Abstract
Public health surveillance systems for COVID-19 are multifaceted and include multiple indicators reflective of different aspects of the burden and spread of the disease in a community. With the emergence of wastewater disease surveillance as a powerful tool to track infection dynamics of SARS-CoV-2, there is a need to integrate and validate wastewater information with existing disease surveillance systems and demonstrate how it can be used as a routine surveillance tool. A first step toward integration is showing how it relates to other disease surveillance indicators and outcomes, such as case positivity rates, syndromic surveillance data, and hospital bed use rates. Here, we present an 86-week long surveillance study that covers three major COVID-19 surges. City-wide SARS-CoV-2 RNA viral loads in wastewater were measured across 39 wastewater treatment plants and compared to other disease metrics for the city of Houston, TX. We show that wastewater levels are strongly correlated with positivity rate, syndromic surveillance rates of COVID-19 visits, and COVID-19-related general bed use rates at hospitals. We show that the relative timing of wastewater relative to each indicator shifted across the pandemic, likely due to a multitude of factors including testing availability, health-seeking behavior, and changes in viral variants. Next, we show that individual WWTPs led city-wide changes in SARS-CoV-2 viral loads, indicating a distributed monitoring system could be used to enhance the early-warning capability of a wastewater monitoring system. Finally, we describe how the results were used in real-time to inform public health response and resource allocation.
Keywords: Wastewater-based epidemiology, COVID-19, SARS-CoV-2, Syndromic surveillance, Lead time, Hospitalizations, Positivity rate
Graphical abstract
1. Introduction
Wastewater disease monitoring is an effective and resource-efficient tool for tracking community infection dynamics of the severe acute respiratory syndrome coronavirus (SARS-CoV-2) (Xiao et al., 2022; Olesen et al., 2021; Kaplan et al., 2021; Galani et al., 2022; Peccia et al., 2020; D’Aoust et al., 2021; Wurtzer et al., 2022; Wurtz et al., 2021). It has advantages as an independent surveillance tool because it captures symptomatic and asymptomatic infections, it does not depend on access to healthcare or testing capacity, and is not biased by individual health-seeking behavior. These advantages, combined with early demonstrations of detecting SARS-CoV-2 in wastewater, sparked implementation of SARS-CoV-2 wastewater monitoring systems across the globe (Naughton et al., 2021). Numerous studies have since demonstrated that wastewater monitoring of SARS-CoV-2 provides a useful, independent, quantitative estimate of community infection trends (Xiao et al., 2022; Olesen et al., 2021; Kaplan et al., 2021; Galani et al., 2022; Peccia et al., 2020; D’Aoust et al., 2021; Fernandez-Cassi et al., 2021).
For public health intervention, wastewater surveillance's main strength is not use in isolation or in place of other metrics, but instead as a complementary metric for evaluating trends alongside a suite of key disease metrics. Other key disease metrics include clinical case counts or positivity rate, and percent of emergency department visits and hospital bed use rates (general and ICU) that are COVID-19 related. State and local health departments can use wastewater surveillance alongside these other metrics to understand the level of severity of COVID-19 in their communities over time. Public health departments rely on a suite of indicators as opposed to a single metric because they represent different aspects of disease surveillance. The wastewater viral load indicates the level of the virus in the community where symptomatic and asymptomatic are represented, with no specificity of illness. The positivity rate or case counts, particularly in early waves of the pandemic, was indicative of the cases in the community but was biased toward symptomatic infections (Symanski et al., 2021). The percent of emergency department visits that are COVID-19-related indicates the percent of people seeking treatment for the illness. The percent of general hospital bed use that are COVID-19 related and then the percent of ICU hospital bed use that are COVID-19 indicate progressively increasing severity of illness.
To establish wastewater monitoring as a part of routine disease surveillance, wastewater information needs to be integrated with existing disease surveillance systems. For interpretation, an understanding of how wastewater levels relate to other disease indicators and how those relationships shifted over the pandemic is needed. This is necessary because the representativeness of the traditional disease indicators and their relationship to wastewater levels changed as the pandemic progressed in time. For example, as the pandemic progressed, the bias in the positivity rate increased likely due to the decline of the clinical PCR testing rate and wide availability of at-home rapid tests which are not picked up by surveillance. Also, as the pandemic progressed, vaccines and new treatments became available, and new variants emerged, which impacted the severity of the disease at the population-level (Christensen et al., 2022). Wastewater, in theory, represents a more consistent disease indicator as it does not depend on health-seeking behavior or available resources. But even the representativeness of the viral load in the wastewater may have changed over time as variants of the virus and vaccination resulted in different shedding rates (Siedner et al., 2022; Prasek et al., 2022). The changing landscape of metrics and what they represent underscores the need for a better understanding of how to use multiple disease indicators to triangulate an understating of the level of concern and inform planning and resource allocation accordingly.
In this study we present the wastewater viral load and other key indicator data used for COVID-19 surveillance in Houston. We examine the relationship between the city-wide wastewater viral load and other indicators including positivity rate, emergency department COVID-related visit rates, and COVID-related general bed and ICU bed use rates. We perform this analysis on data collected 86 weeks between September 1, 2020 and April 25, 2022 which included three major waves of COVID-19 due to the Alpha, Delta, and Omicron SARS-CoV-2 variants and discuss how wastewater data is used to threshold city-wide COVID concern levels. Further, we identify instances where changes in the viral load at individual wastewater treatment plants led city-wide changes in viral load. Our results demonstrate how wastewater monitoring can complement existing disease surveillance systems, and how having numerous distributed monitoring sites may enable early detection of city-wide COVID-19 surges.
2. Methods
2.1. Wastewater samples and wastewater treatment plant flow data
Samples were collected from 39 different wastewater treatment plants (WWTPs) within Houston serving approximately 2.3 million people and covering a total service area of approximately 580 mile2. Details for each WWTP including average daily flow rate, sewershed area, and service population are provided in Table S1.
Raw wastewater samples were collected from the influent channel of each WWTP using refrigerated 24-hour composite samplers. Influent flowrates per WWTP was also provided by Houston Water personnel for each 24-hour sampling period. After sample collection, samples were stored on ice and transported to Houston Water's laboratory, aliquoted into 250 mL and 500 mL bottles and transported on ice for further processing to Rice University (Rice) for the entire study period, and to Rice and the Houston Health Department (HHD) laboratory starting June 7, 2021. Methods used for sample concentration, RNA extraction, and quantification differed slightly between laboratories and evolved over time. Sample processing methods and procedures for calibrating results across laboratories and method changes are described in the following sections.
