SUMMARY
Extended-spectrum β-lactamase (ESBL) production has been very rare in serotype K1 Klebsiella pneumoniae ST23 strains, which are well-known invasive community strains. Among 92 ESBL-producing strains identified in 218 isolates from nine Asian countries, serotype K1 K. pneumoniae strains were screened. Two ESBL-producing K. pneumoniae isolates from Singapore and Indonesia were determined to be serotype K1 and ST23. Their plasmids, which contain CTX-M-15 genes, are transferable rendering the effective transfer of ESBL resistance plasmids to other organisms.
Key words: Extended-spectrum β-lactamase, Klebsiella pneumoniae, serotype K1
Klebsiella pneumoniae is an important pathogen causing hospital-acquired infections and multidrug resistance in these bacteria has increased [1, 2]. Treatment of multidrug-resistant K. pneumoniae infections has been a big challenge and there is an increasing concern about the clinical impact of these bacteria [3]. Invasive K. pneumoniae strains belonging to serotype K1 have emerged as important causes of community-acquired infections including liver abscess in many Asian countries [4–6]. Despite a high prevalence of community infections by these invasive K. pneumoniae strains has been characterized by ST23, which appears to be susceptible to most antibiotic classes except ampicillin and piperacillin [6]. Only a few case reports of Extended-spectrum β-lactamase (ESBL)-producing serotype K1 K. pneumoniae have been reported [7–9]. In this study, we molecularly characterized two serotype K1 ESBL-producing K. pneumoniae strains and investigated the transferability of plasmid DNA from these isolates.
As part of a multinational Asian Network for Surveillance of Resistant Pathogens (ANSORP) surveillance study on hospital-acquired pneumonia during 2008 and 2009 [10], a total of 218 K. pneumoniae isolates were collected from nine Asian countries. Of the 218 strains, 92 isolates were identified as ESBL producers [11]. In this study, the 92 ESBL-producing K. pneumoniae isolates were tested for serotype K1, and serotype K1 ESBL-producers were included for further characterization. In vitro antimicrobial susceptibility testing was performed by a broth microdilution method and double-disk synergy test following Clinical and Laboratory Standards Institute (CLSI) guidelines. Double-disk synergy test-positive isolates were further tested by polymerase chain reaction (PCR) and sequence analyses to determine the gene responsible for the ESBL phenotype. PCR for blaTEM, blaSHV, and blaCTX-M was conducted using previously described PCR primers and conditions [12]. All PCR products were sequenced and the sequences were compared using the database available at http://www.lahey.org/studies/.
K1 serotyping was tested by PCR using a primer pair specific for magA, which is specific for the K1 antigen. The primers used were as described previously [13]. Multilocus sequence typing was performed on serotype K1 K. pneumoniae by determining the nucleotide sequences of seven housekeeping genes as described previously [14]. For pulsed-field gel electrophoresis (PFGE), agarose embedded bacterial genomic DNA was digested with 20 U XbaI. The restriction fragments were separated by electrophoresis in 0·5× Tris-borate-EDTA buffer. The PFGE patterns were analysed using GelCompar II v. 6.1 (Applied Maths, Belgium). Conjugation experiments were performed using azide-resistant E. coli J53 as a recipient strain. Mixed strains at a ratio of 1:10 (donor:recipient) were used for broth and filter mating assays. Transconjugants were selected on lysogeny broth medium containing cefotaxime (4 mg/l) and sodium azide (200 mg/l). Isolated DNA plasmids from serotype K1 ESBL transconjugants were used for transformation in the E. coli DH5α strain. All transconjugants and transformants were tested for ESBL production using the disk and broth microdilution method followed by PCR amplification of the ESBL genes.
