SUMMARY
We investigated the first laboratory-confirmed human case of cowpox virus infection in Russia since 1991. Phylogenetic studies of haemagglutinin, TNF-α receptor-like protein and thymidine kinase regions showed significant differences with known orthopoxviruses, including unique amino-acid substitutions and deletions. The described cowpox virus strain, taking into account differences, is genetically closely related to strains isolated years ago in the same geographical region (European part of Russia and Finland), which suggests circulation of viral strains with common origin in wild rodents without spread over long distances and appearance in other parts of the world.
Key words: Cowpox, phylogenetic studies, Russia, vaccine-like
On 1 September 2015, a 13-year-old boy was admitted to the regional hospital of Kostroma, Russia, with complaints for a cutaneous lesion on the lower third of the right shin that had evolved to four crater-shaped ulcers with black necrotic scabs (Fig. 1a), an enlarged lymph node 5·0 × 2·5 cm in the right groin and haemorrhage in the sclera of the right eye. The child was placed in a children's infectious disease department for suspected especially dangerous infections, i.e. anthrax or tularemia. The battery of laboratory studies of biological material from the skin lesions of the patient allowed us to exclude these dangerous infections. The specimen was sent to State Research Centre of Virology and Biotechnology ‘Vector’ (SRC VB VECTOR), the World Health Organization Collaborating Centre for Orthopoxvirus Diagnosis and Repository for Variola Virus Strains and DNA, for diagnosis of possible orthopoxvirus (OPV) infection. Ethical approval was granted by the Institutional Review Board at SRC VB VECTOR. The authors assert that all procedures contributing to this work comply with the ethical standards of the relevant national and institutional committees on human experimentation and with the Helsinki Declaration of 1975, as revised in 2008.
Fig. 1.
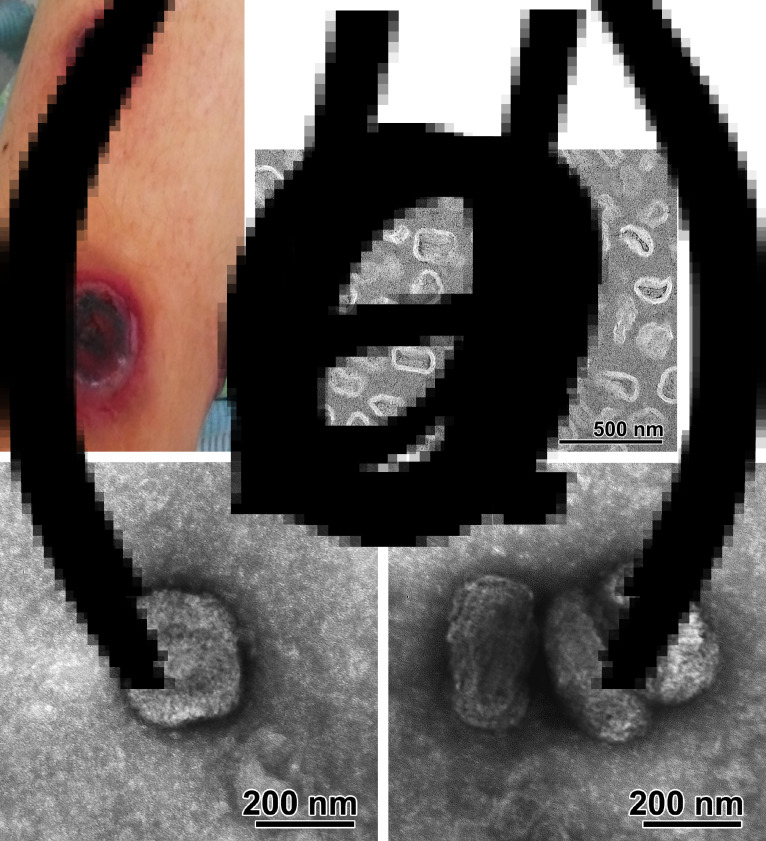
Cowpox virus infection in a 13-year-old boy. (a) Cutaneous lesion on the lower third of the right shin; (b) ultrathin section of the scabs fragments, electron microscopy image; scale bar, 500 nm; (c, d) virions in the cell lysate, negative staining electron microscopy image; scale bar, 200 nm.