2.2. Wastewater concentration, nucleic acid extraction, and SARS-CoV-2 quantification
Rice methods. Concentration was performed in duplicate for each wastewater sample. For each replicate, 50 mL of wastewater was concentrated via filtration using electronegative filters. Between September 1, 2020 and January 25, 2021, nucleic acid extraction was performed on a Maxwell 48 RSC automated platform (AS8500, Promega) using a modified protocol for the Maxwell RSC PureFood GMO and Authentication Kit (AS1600, Promega). From February 1, 2021 to April 25, 2022, extraction was performed on a chemagic 360 automated platform using the Viral DNA/RNA 300 Kit H96 (CMG-1433, PerkinElmer) following the manufacturer's protocol. The extraction method was changed to adjust for the increase in weekly samples. SARS-CoV-2 N1 and N2 gene targets were quantified in wastewater samples using a one-step RT-ddPCR assay. RT-ddPCR was performed on a QX200 AutoDG Droplet Digital PCR System (Bio-Rad) and a C1000 Thermal Cycler (Bio-Rad). Further details on concentration, extraction, and quantification including information on assay setup and thermocycling conditions, controls, and limit of detection, are provided in the Supporting Information.
2.2.1. HHD methods
xConcentration and extraction were performed identically to the Rice method using the chemagic 360 extraction platform with the exception of the volume of lysate supernatant transferred into the RNA extraction plate (600 μL instead of 300 μL) and the final eluate volume (60 μL instead of 50 μL). Quantification of N1 and N2 were performed by RT-qPCR using the Water SARS-CoV-2 RT-PCR test kit (IDEXX Laboratories, Inc., Westbrook, ME) following the manufacturer's instructions. Further details are provided in the Supporting Information.
2.3. Diagnostic testing, syndromic surveillance, and hospital bed use data
Positive and negative COVID-19 diagnostic (PCR) testing results were extracted from the Houston Electronic Disease Surveillance System (HEDSS). The daily positivity rate was calculated as follows:
Results were limited to patients located within one of the 105 Houston-area ZIP codes. Test records were deduplicated by person by specimen date (i.e., if a person is tested multiple times in a single day, they are counted once for that day; if a person is tested across multiple days, they are counted once per each of those days). Results were aggregated by specimen date.
COVID-related emergency department (ED) visit data were extracted from the Southeast Texas Electronic Syndromic Surveillance System for the Early Notification of Community-Based Epidemics (HHD-ESSENCE) and includes data from over 130 healthcare facilities across the Public Health Region 6/5 South in Texas. Syndromic surveillance via the HHD-ESSENCE system is routinely used for monitoring other influenza-like illnesses. A COVID-related ED visit was defined as an emergency-related visit with a COVID-19 diagnosis code in the discharge diagnosis field. The daily COVID ED Visit Rate was calculated as follows:
The ED data was not limited geographically and reflects data from all facilities currently reporting to HHD-ESSENCE for the Texas Public Health Region 6/5 South. Results were aggregated by visit date.
Hospital bed usage data were extracted from the Southeast Texas Regional Advisory Council (SETRAC) COVID Executive Hospital Summary dashboard (Covid Executive Hospital Summary; SENTRAC). The daily COVID ICU and general bed usage rates were calculated as follows:
The hospital bed usage variables were limited to the county-level and reflect data from Houston's main county, Harris. Results were aggregated to bed usage date.
2.4. Statistical methods
2.4.1. Aggregating data across labs
As mentioned, the 24-hour composite wastewater samples collected each week from the 39 wastewater treatment plants are analyzed by multiple labs using multiple methods (additional details are provided in the SI). Before statistical analysis is performed on the sample results, calibration across labs is performed. This calibration is calculated through regression. Measurements from Rice using Maxwell platform and HHD were adjusted to Rice chemagic 360 levels using two different regressions. The Rice Maxwell regression model for adjustments is a linear regression model that was built on samples from January 25, 2021 in which we extracted replicate samples (n = 24) via Maxwell and chemagic 360 in a head-to-head comparison. The HHD regression model for adjustments is a cubic-polynomial regression model that was built on samples from April 12, 2021 to August 16, 2021 (n = 387). For the HHD regression model, the sample results are converted to the log10 scale prior to the regression, as such the adjusted values are raised to the 10th power after adjustments are applied. The transformation of log10 resolves the issue of extreme values and skewness of the copies per liter measurements. Additional details on the regressions are provided in the Supporting Information.
2.4.2. Spline-smoothing of wastewater viral load
Once the regression adjustments are applied, and the adjusted values are on the copies per liter scale rather than the log10 copies per liter scale, they are normalized by flow. The flow for each of the 39 wastewater treatment plants is reported in liters per day. After flow normalization, the measurements are in copies per day. The flow normalized measurements are calculated as follows:
A regression spline model is used to fit the time series of the log10 copies per day for each of the 39 wastewater treatment plants. The transformation of log10 again resolves the issue of extreme values and skewness. The knots in the spline are chosen optimally based on the quantiles of the copies per day. The number of knots or degrees of freedom is a sensitivity parameter that is checked each week. To estimate the city-wide total copies per day, the log10 estimated copies per day at each wastewater treatment plant are raised to the 10th power, summed and then again transformed to log10. The total estimated copies per day is an estimate of the median of the probability distribution for copies per day.
2.4.3. Weekly percent change
The relative week-to-week percent change is calculated for each of the 39 wastewater treatment plants as well as the city-wide total. The weekly relative percent change for a certain week is calculated using the below formula:
The estimated viral load values used in the relative week-to-week percent change calculations for each of the 39 wastewater treatment plants are the log10 copies per day estimates produced by the spline regressions described in the prior section. The estimated viral load values used in the relative week-to-week percent change calculations for the city-wide total is the summed total log10 copies per day estimate described in the prior section.
2.4.4. Cross-correlation analysis
To assess whether there was a leading or lagging relationship between the wastewater viral load and other metrics, we performed a time-step cross-correlation analysis where the city's wastewater viral load was lagged −7 to +21 days against each raw daily rate: positivity, COVID ED visits, COVID ICU beds, and COVID general beds rates. The Pearson correlation coefficient was calculated for each day interval lag in wastewater viral load for each of the aforementioned daily rates. The cross-correlation analysis was performed for each of the COVID-19 waves individually. The first (Alpha) wave corresponded to September 1, 2020 to January 20, 2021; the second wave ran from June 15, 2021 to August 31, 2021; and the third wave was November 20, 2021 to January 15, 2022. These date ranges were selected to include the lead time before the peak, i.e., the date of the minimum before the peak, to the date of the peak of the most lagging metric. Confidence intervals for the correlation coefficients were calculated where the lower confidence bound for each cross-correlation took into account the fact that each series is autocorrelated (Shumway et al., 2000). Details on how the cross-correlation analysis was performed are provided in the Supporting Information.
