Abstract
Vibrio cholerae synthesizes the catechol siderophore vibriobactin. In this report, we present the complete map of a vibriobactin gene region containing two previously unreported vibriobactin biosynthetic genes. vibD encodes a phosphopantetheinyl transferase, and vibH encodes a novel nonribosomal peptide synthase. Both VibD and VibH are required for vibriobactin biosynthesis.
Vibrio cholerae, like most other bacterial pathogens, requires iron for growth and survival, and it possesses multiple systems for iron acquisition (11, 14, 17, 20). One mechanism by which V. cholerae acquires iron is the synthesis and transport of the catechol siderophore vibriobactin (11). Vibriobactin is synthesized and secreted into the environment, where it binds ferric iron with high affinity. The ferri-vibriobactin complex is then transported into the cell by a process that requires the outer membrane receptor ViuA (6, 27), a functional TonB system (17), and an inner membrane permease system (31). The ViuB protein then removes the iron from the ferri-siderophore complex (4).
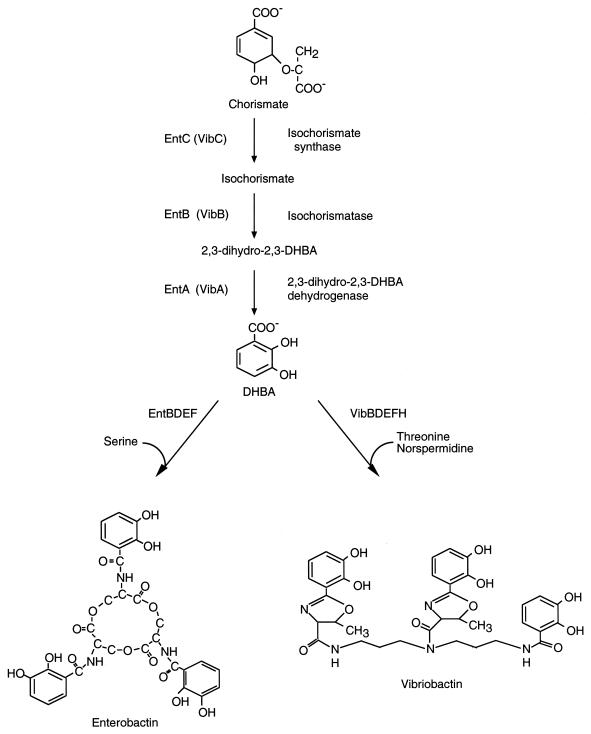
Vibriobactin contains three molecules of 2,3-dihydroxybenzoic acid (DHBA) linked either directly or through threonine residues to the polyamine norspermidine (Fig. 1) (11). Norspermidine is rarely synthesized by bacteria, but it is a common polyamine in members of the family Vibrionaceae (32, 33). Although the structure of vibriobactin is unique, its biosynthesis shares features with the synthesis of the prototype catechol siderophore enterobactin. The first committed steps in the synthesis of enterobactin lead to the synthesis of DHBA from chorismate (8, 29). Vibriobactin biosynthesis also requires the synthesis of DHBA from chorismate (11), and the pathway for DHBA synthesis appears to be the same in V. cholerae and in Escherichia coli. V. cholerae contains genes homologous to entA, entB, and entC, the three genes required for DHBA synthesis in E. coli (8, 29), and each of these genes in V. cholerae, vibABC, complements a defect in the homologous ent gene (30). In addition, a V. cholerae vibA mutant produces no DHBA, confirming that the role of these genes in V. cholerae is DHBA synthesis (30).
FIG. 1.
Biosynthetic pathways and structures of the catechol siderophores enterobactin and vibriobactin (11).
The mechanism of vibriobactin biosynthesis from DHBA, threonine, and norspermidine is different from the mechanism of enterobactin synthesis from DHBA and serine, but some aspects of the synthesis are conserved. In a recently proposed model of enterobactin biosynthesis (9), EntD is the phosphopantetheinyl transferase (15) which catalyzes the transfer of 4′-phosphopantetheine (pPant) to the side chain hydroxyl of a conserved serine residue within EntB. This posttranslational modification allows EntB to serve as the acyl carrier protein for DHBA. EntE catalyzes the adenylation of DHBA and transfer of the activated DHBA to the pPant moiety on EntB (10). EntF is a 142-kDa protein with four distinct domains (Fig. 2). The peptide carrier domain of EntF is covalently modified by the addition of a pPant moiety that allows it to act as the carrier protein for the serine moiety. This modification is catalyzed by EntD. All subsequent enzymatic reactions are catalyzed by EntF, including adenylation of serine and transfer of the activated serine to the endogenous pPant moiety (adenylation domain), formation of the amide bonds joining three DHBA molecules with three serines (condensation domain), and formation of the ester bonds which join the three serine-DHBA moieties to form the cyclic enterobactin molecule (thioester domain) (10, 22).
