SUMMARY
It has been suggested that zoonotic transmission of Staphylococcus aureus (SA) and methicillin-resistant S. aureus (MRSA) can occur between owners and their pets within the same household. However, the influence that pet-ownership could have in the biodiversity of SA/MRSA strains circulating among owners is not fully understood. The objective of this study was to perform a molecular epidemiological analysis to evaluate and compare the biodiversity of SA/MRSA strains in dog-owning and non-dog-owning healthy households within the same community. Antimicrobial resistance, SCCmec type, USA type and clonality were assessed. Overall, 33·1% (165/499) of human subjects carried SA and 2·8% (14/499) carried MRSA. Among dogs, 7·1% (8/113) carried SA but none were MRSA positive. No difference was detected in the diversity index of SA/MRSA pulsotypes between dog-owning and non-dog-owning households; but, a marked variation was still observed in the pulsotypes circulating in each type of household. Additionally, simultaneous carriage of the same SA pulsotype in owner(s) and dog was observed in 57% of households with positive humans and pets. These results demonstrate that dogs can indeed participate in the circulation of SA/MRSA pulsotypes within a home and that the presence of a pet does not seem to favour certain strains within their household.
Key words: Biodiversity, household, molecular epidemiology, pet-ownership, Staphylococcus aureus
INTRODUCTION
In 2012, the American Veterinary Medical Association estimated that the dog population in the United States alone was about 70 million with over 43 million households (roughly 36·5%) owning one or more dogs [1]. Some studies have suggested that the human–animal bond brings physiological, psychological, social and emotional health benefits for pet-owners, including improved physical health (perhaps through increased physical activity) and emotional comfort and companionship for elderly and individuals with special needs [2, 3]. Additionally, 66·7% of dog-owners consider their dogs as another family member [1]. However, there are always two sides to every story.
At the same time, human–animal interactions could also be associated with health risks, including the transmission of zoonotic pathogens such as Staphylococcus aureus and methicillin-resistant S. aureus (MRSA). At the household level, it is suspected that S. aureus/MRSA transmission occurs through direct contact during owner–pet interactions (petting, bathing, licking, sharing couch/bed space, among others) or through indirect contact with contaminated household surfaces [4]. Thus, it is not surprising to observe that MRSA clones found in pets reflect those strains that are carried by colonized/infected people in close contact with them [5–7].
Since pets are believed to be a potential reservoir for S. aureus/MRSA [8, 9] and some strains tend to prefer a particular animal species [10–12], it is feasible to assume that the diversity of S. aureus/MRSA strains in household members could vary depending on the presence and contact with certain companion animal species. Unfortunately, only a small number of reports and studies have described S. aureus/MRSA carriage and zoonotic transmission within households, and in most cases the owner, dog, or both were known to be colonized/infected with MRSA [5, 13–18]. Only a few publications involved a selection of subjects (humans and dogs) with unknown carriage or no active infections [7, 19–22]. Moreover, no previous studies have compared the epidemiology and molecular characteristics of S. aureus/MRSA strains between dog-owning and non-dog-owning households within the same community. Consequently, little is known about how the presence of a pet in a ‘healthy’ household could potentially change or shift the type and diversity of S. aureus/MRSA strains circulating among family members.
Previous publications have identified dogs colonized or infected with MRSA strains typically classify as hospital-acquired strains (e.g. USA100) [22–25] that are considered opportunistic pathogens and important causes of morbidity and mortality. If the presence of pets could promote or favour the presence of certain S. aureus/MRSA strains then compared to non-dog-owners, dog-owning households could have an increased risk of exposure to potentially pathogenic or virulent strains (e.g. pandemic strains that could cause infections in humans).
In order to address this gap in current knowledge, the objective of this study was to screen members of dog-owning and non-dog-owning healthy households within the same community for S. aureus/MRSA to perform a molecular epidemiological analysis to evaluate and compare the biodiversity of S. aureus/MRSA strains and pulsotypes in the presence or absence of pets. In dog-owning households, owner–pet interactions were evaluated to determine if certain behaviours were more likely to be associated with the presence of specific S. aureus/MRSA strains or pulsotypes.
MATERIALS AND METHODS
Study population, sample size and recruitment
The study was conducted between May 2012 and May 2013 in the Midwestern United States in the Central Ohio region. Households with and without dogs were considered for the study. The sample size of this study was calculated assuming a baseline of S. aureus colonization prevalence of 33% in a weighted human population, with 65% non-dog-owners and 35% dog-owners, and an alpha of 0·05.
Families were recruited using three different methods. First, potential volunteers were located through the Research Match™ (NIH-sponsored registry) by selecting individuals that indicated an interest in research projects. Second, participants already enrolled in the study were asked if they would consider referring other households. Third, recruitment material (leaflet) was available to clients at The Ohio State University Veterinary Medical Center asking for volunteers to participate in this study. Households were excluded from the study if they participated in commercial breeding operations or boarding of dogs at their primary residence. The study excluded participants that engaged in livestock operations such as farming and animal rescue activities. Moreover, a subject was not included if he/she refused to participate in the study, or if he/she was not present in the house at the time of sampling. The presence of other non-dog pets in the household was recorded (number and animal species) and considered as a variable for further analyses. Once enrolled, consent was obtained in writing from each of the participants in the household. Assent was obtained from children (under the age of 18 years) and formal consent from parents or guardians. Informed consent was obtained from owners to collect samples from dogs.
Alhough not reported here, epidemiological data was collected from each household member including general household data and human health/risk factors. For dog-owners, information was collected including general information about the family's pets (age, sex, breed, health factors), and the types and frequencies of interaction between the family members and their pets. For the purposes of this manuscript, only the information regarding the owner–pet interaction will be included in the data analysis (Table 1).