3. Results
3.1. City-wide wastewater SARS-CoV-2 viral loads were strongly correlated with positivity rate, ED visits, hospital general bed use rates
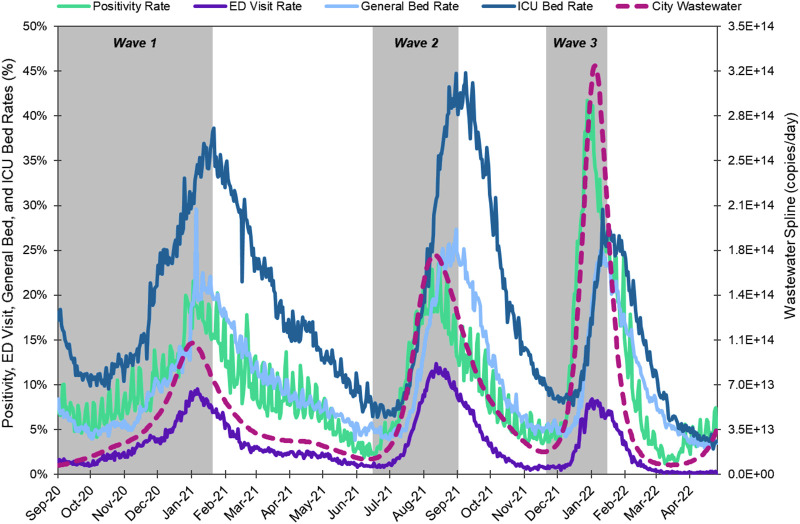
We quantified SARS-CoV-2 RNA concentrations in wastewater samples collected at 39 wastewater treatment plants that serve approximately 2.3 million people over 86 weeks. The study period captured three major infection waves of COVID-19 that corresponded to the emergence of Alpha (B.1.1.7 and Q lineages), Delta (B.1.617.2 and AY lineages), and Omicron (B.1.1.529, BA.1 lineages) variants. We used a flow-based normalization approach to calculate the viral load at each WWTP by multiplying the SARS-CoV-2 RNA concentration by the influent flow rate for each WWTP, an approach that numerous other studies have applied for normalization and to account for impacts of dilution (Acosta et al., 2022; van Boven et al., 2022; Yaniv et al., 2021; Nagarkar et al., 2021; Isaksson et al., 2022; Tiwari et al., 2022). Fig. 1 shows the time series data for the city-wide aggregated SARS-CoV-2 RNA viral load, disease indicators including positivity rate, COVID ED visit rate, COVID ICU bed use rate, and COVID general bed use rate. Wastewater viral load and all other conventional metrics experienced peaks corresponding to the three waves caused by different SARS-CoV-2 variants. Across the entire 86-week study period (September 1, 2020 – April 25, 2022), the wastewater viral load was strongly correlated with positivity rate (ρ = 0.91), ED visit rate (ρ = 0.80), general bed use rate (ρ = 0.71), and moderately correlated with ICU bed use rate (ρ = 0.47) (Table S2).
Fig. 1.
Positivity rate, COVID-diagnosis ED visit rate, hospital general bed use rate, and ICU bed use rate (left y-axis), and smoothed city-wide wastewater viral load (right y-axis) time series for September 1, 2020 through April 25, 2022. Positivity rate is the daily positivity rate for the City of Houston's 105 zip codes. ED visit rate is the daily rate of COVID-related emergency department visits for the Texas PH Region 6/5S. General and ICU bed rates are the daily rates of general and ICU beds in use for COVID patients for Harris County, respectively. City wastewater is the daily spline-smoothed aggregate viral load for the city of Houston. Grey shading indicates the time period included for each wave in the time-step correlation analysis.
3.2. The leading indicator metric for city-wide SARS-CoV-2 infection burden varied across COVID-19 waves
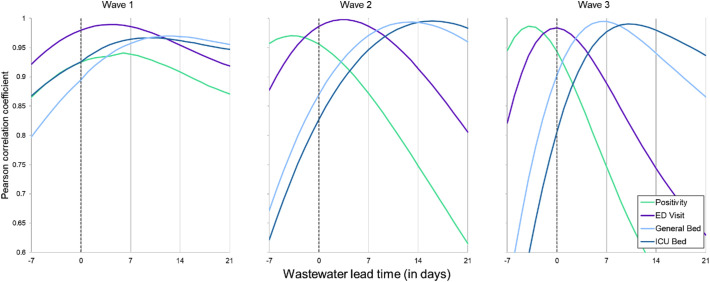
We next compared the relative timing of the wastewater viral load to the other disease indicators for each wave individually. In this analysis, we used the data corresponding to the rising edge of the peak for each wave (Fig. 1, grey shaded time periods) and computed the correlation coefficients between the wastewater and indicators. To evaluate if wastewater viral load lead or lagged the other timeseries datasets, we performed a cross-correlation analysis and compared the correlation between the wastewater viral load and other indicators after offsetting by a period of −7 to +21 days. The leading metric differed for each COVID-19 wave, as determined by comparing lead/lag offset with the strongest correlation coefficient between the wastewater viral load and other disease indicators. In the first (Alpha) wave, all other indicators were strongly correlated with wastewater viral load, over lag windows between −7 and 21 days. In this wave, the wastewater viral load, COVID ED visit rate, and positivity rate generally moved together and led COVID ICU and general bed use rates (Fig. 2A and Table 1 ). In the second (Delta) wave, the strongest correlation coefficients were observed when positivity rate led, followed by wastewater viral load, COVID ED visit, COVID general bed use, and COVID ICU bed use rates. The third (Omicron) wave was more similar to the second wave than the first in terms of the temporal patterns of each of the metrics, with the exception that the strongest correlation between wastewater viral load and COVID ED visit rates occurred when there was no lag between the datasets.
Fig. 2.
Pearson correlation coefficients for the cross correlation between disease metrics and daily spline-smoothed wastewater viral load for the city offset between −7 (lagging) and +21 (leading) days. The correlation analyses were performed using the raw daily rates for positivity, ED visit, general bed use, and ICU bed use rates. Triangles indicate correlation coefficients significantly different from zero.
Table 1.
Lead time of wastewater relative to other disease indicator metrics based on maximum correlation coefficient from time-step analysis. Positive values indicate wastewater led, while negative values indicate wastewater lagged other metrics.
