SUMMARY
A subtyping methodology for Campylobacter, Comparative Genomic Fingerprinting (CGF40), has been described recently. The objective of this study was to assess the utility of CGF40 as a tool to enhance routine public health surveillance of campylobacteriosis. Isolates of Campylobacter from across the province were requested and sent for CGF40 subtyping. Epidemiological data from cases reported to public health officials in Nova Scotia, Canada, from January 2012 to March 2015 were linked with blinded CGF40 subtyping results. CGF40 was epidemiologically valid; subtyping discerned known epidemiologically related isolates and augmented case-finding. Predominant sources and locations of subtype detection from the national reference database showed some study subtypes were rare and even novel to the database, while others were more commonly identified over multiple years and with exposures locally and internationally. A case-case study design was applied to examine risk factors for the most common CGF40 subtypes detected. Differences in the epidemiology of different CGF40 subtypes were observed. Statistically significant associations were noted for specific subtypes with rural residence, local exposure, contact with a pet dog or cat, contact with chickens, and drinking unpasteurized milk. With prospective use, CGF40 could potentially identify unrecognized outbreaks and contribute to epidemiological investigations of case clusters.
Key words: Campylobacter, molecular epidemiology, outbreaks, surveillance
INTRODUCTION
Foodborne diseases remain a major cause of illness in Canada and worldwide. In Canada, Campylobacter is the leading reportable bacterial cause of gastroenteritis, corresponding to an estimated burden of 145 000 cases per year [1]. While gastroenteritis caused by Campylobacter is typically a self-limiting illness, there can be serious sequelae of infection. A number of pathogenic Campylobacter species have been associated with gastrointestinal as well as extra-gastrointestinal diseases, including Guillain–Barré syndrome, Miller Fisher syndrome, brain abscesses and meningitis, bacteraemia, sepsis, and reactive arthritis [2]. Domestically acquired Campylobacter is estimated to cause 565 hospitalizations and five deaths each year in Canada [3]. This estimate does not include morbidity and mortality related to chronic sequelae related to the acute infection.
In Nova Scotia, the incidence of Campylobacter is similar to that of Salmonella [4]. Current routine laboratory characterization in many jurisdictions, including Canada, stops at genus for Campylobacter. In contrast, routine serotyping of Salmonella began to uncover the epidemiology of salmonellosis, and the addition of typing via pulsed-field gel electrophoresis (PFGE) for further discrimination of strains provides a robust surveillance strategy for the rapid identification of outbreaks [5]. In the absence of a routine typing surveillance programme for Campylobacter, the ability to understand case aetiology, monitor trends and detect outbreaks is limited. While serotyping and a number of molecular subtyping methods, including PFGE, restriction fragment-length polymorphism analysis of the flagellin gene, DNA sequencing of the flagellin gene short variable region, and multilocus sequence typing, have been used to describe the molecular epidemiology of Campylobacter, these methods do not lend themselves to large-scale deployment for routine epidemiological surveillance, which has prevented the implementation of any typing strategy for routine Campylobacter surveillance in Canada to date [6]. In part, these challenges stem from resource constraints due to the large numbers of Campylobacter isolates that are produced when clinical specimens from campylobacteriosis cases are systematically subjected to microbiological analysis. Questions have also been raised regarding the utility of real-time molecular typing for case cluster detection using leading subtyping methods [7, 8].
A recently described approach, comparative genomic fingerprinting (CGF), is a high-resolution and rapid typing method which applies standard laboratory technology (polymerase chain reaction, PCR) to discriminate strains on the basis of differences in genome content [9]. The method was developed to overcome some of the technical and logistical hurdles to implementing typing in the context of routine Campylobacter surveillance, which include insufficient discriminatory power of the typing method, required resources, turn-around time for results, and ease of analysis.
The objective of this study was to assess the utility of CGF as a tool to enhance routine public health surveillance of campylobacteriosis through linkage of epidemiological data with blinded CGF subtyping results. First, the epidemiological validity of CGF subtyping was assessed using reported outbreaks and case clusters determined to be related through routine public health case follow-up activities. Secondly, descriptive and analytic epidemiology approaches were applied to the most common CGF40 subtypes detected. Significant associations from these analyses may provide a starting point for outbreak hypothesis generation for specific CGF40 subtypes. This study is the first to quantify associations between specific CGF40 subtypes and individual risk factors reported by cases.
METHODS
Case reports and populations
As per the Health Protection Act of Nova Scotia, cases of campylobacteriosis are notifiable to provincial public health authorities. Laboratories report positive Campylobacter culture results to local public health officials, and through case management activities, public health nurses document demographic and exposure information on case report forms. For this study, the Population Health Assessment and Surveillance unit of the Nova Scotia Department of Health and Wellness, provided access to case report forms. Demographic and exposure data were entered into a database and linked to subtyping results by unique laboratory accession numbers. Specimen collection date was substituted for onset date when the date of symptom onset was unavailable. In Nova Scotia, the number 0 is assigned to the second position in postal codes for rural areas and numbers 1–9 are assigned to the second position for urban areas. Cases were classified as urban or rural based on postal code of residence.
