Abstract
A 30-kb region surrounding the replication origin in Caulobacter crescentus was analyzed. Comparison to the genome sequence of another α-proteobacterium, Rickettsia prowazekii, revealed a conserved cluster of genes (RP001, hemE, hemH, and RP883) that overlaps the established origin of replication in C. crescentus and the putative origin of replication in R. prowazekii. The genes flanking this cluster differ between these two organisms. We therefore propose that this conserved gene cluster can be used to identify the origin of replication in other α-proteobacteria.
Caulobacter crescentus and Rickettsia prowazekii behave very differently despite being members of the α-subdivision of the gram-negative proteobacteria group. Unlike the free-living and nonpathogenic C. crescentus, R. prowazekii is an obligate intracellular parasite that causes epidemic louse-borne typhus in humans (14). Phylogenetic studies indicate that R. prowazekii is the closest living relative to the eukaryotic mitochondrion, thus suggesting that the mitochondrion may have originated from the ancestral group which spawned the α-proteobacteria group (2, 21).
C. crescentus divides asymmetrically to produce two cell types that differ in both their morphological and developmental programs (23). In the sessile stalked cell, chromosome replication initiates immediately after cell division, whereas in the chemotatic flagellated swarmer cell, replication is repressed until the swarmer cell differentiates into the stalked-cell type (18). However, R. prowazekii does not have a complex developmental cycle. Instead, the nonflagellated pleomorphic organism divides by binary fission (15).
The C. crescentus replication origin and the putative origin region of R. prowazekii have been determined by previous studies (2, 17). C. crescentus is a model organism for replication studies since synchronized populations can be isolated on a density gradient (9). The C. crescentus replication origin (Cori) was initially identified by in vivo 32P labeling of the earliest replicating DNA and by autonomous plasmid replication assays (17). Sequence analysis of Cori suggest both similarities and differences with the Escherichia coli replication origin (oriC). Cori possess elements (AT-rich region, 13-mers, and DnaA boxes) that are similar to those of oriC in E. coli. However, Cori also possesses unique sequence features. For example, five iterons (TTAA-N7-TTAA) identified as binding sites for the response regulator CtrA (cell cycle transcription regulator) are implicated in repressing chromosome replication (25, 28). A second feature is the hemE (uroporphyrinogen decarboxylase) homolog (17), whose 5′ end with its weak promoter (Pw) overlaps the essential Cori autonomous replication sequences (18). A strong transcription promoter (Ps) is located 5′ to hemE (Fig. 1C) and overlaps the AT-rich region and CtrA binding sites a and b (Fig. 2B). This Ps promoter produces transcripts that are poorly translated, and most hemE transcription is directed from the weak promoter (Pw; Fig. 1C and 2B). Interestingly, Ps transcription coincides with DNA replication in stalked cells and may play a role in Cori replication initiation (18). The organizational differences observed between C. crescentus and E. coli replication origins suggests an alternative class of replication origins in the α-subdivision of proteobacteria.
FIG. 1.
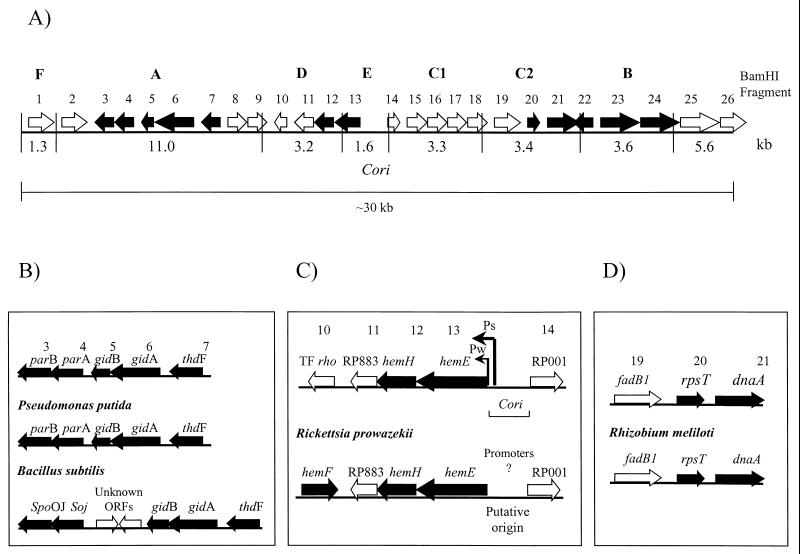
Gene clusters in the ∼30-kb region of earliest replicating DNA in C. crescentus. (A) Solid boxes represent previously described C. crescentus genes, and open boxes represent potential genes with high homology to those found in genomes of closely related bacteria (Tables 1 and 2). (B) Comparative parB-parA-gidB-gidA-thdF gene clusters of C. crescentus, P. putida, and B. subtilis. (C) Comparative RP883-hemH-hemE-RP001 gene cluster around the replication origins of C. crescentus and R. prowazekii. The strong promoter (Ps), the weak promoter (Pw), and the RP001 promoter (P3) are indicated for Cori; the rho ORF is an E. coli homolog of transcription termination factor rho also observed in R. sphaeroides (12) and R. prowazekii (α-proteobacteria). (D) Comparative fadB1-rpsT-dnaA gene cluster of C. crescentus and R. meliloti.
