SUMMARY
HBsAg reappearance may constitute not only a risk for liver disease but also an infectious source. We aimed to determine whether HBsAg may reappear after spontaneous HBsAg seroclearance. A cohort of 2999 HBsAg-positive subjects aged 30–55 years was recruited in Guangxi, China in 2004. HBsAg was tested every 6 months from July 2004 to June 2007, then, one more time in December 2013. The results showed that spontaneous HBsAg seroclearance occurred in 41 subjects in the first 3 years, giving a 0·54% annual seroclearance rate. Thirteen of the 41 subjects were randomly tested for HBsAg in 2013. Four subjects became HBsAg positive. S gene sequences of HBV were analysed from serum collected before seroclearance and after reappearance, respectively, for subject QS840 (11 and 12 clones), subject TN98 (13 and 13 clones) and subject WX227 (10 and 8 clones). Serotype, subgenotype and amino-acid substitution pattern in each sample collected after reappearance was observed in the sample collected before HBsAg seroclearance. Nucleotide similarity between the two sequences from each subject was >99% and five sequences from subject TN98 were the same. In conclusion, following reactivation, HBsAg may reappear in individuals with spontaneous HBsAg seroclearance many years previously.
Key words: Genotypes, HBsAg, hepatitis B virus (HBV), mutations, reappearance, seroclearance, serotype
INTRODUCTION
Although the implementation of a universal hepatitis B vaccination programme has proven successful in preventing acute hepatitis B virus (HBV) infection [1], persistent infection with HBV remains a major global public health problem. There are about 240 million hepatitis B surface antigen (HBsAg) carriers worldwide [2] who are in the risk of acute and chronic liver diseases including cirrhosis and hepatocellular carcinoma (HCC) [3].
HBsAg is an important serological marker for the diagnosis and monitoring of chronic hepatitis B patients [4]. HBsAg seroclearance during the clinical course can be considered as resolved hepatitis B infection [5]. HBsAg seroclearance may occur during the natural history of HBV infection and following antiviral therapy [6].
The incidence of spontaneous HBsAg seroclearance, defined as the absence of serum HBsAg on two occasions at least 6 months apart and remaining negative up to the last visit [7], varies with geographical area. It is estimated that the annual incidence is about 0·7–1·7% in regions where the prevalence of HBsAg is low, such as Western countries [8–10], while that in regions where HBV is endemic, such as Asian countries, is about 0·5–3·08% [7, 11, 12].
Spontaneous HBsAg seroclearance is a rare event in chronic hepatitis B. However, most patients with HBsAg seroclearance have an excellent outcome and are in a state closest to a ‘cure’ [13], although some of them remain at risk of HCC [14]. Therefore, much effort has been expended to identify factors associated with HBsAg seroclearance with the objective of increasing the rate of HBsAg seroclearance. Older age, HBeAg seronegativity, clinical remission and cirrhosis have been reported to be associated with HBsAg seroclearance [13]. It also has been reported that HBV viral load and quantitative serum HBsAg levels can accurately predict HBsAg seroclearance [15]. However, little is known about the reappearance of HBsAg after spontaneous HBsAg seroclearance, which may constitute not only an increased risk of liver disease but also an infectious source.
In this study, based on our long-term cohort study [16], we report some asymptomatic HBsAg carriers who experienced spontaneous HBsAg seroclearance, with HBsAg reappearance many years later. We also determined whether the reappearance was caused by reinfection or reactivation using sequence analysis.
MATERIALS AND METHODS
Study subjects
All of the study subjects were recruited from the Long An cohort, which has been described previously [16]. The cohort was established in early 2004 from 27 340 agricultural workers aged 30–55 years living in the rural area of Long An county, Guangxi, China, using stratified sampling. It consists of 2998 HBsAg carriers. We followed the study subjects once every 6 months from 1 July 2004 to 1 July 2007. Then, we continued to follow-up some of the study subjects randomly once in 2013 when we randomly recruited 300 subjects with wild type of HBV core promoter and 300 subjects with HBV core promoter double mutations (1762T1764A) from the Long An cohort with the purpose of searching for human genes related to the double mutations. Each study subject provided a serum sample for the assessment of virological parameters and alpha fetoprotein concentrations.
Informed consent in writing was obtained from each individual. The study protocol conforms to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Guangxi Institutional Review Board.
Serological testing
Sera were tested for HBsAg, HBeAg/anti-HBe, and anti-HBc using enzyme immunoassays (Beijing Wantai Biological Pharmacy Enterprise Co. Ltd, China). Alanine aminotransferase (ALT) levels were determined using a Reitman kit (Shanghai Fortune Long March Medical Science, China).
Measurement of viral loads
Serum HBV DNA concentrations were quantified by real-time polymerase chain reaction (PCR) using commercial reagents (Sansure Biotech Inc., China) in an ABI Prism 7500 sequence detection system (Applied Biosystems, USA) with a dynamic range of 1×102 –5 × 109 IU/ml.