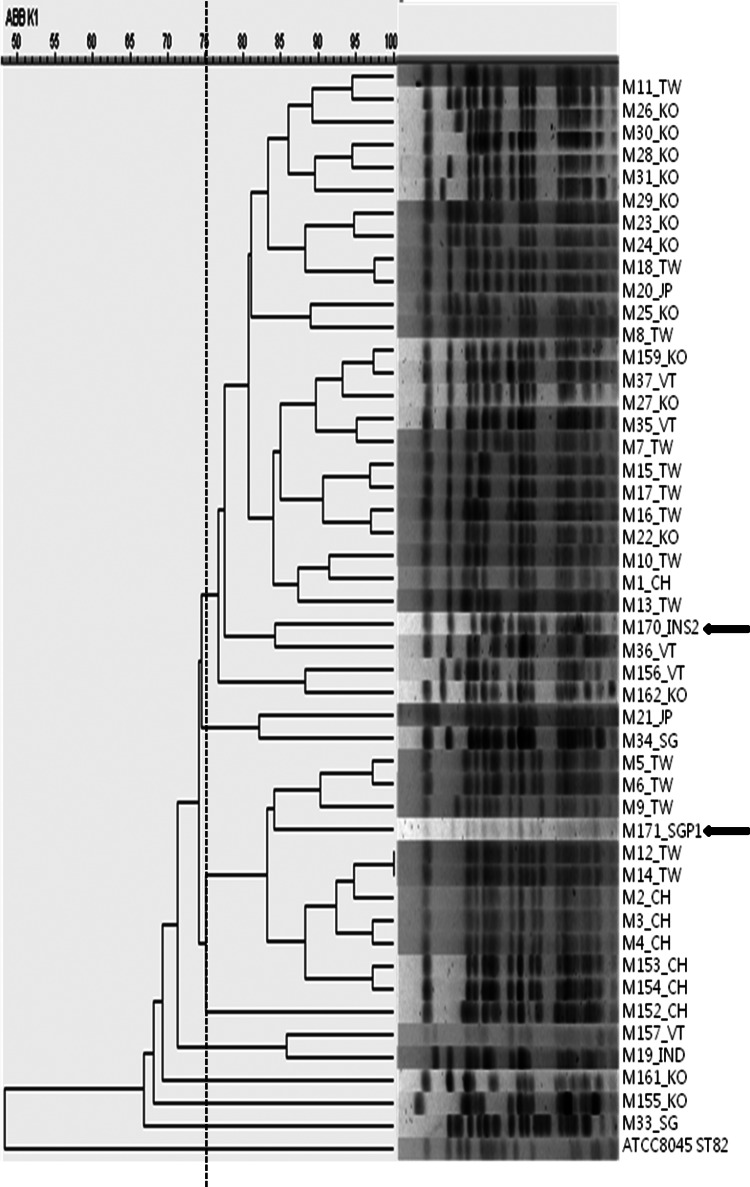
Among the 92 ESBL-producing K. pneumoniae isolates, only two were determined as serotype K1. The two isolates were from patients in Singapore and Indonesia named, SGP-1 and INS-2, respectively. The results of antimicrobial susceptibility testing of these isolates are shown in Table 1. Both SGP-1 and INS-2 belonged to ST23 and carried the blaCTX-M-15 gene. Moreover, those isolates were negative for TEM and SHV ESBL genes and fully susceptible to carbapenems. SGP-1 was isolated in a patient with ventilator-associated pneumonia (VAP) after neurosurgery. The patient was treated with meropenem and discharged. INS-2 was isolated in a patient with VAP after surgery. Ceftazidime was given empirically and replaced by meropenem. Although VAP improved, the patient died due to cardiac arrest 77 days after admission. The PFGE analysis showed that the PFGE patterns of these strains and serotype K1 non-ESBL K. pneumoniae isolates of ST23 from Asian countries clustered into a major group with a similarity of >75% (Fig. 1). To determine whether ESBL resistance in serotype K1 ST23 K. pneumoniae strains was transferable, a conjugation experiment was performed. The plasmids carrying blaCTX-M-15 from SGP-1 and INS-2 were successfully transferred into recipient J53 E. coli isolates. The transfer of the plasmid carrying blaCTX-M-15 from the JI-14 transconjugant was accomplished using an E. coli DH5α strain. All transconjugants (JS-1, JS-2, JS-4, JS-5, JS-6, JS-7, and JS-8 from SGP-1; JI-2, JI-3, JI-5, JI-10, JI-11, and JI-15 from INS-2) and transformants (FI14-1 and FI14-2) were tested for ESBL production using the disk and broth microdilution method followed by PCR amplification of ESBL genes. All transconjugants and transformants were found to express blaCTX-M-15 genes. Antimicrobial resistance patterns did not differ between the donors and transconjugants; however, all of the transformants showed decreased minimum inhibitory concentrations for tested antimicrobials (Table 1).
Table 1.