Table 1.
Questionnaire used to evaluate owner–pet interactions in dog-owning households in the central Ohio region
| |
|
|
| |
| |
|
|
| |
|
|
| |
|
|
| |
|
|
| |
| |
| |
|
|
| |
|
|
| |
|
|
| |
|
|
| |
|
|
Ethical approval
The study was reviewed and approved by the Institutional Review Board Committee (FWA#00006378, protocol 2011H0173) and the Institutional Animal Care Committee (protocol 2010A00000173).
Sample collection and bacterial isolation
A sample from the nostrils of all human subjects was collected using a sterile cotton-tip swab [BD BBL™ CultureSwab, Becton Dickinson and Company (BD), USA] after adding approximately 2–3 ml of sterile saline solution (SalJet, Winchester Laboratories, USA) into each nostril. Additionally, in households with pets, two samples were collected from each animal. Sterile cotton-tip swabs pre-moistened in sterile trypticase soy broth (TSB; BD BBL, BD) with 2·5% NaCl were used to collect individual samples from the nose and the perianal region. All samples were immediately taken to The Ohio State University Diagnostic and Research Laboratory for Infectious Diseases (DRLID) and incubated in TSB 2·5% NaCl for 24 h at 35°C. Samples were then streaked onto mannitol salt agar (MSA; BD BBL, BD). As previously described [26], colonies were selected based on typical staphylococcal morphology and mannitol reaction to be transferred onto blood agar plates (TSA with 5% sheep blood; Remel®, USA). Identification of S. aureus was performed by standard colony morphology (including size, pigmentation and haemolysis pattern), and biochemical tests reactions that included: mannitol fermentation, Gram stain, catalase, tube coagulase, anillin fermentation, polymyxin B susceptibility, acetoin production (Vogues–Proskauer test), and latex agglutination (Sure-Vue® Color Staph ID, Biokit USA Inc., USA). All S. aureus isolates were streaked onto OSA plates (BD BBL Oxacillin Screen Agar®, BD) containing 6 μg/ml oxacillin and supplemented with NaCl for phenotypic confirmation of MRSA, following the Clinical Laboratory Standards Institute protocols [27].
Antimicrobial susceptibility testing
All S. aureus isolates were phenotypically tested against 16 antimicrobial drugs used in veterinary and human medicine. The Kirby–Bauer disc diffusion technique was used to test amikacin 30 μg, ampicillin 10 μg, amoxicillin with clavulanic acid 20/10 μg, cefpodoxime 10 μg, cephalotin 30 μg, chloramphenicol 30 μg, ciprofloxacin 2 μg, clindamycin 2 μg, doxycycline 30 μg, enrofloxacin 5 μg, erythromycin 15 μg, gentamicin 1 μg, oxacillin 1 μg, sulfamethoxazole trimethoprim 1·25/23·75 μg, and tetracycline 30 μg. Additionally, vancomycin was tested using Vancomycin Screen Agar plates [VSA(6 mg/l); BD BBL, BD]. The D test was performed to detect inducible clindamycin resistance [28]. The following control strains were used: S. aureus (ATCC 43300), S. aureus (ATCC 29213), S. aureus (ATCC 25923), Enterococcus faecalis (ATCC 23212), Escherichia coli (ATCC 25922), and Pseudomonas aeruginosa (ATCC 27853). Isolates resistant to three or more classes of antimicrobials (including beta-lactams after confirmation of the mecA gene) were considered multidrug resistant.
Staphylococcal chromosome cassette mec (SCCmec) typing and pulsed-field gel electrophoresis
Methicillin-resistant status was confirmed by detection of the mecA gene and typing of the SCCmec by performing a multiplex PCR [29] with few modifications [26]. Additionally, clonality between S. aureus isolates was evaluated by digestion of chromosomal DNA with SmaI following the Centers for Disease Control and Prevention (CDC) protocol [30]. Restriction fragments were separated using a CHEF mapper system (Bio-Rad Laboratories, Belgium). The resulting band patterns were analysed by BioNumerics® software v. 6·6, (Applied Maths, Belgium) to determine the relatedness between strains by using Dice coefficient and Unweighted Pair Group Method using Arithmetic averages (UPGMA) to achieve dendrograms with a 1% band position tolerance [31]. Dendrogram interpretation was performed as described previously [26]: each band pattern represented a pulsotype; isolates with ⩾98% similarity were considered indistinguishable and characterized as the same pulsotype. These types were grouped in clusters of ⩾80% similarity that were considered to be closely related. A CDC database containing 100 S. aureus strains with the most typical band patterns was used to designate USA types (strain type), using ⩾80% similarity as the cut-off point.
Data analysis
The data was divided in two groups (dog-owners and non-dog-owners) and later analysed at the individual and household level. A household was considered positive when at least one member (human or canine) was positive for S. aureus/MRSA. Diversity between bacterial strains found in dog-owning and non-dog-owning households was established based on Simpson's Diversity Index [32]. Owner–pet interactions were evaluated in positive dog-owning households to establish concurrent S. aureus/MRSA carriage with a particular strain. Unadjusted odds ratios (OR) and their 95% confidence intervals (CI) were assessed using Pearson's χ2 (or Fisher's exact test when appropriate) using Stata (Small Stata v. 12.0, StataCorp LP, USA). Independent variables that were associated with S. aureus/MRSA pet carrier status upon this initial screening at P < 0·2 were tested for inclusion in a multivariable logistic regression model using the same software. The final logistic regression model was obtained using a forward selection procedure with a P < 0·05 based on the likelihood ratio χ2 statistic required to remain in the model.