Nested PCR for HBV DNA and nucleotide sequencing
DNA was extracted from 85 µl serum by pronase digestion followed by phenol/chloroform extraction. The first round of PCR was carried out in a 50 µl reaction using primers MD14 (nt 418–433, 5'-GCGCTGCAGCTATGCCTCATCTTC-3') and HCO2 (nt 761–776, 5'-GCGAAGCTTGCTGTACAGACTTGG-3'), with 5 min hot start followed by 35 cycles of 94 °C for 45 s, 45 °C for 45 s, and 72 °C for 60 s. The second round of PCR was carried out on 5 µl of the first-round products in a 50 µl reaction using primers ME15 (nt 455–470, 5'- GCGCTGCAGCAAGGTATGTTGCCCG-3') and HDO3 (nt 734–748, 5’-GCGAAGCTTCATCATCCATATAGC-3’), with 5 min hot start followed by 30 cycles of 94 °C for 45 s, 55 °C for 45 s, and 72 °C for 60 s. Products from the second round were confirmed by agarose gel electrophoresis. HBV DNA-positive products were sent to Sangon Biotech (China) for cloning sequencing. Amplicons were cloned into the vector pUCm-T (Sangon Biotech). Plasmid DNA was extracted using a SK1191 UNIQ-10 kit (Sangon Biotech) and the purified DNA was sequenced using a BigDye Terminator v. 3.1 Cycle Sequencing kit (Applied Biosystems) with sequencing primer ME15.
HBV serotyping
Serotypes were determined according to a single amino acid at the following positions: adrq- (122 K+127P+134 F+159 V+160R+177 A+178P), adrq+ (122 K+127P+134 F+159A+160R+177 V+178P), adw2 (122 K+127P+134 F+159A+160 K+177 V+178P), adwq+ (122 K+127 L+134 F+159A+160 K+177 V+178P), ayr (122R+127P+134 F+159A+160R+177 V+178P), ayw1 (122R+127P+134 F+159A+160 K+177 V+178P), ayw2 (122R+127P+134Y+159G+160 K+177 V+178P), ayw3 (122R+127 T+134 F+159G+160 K+177 V+178P) [17–19].
HBV genotyping
HBV genotypes were determined using phylogenies reconstructed on the basis of the complete S region (294 nt) of the viruses. The sequences were aligned to 16 HBV sequences of all known genotypes retrieved from GenBank using Clustal W and visually confirmed with the sequence editor BioEdit [20]. These sequences include M57663-Philippines, D23677-Japan, KT347089-South Africa, KM359441-Argentina, X75656-Polynesia, KF873543-Australia, KM999992-Philippines, EU670263-Philippines, Y07587-Germany, AB032431-Liberia, AY090456-Nicaragua, AF160501-USA, AY090460-USA, AF241407-Vietnam, AB486012-Japan and AY226578-Woolly monkey. Neighbour-Joining trees were reconstructed under the Kimura two-parameter substitution model with the program MEGA [21]. The reliability of clusters was evaluated using interior branch test with 1000 replicates, and the internal nodes with over 95% support were considered reliable.
Statistical analysis
Statistical comparison of variables was performed using the χ2 test. The 95% confidence intervals (CI) for the prevalence of variables were estimated. Logistic regression analysis was carried out to identify factors associated with spontaneous HBsAg seroclearance. All P values were two-tailed and P < 0·05 was considered to be significant. Statistical analysis was performed using SPSS v. 16.0 software (SPSS Inc., USA).
RESULTS
Baseline characteristics
In this study, we carried out seven rounds of sampling. Sampling of the study subjects depended on their consent. The seventh sampling, incorporated in another study, included a subset of the subjects in 2013. Again, the study subjects provided samples after giving consent. The total rate of HBsAg negativity was 2·0% (201/12355, 95% CI 1·4–1·8). The rate of HBsAg negativity in males (1·6%, 101/6413) was not significantly different from that in females (1·7%, 100/5942) (χ2 = 0·225, P > 0·05). The rate of HBsAg negativity in the group originally HBeAg positive (0·3%, 5/1821) was significantly higher than that in the HBeAg-negative group (0·18%, 179/10134) (χ2 = 22·668, P < 0·05). The difference in the rate of HBsAg negativity between the abnormal ALT at baseline group (1·4%, 23/1691) and normal ALT at baseline group (1·6%, 161/10264) was not significant (χ2 = 0·416, P > 0·05) (Table 1). Clearly, HBeAg status is associated with HBsAg negativity.
Table 1.
