SUMMARY
Human parainfluenza virus (HPIV) infections are one of the commonest causes of upper and lower respiratory tract infections. In order to determine if there have been any recent changes in HPIV epidemiology in England and Wales, laboratory surveillance data between 1998 and 2013 were analysed. The UK national laboratory surveillance database, LabBase, and the newly established laboratory-based virological surveillance system, the Respiratory DataMart System (RDMS), were used. Descriptive analysis was performed to examine the distribution of cases by year, age, sex and serotype, and to examine the overall temporal trend using the χ2 test. A random-effects model was also employed to model the number of cases. Sixty-eight per cent of all HPIV detections were due to HPIV type 3 (HPIV-3). HPIV-3 infections were detected all year round but peaked annually between March and June. HPIV-1 and HPIV-2 circulated at lower levels accounting for 20% and 8%, respectively, peaking during the last quarter of the year with a biennial cycle. HPIV-4 was detected in smaller numbers, accounting for only 4% and also mainly observed in the last quarter of the year. However, in recent years, HPIV-4 detection has been reported much more commonly with an increase from 0% in 1998 to 3·7% in 2013. Although an overall higher proportion of HPIV infection was reported in infants (43·0%), a long-term decreasing trend in proportion in infants was observed. An increase was also observed in older age groups. Continuous surveillance will be important in tracking any future changes.
Key words: England and Wales, epidemiology, laboratory surveillance, parainfluenza, seasonality, temporal trend
INTRODUCTION
Human parainfluenza viruses (HPIVs) are among the commonest causes of upper and lower respiratory tract infections, especially in young children [1–3]. Some literature has reported that in children aged <5 years, HPIV infections were the second most common cause of acute respiratory illness resulting in hospitalization, surpassed only by respiratory syncytial virus but ahead of influenza viruses [4]. Immunity to HPIV is incomplete, and infection and reinfection occur throughout life [5, 6]. There are four currently recognized HPIV types (1–4), with all causing a full spectrum of respiratory illness, including the common cold, upper respiratory tract illness (URTI), croup, and severe lower respiratory tract illness (LRTI), such as bronchiolitis and pneumonia [2]. Studies suggest that 2–17% of hospitalizations for acute respiratory infections in children aged <5 years are due to HPIV infection [7–9]. HPIV-3 is the most virulent of the HPIVs and is associated with significant morbidity and mortality [10]. A modelling study based on all hospitalizations for lower respiratory tract infection in children aged <5 years in England between 1995 and 1998 estimated that 12·9% of unspecified pneumonia hospitalizations were due to parainfluenza infections and 12·7% of unspecified bronchiolitis hospitalizations [11]. A study found that HPIV infections accounted for 6·8% of all hospitalizations for fever, acute respiratory illness, or both in children aged <5 years [12]. Among adults, most HPIV infections cause mild disease manifesting as upper respiratory tract symptoms [2]. Adult patients more frequently present with influenza-like symptoms compared to children [13]. HPIV infections, however, also cause more severe disease. One study [14] reported that at least 2% of cases of community-acquired pneumonia requiring hospitalization in adults were associated with an increase in HPIV antibodies. HPIV infections have also been identified as a cause of nosocomial outbreaks involving paediatric, geriatric and bone marrow transplant wards [15–18]. In addition, HPIV infections can cause outbreaks of acute respiratory illness in institutionalized elderly people [19, 20].
Specific antiviral therapy of proven value for HPIV infection is not yet established although immunosuppressed patients have been given oral ribavirin for treatment which showed that use of ribavirin early and at higher doses might improve the outcome [3, 21]. Despite decades of research, there are currently no licensed vaccines available to protect against infection caused by any of the HPIV types [4, 10]. Prevention thus relies on more traditional non-pharmaceutical respiratory infection control and prevention packages such as isolation, cohorting, hand hygiene, cough etiquette and face masks.
The last detailed surveillance report [22] on HPIV in England and Wales was published in 1999 based on the routine laboratory infection reports between 1975 and 1997, which reported that HPIV-3 was the main HPIV type with a clear annual peak occurring between late spring and summer. The seasonality and circulation for other types of HPIV were also described. Since then no further in depth publication has been reported. In recent years, routine clinical surveillance data [23] have shown secular declining trends in consultation rates in primary-care setting due to respiratory infections such as influenza-like illness, acute bronchitis, pneumonia, upper and lower respiratory tract infections in England and Wales. Therefore, this analysis of laboratory surveillance data on HPIV infection has been undertaken to examine the recent situation in relation to HPIV and determine if there have been any changes in the clinical and epidemiological characteristics of HPIV infections between 1998 and 2013 in England and Wales.