RESULTS
A total of 135 households (represented by 499 human subjects and 113 canines) were sampled during the study period. Of those, 59·3% (80/135, represented by 155 adults, 129 children and 113 canines) were dog-owners and, 40·7% (55/135, represented by 107 adults and 108 children) were non-dog-owners. The average number of members per household was 4 (range 1–11); with an average of 93% of the household members actively participating in the study. The average number of dogs per household was 2·5 (range 1–4).
Dog-owning households/individuals
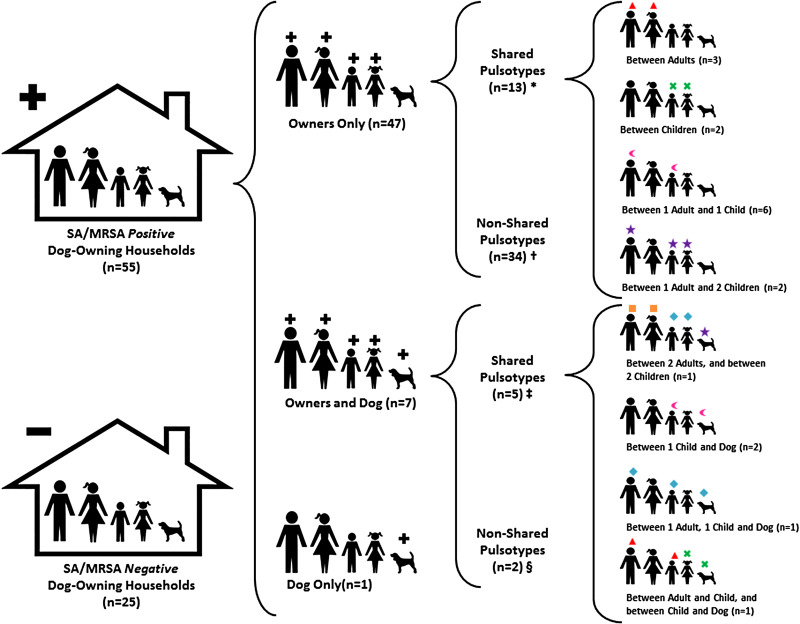
In 68·8% (55/80, 109 positive individuals) of the dog-owning households, one or more family members were found to be S. aureus carriers; of those, 10% (8/80, 12 individuals) were MRSA positive (Table 2). Of these households, 85·5% (47/55) had only human positive members, 12·7% (7/55) had positive humans and dogs, and 1·8% (1/55) had only positive pets (Fig. 1). With regard to individuals, 35·6% (101/284) were positive to S. aureus and 4·2% (12/284) to MRSA, with children showing the highest carriage rates (41·9% and 5·4%, respectively) (Table 2). Additionally, 7·1% (8/113) of the pets carried S. aureus, but all of them were negative for MRSA. Among the eight households with S. aureus-positive pets only one dog was found positive per house, regardless of the total number of canines present at home (ranging from 1 to 2). Statistical analyses performed to determine if multi-dog (⩾2 pets) households were more likely to be S. aureus positive showed no significant association (P = 0·26). Additionally, no significant association was observed between the presence of non-dog pets [cats (P = 0·18), reptiles/amphibians (P = 0·78), small mammals (P = 0·66) and fish (P = 0·76)] and S. aureus/MRSA carriage. As expected, S. aureus/MRSA pulsotypes were shared among family members and between humans and their pets. In fact, in households where both humans and pets were positive, owners and their dog(s) shared the exact same pulsotype in 57·1% (4/7) of the occasions (Fig. 1).
Table 2.
Staphylococcus aureus and MRSA prevalence in dog-owning and non-dog-owning healthy households in the central Ohio region
| Households | Individual subjects | ||||||
|---|---|---|---|---|---|---|---|
| Total N | S. aureus (%)* | MRSA (%) | Total N | S. aureus (%)* | MRSA (%) | ||
| Dog-owning | 80 | 55 (68·8)† | 8 (10·0)‡ | Adult | 155 | 47 (30·3) | 5 (3·2) |
| Child | 129 | 54 (41·9) | 7 (5·4) | ||||
| People | 284 | 101 (35·6) | 12 (4·2) | ||||
| Dog | 113 | 8 (7·1) | 0 (0·0) | ||||
| Non-dog-owning | 55 | 30 (54·5)† | 2 (3·6)‡ | Adult | 107 | 34 (31·8) | 1 (0·9) |
| Child | 108 | 30 (27·8) | 1 (0·9) | ||||
| People | 215 | 64 (29·8) | 2 (0·9) | ||||
| Dog | − | − | − | ||||
| Overall | 135 | 85 (63·0) | 10 (7·4) | People | 499 | 165 (33·1) | 14 (2·8) |
MRSA, Methicillin-resistant S. aureus; MSSA, methicillin-susceptible S. aureus.
* This category includes both MSSA and MRSA.
† A household was considered positive when at least one member (human or dog) was positive for S. aureus (either MSSA or MRSA).
‡ A household was considered positive when at least one member was positive for MRSA.
Fig. 1.
Distribution of Staphylococcus aureus and MRSA pulsotypes between owners and their pets in dog-owning households in the central Ohio region. Each S. aureus (SA)/MRSA pulsotype is represented with a unique geometrical symbol above the individual (owner or dog) colonized with it. Numbers in parentheses represent the number of households. + Denotes positive household or individual (owner and/or dog). – Denotes negative household. * Positive individuals (two or more owners) within the same household, colonized with the same SA/MRSA pulsotype. † Positive individuals (one or more owners) within the same household, each colonized with a different SA/MRSA pulsotype. ‡ Positive individuals (one or more owners and one dog) within the same household, colonized with the same SA/MRSA pulsotype. § Positive individuals (one or more owners and one dog) within the same household, each colonized with a different SA/MRSA pulsotype.
