diseases in wild swine in Texas. Journal of the American Veterinary Medical Association 1986; 189: 1029–1032. [PubMed] [Google Scholar]
- 31.Ebani VV, et al. Prevalence of Leptospira and Brucella antibodies in wild boars (Sus scrofa) in Tuscany, Italy. Journal of Wildlife Diseases 2003; 39: 718–722. [DOI] [PubMed] [Google Scholar]
- 32.Ridoutt C, et al. Detection of brucellosis and leptospirosis in feral pigs in New South Wales. Australian Veterinary Journal 2014; 92: 343–347. [DOI] [PubMed] [Google Scholar]
- 33.Shotts J, et al. Leptospirosis in cottontail and swamp rabbits of the Mississippi delta. Journal of Wildlife Diseases 1971; 7: 115–117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://doi.org/10.1017/S0950268816002247.
click here to view supplementary material