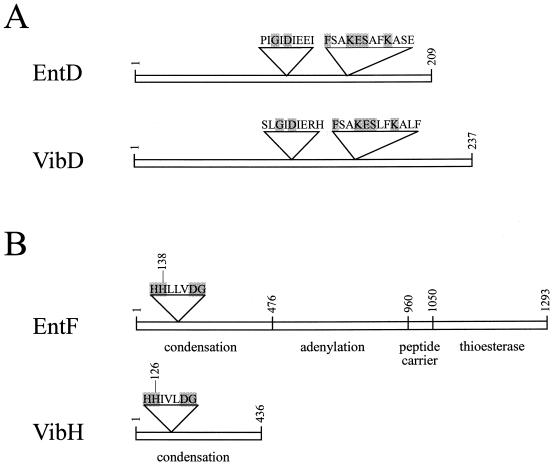
FIG. 2.
(A) Schematic representation of EntD and VibD. The proposed phosphopantetheinyl transferase consensus regions are shown (15). The most highly conserved amino acid residues are shaded. (B) Domain structure of EntF and VibH. The EntF protein contains condensation (amino acids 1 to 475), adenylation (amino acids 476 to 960), peptide carrier (961 to 1049), and thioesterase (amino acids 1050 to 1293) domains (10, 22), while VibH contains only a condensation domain. The proposed catalytic region for the condensation domain is shown, and the most highly conserved amino acids are shaded. The approximate amino acid number at the junctions of the domains is indicated.
In vibriobactin biosynthesis, the mechanism to form the amide bonds that join the DHBA molecules to threonine or norspermidine and the threonine to the norspermidine backbone could be similar to the mechanism for transferring DHBA to serine in enterobactin. Genetic evidence also suggests that the mechanism of the late steps of vibriobactin and enterobactin biosynthesis may be similar, in that V. cholerae has homologues of entBDEF, the genes required for late steps in enterobactin biosynthesis (5, 30).
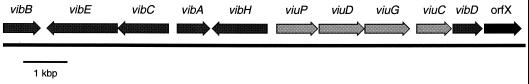
Unlike enterobactin, for which the biosynthetic and transport genes are located in a single 22-kbp genetic locus (8), vibriobactin genes are located in two separate genetic clusters (4–6, 27, 30, 31). Both gene clusters are located on V. cholerae replicon 1 but are separated by approximately 106 bp (13, 28). Each cluster contains both biosynthetic genes and genes for vibriobactin utilization. One of these clusters contains the vibriobactin transport and utilization genes viuA and viuB (4, 6) and the biosynthetic gene vibF (5). The second region (Fig. 3), which is the subject of this report, includes the previously described genes for the synthesis of DHBA from chorismate (vibABC) and a gene for the activation of DHBA (vibE) (30). The region also contains genes for a periplasmic binding protein-dependent ABC transport system, which transports vibriobactin and enterobactin through the periplasm and across the inner membrane.
FIG. 3.
Organization of the second vibriobactin gene region. The arrows indicate the direction of transcription of the vibriobactin genes. Vibriobactin biosynthetic genes are indicated with dark arrows, and the transport genes are shown with light gray arrows. OrfX is closely linked to vibD, but a role in vibriobactin transport or utilization has not been shown. The DNA sequence of this region is posted at GenBank accession number U52150.
Vibriobactin gene cluster contains an entD homologue that is required for vibriobactin biosynthesis.
To identify all of the genes required for vibriobactin biosynthesis and transport, the DNA sequence of the entire vibriobactin region was determined. Two genes, vibD and vibH, that have not been described previously were identified, and their locations relative to the other vibriobactin genes are shown in Fig. 3.
One of these genes, vibD, encodes a protein with sequence homology to E. coli EntD (2) and other phosphopantetheinyl transferase proteins (15). A ClustalW alignment (18) of VibD and EntD sequences shows 31% amino acid identity and 16% conservative substitutions. VibD also contains regions with sequence similarity to each of the two phosphopantetheinyl transferase superfamily consensus motifs (15) (Fig. 2). The assignment of vibD as the V. cholerae entD homologue is supported by the observation that vibD complements an E. coli entD mutation (33 and data not shown).