Case clusters were defined in two ways: (a) public health-reported clusters; clusters of cases reported by public health officials as being epidemiologically linked or (b) temporal CGF40 subtype clusters: clusters of ⩾2 isolates with matching CGF40 results with associated case symptom onset dates falling within a 30-day period of each other. Cluster definitions are not mutually exclusive.
A case-case study design [10] was used to compare exposures within CGF40 subtype clusters of ⩾5 cases to exposures among sporadic cases. Sporadic cases were defined as CGF subtypes that were detected only once in the study period. Exact odds ratios were calculated to assess the significance of the exposures using a cut-off value of P = 0·05. Exposures assessed were: urban residence, acquisition of illness in Nova Scotia, age, sex, work in chicken or mink farming/processing, contact with chickens through owning or visiting a farm or working in chicken farming/processing, drinking unpasteurized milk, contact with a pet dog and/or cat, contact with recreational water, contact with farm/zoo/wildlife animals. Statistical analyses were conducted using SAS v. 9.4 software (SAS Institute Inc., USA).
CGF
Case isolates were forwarded from local laboratories to the Provincial Public Health Laboratory and onward to the National Microbiology Laboratory at Lethbridge (NML), Public Health Agency of Canada (Lethbridge, Alberta). Some isolates were identified retrospectively and isolates stored at −70 °C were sent for typing. Laboratory staff were blinded to epidemiological information. Species was determined by PCR (NML) as previously described [11] and/or matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (Nova Scotia). CGF40 fingerprints were generated from eight multiplex PCRs for each isolate using primer sets and methodology described previously [9]. PCR products were scored positive (1) or negative (0) based on presence or absence of each target amplicon in order to create a binary CGF40 fingerprint. For ease of comparison, CGF40 fingerprints were assigned a three-digit CGF subtype derived from cluster membership in the national reference CGF database. This database is comprised of a pan-Canadian collection of isolates (n = 22 011) obtained from human clinical, animal, and environmental sources from 1998 to date, representing samples obtained from several national and regional surveillance programmes and a range of ad hoc research activities. Isolates with patterns identical to existing fingerprints were assigned the appropriate CGF subtype, while novel patterns were assigned a CGF type based on its hierarchical position in the clustered database.
For all CGF40 subtypes related to cases in the study, a query was conducted on the reference database for frequencies, sources and regions from which the strains were isolated.
Ethics statement
The research ethics board of Capital District Health Authority approved this study, which involved the secondary use of clinical isolates and epidemiological data.
RESULTS
There were 581 cases reported to public health officials with onset dates ranging from 1 January 2012 to 31 March 2015. Speciation and CGF subtyping was completed for 299 (52%) cases. There were 265 Campylobacter jejuni, 25 C. coli, and nine other Campylobacter species isolated. There were a total of 141 distinct CGF40 subtypes, with 70% of isolates sharing fingerprints with one or more isolates (Table 1). There were 89 CGF40 subtypes isolated only once in Nova Scotia in the study period.
Table 1.
CGF40 subtypes by frequency of detection, Nova Scotia, January 2012–March 2015 (n = 299)
| Frequency of detection of CGF40 subtype (no. of cases) | No. of CGF40 subtypes |
|---|---|
| 1 | 89 |
| 2 | 26 |
| 3 | 5 |
| 4 | 6 |
| 5 | 1 |
| 6 | 7 |
| 7 | 3 |
| 8 | 1 |
| 12 | 1 |
| 14 | 1 |
| 17 | 1 |
Significantly more isolates from cases residing in urban areas were available for CGF subtyping compared to rural areas (Table 2), due to differences in isolate storage practices among laboratories. The subset of cases for which isolates were subtyped differed in distribution by episode year when compared to those not subtyped, with a greater proportion of cases subtyped in 2014 and 2015, due to increased availability of isolates for testing over time. Subtyped cases did not differ significantly from non-subtyped cases in age or sex distribution (Table 2, P = 0·07 and P = 0·06).
Table 2.