Antimicrobial susceptibilities and molecular characteristics of recipient, donor and transconjugants
| Strain | Description | ESBL | Molecular characteristics | MICs and antimicrobial susceptibility | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | CAZ | CPM | P/T | CIP | AZT | |||||||||||
| Recipient | J53 | E. coli J53 | − | − | 0·12 | S | 0·12 | S | 0·12 | S | 2/4 | S | 0·12 | S | 0·06 | S |
| Donor 1 | SGP-1 | ST23 serotype K1 K. pneumoniae |
+ | CTX-M-15 | >128 | R | 64 | R | >128 | R | 8/4 | S | 2 | I | >64 | R |
| Transconjugants | JS-1 |
E. coli J53/ESBL K. pneumoniae SGP-1 |
+ | CTX-M-15 | >128 | R | 64 | R | 128 | R | 32/4 | I | 1 | S | >64 | R |
| JS-2 | + | CTX-M-15 | >128 | R | 64 | R | >128 | R | 32/4 | I | 0·5 | S | >64 | R | ||
| JS-4 | + | CTX-M-15 | >128 | R | 64 | R | 32 | R | 32/4 | S | 1 | S | >64 | R | ||
| JS-5 | + | CTX-M-15 | >128 | R | 64 | R | 32 | R | 64/4 | I | 1 | S | >64 | R | ||
| JS-6 | + | CTX-M-15 | >128 | R | 128 | R | 128 | R | 64/4 | I | 2 | I | >64 | R | ||
| JS-7 | + | CTX-M-15 | >128 | R | 32 | R | 32 | R | 16/4 | S | 1 | S | >64 | R | ||
| JS-8 | + | CTX-M-15 | >128 | R | 64 | R | 128 | R | 32/4 | I | 1 | S | >64 | R | ||
| Donor 2 | INS-2 | ST23 serotype K1 K. pneumoniae |
+ | CTX-M-15 | >128 | R | 64 | R | 128 | R | 8/4 | S | 2 | I | >64 | R |
| Transconjugants | JI-2 |
E. coli J53/ESBL K. pneumoniae INS-2 |
+ | CTX-M-15 | >128 | R | 32 | R | 128 | R | 8/4 | S | 2 | I | >64 | R |
| JI-3 | + | CTX-M-15 | >128 | R | 16 | R | 64 | R | 8/4 | S | 2 | I | >64 | R | ||
| JI-5 | + | CTX-M-15 | >128 | R | 64 | R | 16 | I | 16/4 | S | 0·12 | S | 64 | R | ||
| JI-10 | + | CTX-M-15 | >128 | R | 16 | R | 128 | R | 8/4 | S | 2 | I | >64 | R | ||
| JI-11 | + | CTX-M-15 | >128 | R | 16 | R | 128 | R | 8/4 | S | 2 | I | >64 | R | ||
| JI-15 | + | CTX-M-15 | >128 | R | 32 | R | 128 | R | 8/4 | S | 2 | I | >64 | R | ||
| Recipient | DH5α | E. coli DH5α | − | − | 0·12 | S | 0·12 | S | 0·12 | S | 0·5/4 | S | 0·12 | S | 0·06 | S |
| Transformants | FI14-1 |
E. coli DH5α/ESBL JI14 plasmid |
+ | CTX-M-15 | 64 | R | 16 | R | 4 | S | 4/4 | S | 0·25 | S | 16 | R |
| FI14-2 | + | CTX-M-15 | 64 | R | 16 | R | 2 | S | 4/4 | S | 0·25 | S | 16 | R | ||
MIC, Minimum inhibitory concentration; CTX, cefotaxime; CAZ, ceftazidime; CPM, cefepime; P/T, piperacillin-tazobactam; CIP, ciprofloxacin; AZT, aztreonam; R, resistant; I, intermediate; S, susceptible.
Fig. 1.
PFGE dendrogram of ST23 serotype K1 K. pneumoniae isolates from Asian countries.
In this study, we investigated the molecular characteristics of two serotype K1 ESBL-producing K. pneumoniae strains belonging to ST23 and the transferability of plasmid DNA from these isolates. The increasing prevalence of ESBL-producing K. pneumoniae is becoming a serious worldwide problem. Over the past decade, the global emergence of multidrug-resistant strains of K. pneumoniae that produce ESBL of CTX-M types has been of considerable concern. In particular, CTX-M-15-producing K. pneumoniae is highly prevalent in Asian counties and worldwide [11, 15]. Although serotype K1 K. pneumoniae strains have emerged as important pathogens causing community-acquired infections in many countries, those strains have shown good susceptibility to most antimicrobials to date. However, our report of two serotype K1 ESBL-producing K. pneumoniae strains carrying blaCTX-M-15 on transferrable plasmids suggests that dissemination of CTX-M-15 producers in highly pathogenic serotype K1 K. pneumoniae strains may become a serious threat to the treatment of community-acquired K. pneumoniae infections in the near future. Furthermore, two isolates in this study were found in the Asian Hospital Acquired Pneumonia surveillance study in the absence of antimicrobial pressure. The results presented here are in agreement with the observation from Shin & Ko [16]. Moreover, we found that ST23 K1 K. pneumoniae showed a similar PFGE pattern regardless of ESBL. Previous reports support this result that the ST23 ESBL K. pneumoniae might have disseminated from a single strain [16, 17].