To determine whether vibD was required for vibriobactin biosynthesis, a vibD mutant, EWV101, was constructed by marker exchange as previously described (30). This strain and the other bacterial strains and plasmids used in this study are described in Table 1. EWV101 was positive for the synthesis of catechols (Table 2), indicating that the mutant had no defect in DHBA biosynthesis. To determine whether EWV101 could synthesize vibriobactin, the ability of EWV101 to cross-feed V. cholerae vibD, vibB, vibH, and vibA mutants was determined (Table 2). The vibD mutant failed to stimulate the growth of the vibD and vibH mutant strains, indicating that it was not secreting vibriobactin. The vibriobactin synthesis defect in this strain was complemented by either vibD or entD encoded on a plasmid (Table 2 and data not shown). Taken together, these data indicate that VibD is likely to provide the phosphopantetheinyl transferase activity required for vibriobactin synthesis. Sequences similar to the phosphopantetheinylation consensus sequence (9) are found in the potential target proteins VibB (FLGLDSI, amino acids 243 to 249) (30) and VibF (DFGGHSL, amino acids 1886 to 1892) (5). The underlined serine residue within these sequences is the likely site of pPant addition by VibD.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| Lou15 | V. cholerae E1 Tor | 24 |
| EWV100 | V. cholerae Lou15 vibA::Tn5 | 30 |
| EWV101 | V. cholerae Lou15 vibD::mini-Tn10kana | This study |
| EWV104 | V. cholerae Lou15 vibB::camb | This study |
| SSV119 | V. cholerae Lou15 vibH::camc | This study |
| DH5α | E. coli host for subcloning | 12 |
| Plasmids | ||
| pBluescript SK− | Cloning vector | Strategene |
| pJSV78 | 7.8-kbp SalI fragment with vibCAH | 30 |
| pJSV90 | 9.0-kbp EcoRI fragment with vibH, viuPDGC, and vibD in pACYC184 | 30 |
| pVIB115 | 1.9-kbp HindIII-StuI fragment with vibD in pBluescript SK− | This study |
The site of the mini-Tn10kan insertion in vibD is indicated in GenBank accession number U52150.
The camcassette is inserted into the NsiI site in vibB.
The camcassette is inserted into the BglII site in vibH.
TABLE 2.
Effect of vibD and vibH mutations on catechol and vibriobactin synthesisa
| Indicator strain (genotype) | Catechol synthesisb | Zone of stimulation (mm) with:
|
||||
|---|---|---|---|---|---|---|
| Lou15 (wild type) | EWV101 | EWV101 (pVIB115) | SSV119 | SSV119 (pJSV78) | ||
| EWV101 (vibD) | + | 30 | 0 | 16 | 0 | 24 |
| EWV104 (vibB) | − | 31 | 0 | 13 | 0 | 18 |
| SSV119 (vibH) | + | 31 | 0 | 15 | 0 | 23 |
| EWV100 (vibA) | − | 33 | 22 | 25 | 22 | 25 |
Cultures of the indicator strains were seeded into L agar containing EDDA (100 μg/ml). The indicated V. cholerae strains were spotted onto the medium. The zone of growth was measured 18 h after inoculation. pVIB115 encodes vibD, and pJSV78 encodes vibH.
Catechol synthesis was determined by the method of Arnow (3).
EWV101 stimulated the growth of the vibA mutant (Table 2). This is likely due to secretion of DHBA by this vibD mutant. DHBA could be taken up and converted to vibriobactin by the vibA mutant, which has no defect in the genes required to convert DHBA to vibriobactin. In contrast, the vibD mutant did not stimulate growth of the vibB mutant EWV104 (Table 2). A role for VibB in the late steps of vibriobactin synthesis suggests that VibB, like its E. coli homologue EntB, is bifunctional, with the amino-terminal region of the protein containing the isochorismatase activity required for the synthesis of DHBA (30), while the carboxy-terminal region functions as the carrier protein for DHBA (9). This is supported by the observation that the VibB amino acid sequence contains the carrier protein consensus sequence FLGLDSI at amino acids 243 to 249 (9).
VibH is required for vibriobactin biosynthesis.