Results of the follow up of the cohort
| Visiting date | Status of HBsAg | No. | Age, years (mean±s.d.) | Male (%) (95% CI) | HBeAg(+) (%) (95% CI) | ALT (%) (95% CI) | HBsAg(-) (%) (95% CI) |
|---|---|---|---|---|---|---|---|
| July 2004 | HBsAg(-) | 11 | 42·4 ± 8·4 | 5 (45·5) (16·1–74·9) | 0 | 1 (9·1) (−7·9 to 26·1) | 0·4 (0·2–0·6) |
| HBsAg(+) | 2547 | 38·7 ± 7·0 | 1105 (43·4) (41·5–45·3) | 402 (15·8) (14·4 to 17·2) | 336 (13·2) (11·9 to 14·5) | ||
| Feb. 2005 | HBsAg(-) | 21 | 41·7 ± 7·5 | 9 (42·9) (21·7–64·1) | 2 (9·5) (−3·0 to 22·0) | 10 (4·8) (−4·3 to 13·9) | 1·1 (0·6–1·6) |
| HBsAg(+) | 1831 | 39·4 ± 6·9 | 1027 (56·1) (53·8–58·4) | 329 (18·0) (16·2 to 19·8) | 263(14·4) (12·8 to 16·0) | ||
| Aug. 2005 | HBsAg(-) | 33 | 42·9 ± 6·9 | 10 (30·3) (14·6–46·0) | 2 (6·1) (−2·1 to 14·3) | 6 (18·2) (5·0 to 31·4) | 1·8 (1·2–2·4) |
| HBsAg(+) | 1802 | 39·6 ± 6·9 | 996 (55·3) (53·0–57·6) | 313 (17·4) (15·6 to 19·2) | 318 (17·7) (15·9 to 19·5) | ||
| Mar. 2006 | HBsAg(-) | 32 | 43·4 ± 7·4 | 15 (46·9) (29·6–64·2) | 1 (3·1) (−2·9 to 9·1) | 2 (6·3) (−2·1 to 14·7) | 1·7 (1·1–2·3) |
| HBsAg(+) | 1840 | 39·0 ± 6·9 | 1010 (54·9) (52·6–57·2) | 283 (15·4) (13·8 to 17·0) | 257 (14·0) (12·4 to 15·6) | ||
| Sept. 2006 | HBsAg(-) | 29 | 42·9 ± 6·8 | 15 (51·7) (33·5–70·0) | 0 | 1 (3·5) (−3·2 to 10·2) | 1·6 (1·0–2·2) |
| HBsAg(+) | 1766 | 40·5 ± 6·9 | 943 (53·4) (51·1–55·7) | 227 (12·9) (11·3 to 14·5) | 254 (14·4) (12·8 to 16·0) | ||
| June 2007 | HBsAg(-) | 58 | 41·1 ± 6·8 | 33 (56·9) (44·2–69·6) | 0 | 3 (5·2) (−0·5 to 10·9) | 2·8 (2·1–3·5) |
| HBsAg(+) | 1985 | 39·3 ± 7·0 | 1081 (54·5) (52·3–56·7) | 262 (13·2) (11·7 to 14·7) | 240 (12·1) (10·7 to 13·5) | ||
| Dec. 2013 | HBsAg(-) | 17 | 51·9 ± 7·5 | 14 (82·4) (64·3–100·5) | n.d. | n.d. | 4·4 (2·4–6·4) |
| HBsAg(+) | 383 | 50·7 ± 6·3 | 150 (39·4) (34·5–44·3) | n.d. | n.d. | ||
| Total | HBsAg(-) | 201 | 101 | 5 | 23 | 1·6 (1·4–1·8) | |
| HBsAg(+) | 12 154 | 6312 | 1816 | 1668 |
CI, Confidence interval; ALT, alanine aminotransferase; n.d., not done.
Spontaneous HBsAg seroclearance and its factors in the first six samples
According to the definition of spontaneous HBsAg seroclearance [7], we found that this occurred in 41 subjects during 7583 person-years of follow-up evaluation, giving a 0·54% annual seroclearance rate. Multivariable logistic regression analysis was performed to identify factors associated with spontaneous HBsAg seroclearance. The independent variables included gender, age, genotype, viral load, HBeAg and ALT. Information for these variables was based on the baseline data. The results showed that older age (> 40 years) was associated with spontaneous HBsAg seroclearance [odds ratio (OR) 6·0, 95% CI 1·3–21·4, P = 0·022]. Low viral load (<103 copies/ml in serum) was also a factor for spontaneous HBsAg seroclearance (OR 6·3, 95% CI 1·4–28·8, P = 0·018) (Table 2).
Table 2.