FIG. 2.
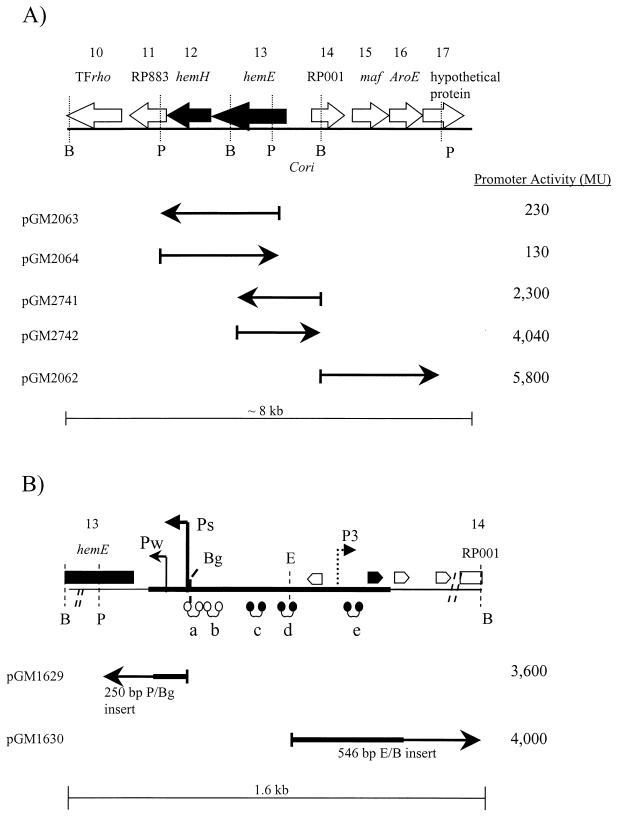
β-galactosidase transcription assays of pRK290lacZ plasmids containing selected fragments of the 30-kb region of earliest replicating DNA. Transcription strengths of each plasmid construct are expressed in Miller units (MU). Background level of the vector alone control is considered to be approximately 200 to 300 MU (data not shown). (A) Transcription assays focused on DNA involving ORFs 10 to 17 described in Tables 1 and 2. (B) Representative region of Cori (18). Thick line, minimal fragment for autonomous plasmid replication; pointed boxes, potential DnaA boxes; filled box, the closest sequence (TGATCCACA [8- or 9-bp match]) to the E. coli consensus; open boxes, weaker matches to the E. coli consensus sequence; dumbbells, CtrA binding sites; closed dumbbells, the CtrA consensus sequence, GTTAA-N7-TTAA; open dumbbells, the 8- or 9-bp matches to this consensus sequence. Abbreviations of restriction enzymes: B, BamHI; Bg, Bg/II; E, EcoRI, P, PstI.
The putative replication origin in R. prowazekii inferred by a switch in GC bias (16) possesses DnaA boxes based on comparison to the E. coli consensus sequence (2); however, the method of initiation or control of replication has not been determined.
This study reports the gene clusters and preliminary analysis of the 30-kb region of earliest replicating DNA in the C. crescentus genome. Based on the identical gene cluster spanning Cori and the R. prowazekii putative replication origin, we propose that this particular gene cluster be utilized to aid in identification and characterization of replication origins in other α-proteobacteria.
DNA sequence analysis of the earliest replicating region in C. crescentus.