METHODS
The LabBase database
All parainfluenza infection reports from England and Wales between specimen weeks 1 (1998) and 52 (2013) were extracted from the Public Health England (PHE) national laboratory reporting scheme, the LabBase database [24]. This database collects routine laboratory clinical test results of all microorganism detections from public health and non-public health laboratories throughout England, Wales and Northern Ireland. All laboratories were required to submit their data to the system as soon as and as often as they could. Indeed, some laboratories do report their data daily. As LabBase has been replaced by a new surveillance system called the Second Generation Surveillance System (SGSS) since 2014, to avoid any possible inconsistency this change might bring in, only data up to 2013 were included in this study. Each record in the database corresponds to an individual episode of illness and contains demographic and certain clinical information of patients. A unique organism-patient-illness-episode identification (OPIE ID) was assigned to each unique record. This OPIE ID was used to identify any duplicate records defined as infections within a 6-week period in an individual within the study dataset. De-duplication was carried out before the analysis. The following data fields were obtained from the database: earliest specimen date, age, sex, serotype, testing method, specimen type, specimen source and clinical features.
Due to multiple testing methods, different specimen types and specimen sources, and various clinical features were reported in the same patients’ samples; 1826 individual patients in the extracted dataset had multiple records. This in total generated 2715 extra records. These extra records were removed when analysing by age, sex and serotype as there should only be one value for each patient episode. However the additional records for each patient were included in the analyses of testing methods, clinical features and specimen source in order to understand the complete picture.
The Respiratory DataMart System (RDMS)
The recently established sentinel laboratory-based virological surveillance system for multiplex-respiratory virus PCR testing of respiratory samples, the RDMS [25] was also used to extract HPIV test data collected from a network of 14 major regional PHE and local National Health Service (NHS) laboratories in England only. RDMS includes both positive and negative test results and therefore the proportion of positive samples can be calculated. The RDMS for non-influenza respiratory viruses was established from the end of 2009. Data de-duplication was also carried out for this system. The complete dataset between specimen week 1 (2010) to week 52 (2013) was used in this analysis. Patients’ demographic data and test data are collected but no clinical data. Positive HPIV test data from 12 out of the 14 RDMS laboratories were also submitted for inclusion in the PHE LabBase database during the study period (the reasons for those two laboratories not submitting data to LabBase were not clear).
Statistical analaysis
Descriptive analysis was undertaken to examine the case distribution by year, age, sex and serotypes, and to examine the overall trend in proportion of cases in different age groups using χ2 test for trend.
The random-effects model in a Bayesian framework was used to model the number of positive detections according to age group and year. Changes in the sampled laboratories over time were accounted for via a fixed effect for the baseline rate in each laboratory. The model included fixed effects for age groups and age group-specific linear trends. Overall trends were allowed to vary between laboratories via a random effect. Finally, overdispersion (excess variability in observed detections) was accounted for via random effects at two levels: in the overall annual number of detections; and specific laboratory-years. This structure allows an appropriate level of uncertainty in the age-specific trends, given the stochasticity of observed counts at different levels of aggregation. Overall numbers in each age group in the population over time were included as a denominator in the model, allowing for changes in demographics over time, in order to produce incidence rate ratios (IRR) between age groups and age-specific trends. Patients with unknown age and laboratories with fewer than 10 reports over the study period were excluded from these models. The model accounts for the varying number of reporting laboratories and changes in demographics in the population.