Logistic regression analysis for factors associated with spontaneous HBsAg seroclearance
| Analysis | Variables | B* | s.e. | P value | OR | 95% CI |
|---|---|---|---|---|---|---|
| Univariate analysis | Sex (male vs. female) | 0·581 | 0·416 | 0·163 | 1·788 | 0·791–4·043 |
| Age (⩾40 years) | 1·356 | 0·473 | 0·004 | 3·881 | 1·535–9·814 | |
| Genotype (B vs. C) | −0·189 | 0·501 | 0·706 | 0·828 | 0·310–2·211 | |
| Viral loads (<103 copies/ml in serum) | 2·003 | 0·549 | 0·0001 | 7·411 | 2·525–21·754 | |
| HBeAg positive | 1·640 | 1·024 | 0·109 | 5·154 | 0·693–38·356 | |
| ALT (abnormal) | 0·039 | 0·623 | 0·951 | 1·039 | 0·307–3·521 | |
| Multivariate analysis | Age | 1·786 | 0·778 | 0·022 | 5·966 | 1·299–27·397 |
| Viral loads | 1·835 | 0·778 | 0·018 | 6·262 | 1·364–28·753 |
OR, Odds ratio; CI, confidence interval; ALT, alanine aminotransferase.
* B, regression coefficient.
Reappearance of HBsAg after spontaneous seroclearance
Thirteen of the 41 individuals who experienced spontaneous HBsAg seroclearance were included in another study and therefore tested for HBsAg in 2013. Surprisingly, we found that four of these subjects had become positive for HBsAg after spontaneous seroclearance more than 8 years previously, making the rate of HBsAg reappearance 30·8% (4/13). None of the 13 individuals had a risk factor for reactivation, such as immunosuppression, steroid use, HIV infection, etc. The four subjects were QS840, TF336, TN98 and WX227; subjects QS840, TF336 and TN98 were positive for anti-HBs when HBsAg reappeared. Subjects QS840 and WX227 had abnormal ALT levels. It is difficult to explain this phenomenon by sex, age, genotype, viral load, HBeAg and ALT (Table 3). In order to determine whether the reappearance of HBsAg is due to reinfection or reactivation, we randomly selected three of the four with HBsAg reappearance for sequencing of the HBV genome.
Table 3.
Characteristics of subjects with HBsAg seroclearance and reappearance
| Subject | Sex | Age | Genotype | Baseline viral load | HBeAg | ALT | July 2004 | Feb. 2005 | Aug. 2005 | Mar. 2006 | Sept. 2006 | June 2007 | Dec. 2013 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| BB255 | F | 53 | C | 737·7 | − | − | + | − | − | − | n.d. | n.d. | − |
| GA102 | F | 36 | U | 1933 | − | − | + | n.d. | n.d. | − | − | − | − |
| NF120 | F | 50 | B | 3509 | − | − | + | + | + | − | − | − | − |
| QS840 | M | 39 | C | 41·92 | + | − | + | − | − | − | − | − | + |
| QZ550 | M | 42 | C | 11 482 | − | − | + | n.d. | − | − | − | − | − |
| TF336 | F | 34 | C | 9·5 × 105 | n.d. | n.d. | + | − | − | n.d. | − | n.d. | + |
| TF405 | M | 30 | C | 701·5 | − | + | + | − | n.d. | − | − | − | − |
| TL203 | M | 35 | n.d. | 168·9 | − | − | + | − | − | n.d. | − | − | − |
| TM015 | F | 41 | C | 769·1 | − | − | + | + | − | n.d. | − | − | − |
| TN98 | F | 40 | C | 135·1 | − | − | + | − | − | − | − | − | + |
| TS117 | F | 50 | C | 57. 9 | − | − | + | + | − | − | − | − | − |
| TW126 | F | 39 | n.d. | 4·8 | − | − | + | − | − | − | n.d. | − | − |
| WX227 | M | 40 | C | 4816 | − | + | + | + | + | − | − | − | + |
ALT, Alanine aminotransferase; n.d., not done.