From the 109 S. aureus-positive individuals (humans and pets) belonging to dog-owning households, one human subject was positive for both MSSA and MRSA. Therefore, 110 isolates [98 methicillin-susceptible S. aureus (MSSA) and 12 MRSA] were obtained and further analysed. Their genotypic characteristics are described in Table 3. The majority of the methicillin-susceptible isolates were classified as USA200 (32/98, 32·7%), USA600 (15/98, 15·3%) and USA900 (14/98, 14·3%), while most of the methicillin-resistant isolates were characterized as USA300 (5/12, 41·7%) and USA100 (3/12, 25·0%). In the case of MRSA strains, three isolates (3/12, 25·0%) were classified as hospital-acquired strains (SCCmec type II/USA100); the rest (9/12, 75·0%) were classified as community-acquired [SCCmec type IV/USA300 (five isolates), and one isolate each for type IV/USA800, type IV/Iberian, type IV/not typable, type V/USA700]. Further genotypic characteristics (dendrogram analysis) are discussed below.
Table 3.
Molecular epidemiological characteristics of Staphylococcus aureus isolates from family members (owners and pets) in dog-owning (n = 110*) and non-dog-owning households (n = 64) in the central Ohio region
| MSSA isolates | MRSA isolates | |||||
|---|---|---|---|---|---|---|
| Type | n | % | Type | n | % | |
| Dog-owning households | USA200 | 32 | 32·7 | USA300 | 5 | 41·7 |
| USA600 | 15 | 15·3 | USA100 | 3 | 25·0 | |
| USA900 | 14 | 14·3 | USA700 | 1 | 8·3 | |
| USA100 | 6 | 6·1 | USA800 | 1 | 8·3 | |
| USA300 | 6 | 6·1 | Iberian | 1 | 8·3 | |
| Other types† | 25 | 25·5 | Not typable | 1‡ | 8·3 | |
| Sub-total | 98 | 100·0 | 12 | 100·0 | ||
| Non-dog-owning households | USA200 | 22 | 35·5 | USA100 | 1 | 50·0 |
| USA600 | 6 | 9·7 | USA300 | 1 | 50·0 | |
| USA100 | 4 | 6·5 | ||||
| USA300 | 3 | 4·8 | ||||
| USA900 | 3 | 4·8 | ||||
| Other types§ | 24 | 38·7 | ||||
| Sub-total | 62 | 100·0 | 2 | 100·0 | ||
MSSA, Methicillin-susceptible S. aureus; MRSA, methicillin-resistant S. aureus.
* Two isolates (one MRSA and one MSSA) were obtained from one particular subject. Therefore, we obtained 110 isolates from 109 subjects (owners and dogs).
† Includes USA700 (n = 3 isolates), EMRSA15 (3), Iberian (1), USA1200 (1), novel type (1), group C (1) and not typable (15, which did not match any USA type from the database used during the analysis)
‡ Isolates that did not match any USA type from the database used during the analysis
§ Includes USA1200 (n = 3 isolates), novel type (3), USA700 (2), Iberian (1), EMRSA15 (1), and not typable (14, which did not match any USA type from the database used during the analysis).
Phenotypically, 14 distinct antimicrobial susceptibility profiles were identified (seven for MSSA and seven for MRSA isolates). In the case of methicillin-susceptible profiles, 71·4% (5/7) were resistant to ampicillin and erythromycin, 57·1% (4/7) were resistant to clindamycin (all inducible) and, 14·3% (1/7) were resistant to enrofloxacin, ciprofloxacin and tetracycline. For methicillin-resistant profiles, besides the expected beta-lactam resistance, 42·9% (3/7) were resistant to erythromycin and ciprofloxacin, 28·6% (2/7) were resistant to enrofloxacin, and 14·3% (1/7) were resistant to clindamycin (all inducible). Only one phenotypic profile (represented by the three hospital-acquired MRSA isolates) was classified as multidrug resistant. All S. aureus isolates were susceptible to gentamicin, amikacin, doxycycline, sulfamethoxazole trimethoprim, chloramphenicol, and vancomycin.
Non-dog-owning households/individuals
One or more family members were found positive for S. aureus in 54·5% (30/55, representing 64 positive individuals) of the non-dog-owning households, with only 3·6% of them positive to MRSA (2/55) (Table 2). With regard to individuals, 29·8% (64/215) were positive to S. aureus and 0·9% (2/215) to MRSA (Table 2). The presence of other non-dog pets within the household was not associated with S. aureus/MRSA carriage in human family members [cats (P = 0·87), reptiles/amphibians (P = 0·85), small mammals (P = 0·66) and fish (P = 0·88)] in this type of household. Forty percent (12/30) of the non-dog-owning households shared pulsotypes between family members (6/12 between adults, 3/12 between children and, 6/12 between adults and children). There was a significant difference between households owning dogs vs. those that do not in regards to S. aureus and MRSA colonization; however, such epidemiological analysis will be discussed elsewhere (data not published).
Only one S. aureus isolate was obtained from each positive subject; therefore, 64 isolates (62 MSSA and two MRSA) were further analysed (Table 3). The majority of the methicillin-susceptible strains were characterized as USA200 (22/62, 35·5%), USA600 (6/62, 9·7%) and USA100 (4/62, 6·5%). The two methicillin-resistant strains, one was classified as a community-acquired strain (SCCmec type IV/USA300) and one was a hospital-acquired strain (type II/USA100). Further genotypic characteristics (dendrogram analysis) are discussed below.