RESULTS
Serotype and seasonality
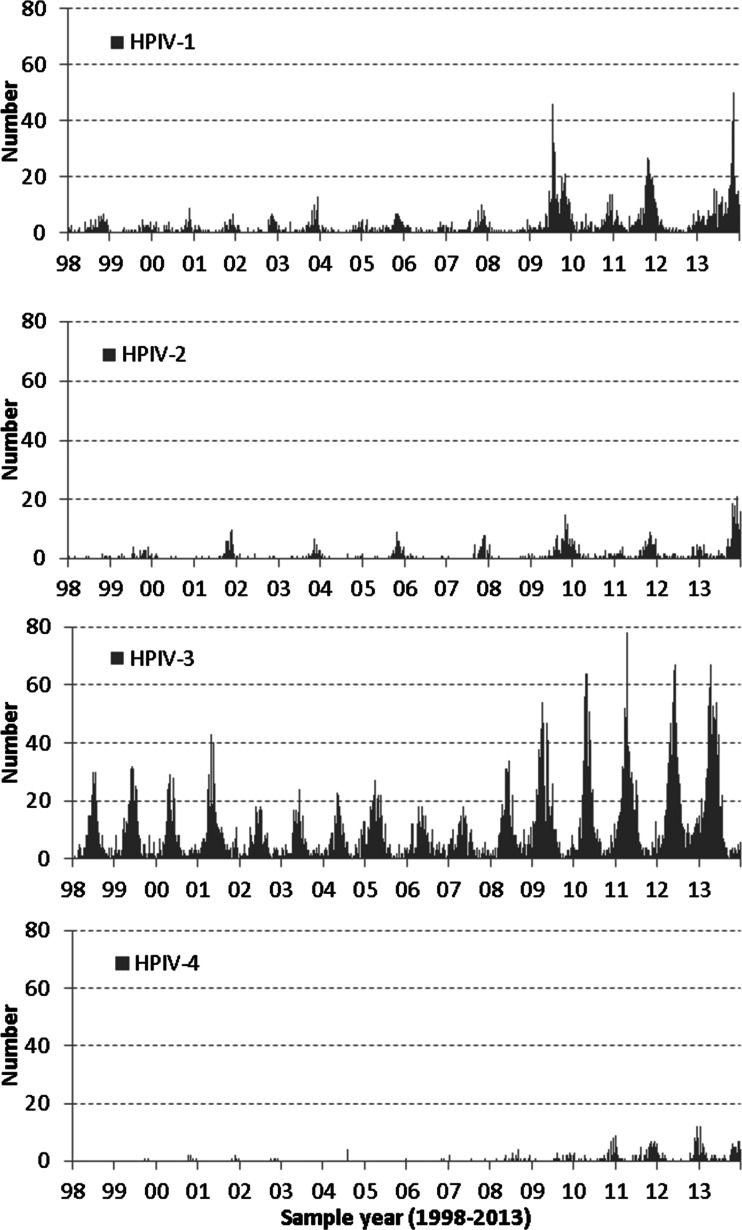
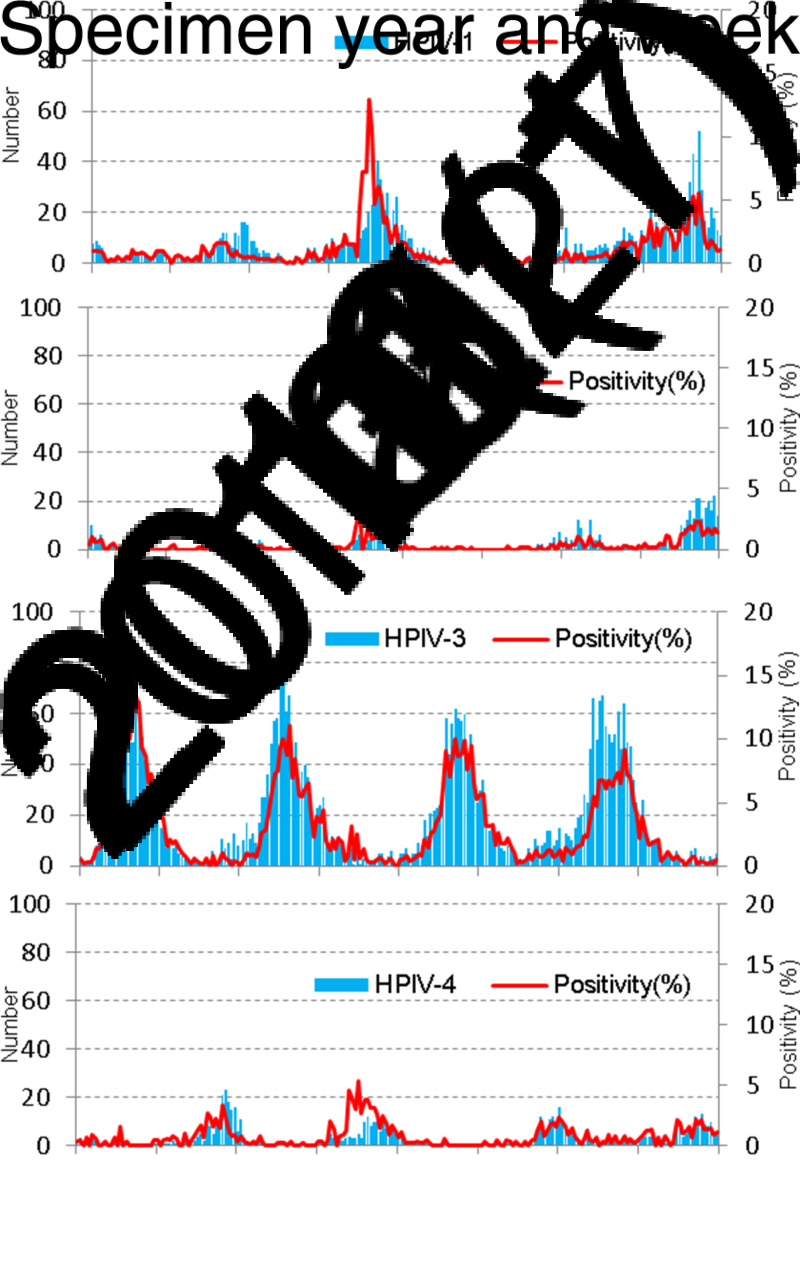
A total of 17 982 unique parainfluenza positive test reports were reported from 158 laboratories in England and Wales over the 16-year study period between specimen week 1 (1998) and week 52 (2013) in the LabBase dataset. HPIV-3 was the major serotype accounting for 68·3% of all positive reports with typing information available (8478/12 420), followed by HPIV-1, 20·3% (2526/12 420), HPIV-2 (7·8%, 963/12 420), and HPIV-4 (3·6%, 453/12 420). The unknown type reports accounted for 30·9% (5562/17 982) of all positive reports. The weekly distribution of HPIV infections by serotype during the study period is shown in Figure 1. HPIV-3 infections have a regular annual seasonal peak activity period between March and June. More detections have been reported for all types of parainfluenza viruses since 2009 when the influenza A(H1N1) pandemic first emerged. HPIV-1 also shows fairly regular biennial seasonality with increased activity usually between October and December in odd-numbered years. HPIV-2 had regular biennial peaks occurring also between October and December in odd-numbered years; HPIV-4 infections were less frequently reported until 2009 and much more commonly reported ever since, usually peaking in the last quarter of the year.