The ∼30-kb region of earliest replicating DNA was subcloned as seven BamHI fragments (17) (Fig. 1A), as well as five PstI fragments and four HindIII fragments in pBluescript II plasmid vectors (Stratagene). Both ends of these subclones were sequenced from minipreps (Qiagen) by using the Sequitherm kit (Epicentre Technologies Inc.) and Queen's University Sequencing Centre (Kingston, Ontario, Canada). These sequences were then matched with preliminary sequence data (contig no. 12574) obtained from The Institute for Genomic Research website (http://www.tigr.org). The matched sequences of the ∼30-kb region were assembled on the basis of a restriction endonuclease map of Cori Cosmid I (17) and previously described C. crescentus genes listed in Table 1. Potential open reading frames (ORFs) were determined by using the ORF Finder program at the National Center for Biotechnology Information website (http://www.ncbi.nlm.nih.gov). Significant homologies (≥50%) of the ORFs to genes of published proteins were determined by the BLASTX program (1) with the nr database, as represented in Table 2.
TABLE 1.
Previously described C. crescentus genes near the replication origin and their homologies to R. prowazekii genes
| Box no. | C. crescentus gene | Reference or GenBank accession no. | % Homology to R. prowazekii gene |
|---|---|---|---|
| 3 | parB | 20 | 53 |
| 4 | parA | 20 | 74 |
| 5 | gidB | AAD40694, AAD40695, AAD40696 | 58 |
| 6 | gidA | AAD40694, AAD40695, AAD40696 | 69 |
| 7 | thdFa | AAD40694, AAD40695, AAD40696 | 59 |
| 12 | hemH | AF184071 | 60 |
| 13 | hemE | 17 | 58 |
| 20 | rpsT | 19 | 71 |
| 21 | dnaA | 34 | 63 |
| 22 | alkB | 7 | —b |
| 23 | dnaK | 13 | 82 |
| 24 | dnaJ | 13 | 56 |
thdF encodes a 50-kDa protein.
—, no homolog in R. prowazekii.
TABLE 2.
Potential ORFs near the C. crescentus replication origin and their highest homologies to related organisms
| Box no. | Homolog in C. crescentus | % Homology to R. prowazekii genea | Highest homology (%) to gene (organism [reference or GenBank accession no.]) |
|---|---|---|---|
| 1 | Leucyl-tRNA ligase | 52 | 54 (E. coli [4]) |
| 2 | Tyrosine aminotransferase | 42b | 47 (R. meliloti [26]) |
| 8 | Putative carboxymethylenebutenolidase | —c | 67 (Azospirillum brasilense [33]) |
| 9 | Quinone oxidoreductase | 58 | 66 (P. aeruginosa [29]) 58 (R. capsulatus [24]) |
| 10 | Transcription termination factor rho | 84 | 89 (R. sphaeroides [12]) |
| 11 | RP883 (unknown protein) | 66 | 63 (integral membrane protein; R. sphaeroides [12]) |
| 14 | RP001 (unknown protein) | 63 | 53 (Arabidopsis thaliana [CAA17540]) |
| 15 | maf | 43 | 51 (Thermotoga maritima [22]) |
| 16 | Shikimate 5-dehydrogenase (aroE) | — | 55 (Aquifex aeolicus [8]) |
| 17 | Hypothetical protein | — | 57 (Deinococcus radiodurans [30]) |
| 18 | Delta subunit of DNA polymerase III (dnaQ) | 60 | 56 (E. coli [AAB08637]) 56 (P. aeruginosa [29]) |
| 19 | Enoyl CoA hydratase (fadB1) | — | 70 (R. meliloti [19]) 67 (E. coli [10]) 51 (R. capsulatus [3]) |
| 25 | mutS (DNA mismatch repair protein) | 63 | 60 (E. coli [CAB92351]) |
| 26 | Uridyltransferase | — | 65 (R. meliloti [AAF37852]) 52 (E. coli [4]) |
Homologies are as described by Andersson et al. (2).
This R. prowazekii homolog was identified as an unknown protein.
—, no homolog in R. prowazekii.
Conserved gene clusters.