Phenotypically, nine antimicrobial susceptibility profiles were identified (seven for MSSA isolates and two for MRSA isolates). In the case of methicillin-susceptible profiles, 57·1% (4/7) were resistant to ampicillin and erythromycin, 42·9% (3/7) were resistant to clindamycin (all inducible), 28·6% (2/7) were resistant to tetracycline and, 14·3% (1/7) were resistant to doxycycline. For methicillin-resistant profiles, besides the expected resistance to beta-lactam, 100·0% (2/2) were resistant to erythromycin and, 50·0% (1/2) were resistant to ciprofloxacin and clindamycin (constitutive resistance). Only one phenotypic profile (represented by the hospital-acquired MRSA isolate) was classified as multidrug resistant. All S. aureus isolates were susceptible to enrofloxacin, gentamicin, amikacin, tetracycline, doxycycline, sulfamethoxazole trimethoprim, chloramphenicol and, vancomycin. The overwhelming majority of the isolates present within the central Ohio region were characterized as pansusceptible, and no significant difference was observed between dog-owning and non-dog-owning households in regards to antimicrobial resistance.
Molecular and diversity analyses: comparison between dog-owning and non-dog-owning households
Molecular analysis of a dendrogram constructed with 110 isolates from dog-owning and 64 isolates from non-dog-owning households showed 108 different pulsotypes (P1–P108) compiled into several small groups (19 clusters). Most pulsotypes were represented by 1–4 isolates with the exception of P7 (n = 7 isolates), P14 (6), P16 (6), P22 (5), P32 (6), and P60 (5). The most prevalent strains were USA200 (31·0%, 54/174 isolates), USA600 (12·1%, 21/174) and USA900 (9·8%, 17/174). Detailed phenotypic and genotypic characteristics present in all S.aureus isolates collected during the study period are described in Tables 4 and 5. No significant difference was observed between the proportion of strains (USA types) present in dog-owning and non-dog-owning households.
Table 4.
Phenotypic and genotypic characteristics of methicillin-susceptible Staphylococcus aureus isolates obtained from dog-owning and non-dog-owning healthy households in the central Ohio region
| Genotyping | Phenotyping | Frequency | |
|---|---|---|---|
| USA type | SCCmec | ||
| Human isolates (n = 152) | |||
| USA200 (n = 54) | n.a. | Amp | 31 |
| AmpEryCliInd | 11 | ||
| S | 6 | ||
| EryCliInd | 5 | ||
| AmpEnoCipEryCliInd | 1 | ||
| USA600 (n = 18) | n.a. | Amp | 13 |
| S | 3 | ||
| AmpEryCliInd | 2 | ||
| USA900 (n = 15) | n.a. | Amp | 14 |
| S | 1 | ||
| USA100 (n = 9) | n.a. | AmpEryCliInd | 3 |
| Amp | 2 | ||
| AmpTetDox | 2 | ||
| Ery | 1 | ||
| S | 1 | ||
| USA300 (n = 9) | n.a. | Amp | 4 |
| AmpEry | 2 | ||
| AmpEryCliInd | 2 | ||
| AmpEryTetCliInd | 1 | ||
| USA700 (n = 5) | n.a. | Amp | 4 |
| AmpEryTetCliInd | 1 | ||
| USA1200 (n = 4) | n.a. | AmpEryCliInd | 2 |
| Amp | 1 | ||
| AmpEryTetCliInd | 1 | ||
| EMRSA15 (n = 4) | n.a. | Amp | 4 |
| Novel type (n = 4) | n.a. | Amp | 4 |
| Iberian (n = 2) | n.a. | Amp | 1 |
| AmpEryCliInd | 1 | ||
| Not typable (n = 28) | n.a. | Amp | 12 |
| S | 10 | ||
| Ery | 3 | ||
| AmpEry | 1 | ||
| AmpEryCliInd | 1 | ||
| EryCliInd | 1 | ||
| Dog isolates (n = 8) | |||
| USA600 (n = 3) | n.a. | Amp | 2 |
| AmpEryCliInd | 1 | ||
| USA900 (n = 2) | n.a. | Amp | 2 |
| USA100 (n = 1) | n.a. | Amp | 1 |
| Group C (n = 1) | n.a. | S | 1 |
| Not typable (n = 1) | n.a. | S | 1 |
Amp, Ampicillin; Cli, clindamycin; CliInd, inducible clindamycin; Cip, ciprofloxacin; Dox, doxycycline; Ery, erythromycin; S, pansusceptible; Tet, tetracycline; n.a., not applied.
Table 5.
Phenotypic and genotypic characteristics of methicillin-resistant Staphylococcus aureus isolates obtained from dog-owning and non-dog-owning healthy households in the central Ohio region
| Genotyping | Phenotyping | Frequency | |
|---|---|---|---|
| USA type | SCCmec | ||
| Human isolates (n = 14) | |||
| USA300 (n = 6) | IV | AmpAmcOxaCpdEry | 3 |
| AmpOxaCpdEry | 2 | ||
| AmpOxaCpdEnoCip | 1 | ||
| USA100 (n = 4) | II | AmpOxaCpdEnoCipEryClinInd | 3 |
| AmpOxaCpdCipEryCli | 1 | ||
| USA700 (n = 1) | V | AmpOxaCpd | 1 |
| USA800 (n = 1) | IV | AmpAmcOxaCpd | 1 |
| Iberian (n = 1) | IV | AmpAmcOxaCpd | 1 |
| Not typable (n = 1) | IV | AmpAmcOxaCpdCip | 1 |
| Dog isolates (n = 0) | |||
Amp, Ampicillin; Amc, amoxicillin with clavunate; Cli, clindamycin; CliInd, inducible clindamycin; Cip, ciprofloxacin; Cpd, cefpodoxime; Ery, erythromycin; Oxa, oxacillin.