Reappearance of HBsAg is due to reactivation
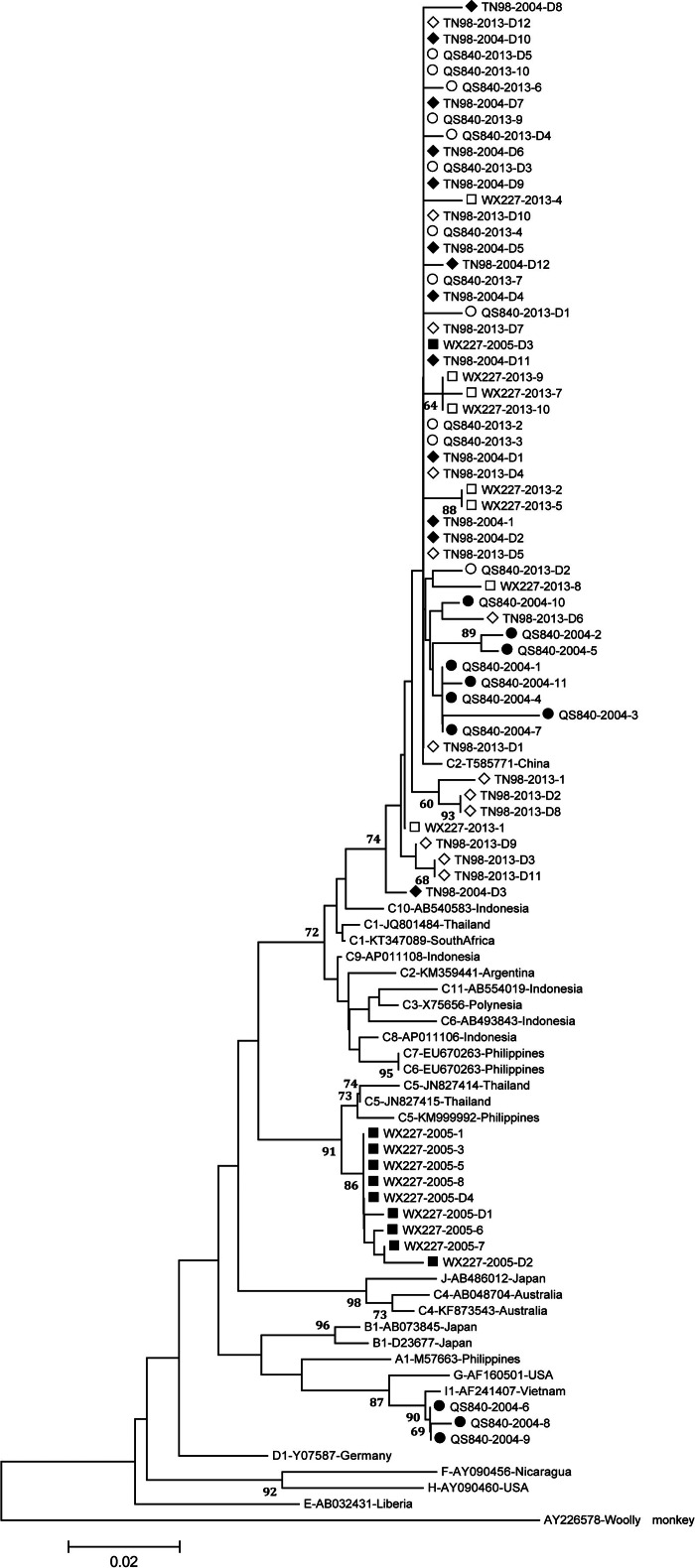
For subject QS840, 11 and 12 clones were selected from serum collected in 2004 and 2013, respectively. Serotypes adrq+ and adw2 were identified in the sample from 2004 but only serotype adrq+ was found in the sample from 2013. Subgenotypes C2 and I1 were identified in the sample from 2004. However, only subgenotype C2 could also be seen in samples collected in 2013. Ten of the 12 sequences from the sample collected in 2013 have the same amino-acid substitution mutation pattern in the major hydrophilic region (T118 K, T123N, G145A) as the sample collected in 2004 (Table 4, Figs 1, 2). All of the sequences from samples collected in 2013 have a similarity rate of 99% to that of 2004 at the nucleotide level. Clearly, the virus in the 2013 samples come from the virus in 2004 because they share the same serotype adrq+, subgenotype C2, mutation pattern and high rate of nucleotide similarity. Therefore, these data suggest that the reappearance of HBsAg in subject QS840 is due to reactivation.
Table 4.
Molecular characteristics of sequences from subject with reappearance of HBsAg
| Sample | Year of follow-up | No. of clones | Serotype | Genotype | Mutations |
|---|---|---|---|---|---|
| QS840 | 2004 | 11 | adrq+ (7*) | C2 (7), | T118 K, T123N, G145A (6) |
| T118 K, T123N, G145A, S174R | |||||
| adw2 (1) | C2 (1) | T118 K, T123N, G145A, F161Y, V168A | |||
| adw2 (3) | I1 (3) | D118N, L129M, K149Q (2) | |||
| D118N, D123G, L129M, K149Q | |||||
| 2013 | 12 | adrq+ (12) | C2 (12) | T118 K, T123N, G145A (10) | |
| T118E, T123N, G145A | |||||
| T118 K, T123N, C124R, G145A, P151H | |||||
| TN98 | 2004 | 13 | adrq+ (13) | C2 (13) | T118 K, T123N, G145A(12) |
| G145A, E164D | |||||
| 2013 | 13 | adrq+ (13) | C2 (13) | T118 K, G145A | |
| T118 K, T123N, G145A(6) | |||||
| T118 K, T123N, G145A, C149Y(2) | |||||
| T118 K, G145A (3) | |||||
| T118 K, T123N, G130A, G145A | |||||
| WX227 | 2005 | 10 | adrq+ (1) | C2 (1) | T118 K, T123N, G145A |
| adw2 (7)† | C5 (9 | Wild type (7) | |||
| K122 | |||||
| K122, S136 T | |||||
| 2013 | 8 | adrq+ (8) | C2(8) | T118 K, G145A | |
| T118 K, T123N, G145A(2) | |||||
| T118 K, G119E, T123N, G145A | |||||
| C107R, T118 K, T123N, G145A, W156L(3) | |||||
| T118 K, T123N, G145A, P151L |
* Number of clones.