Our analysis noted three distinct gene clusters (Fig. 1A) in the C. crescentus ∼30-kb region. One spans the replication origin, and the other two flank the Cori region. The two flanking gene clusters have been previously reported to be similar to those in genomes of related organisms (GenBank accession no. AAD40694, AAD40695, and AAD40696) (11, 19). The first gene cluster is similar to that of Rhizobium meliloti, a member of the α-proteobacteria (19). The reported gene cluster involved the dnaA locus in C. crescentus and R. meliloti and was previously observed to be similar with respect to the neighboring rpsT (ribosomal protein S20) (19). Further sequence analysis of this dnaA gene locus in C. crescentus had led to the observation of an additional ORF upstream of dnaA which has 70% amino acid homology with the ORF in a similar position in R. meliloti identified as fadB1 (enoyl CoA hydratase) (19). (Fig. 1D; Table 2). With the addition of fadB1, the gene cluster fadB1-rpsT-dnaA is now identical to the previously established gene cluster in the respective R. meliloti genome (Fig. 1D). The R. meliloti fadB1 gene, as well as homologs (Table 2) in E. coli (10) and Rhodobacter capsulatus (3), were identified based on homology to the mammalian mitochondrial enoyl-coenzyme A (CoA) hydratase. This suggests that this particular homolog is not necessarily restricted to the higher forms of eukaryotes and may be a carryover from the original ancestral group (2, 21). Sequence analysis beyond the fadB1-rpsT-dnaA gene cluster revealed no further similarities between the dnaA locus in C. crescentus and R. meliloti. Since R. prowazekii is also a member of the α-proteobacteria, it was expected that this gene cluster would be found in the genome. This is not the case, as there is no fadB1 homolog within the R. prowazekii genome (Table 2), and in addition, the rpsT and dnaA homologs are not located close to one another, but rather a considerable distance apart (2). However, there is significant amino acid homology with C. crescentus dnaA and rpsT products (71 and 63%, respectively), as well as with products of other important replication regulatory genes in C. crescentus, E. coli, Pseudomonas aeruginosa, and Rhodobacter sphaeroides (Tables 1 and 2).
The second gene cluster flanking Cori involves the genes parB-parA-gidB-gidA-thdF (Fig. 1B; Table 2). The partial gene cluster parB-parA-gidB-gidA has been previously observed in C. crescentus (GenBank accession no. AAD40694, AAD40695, and AAD40696). This gene cluster is identical to that of Pseudomonas putida and similar to that of Bacillus subtilis, both of which are also located near their respective replication origins (11). The C. crescentus plasmid and/or chromosome partitioning proteins ParA and ParB were identified as homologs to these genes found in P. putida, as well as homologs to the regulators of sporulation and chromosome partitioning identified in B. subtilis, Soj and Spo0J, respectively (20). Homologs of the E. coli glucose inhibition of division proteins GidA and GidB were also identified in C. crescentus (GenBank accession no. AAD40694, AAD40695, and AAD40696), and the parB-parA-gidB-gidA gene cluster (Fig. 1B) is observed in other eubacteria, such as P. putida (gram negative) and B. subtilis (gram positive) (11). We observed an ORF in C. crescentus that was previously identified as a homolog to the 50-kDa thiophene/furan oxidation protein ThdF in E. coli, whose gene is situated upstream of the gidA gene. In comparison, this thdF homolog is also located upstream of gidA in P. putida and B. subtilis (GenBank accession no. AAD40694, AAD40695, and AAD40696) (11). The gene cluster parB-parA-gidB-gidA-thdF in B. subtilis is similar but not identical to those found in P. putida and C. crescentus, as B. subtilis has two additional ORFs between the parBA and gidBA gene sets which have not been identified (Fig. 1B) (11). The products of the C. crescentus parB, parA, gidB, and gidA homologs share significant amino acid homology with the products of corresponding genes in R. prowazekii (Table 1). Surprisingly, the gene cluster parB-parA-gidB-gidA is present in the genome; however, it is located ∼70 kb away from the putative origin in reverse order. It is thought that the actual gene order is of importance for functionality of the proteins transcribed, but the location of the cluster relative to the replication origin or the orientation of the gene order seems to be irrelevant.
Identical gene cluster spans replication origins in C. crescentus and R. prowazekii.
The third gene cluster, which spans the Cori region, is unique, consists of C. crescentus genes as well as homologs to R. prowazekii genes, and is identical to the gene cluster overlapping the uncharacterized putative replication origin in R. prowazekii (Fig. 1C; Table 1). Whole-genome sequence analysis of R. prowazekii determined that a homolog of hemE is present and that the 5′ region may also overlap the putative replication origin. The hemE product has 58% amino acid homology to that of the C. crescentus hemE (Fig. 1C; Table 1), and further analysis of the flanking regions of Cori and the putative R. prowazekii origin revealed striking similarities (Table 1; Fig. 2B). Three additional ORFs demonstrating 60 to 66% amino acid homology were identified and present in the same cluster (Fig. 1C). A homolog of the R. prowazekii ferrochetase hemH gene (60% amino acid homology) is located downstream of hemE in C. cresecentus. Unknown predicted proteins RP883 and RP001 also share a high degree of amino acid homology (66 and 63%, respectively) with the products of the C. crescentus ORFs. RP883 also shares 63% amino acid homology with an integral membrane protein in R. sphaeroides which may aid in identification of the function of the RP883 protein (12) (Table 2). The function of RP001 has yet to be established but it may possess an ATPase activity (2). Two known promoters (Ps and Pw) are located within Cori; however, it is not known as of yet whether comparable promoters are present within the R. prowazekii putative origin (Fig. 1C).