When compared by type of household, 75 pulsotypes were associated with dog-owning houses, while 44 pulsotypes were found in non-dog-owning houses. Only 10·2% (11/108) of the pulsotypes were observed in both types of households. These shared pulsotypes were P7 (USA200), P13 (USA200), P14 (USA200), P15 (USA200), P16 (USA200), P18 (USA200), P22 (USA200), P32 (USA600), P50 (Iberian), P65 (USA300, only MRSA shared pulsotype) and, P85 (not typable). In the case of dog-owning households, no significant difference was observed in the number of pulsotypes present in single-dog households (ranging from 0 to 3 pulsotypes, and one house with five pulsotypes) compared to multi-dog (⩾2 pets, 0–3 pulsotypes) households.
In regards to diversity, Simpson's index of diversity in isolates from dog-owning and non-dog-owning households was 0·99 for each group, indicating a high diversity of isolates in both type of households.
Owner–pet interactions between members of dog-owning households
To determine if owners and their dogs had concurrent S. aureus/MRSA carriage by a particular pulsotype due to a particular owner–pet interaction, sixty variables (Table 1) describing such interactions were evaluated in the S. aureus/MRSA-positive dog-owning households (n = 55). Due to the small numbers in this data subset, only 45 variables were eligible for univariable model calculations. Of those, four variables (do you wash your hands after interacting with your pet?; when at home, is the dog restricted to the basement?; when not at home, is the dog restricted to other areas?; and how often does your dog jump when interacting with family members?) showed P values of < 0·2 and were included in the multivariable model, where none were significantly associated. Therefore, no particular human–pet interaction was associated with simultaneous owner–dog colonization with the same S. aureus/MRSA pulsotype.
DISCUSSION
After sampling 135 households within the central Ohio region, it is evident that S. aureus and MRSA are indeed circulating in the majority of households (63%) in this community, where the overall carriage rate of this population (33·1% for S. aureus and 2·8% for MRSA) was similar to that reported in the United States (28–30% S. aureus, 1·8–2% MRSA) [33]. Other studies performed in healthy individuals in other communities have also shown a similar prevalence (18–42% S. aureus, 0–6% MRSA) [7, 19–22]. Overall, households with dogs had a higher S. aureus/MRSA carriage rate, both at the household level (68·8% dog-owning vs. 54·5% non-dog-owning) and at the individual subject level, especially in the case of MRSA (4·2% vs. 0·9%, respectively). In this sample, 7·1% of dogs were positive for S. aureus but none of them carried MRSA at the time of sampling. Previous studies have reported a wide range of prevalence of MRSA in dogs, 1–14% and 0–4% for S. aureus and MRSA, respectively [7, 19–22].
Similarly to what has been reported in the Midwest region and other parts of the United States, most of the S. aureus/MRSA human isolates obtained in this study were susceptible to doxycycline, gentamicin, tetracycline, and sulfamethoxazole trimethoprim [34, 35]. Likewise, even though differences were observed in percentages of resistance to ampicillin, ciprofloxacin, clindamycin and erythromycin, a significant proportion (in some cases the majority) of the isolates described in this and other studies still showed a resistant phenotype to this group of antibiotics [34, 35]. Unfortunately, due to the low prevalence and minor significance of MSSA colonization in dogs, information describing antimicrobial susceptibility patterns in this animal species is mostly limited to methicillin-resistant strains. For this reason, as well as the low number of canine isolates obtained in this study, an accurate comparison with other studies cannot be made.
Considering prior reports of S. aureus strain(s) similarity within a particular animal species [10–12] (i.e. dogs are frequently associated to USA100, which are considered hospital-acquired MRSA strains) [22–25], it was expected that dog-owning households would have less diverse populations of S. aureus/MRSA strains than non-dog-owning households. However, when comparing the results from both types of houses, this study found that strains commonly described in the United States [22, 36] were not only present in both groups, but also similarly distributed (Table 3). In particular, USA200 and USA600 have been described as the most prevalent MSSA strains present the Midwest region (including Ohio), and USA100 and USA300 as the most prevalent MRSA strains within the same area [34, 37]. Since there was not a predominant strain in dog-owning households, it was concluded that dogs, at least in this community, do not appear to play a major role in establishing, maintaining, or favouring the presence of specific S. aureus/MRSA strains within healthy homes.
When analysing at the pulsotype level, Simpson's diversity index revealed that both types of households were highly and equally diverse (index of 0·99 for each). In short, even though there was no significant difference in the proportions of S. aureus/MRSA strains present in dog-owning vs. non-dog-owning houses, the pulsotypes present on each group were very different. In fact, only 10·2% (11/108) of the pulsotypes obtained during the study were found in both groups of households. Unfortunately, based on the information gathered in this study, it is unclear if this effect is completely or partially due to the high variety of pulsotypes that appears to be present in the targeted community. Or, it might be due to other confounding factors such as household routine (including cleaning), geographical location, presence of other animal species at home, human and animal demographics/interactions no recorded in this survey, among others.
In regards to zoonotic transmission, in 85·5% of the dog-owning households, in which the owners tested positive, the pet(s) were negative at time of sampling. This suggests that zoonotic transmission (within healthy households) and persistent colonization with S. aureus/MRSA in dogs is infrequent. However, in households where humans and animals were positive, owners and pets frequently (57%) shared the same S. aureus pulsotype, showing that indeed zoonotic transmission occurs even in healthy households. Comparable results were described in studies performed in Missouri (USA) [22] and Canada [7], with owners and pets sharing the same S. aureus pulsotype in 59·1% and 50·0% of the observations, respectively. However, lower (23·5%) and higher (80·0%) rates have been described in Spain [21] and China [19]. Additionally, scenarios in which dogs carried S. aureus without sharing it with their owners, or were dogs carried a different pulsotype and/or strain from those isolated in their owners, were also observed. The authors believe that the interchange of pulsotypes in these households is dynamic and future studies should be conducted longitudinally to identify if the scenarios described above continuously occur and cycle in the same home (people positive-dog negative, people negative-dog positive, all positive, etc.). Regardless, based on the current data obtained in this study, dogs are indeed involved in the ecology and circulation of S. aureus in the household, but they do not appear to be the main drivers introducing and spreading this pathogen in healthy homes.