† Two sequences could not be serotyped because they have a stop codon at aa122.
Fig. 1.
Frequency and distribution of amino-acid substitutions in the MHR of HBsAg from each clone of (a) subject QS840, (b) subject TN98, (c) subject WX227.
Fig. 2.
Neighbour-joining trees. The trees were reconstructed on the basis of the S region (294 nt) of the viruses under the Kimura two-parameter substitution model with the program MEGA [21]. The branch lengths represent the number of substitutions per site. The reliability of clusters was evaluated using the interior branch test with 1000 replicates and the internal nodes with over 75% support are considered reliable.
For subject TN98, 13 and 13 clones were selected from serum collected in 2004 and 2013, respectively. Serotype adrq+ was the only serotype in both samples. Subgenotype C2 was also the only genotype in both samples. Most sequences from 2004 and 2013 share the same amino-acid substitution mutation pattern in the major hydrophilic region: T118 K, T123N, G145A (Table 4, Figs 1, 2). Five of the 13 DNA sequences from 2013 are the same as that from 2004. The remaining sequences have a similarity rate of 99% to that from 2004. Clearly, the reappearance of HBsAg in subject TN98 is also due to reactivation.
For subject WX227, 10 and eight clones were selected from serum collected in 2005 and 2013, respectively. There are two serotypes in the sample from 2005: adrq+ and adw2. However, serotype adrq+ was the only serotype in samples from 2013. There are two subgenotypes in samples from 2005: C2 and 5. However, only subgenotype C2 could be found in one clone in samples from 2013. Most sequences from 2005 were wild type while none of the sequences from 2013 were wild type in the major hydrophilic region (Table 4, Figs 1, 2). All DNA sequences from 2013 had a similarity rate of 99% to that from the only clone with subgenotype C2 from samples of 2005 and share the same mutation (T118 K and G145A). Their serotypes are all adrq+. These data also suggest that the reappearance of HBsAg in subject WX227 was due to reactivation.
Clearly, based on serotype, genotype, amino-acid substitution mutation pattern in the major hydrophilic region and similarity of nucleotide sequence in the S gene, the reappearance of HBsAg in this study is due to reactivation, suggesting that the definition of spontaneous HBsAg seroclearance [7] has been challenged. Interestingly, all sequences involved in HBsAg reappearance encoded a G145A substitution before HBsAg seroclearance. However, none of sequences without G145A in samples QS840 and WX227 appear after reactivation (Table 4).
DISCUSSION
This study, based on the Long An cohort, is the first to report that reappearance of HBsAg may occur in individuals with HBsAg spontaneous seroclearance many years previously. The reappearance resulted from reactivation, rather than reinfection. We also found that the rate of spontaneous HBsAg seroclearance varies with cohort, not geographical area. Older age (>40 years) and low viral load (<103copies/ml in serum) were associated with spontaneous HBsAg seroclearance. However, factors that explain the reappearance of HBsAg remain unclear. The strength of this study is that it was based on a long-term followed-up cohort, which allowed us to obtain information from asymptomatic HBsAg carriers to spontaneous HBsAg seroclearance, and then HBsAg reappearance many years later. Furthermore, none of our study subjects were treated with antiviral drugs, which should avoid the influence of these drugs on HBsAg seroclearance and reappearance. The weakness of this study is that we did not test all of incidences of spontaneous HBsAg seroclearance, which may provide more information about reappearance and make the conclusion stronger.
Although HBsAg seroclearance was considered to be the state closest to a ‘cure’ [13], reactivation after seroclearance remains possible. This phenomenon is not a rare event in individuals with HBsAg seroclearance through antiviral therapy [22, 23]. It could also be observed in people with other diseases, such as HCC [24], haematological malignancies [25] and HIV infection [26, 27]. However, HBsAg reappearance in the general population with spontaneous HBsAg seroclearance has not been reported. Furthermore, none of the studies determined whether reinfection or reactivation was responsible for the reappearance of HBsAg. In this study, we are the first to report that HBsAg reappearance occurred in the general population and found that reactivation was the cause of its reappearance.
The natural course of chronic HBV infection can be divided into four phases, based on the virus–host interaction: immune tolerance, immunoreactive, low or no replication, and reactivation. The third phase is characterized by undetectable or low levels of HBV DNA [28]. In general, serum HBsAg titre has a positive correlation with HBV DNA concentrations [29]. It has been reported that patients with HBsAg seroclearance still harbour HBV within the liver (mainly in the form of cccDNA) up to 4 years after seroclearance, albeit at a very low replicative level and in a transcriptionally inactive phase [13]. It is possible that HBV exists within the liver for longer and causes reactivation when the host becomes disadvantaged, such as through immune suppression. If so, this could explain not only the reappearance of HBsAg but also why those experiencing HBsAg seroclearance remain at risk for HCC [14]. Therefore, our findings are important to remind clinicians to continue to monitor cirrhosis and HCC for individuals with HBsAg seroclearance, because spontaneous HBsAg seroclearance during the course of HBV infection does not mean resolved hepatitis B infection. It is also important for medical workers to monitor HBsAg because its reappearance constitutes a potential source of infection.