It is proposed that the presence of the identical RP883-hemH-hemE-RP001 gene cluster surrounding Cori and the R. prowazekii putative replication origin supports selection of this region as the R. prowazekii origin. This proposal is supported by the genetic and radiolabel evidence identifying Cori as a replication origin (17, 18), as well as evidence via Brewer & Fangman two-dimensional DNA gel electrophoresis analysis (5), which has ascertained bidirectional replication initiation in Cori (A. K. C. Brassinga and G. T. Marczynski, unpublished data). It is also suggested that the RP883-hemH-hemE-RP001 gene cluster can be utilized to aid in identification of replication origins in related members of the α-subdivision group of proteobacteria via prediction of the location between hemE and the RP001 gene.
Molecular analysis of the RP883-hemHE-RP001 gene cluster in C. crescentus.
Inferences from our sequence analysis were confirmed by gene reporter assays. DNA fragments from Cosmid I (17) (Fig. 1A) were subcloned into a broad-host-range lacZ transcription reporter plasmid, pRK2901acZ (18), were introduced into wild-type C. crescentus strain NA1000, and were assayed for β-galactosidase as previously described (18). Previous analysis of hemE expression indicated that both transcription and translation of hemE are primarily directed by the constitutive Pw transcription promoter (18). Paradoxically, the cell cycle-regulated Ps transcription promoter contributes relatively little towards hemE expression, presumably due to RNA instability or secondary structure (18).
The plasmid construct pGM2063 PstI-PstI fragment included most of the proposed hemEH-RP883 operon (Fig. 2A), but did not include the Pw and Ps promoters. Below-promoter activity (230 Miller units [MU]) (Fig. 2A) clearly indicated that no other promoters are present within this region, which strongly supports our proposal of the hemEH-RP883 operon. When promoter activity was assayed in the opposite orientation, pGM2064 demonstrated below-background-level activity (130 MU) (Fig. 2A), thereby confirming the assumption that there are no promoters present in the region in the opposite direction relative to Ps and Pw. Construct pGM2741 (Fig. 2A) included Cori the upstream region flanking Cori, and the Pw promoter. This pGM2741 construct did not include the Ps promoter, as this had been deleted from the fragment. This construct expressed significantly higher activity (2,300 MU) at the hemE 5′ region (Fig. 2A). These results argue that hemE, hemH, and the RP883 gene form an operon transcribed only by the Pw promoter from inside Cori. This is consistent with DNA sequence analysis suggesting translation coupling between these ORFs, since the stop and start codons between hemE and hemH are spaced by 3 nucleotides and no nucleotide spacing is present between the stop and start codons of hemH and RP883.
Promoter activity of the Cori fragment pGM2742 assayed in the opposite direction yielded significant transcription (4,040 MU) at the RP001 gene 5′ region (Fig. 2A). Significant transcription was also observed from pGM2062 BamHI-PstI fragment expressing 5,800 MU in the proper orientation. The increased transcriptional activity indicates the presence of at least one additional promoter, which was designated P3, that may direct RP001 transcription (Fig. 2B). In order to determine the approximate location of the P3 promoter, two smaller and more specific lacZ constructs were devised (Fig. 2B). Construct pGM1629, a 250-bp PstI-Bg/II fragment, was utilized as a comparison control since it included only the Pw promoter. In agreement with previous studies (18), the PstII-Bg/II fragment expressed 3,600 MU (Fig. 2B), since it contains Pw, but not Ps, and ends inside hemE coding sequences. The second construct, pGM1630 with a 546-bp EcoRI-BamHI fragment, retained transcription activity of 4,000 MU, thus indicating that the approximate location of the RP001 (P3) promoter lies within this 546-bp region (Fig. 2B). Sequence analysis of the 546-bp region determined that the start codon of the RP001 ORF lies 88 bp inside the fragment from the BamHI site of the 546-bp EcoRI-to-BamHI fragment. Therefore, the location of the P3 promoter is deduced to be within a 458-bp segment of this fragment upstream of the RP001 start codon (Fig. 2B). These results demonstrate that hemEH-RP883 form an operon and that RP001 is transcribed and translated from the right region of Cori (Fig. 2A and B).