| Wave | Positivity | ED visit | General bed | ICU bed |
|---|---|---|---|---|
| 1 | +6 | +4 | +12 | +10 |
| 2 | −4 | +3 | +13 | +16 |
| 3 | −4 | 0 | +7 | +10 |
3.3. Individual wastewater treatment plant viral loads had greater lead times for city-wide positivity rates, ED visits, and hospital bed use rates
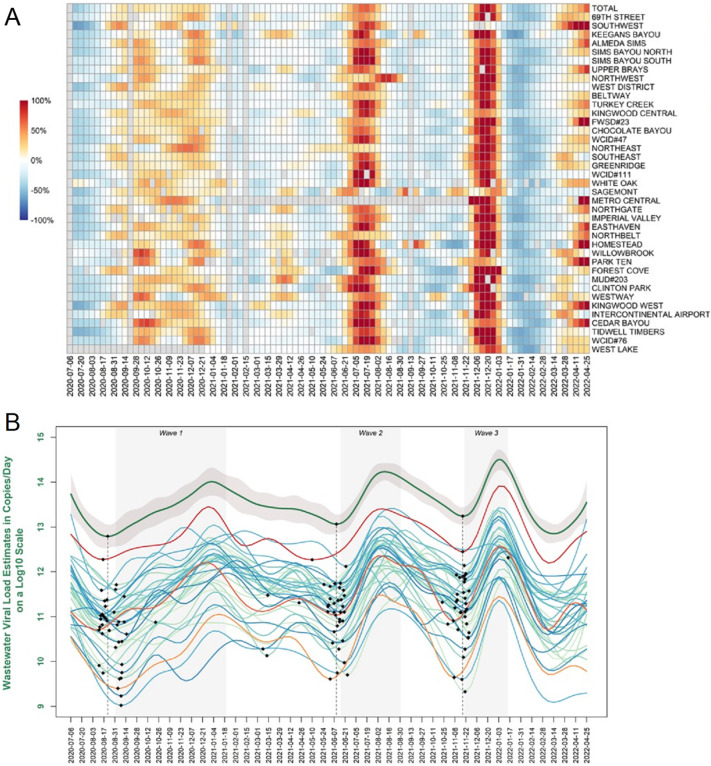
We next asked whether changes in viral load at individual wastewater treatment plants led changes in the city-wide viral load. For each wave, there was some spatial variation in timing and rate of change of the viral load observed at each individual wastewater treatment plants (Fig. 3A). Certain wastewater treatment plants experienced earlier increases in weekly percent change in viral loads than the city-wide total, serving as early indicators of a city-wide surge. To identify sewersheds that were early indicators of the city-wide surges, we compared the relative timing of the trough (minimum) leading up to each wave for each individual wastewater treatment plant versus the city-wide minimum (Fig. 3B, minimums indicated as black diamonds). For example, in wave 1, the 69th wastewater treatment plant's wastewater viral load began increasing prior to the city-wide total (Fig. 3B, red line), with the minimum for the 69th wastewater treatment plant occurring 6 days before the city-wide minimum. In contrast, other wastewater treatment plants lagged behind the city-wide trend in wave 1, such as Clinton Park (Fig. 3B, orange line), for which the minimum was 13 days after the city-wide minimum. In total, there were 17, 18, and 14 wastewater treatment plants whose minimums led the city-wide minimum for waves 1, 2 and 3, respectively. There were a three wastewater treatment plants that led the citywide total waves across all three waves: Beltway, Chocolate Bayou, and Intercontinental Airport. The Intercontinental Airport wastewater treatment plant serves the George Bush Intercontinental Airport, and these results suggest that it may be a good sentinel site for early detection of infection surges.
Fig. 3.
(A) Heatmap of the week-to-week percent change in wastewater viral load for the city-wide total and each of the 39 wastewater treatment plants. Wastewater treatment plants are ordered by population on the y-axis. Grey boxes indicate no sample was collected. (B) Wastewater viral load trends (splines) for city-wide total (top green line with 95 % confidence band in grey) and individual wastewater treatment plants. As examples, 69th Street is shown in red, Intercontinental Airport in dark orange, and Clinton Park in light orange. Black markers indicate the minimums for the city total and each site leading up to each wave. Dotted line indicates the date of the city-wide total minimum. Grey shaded areas indicate the date ranges corresponding to each wave.
4. Discussion
In this study, we showed that SARS-CoV-2 RNA viral loads in wastewater were positively correlated to all other disease metrics used for city-wide COVID-19 surveillance: positivity rate, syndromic surveillance of ED visits, and hospital general and ICU bed use rates. The strong relationship between these metrics and wastewater viral loads across three COVID-19 surges demonstrates that wastewater can provide an independent, complementary source of surveillance information for a major city that is a relatively inexpensive, rapid, and broad as compared to diagnostic testing. Our findings are consistent with other studies reporting correlations between SARS-CoV-2 wastewater levels and new cases, positivity rate, and/or hospitalizations (Galani et al., 2022; Peccia et al., 2020; D’Aoust et al., 2021; Krivoňáková et al., 2021; Graham et al., 2021; Feng et al., 2021; Hoar et al., 2022; Wang et al., 2021; Stadler et al., 2020; Pileggi et al., 2022; Zhao et al., 2022), and underscores its value as a tool for public health action (Medema et al., 2020; McClary-Gutierrez et al., 2021; Farkas et al., 2020; Kirby et al., 2021; Deng et al., 2022; Sharara et al., 2021). The strong correlations between wastewater and metrics such as positivity rate, syndromic surveillance, and hospitalizations occurred across three major waves of COVID-19, despite an evolving virus and new variants, widespread vaccination and boosters, and natural immunity. This highlights the strength of wastewater as a reliable metric of community infections. Wastewater surveillance will become more valuable as diagnostic testing declines and the use of at-home tests increases. Further, as SARS-CoV-2 evolves and medical treatments improve, resulting in overall less severe disease and fewer hospitalizations, wastewater can still serve as a reliable metric of community spread, and even provide information on the relative severity of the a given variant by comparing it against hospitalizations.
Beyond using wastewater as an independent, quantitative measure of community infection trends, there is interest in applying wastewater monitoring as an early-warning system that can be used to forecast surges in infections that stress healthcare resources. Previous studies have claimed that concentrations of SARS-CoV-2 in wastewater can lead reported case counts (Xiao et al., 2022; Olesen et al., 2021; Kaplan et al., 2021; Peccia et al., 2020; D’Aoust et al., 2021). However, many of these reports were from the initial surge of COVID-19, when there were no or very few known infections, and testing rates were low and/or testing resources were scarce. However, in later infection waves, where there was more availability and access to testing resources, lead times were generally nonexistent or even found to lag behind reported cases (dated by specimen date) (Xiao et al., 2022; Feng et al., 2021). One explanation for why wastewater may lead case counts is that individuals start shedding virus in their stool before they seek testing, which typically occurs after symptoms present. However, many other factors contribute to the relative timing between wastewater viral loads and case counts, such as health-seeking behavior, disease severity and progression, and access to testing resources, which are difficult to separate during analysis.