Statistical analyses showed that none of the variables describing the owner–pet interactions were associated with simultaneous carriage of the same S. aureus pulsotype in human and animal family members, or with the predominance of a strain within a household. Similar results were found in a study performed in healthy households in China, where the authors reported that no higher risk of carriage was observed between owners that admitted frequent close contact with their animals (including kissing, allowing face licking, and sleeping on their bed) and those who did not [19]. Nevertheless, an association could be found with other risk factors (e.g. general household data, human/animal demographics, human/animal health factors, among others) that were outside the scope of this manuscript.
Last, this study has limitations that need to be taken into consideration. First, due to study design (cross-sectional), long-term colonization and directionality of bacterial transmission could not be assessed, even though we were able to see the sharing of common pulsotypes. It is also important to recognize that differences among communities in the United States and other countries may reduce the ability to extrapolate the results presented in this manuscript. In addition, since very few households had humans and pets test positive for the same pulsotype, the evaluation of risk factors associated with owner-pet interactions was limited. Finally, other variables (e.g. household cleaning routine, general hygiene practices of family members, among others) should be assessed in future studies as plausible factors that can explain the presence of certain strains/pulsotype in the household. Regardless, this is the first study that evaluates the influence that dogs may have in the diversity of S. aureus/MRSA strains circulating in households, and could be used as a reference if the same effect is observed in other communities.
In conclusion, this study showed that several S.aureus/MRSA strains were present and similarly distributed in both, dog-owning and non-dog-owning healthy households. Therefore, it appears that the presence of a pet does not play a major role in establishing, maintaining or promoting the proliferation of a particular S. aureus/MRSA strain within healthy households in this community. However, a very different set of pulsotypes was observed in each household group. The role that dogs might have played in this selection remains uncertain, especially since no risk factors were associated with concurrent carriage of the same pulsotype in owners and pets. Future studies will be needed in other communities, regions and/or countries to determine if indeed dogs contribute in the ‘selective pressure’ of S. aureus/MRSA strains and/or pulsotypes at the household level.
ACKNOWLEDGEMENTS
The authors thank students Rhiannon Marcello, Cory Berliner, and Madeline Anna for their aid during the sampling period, and Dr Dimitria Mathys who assisted with data analysis. The Center for Disease Control and Prevention facilitated the S. aureus database for PFGE characterization. Dr Herminia de Lencastre, from the Universidade Nova de Lisboa in Portugal provided MRSA controls. The Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) program provided control strains. NARSA is supported under NIAID, NIH contract number HHSN272200700055C. We acknowledge Kevin Grandfield for editing and reading this manuscript. Assistance with study design and recruitment was provided by The Ohio State University Center for Clinical and Translational Science (NIH UL1TR001070). The project described was supported by grant number 1R03HD070633-01 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development and Mars-WALTHAM®. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Centers for Disease Control or the Eunice Kennedy Shriver National Institute of Child Health & Human Development, the National Institutes of Health, or Mars-WALTHAM®.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.American Veterinary Medical Association U.S. Pet Ownership Statistics (https://www.avma.org/KB/Resources/Statistics/Pages/Market-research-statistics-US-pet-ownership.aspx). Accessed 24 October 2016.
- 2.O'Haire M. Companion animals and human health: benefits, challenges, and the road ahead. Journal of Veterinary Behavior: Clinical Applications and Research 2010; 5: 226–234. [Google Scholar]
- 3.McConnell A, et al. Friends with benefits: on the positive consequences of pet ownership. Journal of Personality and Social Psychology 2011; 101: 1239–1252. [DOI] [PubMed] [Google Scholar]
- 4.Guardabassi L, Schwarz S, Lloyd D. Pet animals as reservoirs of antimicrobial-resistant bacteria. Journal of Antimicrobial Chemotherapy 2004; 54: 321–332. [DOI] [PubMed] [Google Scholar]
- 5.Faires M, Tater K, Weese J. An investigation of methicillin-resistant Staphylococcus aureus colonization in people and pets in the same household with an infected person or infected pet. Journal of the American Veterinary Medical Association 2009; 235: 540–543. [DOI] [PubMed] [Google Scholar]
- 6.Weese JS, van Duijkeren E. Methicillin-resistant Staphylococcus aureus and Staphylococcus pseudintermedius in veterinary medicine. Veterinary Microbiology 2010; 140: 418–429. [DOI] [PubMed] [Google Scholar]
- 7.Hanselman B, et al. Coagulase positive staphylococcal colonization of humans and their household pets. The Canadian Veterinary Journal 2009; 50: 954–958. [PMC free article] [PubMed] [Google Scholar]
- 8.Paterson G, et al. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nature Communications 2015; 6: 6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrison E, et al. A shared population of epidemic methicillin-resistant Staphylococcus aureus 15 circulates in humans and companion animals. mBio 2014; 5: e00985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herron-Olson L, et al. Molecular correlates of host specialization in Staphylococcus aureus. PLoS ONE 2007; 2: e1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pantosti A. Methicillin-resistant Staphylococcus aureus associated with animals and its relevance to human health. Frontiers in Microbiology 2012; 3: 127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarthy A, Lindsay J, Loeffler A. Are all meticillin-resistant Staphylococcus aureus (MRSA) equal in all hosts? Epidemiological and genetic comparison between animal and human MRSA. Veterinary Dermatology 2012; 23: 267–275. [DOI] [PubMed] [Google Scholar]