To confirm that the RP001 homolog is translated in C. crescentus, we created a RP001::TEM β-lactamase protein fusion in the predicted and alternate reading frames (data not shown). The EcoRI-BamHI Cori RP001 fragment (Fig. 2B) was ligated into the pJAMY30, -31, and -32 plasmid vectors (32), which were originally derived from TEM β-lactamase plasmid pJBS633 (6), and these were likewise introduced into C. crescentus β-lactamase-deficient strain CB15N Δbla (17). Of the constructs tested, only the predicted in-frame fusion protein allowed this C. crescentus strain to grow in the presence of ampicillin. Similar protein fusion experiments using HemE and β-galactosidase (18) and HemE-TEM β-lactamase fusion proteins (32) demonstrated that hemE is actively translated in C. crescentus.
R. prowazekii homologs RP883 and RP001 have been identified in C. crescentus; they comprise part of an identical gene cluster spanning the replication origins in C. crescentus and R. prowazekii. Based on the pRK290lacZ construct assays, it is suggested that RP883-hemH-hemE form a operon and are driven by the Pw promoter. In addition, the pRK290lacZ construct and protein fusion assays support the existence of the ORF identified as an RP001 homolog in C. crescentus. Therefore, the genes flanking Cori are transcribed and translated in C. crescentus and the identical RP883-hemH-hemE-RP001 gene cluster spanning the R. prowazekii putative origin supports the selection of this region as the R. prowazekii origin.
Three distinct gene clusters have been identified in the ∼30-kb region of earliest replicating DNA in C. crescentus. The parB-parA-gidB-gidA-thdF and fadB1-rpsT-dnaA gene clusters flank the Cori region. The parB-parA-gidB-gidA-thdF gene cluster has been found to be identical to that in P. putida and similar to that in B. subtilis, both of which are located relatively close to their respective origins. As both C. crescentus and P. putida are classified as gram negative, it is not surprising that the gene cluster is identical in both organisms. It is reasonable to assume that since B. subtilis is classified as a gram-positive eubacterium, the parB-parA-gidB-gidA-thdF gene cluster may differ due to evolutionary changes. Also, the parB-parA-gidB-gidA grouping has also been found in R. prowazekii but at a much further distance from the putative origin in reverse order. Evidently, there is a loss of the thdF gene and the parB-parA-gidB-gidA grouping has been relocated, but the gene order is of importance presumably for the transcribed protein functionality.
The fadB1-rpsT-dnaA gene cluster in C. crescentus that was determined is identical to that found in R. meliloti, a member of the α-proteobacteria. However, this gene cluster was not determined for R. prowazekii since the fadB1 homolog is not present and the rpsT and dnaA homologs are located separately in the genome. The C. crescentus rpsT and dnaA homologs, as well as other important replication regulatory genes, share significant amino acid homology with those determined for R. prowazekii. However, these genes are not grouped in the gene clusters described above but are dispersed throughout the genome. Interestingly, homologs of the CcrM DNA methyltransferase, which is essential for viability in C. crescentus, R. meliloti and Brucella abortus and is present in other members of the α-proteobacteria (27, 31), has not been found in R. prowazekii (2).
The third gene cluster, RP883-hemH-hemE-RP001, spans the Cori region and is identical to that spanning the R. prowazekii putative region. RP883, hemH, and hemE form an operon that is driven by the Pw promoter in C. crescentus. A third promoter found in Cori, P3, has been determined to direct transcription of the RP001 homolog. This evidence supports the validity of the RP001 homolog in C. crescentus initially identified by sequence comparison. It is proposed that the R. prowazekii putative replication origin be characterized based on the properties of Cori.
R. prowazekii and C. crescentus have retained identical gene clusters around the replication origin. Two gene clusters flanking Cori have not been retained in R. prowazekii, although the individual replication regulatory genes have been retained and presumably deemed essential. The AT-rich R. prowazekii genome is a third of the size of the GC-rich C. crescentus genome, yet they share significant homology in the gene cluster in and around the replication origin. The substantial difference between the two genomes clearly indicates retained essential genetic information for survival and, in the case of R. prowazekii, deleterious mutations of expendable genes. Therefore, we argue that the retained gene cluster spanning the replication origins of C. crescentus and R. prowazekii has functional significance and enhances its predictive value for locating chromosome replication origins in related members of the α-proteobacteria.
Acknowledgments
We thank Urs Jenal and M. R. K. Alley for their interest in our work and Herbert Winkler, David Wood, and William McSween for their discussions regarding R. Prowazekii. We also thank Boris Gorbatyuk, William Spencer, E. C. S. Chan, and G. Matlashewski for critical reading of the manuscript.