An additional open reading frame was located between vibA and viuP (Fig. 3). To determine whether this gene, named vibH, was required for vibriobactin biosynthesis, a vibH mutant of V. cholerae was constructed. Like the vibD mutant, the vibH mutant strain, SSV119, was positive for the synthesis of catechols (Table 2), indicating that the conversion of chorismate to DHBA was not impaired. SSV119 did not cross-feed the vibD, vibB, or vibH mutants, indicating a defect in vibriobactin biosynthesis (Table 2). Providing the vibH gene in trans on plasmid pJSV78 restored the ability to stimulate growth of each of the mutant strains (Table 2). SSV119 did stimulate the growth of the vibA mutant, consistent with the ability of SSV119 to produce the catechol DHBA, as discussed above. Thus, the phenotype of the vibH mutant suggests that VibH, like VibD, is required for the assembly of vibriobactin from DHBA, threonine, and norspermidine.
Analysis of predicted VibH protein sequence.
The predicted VibH protein has a calculated molecular mass of 49.8 kDa and a predicted pI of 5.8. A Blast search (1) revealed that VibH has sequence homology with nonribosomal peptide synthase proteins, including Bacillus subtilis DhbF (GenBank accession no. Z99120) (21), Streptomyces coelicolor A3(2) calcium-dependent antibiotic synthase I (GenBank AL035640) (19), E. coli EntF (22), Streptomyces chrysomallus actinomycin synthetase II (23), and Serratia liquefaciens SwrA (16). These homologies suggest that VibH is a member of the nonribosomal peptide synthase family. However, VibH is much smaller than other nonribosomal peptide synthase proteins. This makes VibH an atypical member of a family in which the proteins generally have molecular weights of greater than 100,000. The unusually small predicted size of VibH cannot be explained by a frameshift or other sequencing error, since vibA is located immediately downstream of the vibH termination codon (Fig. 3).
Alignment of VibH with the best characterized of the closely related proteins, EntF, revealed that VibH protein aligns well with the first 452 amino acids of EntF. A ClustalW alignment shows 24% amino acid identity and 17% conservative amino acid substitutions. This region of EntF is the condensation domain of the protein, suggesting that VibH has a condensation function. This is supported by the observation that the sequence HHIVLDG (VibH amino acids 125 to 131) matches the condensation domain consensus sequence HHXXXDG (7, 26). The second histidine of this sequence is the catalytic residue. An aligned map of the VibH and EntF domain structures is shown in Fig. 2.
Nonribosomal peptide synthases have a modular structure in which a condensation domain is present together with an adenylation domain and a peptide carrier domain, which is the site of pPant attachment. VibH contains only the condensation domain, and no regions of homology to either an adenylation domain (25) or a peptide carrier domain (9) are present (Fig. 2). This unusual protein structure raises questions about the mechanism of action of VibH. Usually the substrate of a condensation domain is the amino acid attached to the pPant moiety of the peptide carrier domain. Since it is not expected that VibH would contain such a covalently attached amino acid, it is unclear how this protein identifies its substrates. The observation that VibH has only one of the domains present in EntF is consistent with our previous observation that plasmid pJSV90, which contains vibH, does not complement an E. coli entF mutation (30).
Conclusions.
Genomic data indicate that all of the genes for vibriobactin synthesis have now been identified (13). At least four distinct coupling reactions must occur during the assembly of vibriobactin from DHBA, threonine, and norspermidine (Fig. 1). One molecule of DHBA is joined directly with a primary amine on norspermidine, the other two DHBA molecules are joined to the cyclized threonines, and the threonine-DHBA conjugates are joined to the norspermidine at either a primary or a secondary amine. It is not known which of these reactions is catalyzed by VibH. The other nonribosomal peptide synthase homologue required for vibriobactin biosynthesis, VibF (5), is a very large protein (269.5 kDa). It is believed that VibF catalyzes all the late steps not performed by VibH that are required for assembly of the vibriobactin molecule.
This work completes the identification of the vibriobactin biosynthesis genes in V. cholerae. It is unclear why the genes for vibriobactin synthesis and transport are divided into two genetic loci, but the separation of genes that usually map together has been observed for other iron acquisition systems in V. cholerae. For example, the heme receptor gene hutA maps at a distance from the other heme transport genes (13, 28). Both of the vibriobactin regions map to chromosome 1, which contains most of the genes required for growth and pathogenicity of V. cholerae (13). This may reflect the central role of vibriobactin synthesis and utilization in the growth and survival of V. cholerae in at least one of its habitats.
Acknowledgments
This work was supported by the Foundation for Research and by grant AI16935 from the National Institutes of Health.