Here, we compared the timing of the city-wide wastewater viral load to other disease indicators using a time-step cross-correlation analysis. We discreetly analyzed each COVID-19 wave's leading edge, and thus can compare the relationship between each metric across three different COVID surges. The lead/lag with the maximum correlation coefficient between wastewater and other metrics shifted across the different waves. Wastewater had the strongest correlation as a leading indicator in wave 1, with the maximum correlation coefficients occurring when it led positivity rate by 6 days, ED visits by 4 days, and hospitalizations by 10–12 days (Table 1). In contrast to the first wave, wastewater lagged behind the positivity rate in the Delta and Omicron surges based on the maximum cross-correlation coefficients. Testing resources were more widely available in the Delta and Omicron surges, which may have resulted in more timely diagnostic testing and a shift in timing as compared to wastewater levels in the first wave (Xiao et al., 2022). The timing of wastewater levels relative to other metrics across each wave could also be due to differences in fecal shedding and symptom onset due to the different viral variants responsible for each wave (Wade et al., 2022; Tlhagale et al., 2022). To understand if wastewater would be expected to lead new cases (independent of testing resources and behavioral factors), a better understanding of typical fecal shedding rates over the course of an infection is needed, and whether different variants result in different fecal shedding patterns. It is important to note that if we account for autocorrelation, the cross-correlation coefficients are only significantly different from zero in the first wave (Table S2). This is because timeseries data is strongly autocorrelated, and only in the first wave is the sample size large enough to overcome the autocorrelation, whereas in waves 2 and 3 the number of days included in the wave are not sufficiently large. This is consistent with a previous study that found that more frequent sampling (twice weekly) resulted in significant associations between wastewater measurements and case counts, but down sampling to fortnightly or weekly measurements resulted in no significant associations when accounting for autocorrelation (Graham et al., 2021). Regardless, because of the overall strong association between the wastewater viral load and other metrics, wastewater could be implemented to serve as an early warning system and reflect community infection prevalence.
In all waves, wastewater viral loads and syndromic surveillance were relatively consistent with respect to timing, and both led changes in COVID-related general and ICU bed use rates by one to two-weeks. This underscores the complementary role that wastewater surveillance can play with syndromic surveillance. Syndromic surveillance data is available in real-time, but may be biased toward severe infections and captures a much more limited portion of the population. However, discharge diagnoses can be omitted or entered into the system after the fact, resulting in incomplete or delayed information. Wastewater surveillance data can have a turnaround time of less than a day (Smruthi et al., 2021), can detect both symptomatic and asymptomatic cases (Schmitz et al., 2021; Gibas et al., 2021), and can represent the population of an entire metropolis in a single sampling event (Hoar et al., 2022; Wang et al., 2021; Stadler et al., 2020). Both wastewater and syndromic surveillance will become increasingly critical for detecting future surges of COVID-19 as diagnostic testing declines. Early detection could mitigate the impact of surges on hospitals, resulting in better patient outcomes and mitigation of large financial losses experienced by hospitals (Kumar et al., 2020; Coughlin et al., 2020). Our results suggest that syndromic and wastewater surveillance could be used together to establish thresholds for when community spread is increasing and trigger hospitals for emergency preparation measures.
Increasing the lead time of wastewater for early detection of future surges could be achieved through a distributed monitoring system. In Houston, the numerous wastewater treatment systems that serve the city enabled a distributed wastewater monitoring system for SARS-CoV-2. We show that monitoring at 39 different wastewater treatment plants could have enabled earlier detection of city-wide surges on the order of two weeks. Our results are aligned with a previous study that used spatial autocorrelation analysis to show that identified neighborhood hot spots led city-wide infection surges (Haak et al., 2022). As expected, these findings suggest that outbreaks of SARS-CoV-2 occurred in clusters, and were not homogeneously distributed across the city. Interestingly, we observed that the Intercontinental Airport viral load led the citywide total across all three waves, indicating airport may serve as sentinel sites for citywide surges of infections. Of note, infected individuals may have contributed to the wastewater signal in multiple sewersheds if they moved throughout the day (i.e., go to the bathroom at home and at work, located in two different sewersheds). This may have contributed to the strong spatial association in the wastewater signal observed across sewersheds. Understanding the impact of wastewater travel time on lead/lag time, and whether decay of signal may be differentially impacting the viral load observed at each wastewater treatment plant is an area that requires further research. We know from previous work that travel times across and within sewersheds in Houston can vary widely (McCall et al., 2022). We did not observe any trends between wastewater treatment plant size or service area and relative timing of each wave. Effective approaches for identifying early hot spots of infection could potentially contain and mitigate a city-wide outbreak if acted upon via public health measures such as targeted communication, deployment of vaccination and testing resources. Integration of wastewater and other disease surveillance with epidemiologic models (Kaplan et al., 2021; Cao and Francis, 2021; Vallejo et al., 2020; Nourbakhsh et al., 2022; McMahan et al., 2021; Huisman Jana et al.) could be used to assess the impact of distributed versus centralized wastewater monitoring systems and the impact of early detection on different outbreak scenarios.
The strong associations between wastewater viral load and positivity rate and hospitalizations led the City of Houston to incorporate information from the wastewater monitoring system into its COVID-19 alert system. Specifically, each week, the (1) percent of population served by a wastewater treatment plant with an increasing trend in viral load, and (2) weekly increase in the percent of population with an increasing trend in viral load, reported and classified based on thresholds as low, medium, or high concern. These metrics, along with city-wide positivity rates, positivity rates in the lowest income areas, general and ICU hospital bed use rates, and number of new outbreaks in K-12 schools, are used to assess the current level of concern for COVID for the city. This alert system represents one of a growing number as cities and states begin to incorporate wastewater levels into routine public health disease surveillance and response systems (e.g., https://covid19.ncdhhs.gov/dashboard, https://www.cdph.ca.gov).
5. Conclusions
This study advances the field of wastewater disease surveillance as it represents one of the longest longitudinal analyses of wastewater viral levels and four different disease indicators. It is one of the only to include syndromic surveillance data. By performing a cross correlation analysis over the entire study period, as well as each of the three waves driven by different variants, we were able to compare how the relative timing of wastewater levels shifted across the pandemic. Our results and interpretation are limited by the fact that numerous factors changed across each wave, including the dominant variant, testing availability, and human behavior, which are difficult to disentangle. Regardless, we should expect that the relative timing and representativeness of the disease indicators will continue to shift as the virus evolves, attitudes toward the pandemic and behaviors change over time, and updated vaccines become available. This again underscores the need for multiple, complementary disease surveillance metrics to understand disease burden in our communities and guide public health response. Our results are also impacted by the resolution of wastewater sampling, which occurred weekly, and the inherent variability in the measurements of SARS-CoV-2 in wastewater samples. To overcome these limitations, we used splines to smooth the wastewater viral load data and interpolate daily values, an approach used and recommended by several other studies (van Boven et al., 2022; Arabzadeh et al., 2021; Aberi et al., 2021; Karthikeyan et al.). Further research should help to establish methods for assessing the optimal sampling frequency and sample locations given resource constraints and better understand and minimize the factors contributing to measurement variability. In addition, incorporating wastewater surveillance information into existing epidemiological disease surveillance systems used by local, state, federal, and international systems will be critical to integrating as a routine surveillance metric.
CRediT authorship contribution statement
K.C. and R.S. performed data acquisition, curation, analysis, visualization of the results and contributed to the drafting the original manuscript. L.H. supervised and administered the project, directed the analysis, and contributed to the writing and editing of the manuscript. D.P. supervised and administered the project, and contributed to the analysis. K.E. supervised and contributed to the analysis and editing of the manuscript. C.M. contributed to the writing and editing of the manuscript. L.B.S. supervised the project and analysis, wrote and edited the manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Houston Water for their assistance in the collection of wastewater samples and providing the wastewater flow rate data. We acknowledge Kaavya Domakonda, Roger Sealy and Jennifer Myers from the Houston Health Department, and Lauren Bauhs, Madeline Wolken, Russell Carlson-Stadler, Kyle Palmer, and Whitney Rich from Rice for analyzing wastewater samples. This work was supported by funds from CDC-COVID-19 ELC enhancing detection expansion. L.B.S. was supported in part by the National Science Foundation (CBET 2029025), and seed funds from Rice University.