Fig. 1.
Weekly numbers of human parainfluenza virus (HPIV) detection by serotype in England and Wales, 1998–2013 (LabBase System).
Age and sex
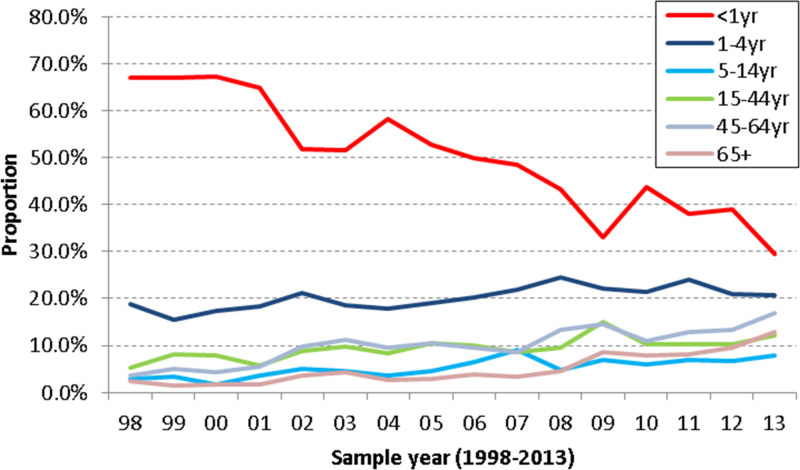
Information on patients’ age was available for 17 717 (98·5%) out of the 17 982 reports in the dataset. Of these HPIV reports with known age, 7620 (43·0%) were from infants aged <1 year, 3715 (21·0%) from children aged 1–4 years, 1089 (6·1%) from ages 5–14 years, 1861 (10·5%) from ages 15–44 years, 2148 (12·1%) from ages 45–64 years and 1284 (7·2%) from patients aged ⩾65 years. The proportions of HPIV infections in different age groups are shown in Figure 2, showing that overall, the most commonly affected age group was children aged <1 year followed by 1- to 4-year-olds. The differences in proportions between the age groups were gradually becoming much smaller over the study period. The proportions from infants aged <1 year have shown a significant long-term decreasing trend (P < 0·001), while the proportions for other age groups show a slow but statistically significant long-term increasing trend: 1–4 years (P < 0·01), 5–14 years (P < 0·001), 15–44 years (P < 0·01), 45–64 years (P < 0·001) and ⩾65 years (P < 0·001). The group with the lowest proportion infected was the 5–14 years age group in recent years.