Sequencing of C. crescentus was accomplished by The Institute of Genomic Research (TIGR) with support from the U.S. Department of Energy. This work was supported by a Fonds pour la Formation de Chercheurs et l'Aide à la Recherche (FCAR) Ph.D. Fellowship and the Department of Microbiology and Immunology F. C. Harrison Fellowship to A.K.C.B. and R.S. and Medical Research Council of Canada (MRC) Grant MT-13453 and MRC Scholarship Award SH-50791-AP007403 to G.T.M.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Andersson S G E, Zomorodipour A, Andersson J O, Sicheritz-Pontén T, Alsmark U C M, Podowski R M, Näslund A K, Eriksson A-S, Winkler H H, Kurland C G. The genome sequence of Rickettsia prowazekiiand the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- 3.Beckman D L, Kranz R G. A bacterial homolog to the mitochondrial enoyl-CoA hydratase. Gene. 1991;107:171–172. doi: 10.1016/0378-1119(91)90313-z. [DOI] [PubMed] [Google Scholar]
- 4.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coliK-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 5.Brewer B J, Fangman W L. The localization of replication origins on ARS plasmids in S. cerevisiae. Cell. 1987;51:463–471. doi: 10.1016/0092-8674(87)90642-8. [DOI] [PubMed] [Google Scholar]
- 6.Broome-Smith J K, Spratt B G. A vector for the construction of translational fusions to TEM β-lactamase and the analysis of protein export signals and membrane protein topology. Gene. 1986;49:341–349. doi: 10.1016/0378-1119(86)90370-7. [DOI] [PubMed] [Google Scholar]
- 7.Colombi D, Gomes S L. An alkB gene homolog is differentially transcribed during the Caulobacter crescentuscell cycle. J Bacteriol. 1997;179:3139–3145. doi: 10.1128/jb.179.10.3139-3145.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deckert G, Warren P V, Gaasterland T, Young W G, Lenox A L, Graham D E, Overbeek R, Snead M A, Keller M, Aujay M, Huber R, Feldman R A, Short J M, Olson G J, Swanson R V. The complete genome of the hyperthermophilic bacterium Aquifex aeolicus. Nature. 1998;392:353–358. doi: 10.1038/32831. [DOI] [PubMed] [Google Scholar]
- 9.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentusstalked and swarmer cells. J Bacteriol. 1977;132:294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrandez A, Prieto M A, Garcia J L, Diaz E. Molecular characterization of PadA, a phenylacetaldehyde dehydrogenase from Escherichia coli. FEBS Lett. 1997;406:23–27. doi: 10.1016/s0014-5793(97)00228-7. [DOI] [PubMed] [Google Scholar]
- 11.Gal-Mor O, Borovok I, Av-Gay Y, Cohen G, Aharonowitz Y. Gene organization in the trxA/B-oriC region of the Streptomyces coelicolorchromosome and comparison with other eubacteria. Gene. 1998;217:83–90. doi: 10.1016/s0378-1119(98)00357-6. [DOI] [PubMed] [Google Scholar]
- 12.Gomelsky M, Kaplan S. The Rhodobacter sphaeroides 2.4.1 rhogene: expression and genetic analysis of structure and function. J Bacteriol. 1996;178:1946–1954. doi: 10.1128/jb.178.7.1946-1954.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomes S L, Gober J W, Shapiro L. Expression of the Caulobacter heat shock gene dnakis developmentally controlled during growth at normal temperatures. J Bacteriol. 1990;172:3051–3059. doi: 10.1128/jb.172.6.3051-3059.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gross L. How Charles Nicolle of the Pasteur Institute discovered that epidemic typhus is transmitted by lice: reminiscences from my years at the Pasteur Institute in Paris. Proc Natl Acad Sci USA. 1996;93:10539–10540. doi: 10.1073/pnas.93.20.10539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T. Group 9. Rickettsias and Chlamydias. In: Hensyl W R, editor. Bergey's manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. p. 351. [Google Scholar]
- 16.Lobry J R. Asymmetric substitution patterns in the two DNA strands of bacteria. Mol Biol Evol. 1996;13:600–605. doi: 10.1093/oxfordjournals.molbev.a025626. [DOI] [PubMed] [Google Scholar]
- 17.Marczynski G T, Shapiro L. Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J Mol Biol. 1992;226:959–977. doi: 10.1016/0022-2836(92)91045-q. [DOI] [PubMed] [Google Scholar]
- 18.Marczynski G T, Lentine K, Shapiro L. A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 1995;9:1543–1557. doi: 10.1101/gad.9.12.1543. [DOI] [PubMed] [Google Scholar]