Demographic characteristics of cases for which CGF40 subtyping was completed and for which subtyping was unavailable
| Cases | CGF40 unavailable | CGF40 complete | |||||
|---|---|---|---|---|---|---|---|
| n | % | n | % | n | % | P value | |
| Sex | |||||||
| Male | 327 | 56·3 | 172 | 61·0 | 155 | 51·8 | 0·07 |
| Female | 254 | 43·7 | 110 | 39·0 | 144 | 48·2 | |
| Urban/rural* | |||||||
| Urban | 306 | 68·8 | 86 | 52·1 | 220 | 78·6 | 1·1 × 10−10 |
| Rural | 139 | 31·2 | 79 | 47·9 | 60 | 21·4 | |
| Age group, years* | |||||||
| <1 | 6 | 1·1 | 5 | 1·8 | 1 | 0·4 | 0·06 |
| 1–4 | 25 | 4·4 | 15 | 5·3 | 10 | 3·5 | |
| 5–9 | 16 | 2·8 | 10 | 3·5 | 6 | 2·1 | |
| 10–19 | 36 | 6·3 | 23 | 8·2 | 13 | 4·6 | |
| 20–29 | 87 | 15·3 | 32 | 11·3 | 55 | 19·3 | |
| 30–39 | 68 | 12·0 | 31 | 11·0 | 37 | 13·0 | |
| 40–49 | 80 | 14·1 | 40 | 14·2 | 40 | 14·0 | |
| 50–59 | 123 | 21·7 | 57 | 20·2 | 66 | 23·2 | |
| 60–69 | 72 | 12·7 | 37 | 13·1 | 35 | 12·3 | |
| 70–79 | 34 | 6·0 | 22 | 7·8 | 12 | 4·2 | |
| ⩾80 | 20 | 3·5 | 10 | 3·5 | 10 | 3·5 | |
| Year | |||||||
| 2012 | 196 | 33·7 | 124 | 44·0 | 72 | 24·1 | 5·1 × 10−9 |
| 2013 | 173 | 29·8 | 101 | 35·8 | 72 | 24·1 | |
| 2014 | 185 | 31·8 | 51 | 18·1 | 134 | 44·8 | |
| 2015 | 27 | 4·6 | 6 | 2·1 | 21 | 7·0 | |
136 missing postal code information to determine urban/rural; 14 cases missing age information.
Some subtypes were detected in multiple years with exposures in local and overseas locations, while others were detected in only one year and were associated with local exposure (Table 3). There were 42 novel CGF40 subtypes added to the reference database from this study, with 99 (70%) subtypes matching existing subtypes in the database. Of the matching subtypes in the reference database, 82/99 were subtypes isolated from animal and/or environmental sources. The leading animal or environmental source was chicken (58·5%), followed by cow (23·2%), then environmental water (12·2%). The remaining 17/99 subtypes matching to the reference database matched to human isolates from other Canadian provinces.
Table 3.
Frequency and characteristics of common CGF40 subtypes, strains for which CGF40 was unavailable, and sporadic CGF40 subtypes
| Nova Scotia study | CGF reference database | |||||||
|---|---|---|---|---|---|---|---|---|
| CGF40 | n | Species | Years isolated | Travel locations | No. of temporal clusters (no. cases in each cluster) | No. in database | Predominant source type | Predominant animal source |
| 0083·001·002 | 17 | jejuni | 2012–2015 | Within Canada | 1 (9) | 278 | Animal | Chicken |
| 0018·001·002 | 14 | jejuni | 2012–2015 | Multiple countries/ continents | 3 (3, 2, 2) | 71 | Animal | Chicken |
| 0253·004·001 | 12 | jejuni | 2012, 2014, 2015 | Within Canada | 3 (3, 3, 2) | 173 | Animal | Chicken |
| 0044·003·001 | 8 | jejuni | 2012–2014 | None | 2 (2, 2) | 259 | Human | Cow |
| 0082·001·001 | 7 | jejuni | 2012–2015 | Within Canada | 1 (2) | 70 | Animal | Chicken |
| 0633·004·002 | 7 | coli | 2013, 2014 | Multiple countries/ continents | 1 (3) | 182 | Animal | Chicken |
| 0169·001·002 | 7 | jejuni | 2012–2014 | Within Canada | 1 (2) | 593 | Animal | Cow |
| 0173·002·004 | 6 | jejuni | 2012, 2014 | None | 1 (2) | 95 | Animal | Chicken |
| 0882·005·001 | 6 | jejuni | 2012–2014 | Within Canada | 1 (2) | 205 | Animal | Chicken |
| 0960·007·001 | 6 | jejuni | 2014 | None | 1 (2) | 167 | Animal | Chicken |
| 0926·002·001 | 6 | jejuni | 2012, 2014 | Within Canada | 1 (3) | 506 | Animal | Chicken |
| 0922·001·002 | 6 | jejuni | 2012 | Within Canada | 1 (3) | 8 | Human | n.a. |
| 0129·001·002 | 6 | jejuni | 2014 | Cuba | 2 (2, 2) | 53 | Animal | Chicken |
| 0117·001·001 | 6 | jejuni | 2012–2014 | Dominican republic | 1 (2) | 168 | Animal | Chicken |
| 0083·007·001 | 5 | jejuni | 2013, 2014 | Multiple countries/ continents | 1 (2) | 38 | Human | Chicken |
The 15 most commonly isolated subtypes (⩾5 cases) encompassed 40% of cases and are described in Table 3. Statistically significant exact odds ratios for exposures were found for some CGF40 subtypes compared to sporadic cases (Table 4). Compared to sporadic cases, cases infected with certain CGF40 subtypes were more likely to live in rural areas, have acquired their infection in Nova Scotia, had contact with chickens, consumed unpasteurized milk, and/or had exposure to a pet dog or cat (Table 4). The odds of exposure to recreational water or to farm/zoo/wildlife animals were not higher for any CGF40 subtype compared to sporadic CGF40 types. Odds ratios for age and sex were not significant for any CGF40 subtype compared to sporadic CGF40 subtypes.
Table 4.