- 13.Weese J, et al. Suspected transmission of methicillin-resistant Staphylococcus aureus between domestic pets and humans in veterinary clinics and in the household. Veterinary Microbiology 2006; 115: 148–155. [DOI] [PubMed] [Google Scholar]
- 14.van Duijkeren E, et al. Human-to-dog transmission of methicillin-resistant Staphylococcus aureus. Emerging Infectious Diseases 2004; 10: 2235–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Duijkeren E, et al. Transmission of a Panton-Valentine leucocidin-positive, methicillin-resistant Staphylococcus aureus strain between humans and a dog. Journal of Clinical Microbiology 2005; 43: 6209–6211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nienhoff U, et al. Transmission of methicillin-resistant Staphylococcus aureus between humans and dogs: two case reports. Journal of Antimicrobial Chemotherapy 2009; 64: 660–662. [DOI] [PubMed] [Google Scholar]
- 17.Manian F. Asymptomatic nasal carriage of mupirocin-resistant, methicillin-resistant Staphylococcus aureus (MRSA) in a pet dog associated with MRSA infection in household contacts. Clinical Infectious Diseases 2003; 36: 26–28. [DOI] [PubMed] [Google Scholar]
- 18.Morris D, et al. Potential for pet animals to harbour methicillin-resistant Staphylococcus aureus when residing with human MRSA patients. Zoonoses and Public Health 2012; 59: 286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boost M, O'Donoghue M, James A. Investigation of the role of dogs as reservoirs of Staphylococcus aureus and the transmission of strains between pet owners and their dogs. Hong Kong Medical Journal 2008; 14: 15–18. [PubMed] [Google Scholar]
- 20.Walther B, et al. Sharing more than friendship – nasal colonization with coagulase-positive staphylococci (CPS) and co-habitation aspects of dogs and their owners. PLoS ONE 2012; 7: e35197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gómez-Sanz E, et al. High diversity of Staphylococcus aureus and Staphylococcus pseudintermedius lineages and toxigenic traits in healthy pet-owning household members. Underestimating normal household contact? Comparative Immunology, Microbiology and Infectious Diseases 2013; 36: 83–94. [DOI] [PubMed] [Google Scholar]
- 22.Kottler S, et al. Prevalence of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus carriage in three populations. Journal of Veterinary Internal Medicine 2010; 24: 132–139. [DOI] [PubMed] [Google Scholar]
- 23.Hoet A, et al. Epidemiological profiling of MRSA positive dogs arriving at a veterinary teaching hospital. Vector Borne and Zoonotic Diseases 2013; 13: 385–393. [DOI] [PubMed] [Google Scholar]
- 24.Loeffler A, et al. Prevalence of methicillin-resistant Staphylococcus aureus among staff and pets in a small animal referral hospital in the UK. Journal of Antimicrobial Chemotherapy 2005; 56: 692–697. [DOI] [PubMed] [Google Scholar]
- 25.Morris D, et al. The prevalence of carriage of meticillin-resistant staphylococci by veterinary dermatology practice staff and their respective pets. Veterinary Dermatology 2010; 21: 400–407. [DOI] [PubMed] [Google Scholar]
- 26.Van Balen J, et al. Presence, distribution and molecular epidemiology of methicillin-resistant Staphylococcus aureus in a small animal teaching hospital: a year-long active surveillance targeting dogs and their environment. Vector Borne and Zoonotic Diseases 2013; 13: 299–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clinical & Laboratory Standards Institute. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated From Animals; Approved Standard. M31-A3, Third Edition. Wayne, Pennsylvania, USA: Clinical and Laboratory Standards Institute (CLSI), 2008, pp. 1–100. [Google Scholar]
- 28.Fiebelkorn KR, et al. Practical disk diffusion method for detection of inducible clindamycin resistance in Staphylococcus aureus and coagulase-negative staphylococci. Journal of Clinical Microbiology 2003; 41: 4740–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrobial Agents and Chemotherapy 2007; 51: 3374–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Centers for Disease Control and Prevention/Pulse-Net Oxacillin-resistant Staphylococcus aureus on PulseNet (OPN). Laboratory protocols for molecular typing of S. aureus by pulsed-field gel electrophoresis (PFGE) (http://www.cdc.gov/mrsa/pdf/ar_mras_PFGE_s_aureus.pdf) Accessed 24 October 2016.
- 31.Murchan S, et al. Harmonization of pulsed-field gel electrophoresis protocols for epidemiological typing of strains of methicillin-resistant Staphylococcus aureus: a single approach developed by consensus in 10 European laboratories and its application for tracing the spread of related strains. Journal of Clinical Microbiology 2003; 41: 1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gardener M. Community Ecology: Analytical Methods using R and Excel, 1st edn. Exeter: Pelagic Publishing Ltd, 2014, p. 425. [Google Scholar]
- 33.Centers for Disease Control and Prevention. Methicillin-resistant Staphylococcus aureus (MRSA) infections (https://www.cdc.gov/mrsa/). Accessed 24 October 2016.
- 34.Davis S, et al. Epidemiology and outcomes of community-associated methicillin-resistant Staphylococcus aureus infection. Journal of Clinical Microbiology 2007; 45: 1705–1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vento T, et al. Staphylococcus aureus colonization of healthy military service members in the United States and Afghanistan. BMC Infectious Diseases 2013; 13: 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tenover F, et al. Characterization of nasal and blood culture isolates of methicillin-resistant Staphylococcus aureus from patients in United States Hospitals. Antimicrobial Agents and Chemotherapy 2012; 56: 1324–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carrel M, Perencevich E, David M. USA300 methicillin-resistant Staphylococcus aureus, United States, 2000–2013. Emerging Infectious Diseases 2015; 21: 1973–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]