Generally, the incidence of spontaneous HBsAg seroclearance is lower in low-endemic areas such as Western countries than in high-endemic areas [30]. Recent findings suggest that the cumulative rate of HBsAg seroclearance in asymptomatic adult carriers from high-endemic areas is higher [11]. In this study, we found that the annual seroclearance rate is lower than that observed in low-endemic areas (European Caucasian subjects, 0·54% vs. 1·43%) [31] and high-endemic areas (Taiwanese subjects 0·54% vs. 1·15%) [11], suggesting that the rate of spontaneous HBsAg seroclearance varies with cohort, not geographical area. We also found that older age and low viral load were associated with spontaneous HBsAg seroclearance, which is in accordance with other studies [30].
Although we found that reappearance of HBsAg was caused by reactivation, we could not reliably identify factors associated with reactivation. In this study, we found serotype adrq+ and adw2 in subjects QS840 and WX227 before seroclearance. However, only serotype adrq+ was found in their samples after reappearance. Furthermore, before seroclearance, adw2 was the predominant serotype in sample WX227 and serotype adrq+ was only found in one of the ten clones. It seems that serotype adrq+ is associated with reactivation. Another interesting observation is that only sequences with the G145A substitution appeared after reappearance. There are other sequences without G145A in the samples from QS840 and WX227 before seroclearance but none of them are present after reactivation. G145A mutation is well known as an escape mutation [32]. The G145A mutation may result in significantly decreased HBsAg expression [33]. When the G145A mutant become predominant, it may reduce the level of HBsAg below the limit of detection so the subjects with this mutant appear to have achieved seroclearance. When the G145A mutant is a minor population, the level of HBsAg is above the limit of detection. However, this hypothesis requires further confirmation.
Reinfection is a possible cause of reappearance of HBsAg. However, none of the three subjects tested in this study had experienced reinfection. This may be attributable to the small sample size. Although 41 individuals with HBsAg spontaneous seroclearance were found in this study, we only tested 13 of them.
In conclusion, HBsAg may reappear in individuals with spontaneous HBsAg seroclearance many years previously. Reactivation is a cause of the reappearance. Therefore, it seems necessary to maintain clinical monitoring and regular surveillance for HBsAg and liver diseases after spontaneous HBsAg seroclearance.
ACKNOWLEDGEMENTS
We are indebted to staff members of Centre for Disease Prevention and Control of Long An and local town hospitals in Long An county, Guangxi, who assisted in recruiting the study subjects and sample collection, and to staff members of the Department of Virology, Centre for Disease Prevention and Control of Guangxi, China for their help in recruiting the study subjects and handling the sera. This study was supported by the Wellcome Trust (WT072058MA) and High Level Academic Backbone 139 Program in Medical Science of Guangxi.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Wen WH, et al. A review of strategies to prevent mother-to-infant transmission of hepatitis B virus infection. Expert Review of Gastroenterology and Hepatology 2015; 17: 1–14. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization. Hepatitis B Fact sheet No. 204, 2014 (http://www.who.int/mediacentre/factsheets/fs204/en/).
- 3.Chen DS. From hepatitis to hepatoma: Lessons from type B. viral hepatitis. Science 1993; 262: 369–370. [DOI] [PubMed] [Google Scholar]
- 4.Cheng HR, et al. Host genetic factors affecting spontaneous HBsAg seroclearance in chronic hepatitis B patients. PLoS ONE 2013; 8: e53008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sorrell MF, et al. National Institutes of Health Consensus Development Conference Statement: management of hepatitis B. Annals of Internal Medicine 2009; 150: 104–110. [DOI] [PubMed] [Google Scholar]
- 6.Lok AS, et al. Chronic hepatitis B. Hepatology 2007; 45: 507–539. [DOI] [PubMed] [Google Scholar]
- 7.Liaw YF, et al. Incidence, determinants and significance of delayed clearance of serum HBsAg in chronic hepatitis B virus infection: a prospective study. Hepatology 1991; 13: 627–631. [PubMed] [Google Scholar]
- 8.Sampliner RE, et al. The liver histology and frequency of clearance of the hepatitis B surface antigen (HBsAg) in chronic carriers. American Journal of the Medical Sciences 1979; 277: 17–22. [DOI] [PubMed] [Google Scholar]
- 9.Villeneuve JP, et al. A long-term follow-up study of asymptomatic hepatitis B surface antigen-positive carriers in Montreal. Gastroenterology 1994; 106: 1000–1005. [DOI] [PubMed] [Google Scholar]