Statistically significant odds ratios and associated 95% confidence intervals for exposures in cases associated with specific CGF40 subtypes vs. sporadic cases (cases with CGF40 subtype detected once) from case-case analysis
| Cases, n | Reported risk factor | Exact OR | ||||
|---|---|---|---|---|---|---|
| CGF40 subtype | Yes/ (yes+no) |
% | OR | 95% CI | Two-sided P value | |
| Urban residence | ||||||
| Sporadic | 89 | 71/85 | 83·5 | Ref. | – | – |
| 0926·002·001 | 6 | 2/6 | 33·3 | 0·102 | 0·008–0·792 | 0·0261 |
| 0129·001·002 | 6 | 2/6 | 33·3 | 0·102 | 0·008–0·792 | 0·0261 |
| Acquired in Nova Scotia | ||||||
| Sporadic | 89 | 35/75 | 46·7 | Ref. | – | – |
| 0083·001·002 | 17 | 12/13 | 92·3 | 13·401 | 1·815–600·55 | 0·0038 |
| 0253·004·001 | 12 | 8/9 | 88·9 | 8·949 | 1·108–415·595 | 0·0354 |
| 004·003·001 | 8 | 7/7 | 100·0 | 10·521* | 1·972-infinity | 0·0142 |
| 0173·002·004 | 6 | 6/6 | 100·0 | 8·947* | 1·631-infinity | 0·0277 |
| Works in chicken or mink farming or processing | ||||||
| Sporadic | 89 | 3/81 | 3·7 | Ref. | – | – |
| 0129·001·002 | 6 | 3/6 | 50·0 | 23·679 | 2·266–277·09 | 0·0067 |
| Contact with chickens | ||||||
| Sporadic | 89 | 6/80 | 7·5 | Ref. | – | – |
| 0882·005·001 | 6 | 3/6 | 50·0 | 11·609 | 1·282–107·827 | 0·0277 |
| 0129·001·002 | 6 | 4/6 | 66·7 | 22·641 | 2·665–298·775 | 0·0026 |
| Drank unpasteurized milk | ||||||
| Sporadic | 89 | 3/72 | 4·2 | Ref. | – | – |
| 0018·001·002 | 14 | 4/12 | 33·3 | 10·91 | 1·554–88·696 | 0·014 |
| Has a pet dog and/or cat | ||||||
| Sporadic | 89 | 40/81 | 49·4 | Ref. | – | – |
| 0083·001·002 | 17 | 13/14 | 92·9 | 13·056 | 1·803–579·77 | 0·0037 |
OR, Odds ratio; CI, confidence interval.
Median unbiased estimate.
Four public health-reported Campylobacter jejuni case clusters were confirmed by CGF40 (Table 5), comprising two case pairs, one cluster of three laboratory-confirmed cases, and one outbreak of six laboratory-confirmed and 12 probable cases. In the cluster of three subtyped cases, one case showed a slight variation in CGF40 subtype with respect to the two other cases (0018·001·003 vs. 0018·001·002), differing at one of the 40 loci analysed by the assay. There were two additional case pairs noted to be epidemiologically linked through public health investigation; however, corresponding isolates speciated as one C. jejuni and one C. coli each. Notably, an unrelated case had C. jejuni isolated from a stool sample and C. fetus isolated from a blood sample.
Table 5.
Temporal clusters of CGF40 subtypes, case onset dates within 30 days, Nova Scotia, January 2012–March 2015 (n = 82)
| Year | CGF40 | No. of cases | Episode date range | Cluster noted by Public Health | Significant association detected for subtype* |
|---|---|---|---|---|---|
| 2012 | 0922·001·002 | 4 | 3 July–4 Aug. | Yes, 3 cases from outbreak with 6 laboratory-confirmed cases and 12 probable cases | No |
| 0018·001·002 | 3 | 9–17 Nov. | No | Unpasteurized milk | |
| 0253·004·001 | 3 | 21 Sept.–21 Oct. | No | Acquired in NS | |
| 0269·004·001 | 2 | 19–23 July | No | n.a. | |
| 0044·003·001 | 2 | 20 Aug.–8 Sept. | No | Acquired in NS | |
| 0891·001·001 | 2 | 20–27 Aug. | No | n.a. | |
| 2013 | 0083·001·002 | 9 | 16 June–10 Oct. | No | Acquired in NS, pet dog or cat |
| 0117·001·001 | 2 | 13–21 Sept. | No | No | |
| 0169·001·002 | 2 | 12 July–1 Aug. | No | No | |
| 0173·010·002 | 2 | 18 May–16 June | No | n.a. | |
| 0027·006·001 | 2 | 26 June –July 10 | Yes, 2 cases | n.a. | |
| 2014 | 0017·001·004 | 3 | 18 May–23 | No | n.a. |
| 0253·004·001 | 3 | 24 July–Aug. 28 | No | Acquired in NS | |
| 0633·004·002 | 3 | 2 Nov.–Dec. 13 | No | No | |
| 0926·002·001 | 3 | 22 June–July 10 | No | Rural residence | |
| 0952·003·002 | 3 | 7 July–Aug. 9 | No | n.a. | |
| 0120·001·007 | 2 | 21–26 Feb. | No | n.a. | |
| 0129·001·002 | 2 | 27–30 May | Yes, 2 cases | Rural residence, contact with chickens | |
| 0129·001·002 | 2 | 1–8 Sept. | No | Rural residence, contact with chickens | |
| 0173·002·004 | 2 | 4–19 Nov. | No | Acquired in NS | |
| 0173·008·002 | 2 | 3–8 Aug. | No | n.a. | |
| 0018·001·002 | 2 | 6–8 June | No | Unpasteurized milk | |
| 0018·001·002 | 2 | 7–12 Aug. | No | Unpasteurized milk | |
| 0253·004·001 | 2 | 5–23 May | No | Acquired in NS | |
| 0044·003·001 | 2 | 19 Nov.–15 Dec. | No | Acquired in NS | |
| 0611·003·001 | 2 | 27 Apr.–5 May | No | n.a. | |
| 0082·001·001 | 2 | 13–29 Nov. | No | n.a. | |
| 0083·007·001 | 2 | 12 Apr.–2 May | No | No | |
| 0882·005·001 | 2 | 8–18 Aug. | No | Contact with chickens | |
| 0904·005·004 | 2 | 16–21 June | No | n.a. | |
| 0933·008·001 | 2 | 16–22 June | No | n.a. | |
| 0960·007·001 | 2 | 18–21 July | No | No | |
| 2015 | 0018·001·002 | 2 | 6–10 Mar. | Yes, 2 cases from cluster of 3 laboratory-confirmed cases in same setting† | Unpasteurized milk |
NS, Nova Scotia; n.a., not applicable.