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.158967.
Appendix A. Supplementary data
Supplementary information
Data availability
Data has been posted to Rice U UDP (DOI provided)
References
- Aberi P., Arabzadeh R., Insam H., Markt R., Mayr M., Kreuzinger N., Rauch W. Quest for optimal regression models in SARS-CoV-2 wastewater based epidemiology. Int. J. Environ. Res. Public Health. 2021;18(20) doi: 10.3390/ijerph182010778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acosta N., Bautista M.A., Waddell B.J., McCalder J., Beaudet A.B., Man L., Pradhan P., Sedaghat N., Papparis C., Bacanu A., Hollman J., Krusina A., Southern D.A., Williamson T., Li C., Bhatnagar S., Murphy S., Chen J., Kuzma D., Clark R., Meddings J., Hu J., Cabaj J.L., Conly J.M., Dai X., Lu X., Chekouo T., Ruecker N.J., Achari G., Ryan M.C., Frankowski K., Hubert C.R.J., Parkins M.D. Longitudinal SARS-CoV-2 RNA wastewater monitoring across a range of scales correlates with Total and regional COVID-19 burden in a well-defined urban population. Water Res. 2022;220 doi: 10.1016/j.watres.2022.118611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabzadeh R., Grünbacher D.M., Insam H., Kreuzinger N., Markt R., Rauch W. Data filtering methods for SARS-CoV-2 wastewater surveillance. Water Sci. Technol. 2021;84(6):1324–1339. doi: 10.2166/wst.2021.343. [DOI] [PubMed] [Google Scholar]
- van Boven M., Hetebrij W.A., Swart A.M., Nagelkerke E., van der Beek R.F.H.J., Stouten S., Hoogeveen R.T., Miura F., Kloosterman A., van der Drift A.-M.R., Welling A., Lodder W.J., de Roda Husman A.M. medRxiv; 2022. Modelling Patterns of SARS-CoV-2 Circulation in the Netherlands, August 2020-February 2022, Revealed by a Nationwide Sewage Surveillance Program. 2022.05.25.22275569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Francis R. On forecasting the community-level COVID-19 cases from the concentration of SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;786 doi: 10.1016/j.scitotenv.2021.147451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen P.A., Olsen R.J., Long S.W., Snehal R., Davis J.J., Ojeda Saavedra M., Reppond K., Shyer M.N., Cambric J., Gadd R., Thakur R.M., Batajoo A., Mangham R., Pena S., Trinh T., Kinskey J.C., Williams G., Olson R., Gollihar J., Musser J.M. Signals of significantly increased vaccine breakthrough, decreased hospitalization rates, and less severe disease in patients with Coronavirus disease 2019 caused by the omicron variant of severe acute respiratory syndrome Coronavirus 2 in Houston, Texas. Am. J. Pathol. 2022;192(4):642–652. doi: 10.1016/j.ajpath.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlin T.A., Ramos C., Samuel-Jakubos H. Wash. DC Urban Inst; 2020. Safety Net Hospitals in the Covid-19 Crisis: How Five Hospitals Have Fared Financially. [Google Scholar]
- COVID Executive Hospital Summary; SETRAC.
- D’Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., Baig A.T., Mayne J., Zhang X., Alain T., Servos M.R., Srikanthan N., MacKenzie M., Figeys D., Manuel D., Jüni P., MacKenzie A.E., Delatolla R. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y., Xu X., Zheng X., Ding J., Li S., Chui H., Wong T., Poon L.L.M., Zhang T. Use of sewage surveillance for COVID-19 to guide public health response: a case study in Hong Kong. Sci. Total Environ. 2022;821 doi: 10.1016/j.scitotenv.2022.153250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas K., Hillary L.S., Malham S.K., McDonald J.E., Jones D.L. Wastewater and public health: the potential of wastewater surveillance for monitoring COVID-19. Environ. Health COVID-19. 2020;17:14–20. doi: 10.1016/j.coesh.2020.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., McLellan S.L. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS EST Water. 2021;1(8):1955–1965. doi: 10.1021/acsestwater.1c00160. [DOI] [Google Scholar]
- Fernandez-Cassi X., Scheidegger A., Bänziger C., Cariti F., Tuñas Corzon A., Ganesanandamoorthy P., Lemaitre J.C., Ort C., Julian T.R., Kohn T. Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high. Water Res. 2021;200 doi: 10.1016/j.watres.2021.117252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galani A., Aalizadeh R., Kostakis M., Markou A., Alygizakis N., Lytras T., Adamopoulos P.G., Peccia J., Thompson D.C., Kontou A., Karagiannidis A., Lianidou E.S., Avgeris M., Paraskevis D., Tsiodras S., Scorilas A., Vasiliou V., Dimopoulos M.-A., Thomaidis N.S. SARS-CoV-2 wastewater surveillance data can predict hospitalizations and ICU admissions. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Roppolo Brazell L., Hinton K., Lontai J., Stark N., Young I., Quach C., Russ M., Kauer J., Nicolosi B., Chen D., Akella S., Tang W., Schlueter J., Munir M. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782 doi: 10.1016/j.scitotenv.2021.146749. 10/gnqvjr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham K.E., Loeb S.K., Wolfe M.K., Catoe D., Sinnott-Armstrong N., Kim S., Yamahara K.M., Sassoubre L.M., Mendoza Grijalva L.M., Roldan-Hernandez L., Langenfeld K., Wigginton K.R., Boehm A.B. SARS-CoV-2 RNA in wastewater settled solids is associated with COVID-19 cases in a large urban sewershed. Environ. Sci. Technol. 2021;55(1):488–498. doi: 10.1021/acs.est.0c06191. [DOI] [PubMed] [Google Scholar]
- Haak L., Delic B., Li L., Guarin T., Mazurowski L., Dastjerdi N.G., Dewan A., Pagilla K. Spatial and temporal variability and data bias in wastewater surveillance of SARS-CoV-2 in a sewer system. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150390. 10/gnqvjq. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoar C., Chauvin F., Clare A., McGibbon H., Castro E., Patinella S., Katehis D., Dennehy J.J., Trujillo M., Smyth D.S., Silverman A.I. Monitoring SARS-CoV-2 in wastewater during New York City’s second wave of COVID-19: sewershed-level trends and relationships to publicly available clinical testing data. Environ. Sci. Water Res. Technol. 2022;8(5):1021–1035. doi: 10.1039/D1EW00747E. [DOI] [Google Scholar]