- 10.Da Silva LC, et al. Spontaneous hepatitis B surface antigen clearance in a long-term follow-up study of patients with chronic type B hepatitis. Lack of correlation with hepatitis C and D virus superinfection. Journal of Gastroenterology 1996; 31: 696–701. [DOI] [PubMed] [Google Scholar]
- 11.Chu CM, et al. HBsAg seroclearance in asymptomatic carriers of high endemic areas: appreciably high rates during a long-term follow-up. Hepatology 2007; 45: 11871–1192. [DOI] [PubMed] [Google Scholar]
- 12.Han ZG, et al. HBsAg spontaneous seroclearance in a cohort of HBeAg-seronegative patients with chronic hepatitis B virus infection. Journal of Medical Virology 2016; 88: 79–85. [DOI] [PubMed] [Google Scholar]
- 13.Chu CM, et al. Hepatitis B surface antigen seroclearance during chronic HBV infection. Antiviral Therapy 2010; 15: 133–143. [DOI] [PubMed] [Google Scholar]
- 14.Kim GA, et al. Incidence of hepatocellular carcinoma after HBsAg seroclearance in chronic hepatitis B patients: a need for surveillance. Journal of Hepatology 2015; 62: 1092–1099. [DOI] [PubMed] [Google Scholar]
- 15.Liu J, et al. A predictive scoring system for the seroclearance of HBsAg in HBeAg-seronegative chronic hepatitis B patients with genotype B or C infection. Journal of Hepatology 2013; 58: 853–860. [DOI] [PubMed] [Google Scholar]
- 16.Fang ZL, et al. HBV A1762 T, G1764A mutations are a valuable biomarker for identifying a subset of male HBsAg carriers at extremely high risk of hepatocellular carcinoma: a prospective study. American Journal of Gastroenterology 2008; 103: 2254–2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Courouce-Pauty AM, et al. Distribution of HBsAg subtype in the world. Vox Sanguinis 1983; 44: 197–211. [DOI] [PubMed] [Google Scholar]
- 18.Okamoto H, et al. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. Journal of General Virology 1988; 69: 2575–2583. [DOI] [PubMed] [Google Scholar]
- 19.Chen X, et al. A description of the hepatitis B virus genomic background in a high-prevalence area in China. Virology Journal 2014; 11: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series 1999; 41: 95–98. [Google Scholar]
- 21.Tamura K, et al. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perrillo R, et al. Entecavir and hepatitis B immune globulin in patients undergoing liver transplantation for chronic hepatitis B. Liver Transplantation 2013; 19: 887–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsu C, et al. Chemotherapy-induced hepatitis B reactivation in lymphoma patients with resolved HBV infection: a prospective study. Hepatology 2014; 59: 2092–2100. [DOI] [PubMed] [Google Scholar]
- 24.Jang JW, et al. Reactivation of hepatitis B virus in HBsAg-negative patients with hepatocellular carcinoma. PLoS ONE 2015; 10: e0122041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han JW, et al. Risk factors and outcomes of hepatitis B virus reactivation in hepatitis B surface antigen negative patients with hematological malignancies. Hepatology Research 2016; 46: 657–668. [DOI] [PubMed] [Google Scholar]
- 26.Waite J, et al. Hepatitis B virus reactivation or reinfection associated with HIV-1 infection. AIDS 1988; 2: 443–448. [DOI] [PubMed] [Google Scholar]
- 27.Vento S, et al. Clinical reactivation of hepatitis B in anti-HBs-positive patients with AIDS. Lancet 1989; 1: 332–333. [DOI] [PubMed] [Google Scholar]
- 28.Fattovich G, et al. Natural history of chronic hepatitis B: special emphasis on disease progression and prognostic factors. Journal of Hepatology 2008; 48: 335–352. [DOI] [PubMed] [Google Scholar]
- 29.Deguchi M, et al. Quantitation of hepatitis B surface antigen by an automated chemiluminescent microparticle immunoassay. Journal of Virology Methods 2004; 115: 217–222. [DOI] [PubMed] [Google Scholar]
- 30.Ferreira SC, et al. Factors associated with spontaneous HBsAg clearance in chronic hepatitis B patients followed at a university hospital. Annals of Hepatology 2014; 13: 762–70. [PubMed] [Google Scholar]
- 31.Lauret E, et al. Long-term outcome in Caucasian patients with chronic hepatitis B virus infection after HBsAg seroclearance. Liver International 2015; 35: 140–147. [DOI] [PubMed] [Google Scholar]
- 32.Lazarevic I, et al. Prevalence of hepatitis B virus MHR mutations and their correlation with genotypes and antiviral therapy in chronically infected patients in Serbia. Journal of Medical Virology 2010; 82: 1160–1167. [DOI] [PubMed] [Google Scholar]
- 33.Martin CM, et al. Mutations associated with occult hepatitis B virus infection result in decreased surface antigen expression in vitro. Journal of Viral Hepatitis 2012; 19: 716–723. [DOI] [PMC free article] [PubMed] [Google Scholar]