In summary, we report two clinical isolates of serotype K1 ESBL-producing K. pneumoniae carrying transferable CTX-M-15 plasmids that could transfer resistance of antimicrobials to other clinical isolates.
ACKNOWLEDGEMENTS
This study received no funding.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Bialek-Davenet S, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerging Infectious Diseases 2014; 20: 1812–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holt KE, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proceedings of the National Academy of Sciences USA 2015; 112: E3574–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pitout JD, Nordmann P, Poirel L. Carbapenemase-producing Klebsiella pneumoniae: a key pathogen set for global nosocomial dominance. Antimicrobial Agents and Chemotherapy 2015; 59: 5873–5884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fang CT, et al. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clinical Infectious Diseases 2007; 45: 284–293. [DOI] [PubMed] [Google Scholar]
- 5.Chung DR, et al. Emerging invasive liver abscess caused by K1 serotype Klebsiella pneumoniae in Korea. Journal of Infection 2007; 54: 578–583. [DOI] [PubMed] [Google Scholar]
- 6.Chung DR, et al. Prevalence and molecular characterization of serotype K1 Klebsiella pneumoniae strains from various clinical specimen sources in 11 Asian countries. Journal of Infection 2012; 64: 622–625. [DOI] [PubMed] [Google Scholar]
- 7.Su SC, et al. Community-acquired liver abscess caused by serotype K1 Klebsiella pneumoniae with CTX-M-15-type extended-spectrum beta-lactamase. Antimicrobial Agents and Chemotherapy 2008; 52: 804–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagano N, Cordevant C, Nagano Y. Upper and lower urinary tract infection caused by Klebsiella pneumoniae serotype K2 and CTX-M-15 beta-lactamase-producing serotype K1: a case report and characterization of serum killing resistance. Journal of Medical Microbiology 2008; 57: 121–124. [DOI] [PubMed] [Google Scholar]
- 9.Hall JM, et al. Temporal flux in beta-lactam resistance among Klebsiella pneumoniae in Western Australia. Journal of Medical Microbiology. Published online: 4 March 2016. doi: 10.1099/jmm.0.000242. [DOI] [PubMed] [Google Scholar]
- 10.Chung DR, et al. High prevalence of multidrug-resistant nonfermenters in hospital-acquired pneumonia in Asia. American Journal of Respiratory and Critical Care Medicine 2011; 184: 1409–1417. [DOI] [PubMed] [Google Scholar]
- 11.Lee MY, et al. High prevalence of CTX-M-15-producing Klebsiella pneumoniae isolates in Asian countries: diverse clones and clonal dissemination. International Journal of Antimicrobial Agents 2011; 38: 160–163. [DOI] [PubMed] [Google Scholar]
- 12.Kim J, et al. CTX-M and SHV-12 beta-lactamases are the most common extended-spectrum enzymes in clinical isolates of Escherichia coli and Klebsiella pneumoniae collected from 3 university hospitals within Korea. FEMS Microbiology Letters 2005; 245: 93–98. [DOI] [PubMed] [Google Scholar]
- 13.Struve C, et al. Investigation of the putative virulence gene magA in a worldwide collection of 495 Klebsiella isolates: magA is restricted to the gene cluster of Klebsiella pneumoniae capsule serotype K1. Journal of Medical Microbiology 2005; 54: 1111–1113. [DOI] [PubMed] [Google Scholar]
- 14.Diancourt L, et al. Multilocus sequence typing of Klebsiella pneumoniae nosocomial isolates. Journal of Clinical Microbiology 2005; 43: 4178–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li B, et al. Analysis of drug resistance determinants in Klebsiella pneumoniae isolates from a tertiary-care hospital in Beijing, China. PLoS ONE 2012; 7: e42280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shin J, Ko KS. Comparative study of genotype and virulence in CTX-M-producing and non-extended-spectrum beta-lactamase-producing Klebsiella pneumoniae isolates. Antimicrobial Agents and Chemotherapy 2014; 58: 2463–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turton JF, et al. Genetically similar isolates of Klebsiella pneumoniae serotype K1 causing liver abscesses in three continents. Journal of Medical Microbiology 2007; 56: 593–597. [DOI] [PubMed] [Google Scholar]