- 19.Margolin W, Bramhill D, Long S R. The dnaA gene of Rhizobium melilotilies within an unusual gene arrangment. J Bacteriol. 1995;177:2892–2900. doi: 10.1128/jb.177.10.2892-2900.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mohl D A, Gober J W. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell. 1997;88:675–684. doi: 10.1016/s0092-8674(00)81910-8. [DOI] [PubMed] [Google Scholar]
- 21.Müller M, Martin W. The genome of Rickettsia prowazekiiand some thoughts on the origin of mitochondria and hydrogenosomes. Bioessays. 1999;21:377–381. doi: 10.1002/(SICI)1521-1878(199905)21:5<377::AID-BIES4>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 22.Nelson K E, Clayton R A, Gill S R, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Nelson W C, Ketchum K A, McDonald L, Utterback T R, Malek J A, Linher K D, Garrett M M, Stewart A M, Cotton M D, Pratt M S, Phillips C A, Richardson D, Heidelberg J, Sutton G G, Fleischmann R D, Eisen J A, Fraser C M, et al. Evidence for lateral gene transfer between Archaea and bacteria from genome sequence of Thermotoga maritima. Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- 23.Newton A, Ohta N. Regulation of the cell division cycle and differentiation in bacteria. Annu Rev Microbiol. 1990;44:689–719. doi: 10.1146/annurev.mi.44.100190.003353. [DOI] [PubMed] [Google Scholar]
- 24.Paoli G C, Vichivanives P, Tabita F R. Physiological control and regulation of the Rhodobacter capsulatus cbboperons. J Bacteriol. 1998;180:4258–4269. doi: 10.1128/jb.180.16.4258-4269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quon K C, Yang B, Domain I J, Shapiro L, Marczynski G T. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc Natl Acad Sci USA. 1998;95:120–125. doi: 10.1073/pnas.95.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rastogi V K, Watson R J. Aspartate aminotransferase activity is required for aspartate catabolism and symbiotic nitrogen fixation in Rhizobium melilotiJ. Bacteriol. 1991;173:2879–2887. doi: 10.1128/jb.173.9.2879-2887.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Robertson G T, Reisenauer A, Wright R, Jensen R, Jensen A, Shapiro L, Roop R M., II The Brucella abortusCcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J Bacteriol. 2000;182:3482–3489. doi: 10.1128/jb.182.12.3482-3489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Siam R, Marczynski G T. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 2000;19:1138–1147. doi: 10.1093/emboj/19.5.1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stover C K, Pham X Q, Erwin A L, Mizoguchi S D, Warrener P, Hickey M J, Brinkman F S, Hufnagle W O, Kowalik D J, Lagrou M, Garber R L, Goltry L, Tolentino E, Westbrock-Wadman S, Yuan Y, Brody L L, Coulter S N, Folger K R, Kas A, Larbig K, Lim R, Smith K, Spencer D, Wong G K, Wu Z, Paulsen I T. Complete genome sequence of Pseudomonas aeruginosaPA01, an opportunistic pathogen. Nature. 2000;406:959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 30.White O, Eisen J A, Heidelberg J F, Hickey E K, Peterson J D, Dodson R J, Haft D H, Gwinn M L, Nelson W C, Richardson D L, Moffat K S, Qin H, Jiang L, Pamphile W, Crosby M, Shen M, Vamathevan J J, Lam P, McDonald L, Utterback T, Zalewski C, Makarova K S, Aravind L, Daly M J, Fraser C M, et al. Genome sequence of the radioresistant bacterium Deinococcus radioduransR1. Science. 1999;286:1571–1577. doi: 10.1126/science.286.5444.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright R, Stephens C, Shapiro L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are observed in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol. 1997;179:5869–6877. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhuang W Y, Shapiro L. CaulobacterFliQ and FliR membrane proteins, required for flagellar biogenesis and cell division, belong to a family of virulence factor export proteins. J Bacteriol. 1995;177:343–356. doi: 10.1128/jb.177.2.343-356.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimmer W, Hundeshagen B. Identification and sequencing of pyrG, the CTP synthetase gene of Azospirillum brasilenseSp7. FEMS Microbiol Lett. 1994;115:273–277. doi: 10.1111/j.1574-6968.1994.tb06650.x. [DOI] [PubMed] [Google Scholar]
- 34.Zweiger G, Marczynski G, Shapiro L. A CaulobacterDNA methyltransferase that functions only in the predivisional cell. J Mol Biol. 1994;235:472–485. doi: 10.1006/jmbi.1994.1007. [DOI] [PubMed] [Google Scholar]