We thank Douglas Henderson and Laura Runyen-Janecky for comments on the manuscript.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Armstrong S K, Pettis G S, Forrester L J, McIntosh M A. The Escherichia coli enterobactin biosynthesis gene, entD: nucleotide sequence and membrane localization of its protein product. Mol Microbiol. 1989;3:757–766. doi: 10.1111/j.1365-2958.1989.tb00224.x. [DOI] [PubMed] [Google Scholar]
- 3.Arnow L E. Colorimetric determination of the components of 3,4-dihydroxyphenylalanine tyrosine mixtures. J Biol Chem. 1937;118:531–537. [Google Scholar]
- 4.Butterton J R, Calderwood S B. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J Bacteriol. 1994;176:5631–5638. doi: 10.1128/jb.176.18.5631-5638.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butterton J R, Choi M H, Watnick P I, Carroll P A, Calderwood S B. Vibrio cholerae VibF is required for vibriobactin synthesis and is a member of the family of nonribosomal peptide synthetases. J Bacteriol. 2000;182:1731–1738. doi: 10.1128/jb.182.6.1731-1738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butterton J R, Stoebner J A, Payne S M, Calderwood S B. Cloning, sequencing, and transcriptional regulation of viuA, the gene encoding the ferric vibriobactin receptor of Vibrio cholerae. J Bacteriol. 1992;174:3729–3738. doi: 10.1128/jb.174.11.3729-3738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crecy-Lagard V, Marliere P, Saurin W. Multienzymatic nonribosomal peptide biosynthesis: identification of the functional domains catalysing peptide elongation and epimerisation. C R Acad Sci Paris. 1995;318:927–936. [PubMed] [Google Scholar]
- 8.Earhart C F. Uptake and metabolism of iron and molybdenum. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: ASM Press; 1996. pp. 1075–1090. [Google Scholar]
- 9.Gehring A M, Bradley K A, Walsh C T. Enterobactin biosynthesis in Escherichia coli: isochorismate lyase (EntB) is a bifunctional enzyme that is phosphopantetheinylated by EntD and then acylated by EntE using ATP and 2,3-dihydroxybenzoate. Biochemistry. 1997;36:8495–8503. doi: 10.1021/bi970453p. [DOI] [PubMed] [Google Scholar]
- 10.Gehring A M, Mori I, Walsh C T. Reconstitution and characterization of the Escherichia coli enterobactin synthetase from EntB, EntE, and EntF. Biochemistry. 1998;37:2648–2659. doi: 10.1021/bi9726584. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths G L, Sigel S P, Payne S M, Neilands J B. Vibriobactin, a siderophore from Vibrio cholerae. J Biol Chem. 1984;259:383–385. [PubMed] [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Heidelberg J F, Eisen J A, Nelson W C, Clayton R A, Gwinn M L, Dodson R J, Haft D H, Hickey E K, Peterson J D, Umayam L, Gill S R, Nelson K E, Read T D, Tettelin H, Richardson D, Ermolaeva M, Vamathevan J, Bass S, Qin H, Dragoi I, Sellers P, McDonald L, Utterback T, Fleishmann R D, Nierman W C, White O, Salzberg S L, Smith H O, Colwell R R, Mekalanos J J, Venter J C, Fraser C M. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–483. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson D P, Payne S M. Cloning and characterization of the Vibrio cholerae genes encoding the utilization of iron from haemin and haemoglobin. Mol Microbiol. 1993;7:461–469. doi: 10.1111/j.1365-2958.1993.tb01137.x. [DOI] [PubMed] [Google Scholar]
- 15.Lambalot R, Gehring A M, Flugel R S, Zuber P, LaCelle M, Marahiel M A, Reid R, Khosla C, Walsh C T. A new enzyme superfamily — the phosphopanthetheinyl transferases. Chem Biol. 1996;3:923–936. doi: 10.1016/s1074-5521(96)90181-7. [DOI] [PubMed] [Google Scholar]
- 16.Lindum P W, Anthoni U, Christophersen C, Eberl L, Molin S, Givskov M. N-Acyl-l-homoserine lactone autoinducers control production of an extracellular lipopeptide biosurfactant required for swarming motility of Serratia liquefaciens MG1. J Bacteriol. 1998;180:6384–6388. doi: 10.1128/jb.180.23.6384-6388.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Occhino D A, Wyckoff E E, Henderson D P, Wrona T J, Payne S M. Vibrio cholerae iron transport: haem transport genes are linked to one of two sets of tonB, exbB, exbD genes. Mol Microbiol. 1998;29:1493–1507. doi: 10.1046/j.1365-2958.1998.01034.x. [DOI] [PubMed] [Google Scholar]