CGF40 clusters with ⩾5 cases were tested for associations with the following risk factors: urban residence, acquisition of illness in Nova Scotia, age, sex, work in chicken or mink farming/processing, contact with chickens through owning or visiting a farm or working in chicken farming/processing, unpasteurized milk, contact with pet dog and/or cat, recreational water, farm/zoo/wildlife animals (see Table 4). These sources may provide leading hypotheses for investigation.
Third outbreak case differed in one PCR result, CGF40 subtype was 0018·001·003; however, this single locus difference was found to be a spurious result upon retesting.
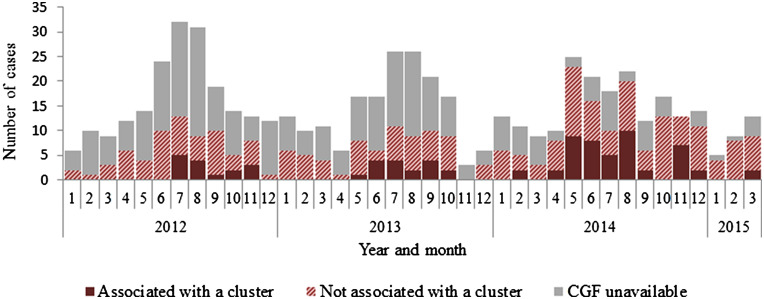
CGF40 identified 33 temporal clusters of cases of a shared CGF40 subtype with onset dates within 30 days of one another (Fig. 1, Table 5). Twenty-seven percent of subtyped cases were associated with clusters (82/299). Due to the small numbers of cases per cluster, analytic epidemiology was only conducted for one temporal cluster of subtype 0083·001·002 (nine cases). For this cluster, the odds of having a pet dog or cat was significantly higher compared to sporadic cases (odds ratio 10·9, 95% confidence interval 2·1–∞, P = 0·01). As such, significant odds ratios for subtypes (inside and outside cluster time period from Table 4) are noted in Table 5.
Fig. 1.
Number of reported cases of campylobacteriosis, associated with or not associated with temporal CGF40 clusters, Nova Scotia, January 2012–March 2015 (n = 581).
DISCUSSION
In this study, blinded CGF40 subtyping was able to discern known epidemiologically related isolates. This subtyping method has been shown to be more discriminatory than PFGE and other molecular typing methods alone and in combination [7]. Here, all reported epidemiologically related cases matched each other at all 40 genes, with the exception of one case in a cluster of three which differed at a single locus. In fact, upon retesting the difference in CGF subtype was found to be a spurious result with the isolate in fact matching at all 40 loci. In an outbreak situation an assessment must be made as to the level of relatedness required to be considered a ‘match’. This issue is not unique to CGF40; it applies to the newest technologies as well, including whole genome sequencing (WGS) [12]. Interpretation criteria for relatedness of strains has been discussed for PFGE [13]. When multilocus variable-number of tandem repeat analysis (MLVA) is included in outbreak case definitions, criteria for allowable deviation from the main outbreak MLVA profile often includes up to three repeats at a single locus or one repeat at two loci [14]. Similarly, CGF40 in outbreak case definitions could potentially include criteria for differences in one or more of the 40 PCR results [9]. Close collaboration between epidemiologists and laboratory colleagues is essential in outbreak investigations occurring in real time. Overall, the discriminatory power of the assay was highly concordant with the epidemiological information.