- Karthikeyan Smruthi Ronquillo Nancy Belda-Ferre Pedro Alvarado Destiny Javidi Tara A. Longhurst Christopher Knight Rob M Cristea Ileana. High-throughput wastewater SARS-CoV-2 detection enables forecasting of community infection dynamics in San Diego County. mSystems 6 (2), e00045-21. doi:10.1128/mSystems.00045-21. [DOI] [PMC free article] [PubMed]
- Isaksson F., Lundy L., Hedström A., Székely A.J., Mohamed N. Evaluating the use of alternative normalization approaches on SARS-CoV-2 concentrations in wastewater: experiences from two catchments in northern Sweden. Environments. 2022;9(3) doi: 10.3390/environments9030039. [DOI] [Google Scholar]
- Kaplan E.H., Wang D., Wang M., Malik A.A., Zulli A., Peccia J. Aligning SARS-CoV-2 indicators via an epidemic model: application to hospital admissions and RNA detection in sewage sludge. Health Care Manag. Sci. 2021;24(2):320–329. doi: 10.1007/s10729-020-09525-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karthikeyan P., Arabzadeh R., Insam H., Markt R., Mayr M., Kreuzinger N., Rauch W. Quest for optimal regression models in SARS-CoV-2 wastewater based epidemiology. Int. J. Environ. Res. Public Health. 2021;18(20) doi: 10.3390/ijerph182010778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby A.E., Walters M.S., Jennings W.C., Fugitt R., LaCross N., Mattioli M., Marsh Z.A., Roberts V.A., Mercante J.W., Yoder J. Using wastewater surveillance data to support the COVID-19 response—United States, 2020–2021. Morb. Mortal. Wkly Rep. 2021;70(36):1242. doi: 10.15585/mmwr.mm7036a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krivoňáková N., Šoltýsová A., Tamáš M., Takáč Z., Krahulec J., Ficek A., Gál M., Gall M., Fehér M., Krivjanská A., Horáková I., Belišová N., Bímová P., Škulcová A.B., Mackuľak T. Mathematical modeling based on RT-QPCR analysis of SARS-CoV-2 in wastewater as a tool for epidemiology. Sci. Rep. 2021;11(1):19456. doi: 10.1038/s41598-021-98653-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P., Kattan O., Broome B., Singhal S. Reassessing Covid-19 needs: how providers can reexamine their surge capacity, supply availability, workforce readiness, and financial resiliency. NEJM Catal. Innov. Care Deliv. 2020;1(3) [Google Scholar]
- McCall C., Fang Z.N., Li D., Czubai A.J., Juan A., LaTurner Z.W., Ensor K., Hopkins L., Bedient P.B., Stadler L.B. Modeling SARS-CoV-2 RNA degradation in small and large sewersheds. Environ. Sci. Water Res. Technol. 2022;8(2):290–300. doi: 10.1039/D1EW00717C. [DOI] [Google Scholar]
- McClary-Gutierrez J.S., Mattioli M.C., Marcenac P., Silverman A.I., Boehm A.B., Bibby K., Balliet M. SARS-CoV-2 wastewater surveillance for public health action. Emerg. Infect. Dis. 2021;27(9) doi: 10.3201/eid2709.210753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan C.S., Self S., Rennert L., Kalbaugh C., Kriebel D., Graves D., Colby C., Deaver J.A., Popat S.C., Karanfil T., Freedman D.L. COVID-19 wastewater epidemiology: a model to estimate infected populations. Lancet Planet. Health. 2021;5(12):e874–e881. doi: 10.1016/S2542-5196(21)00230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema G., Been F., Heijnen L., Petterson S. Implementation of environmental surveillance for SARS-CoV-2 virus to support public health decisions: opportunities and challenges. Environ. Health COVID-19. 2020;17:49–71. doi: 10.1016/j.coesh.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagarkar M., Keely S.P., Jahne M., Wheaton E., Hart C., Smith B., Garland J., Varughese E.A., Braam A., Wiechman B., Morris B., Brinkman N.E. SARS-CoV-2 monitoring at three sewersheds of different scales and complexity demonstrates distinctive relationships between wastewater measurements and COVID-19 case data. Sci. Total Environ. 2021 doi: 10.1016/j.scitotenv.2021.151534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naughton C.C., Roman F.A., Alvarado A.G.F., Tariqi A.Q., Deeming M.A., Bibby K., Bivins A., Rose J.B., Medema G., Ahmed W., Katsivelis P., Allan V., Sinclair R., Zhang Y., Kinyua M.N. medRxiv; 2021. Show Us the Data: Global COVID-19 Wastewater Monitoring Efforts, Equity, and Gaps. 2021.03.14.21253564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nourbakhsh S., Fazil A., Li M., Mangat C.S., Peterson S.W., Daigle J., Langner S., Shurgold J., D’Aoust P., Delatolla R., Mercier E., Pang X., Lee B.E., Stuart R., Wijayasri S., Champredon D. A wastewater-based epidemic model for SARS-CoV-2 with application to three Canadian cities. Epidemics. 2022;39 doi: 10.1016/j.epidem.2022.100560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olesen S.W., Imakaev M., Duvallet C. Making waves: defining the Lead time of wastewater-based epidemiology for COVID-19. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., Warren J.L., Weinberger D.M., Arnold W., Omer S.B. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38(10):1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pileggi V., Shurgold J., Sun J.S., Yang M.I., Edwards E.A., Peng H., Tehrani A., Gilbride K.A., Oswald C.J., Wijayasri S., Al-Bargash D., Stuart R., Khansari Z., Raby M., Thomas J., Fletcher T., Simhon A. Quantitative trend analysis of SARS-CoV-2 RNA in municipal wastewater exemplified with sewershed-specific COVID-19 clinical case counts. ACS EST Water. 2022 doi: 10.1021/acsestwater.2c00058. [DOI] [Google Scholar]
- Prasek S.M., Pepper I.L., Innes G.K., Slinski S., Ruedas M., Sanchez A., Brierley P., Betancourt W.Q., Stark E.R., Foster A.R., Betts-Childress N.D., Schmitz B.W. Population level SARS-CoV-2 fecal shedding rates determined via wastewater-based epidemiology. Sci. Total Environ. 2022;838 doi: 10.1016/j.scitotenv.2022.156535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharara N., Endo N., Duvallet C., Ghaeli N., Matus M., Heussner J., Olesen S.W., Alm E.J., Chai P.R., Erickson T.B. Wastewater network infrastructure in public health: applications and learnings from the COVID-19 pandemic. PLOS Glob. Public Health. 2021;1(12) doi: 10.1371/journal.pgph.0000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shumway R.H., Stoffer D.S., Stoffer D.S. Vol. 3. Springer; 2000. Time Series Analysis and Its Applications. [Google Scholar]