- 18.Olson S A. MacVector: an integrated sequence analysis program for the Macintosh. Methods Mol Biol. 1994;25:195–201. doi: 10.1385/0-89603-276-0:195. [DOI] [PubMed] [Google Scholar]
- 19.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 20.Rogers M B, Sexton J A, DeCastro G J, Calderwood S B. Identification of an operon required for ferrichrome iron utilization in Vibrio cholerae. J Bacteriol. 2000;182:2350–2353. doi: 10.1128/jb.182.8.2350-2353.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rowland B M, Grossman T H, Osburne M S, Tabor H W. Sequence and genetic organization of a Bacillus subtilis operon encoding 2,3-dihydroxybenzoate biosynthetic enzymes. Gene. 1996;178:119–123. doi: 10.1016/0378-1119(96)00349-6. [DOI] [PubMed] [Google Scholar]
- 22.Rusnak F, Sakaitani M, Drueckhammer D, Reichert J, Walsh C. Biosynthesis of the Escherichia coli siderophore enterobactin: sequence of the entF gene, expression and purification of EntF, and analysis of covalent phosphopantetheine. Biochemistry. 1991;30:2916–2927. doi: 10.1021/bi00225a027. [DOI] [PubMed] [Google Scholar]
- 23.Schauwecker F, Pfennig F, Schroder W, Keller U. Molecular cloning of the actinomycin synthetase gene cluster from Streptomyces chrysomallus and functional heterologous expression of the gene encoding actinomycin synthetase II. J Bacteriol. 1998;180:2468–2474. doi: 10.1128/jb.180.9.2468-2474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigel S P, Payne S M. Effect of iron limitation on growth, siderophore production and expression of outer membrane proteins of Vibrio cholerae. J Bacteriol. 1982;150:148–155. doi: 10.1128/jb.150.1.148-155.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stachelhaus T, Marahiel M A. Modular structure of genes encoding multifunctional peptide synthetases required for non-ribosomal peptide synthesis. FEMS Microbiol Lett. 1995;125:3–14. doi: 10.1111/j.1574-6968.1995.tb07328.x. [DOI] [PubMed] [Google Scholar]
- 26.Stachelhaus T, Mootz H D, Bergendahl V, Marahiel M A. Peptide bond formation in nonribosomal peptide biosynthesis: catalytic role of the condensation domain. J Biol Chem. 1998;273:22773–22781. doi: 10.1074/jbc.273.35.22773. [DOI] [PubMed] [Google Scholar]
- 27.Stoebner J A, Butterton J R, Calderwood S B, Payne S M. Identification of the vibriobactin receptor of Vibrio cholerae. J Bacteriol. 1992;174:3270–3274. doi: 10.1128/jb.174.10.3270-3274.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trucksis M, Michalski J, Deng Y K, Kaper J B. The Vibrio cholerae genome contains two unique circular chromosomes. Proc Natl Acad Sci USA. 1998;95:14464–14469. doi: 10.1073/pnas.95.24.14464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh C T, Liu J, Rusnak F, Sakaitani M. Molecular studies on enzymes in chorismate metabolism and enterobactin biosynthetic pathway. Chem Rev. 1990;90:1105–1129. [Google Scholar]
- 30.Wyckoff E E, Stoebner J A, Reed K E, Payne S M. Cloning of a Vibrio cholerae vibriobactin gene cluster: identification of genes required for early steps in siderophore biosynthesis. J Bacteriol. 1997;179:7055–7062. doi: 10.1128/jb.179.22.7055-7062.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wyckoff E E, Valle A-M, Smith S L, Payne S M. A multifunctional ABC transporter system from Vibrio cholerae transports vibriobactin and enterobactin. J Bacteriol. 1999;181:7588–7596. doi: 10.1128/jb.181.24.7588-7596.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamamoto S, Chowdhury M A R, Kuroda M, Nakano T, Koumoto Y, Shinoda S. Further study on polyamine compositions in Vibrionaceae. Can J Microbiol. 1991;37:148–153. doi: 10.1139/m91-022. [DOI] [PubMed] [Google Scholar]
- 33.Yamamoto S, Shinoda S, Kawaguchi M, Wakamatsu K, Makita M. Polyamine distribution in Vibrionaceae: norspermidine as a general constituent of Vibrio species. Can J Microbiol. 1983;29:724–728. [Google Scholar]