Case-case analysis provided evidence of differences in the epidemiology of different CGF40 subtypes. The odds of having had contact with chickens (beyond meat handling and/or consumption) was 23 times higher for subtype 0129·001·002 compared to sporadic cases. The odds of having a pet dog or cat was 13 times higher for cases of subtype 0083·001·002 compared to sporadic cases. These and other detected associations may provide the evidence required to direct further investigation of clinically relevant strains of Campylobacter with a goal of ultimately informing prevention and control measures. This type of evidence could inform further follow up of cases of particular CGF40 subtypes with hypothesis-generating or hypothesis-testing questionnaires and direct environmental sampling. Retrospective data collected for this study included answers to closed-ended interview questions. Further information may be required to assess the likelihood of an exposure as a transmission risk versus a confounder. Recovering a matching strain from the source would be ideal. While case numbers were low, it is believed this is the first published evidence of significant associations with risk factors by subtype of Campylobacter.
Predominant sources and geographical regions from which subtypes have been isolated nationally supported the findings from the analytic epidemiology. All of the subtype clusters found to be likely acquired in Nova Scotia, have been observed in concurrent or previous years from non-human sources in that province. Moreover, non-human isolates from subtype 0129·001·002 in the CGF database are derived exclusively from poultry sources, supporting the association with chicken found in this study. Due to the limited number of isolates from pets or companion animals in the database, we were not able to find evidence of subtype 0083·001·002 association with pet ownership on a broader scale. Thus, while comparisons to a reference database may be useful in providing context for subtype characteristics, these may be limited by the content of the database. When transmission routes involve indirect contact with predominant reservoirs, epidemiologic investigation is required to determine the exposure route. Investigation of a Campylobacter outbreak in Alaska in 2008 found that cases were associated with consumption of raw commercially grown peas that had been contaminated by the feces of wild birds [15]. Both the source of the Campylobacter (cranes) and the exposure of cases (consumption of raw peas) are important findings to inform prevention and control measures.
The national CGF reference database can provide context for subtyping results. Similar to other molecular typing systems, rare subtypes can be flagged for potential investigation. The Campylobacter outbreak in Nova Scotia in 2012 was comprised of six confirmed cases and 12 probable cases. Three isolates from this outbreak were available for subtyping and typed as 0922·001·002. Only one other isolate matching this CGF40 result was found in the reference database, a human clinical isolate from the province of Ontario; there were no isolates from animal or environmental sources. There were three more Nova Scotian Campylobacter cases of this subtype isolated within the four months following the outbreak. It is possible that all cases were related to a common source, for which there may have been the potential to implement control measures. This is an example of how CGF40 can direct investigations and increase our knowledge of Campylobacter epidemiology. Some subtypes were more commonly detected. Campylobacter strains show host specificity [16], with host effects found to be stronger than geographic and temporal effects [17, 18]. Strong host effects support the findings here of some subtypes displaying wide global distribution. Thus, knowledge of the frequency of detection and predominant host source for a subtype can support outbreak investigation and source attribution estimates.
There are limitations to these findings. Epidemiological information and/or CGF data were missing for some cases, and due to low case numbers, case-case analyses were limited to univariate analysis of CGF subtype clusters. A greater proportion of isolates were available for subtyping from the largest urban area of the province where the provincial public health laboratory is situated, due to archiving practices. This affected the ability to identify clusters and to describe the epidemiology of strains circulating in rural areas. Mink farmers are known to be over-represented in reported cases of campylobacteriosis (unpublished observation, Nova Scotia Department of Health and Wellness); however, there were no clusters associated with mink farming detected. Whether the proportion of strains with poultry associations would decrease with more rural strains present is unknown. Referral rates from outside laboratories increased over time and the continuation of isolate referral as routine practice along with sharing summaries of subtyping results with referring laboratories may increase participation and reduce this bias. Another issue is a problem common to all subtyping systems, where some widespread subtypes representing clonal lineages require additional subtyping to further distinguish between strains. Common PFGE patterns for certain Salmonella serotypes lead to the need for other typing techniques, such as MLVA or WGS for differentiation. Here, a number of subtypes were detected in multiple years and/or with multiple countries of likely acquisition. Further typing through other methods may be required for some commonly detected subtypes. Sequencing of the flaA short variable region may provide additional discriminatory power and has been suggested as a secondary method to support CGF [9]. CGF40 was developed for C. jejuni, and this method is less discriminatory of other species. Two C. coli CGF40 clusters had multiple countries of likely acquisition. However, with the small numbers of other species identified in Nova Scotia over the time period, speciation may be sufficient to suggest relatedness of strains clustered in time and space for locally acquired cases of non-jejuni species, saving costs of further typing. Strengths of case-case compared to case-control analysis include reduction in selection and recall biases, timeliness and cost-effectiveness [19, 20].