- Siedner M.J., Boucau J., Gilbert R.F., Uddin R., Luu J., Haneuse S., Vyas T., Reynolds Z., Iyer S., Chamberlin G.C., Goldstein R.H., North C.M., Sacks C.A., Regan J., Flynn J.P., Choudhary M.C., Vyas J.M., Barczak A.K., Lemieux J.E., Li J.Z. Duration of viral shedding and culture positivity with postvaccination SARS-CoV-2 delta variant infections. JCI Insight. 2022;7(2) doi: 10.1172/jci.insight.155483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smruthi Karthikeyan, Andrew Nguyen, Daniel McDonald, Yijian Zong, Nancy Ronquillo, Junting Ren, Jingjing Zou, Sawyer Farmer, Greg Humphrey, Diana Henderson, Tara Javidi, Karen Messer, Cheryl Anderson, Robert Schooley, Martin Natasha K., Rob Knight, John McGrath. Rapid, large-scale wastewater surveillance and automated reporting system enable early detection of nearly 85% of COVID-19 cases on a University Campus. mSystems. 2021;6(4) doi: 10.1128/mSystems.00793-21. e00793-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler L.B., Ensor K.B., Clark J.R., Kalvapalle P., LaTurner Z.W., Mojica L., Terwilliger A., Zhuo Y., Ali P., Avadhanula V., Bertolusso R., Crosby T., Hernandez H., Hollstein M., Weesner K., Zong D.M., Persse D., Piedra P.A., Maresso A.W., Hopkins L. medRxiv; 2020. Wastewater Analysis of SARS-CoV-2 as a Predictive Metric of Positivity Rate for a Major Metropolis. [Google Scholar]
- Symanski E., Ensor K.B., Piedra P.A., Sheth K., Caton K., Williams S.L., Persse D., Banerjee D., Hopkins L. Population-based estimates of SARS-CoV-2 seroprevalence in Houston, Texas as of September 2020. J. Infect. Dis. 2021;224(10):1649–1657. doi: 10.1093/infdis/jiab203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari A., Lipponen A., Hokajärvi A.-M., Luomala O., Sarekoski A., Rytkönen A., Österlund P., Al-Hello H., Juutinen A., Miettinen I.T., Savolainen-Kopra C., Pitkänen T. Detection and quantification of SARS-CoV-2 RNA in wastewater influent in relation to reported COVID-19 incidence in Finland. Water Res. 2022;215 doi: 10.1016/j.watres.2022.118220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tlhagale M., Liphadzi S., Bhagwan J., Naidoo V., Jonas K., van Vuuren L., Medema G., Andrews L., Béen F., Ferreira M.L., Saatci A.M., Alpaslan Kocamemi B., Hassard F., Singer A.C., Bunce J.T., Grimsley J.M.S., Brown M., Jones D.L. Establishment of local wastewater-based surveillance programmes in response to the spread and infection of COVID-19 – case studies from South Africa, the Netherlands, Turkey and England. J. Water Health. 2022;20(2):287–299. doi: 10.2166/wh.2022.185. [DOI] [PubMed] [Google Scholar]
- Vallejo J.A., Rumbo-Feal S., Conde-Pérez K., López-Oriona Á., Tarrío-Saavedra J., Reif R., Ladra S., Rodiño-Janeiro B.K., Nasser M., Cid Á., Veiga M.C., Acevedo A., Lamora C., Bou G., Cao R., Poza M. medRxiv; 2020. Predicting the Number of People Infected with SARS-COV-2 in a Population Using Statistical Models Based on Wastewater Viral Load. 2020.07.02.20144865. [DOI] [Google Scholar]
- Wade M.J., Lo Jacomo A., Armenise E., Brown M.R., Bunce J.T., Cameron G.J., Fang Z., Farkas K., Gilpin D.F., Graham D.W., Grimsley J.M.S., Hart A., Hoffmann T., Jackson K.J., Jones D.L., Lilley C.J., McGrath J.W., McKinley J.M., McSparron C., Nejad B.F., Morvan M., Quintela-Baluja M., Roberts A.M.I., Singer A.C., Souque C., Speight V.L., Sweetapple C., Walker D., Watts G., Weightman A., Kasprzyk-Hordern B. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 2022;424 doi: 10.1016/j.jhazmat.2021.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Zarei-Baygi A., Sauceda C., Iskander S.M., Smith A.L. Long-term surveillance of wastewater SARS-CoV-2 in Los Angeles County. Environ. Sci. Water Res. Technol. 2021;7(12):2282–2294. doi: 10.1039/D1EW00586C. [DOI] [Google Scholar]
- Wurtz N., Lacoste A., Jardot P., Delache A., Fontaine X., Verlande M., Annessi A., Giraud-Gatineau A., Chaudet H., Fournier P.-E., Augier P., La Scola B. Viral RNA in City wastewater as a key indicator of COVID-19 recrudescence and containment measures effectiveness. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.664477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurtzer S., Waldman P., Levert M., Cluzel N., Almayrac J.L., Charpentier C., Masnada S., Gillon-Ritz M., Mouchel J.M., Maday Y., Boni M., Marechal V., Moulin L. SARS-CoV-2 genome quantification in wastewaters at regional and City scale allows precise monitoring of the whole outbreaks dynamics and variants spreading in the population. Sci. Total Environ. 2022;810 doi: 10.1016/j.scitotenv.2021.152213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao A., Wu F., Bushman M., Zhang J., Imakaev M., Chai P.R., Duvallet C., Endo N., Erickson T.B., Armas F., Arnold B., Chen H., Chandra F., Ghaeli N., Gu X., Hanage W.P., Lee W.L., Matus M., McElroy K.A., Moniz K., Rhode S.F., Thompson J., Alm E.J. Metrics to relate COVID-19 wastewater data to clinical testing dynamics. Water Res. 2022;212 doi: 10.1016/j.watres.2022.118070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv K., Shagan M., Lewis Y.E., Kramarsky-Winter E., Weil M., Indenbaum V., Elul M., Erster O., Brown A.S., Mendelson E., Mannasse B., Shirazi R., Lakkakula S., Miron O., Rinott E., Baibich R.G., Bigler I., Malul M., Rishti R., Brenner A., Friedler E., Gilboa Y., Sabach S., Alfiya Y., Cheruti U., Davidovich Nadav, Moran-Gilad J., Berchenko Y., Bar-Or I., Kushmaro A. City-level SARS-CoV-2 sewage surveillance. Chemosphere. 2021;283 doi: 10.1016/j.chemosphere.2021.131194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Zou Y., Li Y., Miyani B., Spooner M., Gentry Z., Jacobi S., David R.E., Withington S., McFarlane S., Faust R., Sheets J., Kaye A., Broz J., Gosine A., Mobley P., Busch A.W.U., Norton J., Xagoraraki I. Five-week warning of COVID-19 peaks prior to the omicron surge in Detroit, Michigan using wastewater surveillance. Sci. Total Environ. 2022;844 doi: 10.1016/j.scitotenv.2022.157040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information
Data Availability Statement
Data has been posted to Rice U UDP (DOI provided)