We conducted molecular testing of the patient's lesion sample using real-time PCR assays for OPV detection [1, 2]. The sample gave a positive result with OPV genus-specific and cowpox virus (CPXV) species-specific probes, and negative results with variola (VARV), monkeypox (MPXV) and vaccinia virus (VACV) species-specific probes. The CPXV strain was designated Kostroma/2015. Moreover, OPV-specific IgG were detected by ELISA in serum samples.
Using electronic microscopy, large concentrations of particles in the shape of an oval or rectangle with rounded corners were found in the scab (Fig. 1b). Maximum outside dimensions of the particles in the section were 255 nm long and 205 nm wide, the majority of the axial ratio of the particles equalled 1·2–1·4. The shell particle had a two-layer structure. Some particles had a biconcave shape flanked by oval bodies (in a concave part). Sometimes circular structures with a diameter of about 40 nm were found. Described particles morphologically correspond to an isolated nucleoid of poxviruses.
The clinical sample was inoculated onto Vero cells for several passages. Non-specific cytopathic effect (CPE) was observed 7 days post-inoculation for the clinical sample. On the second passage a massive OPV-specific CPE was observed on day 5 post-inoculation with earlier plaque formation depending on the passage: plaques were detected from 4 days after infection for the second passage, 2 days for the third and from 1 day for the fourth passage. When studying the cell lysate using negative contrast, brick-shaped particles with sizes of 290–310 nm to 210–255 nm were found (Fig. 1 c, d).
After DNA extraction from cell culture of the first passage, the viral genome was also amplified by PCR using primer pairs to several genomic regions: A58R (haemagglutinin, HA; 1023 nt), D2L (TNF-α receptor-like protein, CrmB; 1313 nt), N5R-O1R (thymidine kinase, TK; 1875 nt) according to CPXV strain GRI-90 nomenclature [3]. The sequences of the corresponding amplicons were determined and were deposited into GenBank under accession numbers: KU861503–KU861505.
The HA and TK sequences are often used for OPV phylogenetic analysis as well as sequences of genes that encode immunomodulatory proteins [4]. Next, phylogenetic studies were conducted by comparing sequences of HA, CrmB and TK regions for the Kostroma/2015 isolate with corresponding sequences of OPVs, including CPXV [strains GRI-90 (X94355), Brighton Red (AF482758), Germany_1991-3 (DQ437593), Finland_2000_MAN (HQ420893), Norway_1994_MAN (HQ420899), Germany_1990_2 (HQ420896), Germany_2002_MKY (HQ420898), France_2001_Nancy (HQ420894), Germany_1980_EP4 (HQ420895), Germany_1998_2 (HQ420897), UK2000_K2984 (HQ420900), Austria_1999 (HQ407377)], VARV (Y16780, L22579), MPXV (DQ011155, DQ011156), taterapox virus (TATV) (DQ437594), camelpox virus (CMPV) (AY009089, AF438165), VACV (AY678276, AM501482, M35027, DQ377945), horsepox virus (HPXV) (DQ792504) and rabbitpox virus (RPXV) (AY484669) [4] (Fig. 2).
Fig. 2.