Notably, two case pairs deemed to be epidemiology-linked differed by CGF40 subtype. Speciation of these isolates determined that they were in fact different species. This finding supports the specificity of CGF subtyping and suggests that speciating and/or CGF40 can provide evidence to rule out linkages. On the other hand, these instances may be evidence supporting the idea that picking single colonies in the routine isolation of Campylobacter may limit the detection of coinfections with multiple species and/or strains. Outbreaks of multiple strains of Campylobacter have been reported [16] and some small studies have shown 7·5–24% of patients with campylobacteriosis to be co-infected with multiple strains [21, 22]. Furthermore, in this study, one patient was known to have C. jejuni isolated from a stool specimen and C. fetus isolated from a blood specimen within a week. While the resources required to routinely identify multiple Campylobacter strains from clinical specimens may not be warranted, epidemiological evidence connecting cases to each other or to a potential source should not be disregarded based on typing differences and multiple isolates from patients and sources may be helpful in investigations with evidence suggesting the presence of multiple strains. The findings here and in the literature serve as a reminder that while subtyping methods enhance outbreak detection overall, these tools must be appropriately applied in the context of an investigation [23, 24].
Evidence is mounting in support of the notion that a larger proportion of cases of campylobacteriosis are related to other cases than was once thought. A national surveillance program (FoodNet) in the United States found 0·5% of campylobacteriosis cases reported in 2011 to be outbreak-related [25]. While this suggests >99% of cases are sporadic, recent studies have suggested that this proportion is likely lower and may be closer to 70–75% [26, 27]. Similarly, the findings of this study suggest 73% of cases were sporadic. Incomplete subtyping and/or related cases beyond the 30 day period may in fact mean that a greater proportion of cases are related. Furthermore, if low level contamination of a widely distributed food product causes illness in a number of jurisdictions, real-time sharing of subtyping results across jurisdictions is required to detect these outbreaks. Protocols could be designed to target public health investigation following the implementation of routine CGF40 subtyping, similar to those in place through PulseNet Canada for Salmonella, Escherichia coli, and Listeria [28, 29]. Our data support other findings that small outbreaks are relatively common [30], beyond those detected through routine public health follow up.
Recent work has shown the potential for WGS in the analysis of Campylobacter isolates in an epidemiological context [31–33] and we anticipate that WGS will play an increasingly prominent role in epidemiological investigations. At the same time, resource constraints typically faced by public health laboratories currently preclude the implementation of WGS for routine analysis of a pathogen as commonly encountered as Campylobacter [6]. A possible short-term strategy could be to leverage subtyping methods such as CGF40, which can be used for rapid screening of large numbers of isolates for the detection of possible case clusters that can be followed-up using WGS.
In summary, routine CGF40 subtyping will identify clusters of genetically related Campylobacter strains which could direct public health investigations and identify potential sources for control. Differences in the epidemiology of different Campylobacter CGF40 subtypes were observed. This level of information is required to understand the epidemiology of this pathogen and in turn inform public health prevention and control activities. The greater the number of isolates undergoing the same subtyping activities across diverse geographic sites, the greater the knowledge base of global epidemiology of Campylobacter. Accessing historical case data from routine enteric disease surveillance for case-case analysis may allow for the swift hypothesis testing critical to outbreak control efforts on a prospective basis. Our findings suggest CGF40 is suited for routine public health surveillance and epidemiological studies of Campylobacter. Further studies of prospective CGF subtyping coupled with investigation of clusters is warranted.
ACKNOWLEDGEMENTS
The authors thank the public health nurses and the laboratory staff across the Province of Nova Scotia, without whom this project and manuscript would not have been possible. We also thank Dr Shiona Glass-Kaastra for the critical review of the statistical approach.
This research received no specific grant from any funding agency, commercial or not-for-profit sectors.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Thomas MK, Murray R, on behalf of the Canadian Burden of Food-borne Illness Estimates Working Group. Estimating the burden of food-borne illness in Canada. Canada Communicable Disease Report 2014; 40: 299–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaakoush NO, et al. Global epidemiology of Campylobacter infection. Clinical Microbiology Reviews 2015; 28: 687–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas KM, et al. Estimates of foodborne illness related hospitalizations and deaths in Canada for 30 specified pathogens and unspecified agents. Foodborne Pathogens and Disease 2015; 12: 820–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nova Scotia Population Health Assessment and Surveillance. Notifiable Diseases in Nova Scotia 2014 Surveillance Report (http://novascotia.ca/dhw/populationhealth/documents/Annual-Notifiable-Disease-Surveillance-Report-2014.pdf). Accessed 4 April 2016.
- 5.Allos BM, et al. Surveillance for sporadic foodborne disease in the 21st century: the FoodNet perspective. Clinical Infectious Diseases 2004; 38: S115–120. [DOI] [PubMed] [Google Scholar]
- 6.Taboada EN, et al. Current methods for molecular typing of Campylobacter species. Journal of Microbiological Methods 2013; 95: 24–31. [DOI] [PubMed] [Google Scholar]
- 7.Clark CG, et al. Comparison of molecular typing methods useful for detecting clusters of Campylobacter jejuni and C. coli isolates through routine surveillance. Journal of Clinical Microbiology 2012; 50: 798–809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michaud S, Ménard S, Arbeit RD. Role of real-time molecular typing in the surveillance of Campylobacter enteritis and comparison of pulsed-field gel electrophoresis profiles from chicken and human isolates. Journal of Clinical Microbiology 2005; 43: 1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taboada EN, et al. Development and validation of a comparative genomic fingerprinting method for high-resolution genotyping of Campylobacter jejuni. Journal of Clinical Microbiology 2012; 50:788–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McCarthy N, Giesecke J. Case-case comparisons to study causation of common infectious diseases. International Journal of Epidemiology 1999; 28: 764–768. [DOI] [PubMed] [Google Scholar]
- 11.Jokinen CC, et al. An enhanced technique combining pre-enrichment and passive filtration increases the isolation efficiency of Campylobacter jejuni and Campylobacter coli from water and animal fecal samples. Journal of Microbiological Methods 2012; 91: 506–513. [DOI] [PubMed] [Google Scholar]
- 12.European Centre for Disease Prevention and Control. Expert Opinion on the introduction of next-generation typing methods for food- and waterborne diseases in the EU and EEA. Stockholm: ECDC, 2015.