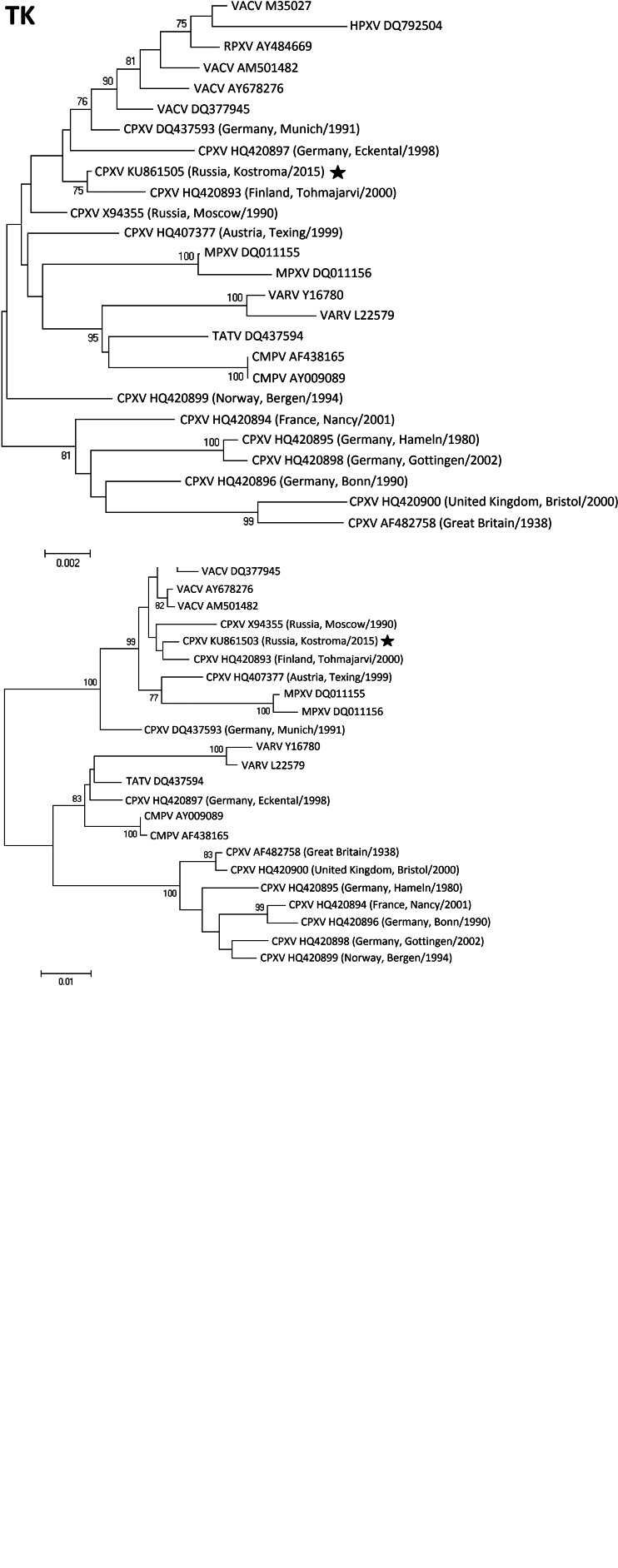
Phylogenetic relationships between three genomic regions of cowpox virus (CPXV) isolates collected in Russia in 2015 and other orthppoxviruses. TNF-α receptor-like protein (CrmB; 1056 nt: positions 2286–3341 of CPXV strain GRI-90); thymidine kinase (TK; 1829 nt: positions 96 315–98 143 of CPXV strain GRI-90); haemagglutinin (HA; 939 nt: positions 178 776–179 720 of CPXV strain GRI-90). Phylogenetic trees were constructed using the neighbour-joining method and MEGA4 software. The numbers at the nodes represent the percent bootstrap support for 1000 replicates. Only values above 75% are shown. Bars at the base of the trees show the genetic divergence. The CPXV strain Kostroma/2015, described in this paper, is indicated by a star (⋆).
In the CrmB region, the Kostroma/2015 isolate showed maximum identity to CPXV strain GRI-90, isolated in 1990 from a 4-year-old girl in Russia [3], with 6 nt changes (out of 1053 nt) leading to two amino acid (aa) substitutions. Next closest to Kostroma/2015 is a group of several CPXV strains: Germany_2002_MKY, Austria_1999 and Finland_2000_MAN with 29–32 nt changes plus six deletions leading to 8–12 aa substitutions and deletion of 2 aa in CrmB sequence.
In the TK region, Kostroma/2015 was closely related to several CPXV and VACV strains, including Finland_2000_MAN (5 nt/0 aa changes) and GRI-90 (6 nt/1 aa changes out of 1829 nt).
Kostroma/2015 has also shown equal identity with several CPXV and VACV strains in the HA region – with minimal 11 nt changes plus nine contiguous nt deletions compared to Finland_2000_MAN, leading to 5 aa substitutions and deletion of 3 aa. Here, the observed deletion of 3 aa (positions 19–21 of HA) is unique and was not found in any other known strains of OPVs. Kostroma/2015 and GRI-90 were found to be relatively distant in the HA region (21 nt changes and 12 deletions/insertions out of 939 nt, leading to 10 aa substitutions, a deletion of 3 aa and an insertion of 1 aa).
Thus Kostroma/2015 isolate, despite the differences, is closest to the two CPXV strains – GRI-90 and Finland_2000_MAN, which in turn belong to the group of ‘Vaccinia-like’ CPXV [5].
Cowpox is transmitted to humans sporadically from cats or rodents [6, 7]. The last cases of cowpox in humans in Russia with a suspected infection from rodents occurred in 1987 (Moscow, source: white rat, vivarium staff aged 31 and 34 years), 1988 (Smolensk region, source: small wild rodents, child aged 10 years), 1990 (Moscow region, source: a mole, child aged 4 years) and 1991 (Moscow, source: white rat, children aged 10 and 12 years) [8]. In Europe the last case of ‘Vaccinia-like’ CPXV was identified in Finland in 2009. This isolate was identical to the CPXV strain Finland_2000_MAN previously identified in Finland [9].
The case of cowpox in a human described in this paper is the first confirmed case in Russia for the last quarter of a century. According to the patient, during the month preceding the start of the disease, he lived in two rural areas of Kostroma region, having contact with domestic animals, namely cats, dogs, cows. He spent a lot of time in the woods, fields and farms. In the course of the epidemiological investigation of the case of cowpox in the Kostroma region blood samples and skin lesions from nine rodents and cats found in places where the patient had visited during the last month before the disease were studied, but no CPXV DNA was amplifiable in the samples and no specific source of infection could be established.
Our observation of the described CPXV strain that it is genetically closely related to strains isolated years ago in the same geographical region (European part of Russia and Finland), which suggests circulation of viral strains with common origin among wild rodents without spread over long distances and appearance in other parts of the world.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Shchelkunov SN, et al. Species-specific identification of variola, monkeypox, cowpox, and vaccinia viruses by multiplex real-time PCR assay. Journal of Virological Methods 2011; 175: 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maksyutov RA, et al. Real-time PCR assay for specific detection of cowpox virus. Journal of Virological Methods 2015; 211: 8–11. [DOI] [PubMed] [Google Scholar]
- 3.Marenikova SS, et al. Biotype and genetic characterization of isolate of cowpox virus having caused infection in a child. Zhurnal mikrobiologii, epidemiologii, i immunobiologii 1996; 4: 6–10. [PubMed] [Google Scholar]
- 4.Dabrowski PW, et al. Genome-wide comparison of cowpox viruses reveals a new clade related to Variola virus. PLoS ONE 2013; 8: e79953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll DS, et al. Chasing Jenner's vaccine: revisiting cowpox virus classification. PLoS ONE 2011; 6: e23086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haller SL, et al. Poxviruses and the evolution of host range and virulence. Infection, Genetics and Evolution 2014; 21: 15–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shchelkunov SN. An increasing danger of zoonotic orthopoxvirus infections. PLoS Pathogens 2013; 9: e1003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shchelkunov SN, Marennikova SS, Moyer RW. Orthopoxviruses Pathogenic for Humans. New York: Springer, 2005, 426 pp. [Google Scholar]
- 9.Kinnunen PM, et al. Severe ocular cowpox in a human, Finland. Emerging Infectious Diseases 2015; 21: 2261–2263. [DOI] [PMC free article] [PubMed] [Google Scholar]