- 13.Tenover FC, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. Journal of Clinical Microbiology 1995; 33: 2233–2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Slayton RB, et al. Outbreak of shiga toxin-producing Escherichia coli (STEC) O157:H7 associated with romaine lettuce consumption, 2011. PLoS ONE 2013; 8: e55300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gardner TJ, et al. Outbreak of campylobacteriosis associated with consumption of raw peas. Clinical Infectious Diseases 2011; 53: 26–32. [DOI] [PubMed] [Google Scholar]
- 16.Cody AJ, et al. Where does Campylobacter come from? A molecular odyssey. Advances in Experimental Medicine and Biology 2010; 659; 47–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sheppard SK, et al. Host association of Campylobacter genotypes transcends geographic variation. Applied and Environmental Microbiology 2010; 76: 5269–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McCarthy ND, et al. Host-associated genetic import in Campylobacter jejuni. Emerging Infectious Diseases 2007; 13: 267–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilson N et al. Case-case analysis of enteric diseases with routine surveillance data: Potential use and example results. Epidemiologic Perspectives and Innovations 2008; 5: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doorduyn Y, et al. Risk factors for indigenous Campylobacter jejuni and Campylobacter coli infections in The Netherlands: a case-control study. Epidemiology and Infection 2010; 138: 1391–1404. [DOI] [PubMed] [Google Scholar]
- 21.Richardson JF, et al. Coinfection with Campylobacter species: an epidemiological problem? Journal of Applied Microbiology 2001; 91: 206–211. [DOI] [PubMed] [Google Scholar]
- 22.Gilpin B, et al. Pulsed-field gel electrophoresis analysis of more than one clinical isolate of Campylobacter spp. from each of 49 patients in New Zealand. Journal of Clinical Microbiology 2012; 50: 457–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osterholm MT. The detection of and response to a foodborne disease outbreak: a cautionary tale. Clinical Infectious Diseases 2015; 61: 910–911. [DOI] [PubMed] [Google Scholar]
- 24.Kwan PSL, et al. Multilocus sequence typing confirms wild birds as the source of a Campylobacter outbreak associated with the consumption of raw peas. Applied and Environmental Microbiology 2014; 80: 4540–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.CDC. Foodborne Diseases Active Surveillance Network (FoodNet): Foodnet Surveillance Report for 2011. Atlanta, Georgia: U.S. Department of Health and Human Services. 2012. (http://www.cdc.gov/foodnet/PDFs/2011_annual_report_508c.pdf). Accessed 29 March 2016.
- 26.Gillespie IA, et al. Point source outbreaks of Campylobacter jejuni infection – are they more common than we think and what might cause them? Epidemiology and Infection 2003; 130: 367–375. [PMC free article] [PubMed] [Google Scholar]
- 27.Castrodale LJ, et al. Calling all Campy – how routine investigation and molecular characterization impacts the understanding of campylobacteriosis epidemiology – Alaska, United States, 2004–2013. Epidemiology and Infection 2016; 144: 265–267. [DOI] [PubMed] [Google Scholar]
- 28.Public Health Agency of Canada. PulseNet Canada (https://www.nml-lnm.gc.ca/Pulsenet/index-eng.htm). Accessed 4 April 2016.
- 29.Rumore JL, Tschetter L, Nadon C. The impact of multilocus variable-number tandem-repeat analysis on PulseNet Canada Escherichia coli O157:H7 laboratory surveillance and outbreak support, 2008–2012. Foodborne Pathogens and Disease 2016; 13: 255–261. [DOI] [PubMed] [Google Scholar]
- 30.Cornelius AJ, et al. Same-day subtyping of Campylobacter jejuni and C. coli isolates by use of multiplex ligation-dependent probe amplification-binary typing. Journal of Clinical Microbiology 2014; 52: 3345–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cody AJ, et al. Real-time genomic epidemiological evaluation of human Campylobacter isolates by use of whole-genome multilocus sequence typing. Journal of Clinical Microbiology 2013; 51: 2526–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kovanen SM, et al. Multilocus sequence typing (MLST) and whole-genome MLST of Campylobacter jejuni isolates from human infections in three districts during a seasonal peak in Finland. Journal of Clinical Microbiology 2014; 52: 4147–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Revez J, et al. Genome analysis of Campylobacter jejuni strains isolated from a waterborne outbreak. BMC Genomics 2014; 15: 768. [DOI] [PMC free article] [PubMed] [Google Scholar]