Fig. 2.
Age group proportions of all types of human parainfluenza virus detection by year in England and Wales, 1998–2013 (LabBase System).
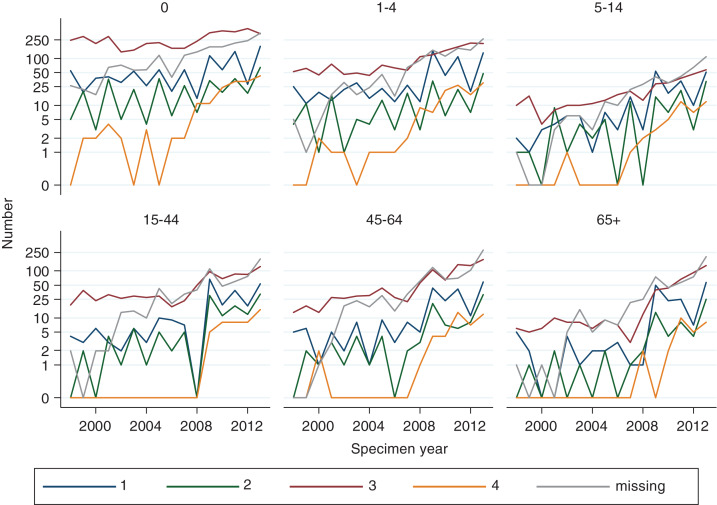
Figure 3 shows the absolute number of HPIV detections in log scale by age group, serotype and specimen year to show their changing trends. The number of detections for all four serotypes increased over time in all age groups, especially HPIV-4, which has shown the fastest increase in recent years. The fastest increase by age in HPIV detections was observed in the ⩾65 years age group for all four serotypes.
Fig. 3.
Human parainfluenza virus detection numbers by age group, serotype (1–4) and specimen year in England and Wales, 1998–2013 (LabBase System).
Sex was known for all but 230 (1·3%) reports. In total, 10 021 cases (55·7%) were male and 7731 (43·0%) female. This slight male dominance was seen in all age groups (especially <1-year-olds) except for the 15–44 and ⩾65 years age groups where similar numbers were found in males and females. The slight male dominance was also observed for all HPIV serotypes except HPIV-4 where a similar number of positive reports were found for males and females.
Laboratory test methods
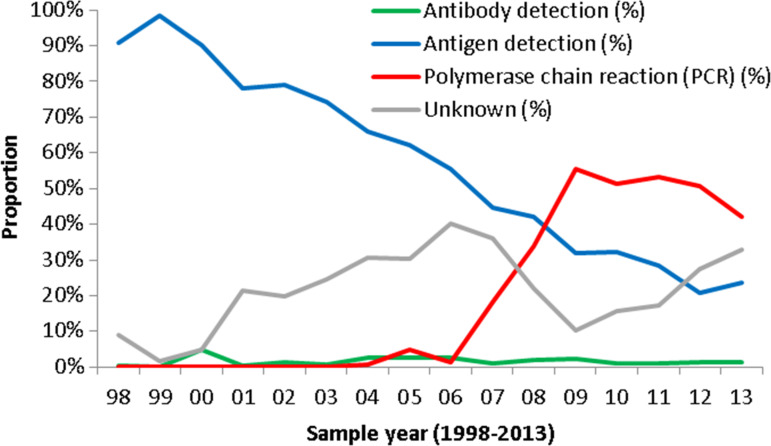
Laboratory testing methods recorded in the database for detecting HPIV infection during the study period included genomic detection (PCR) (34·1%), culture (15·7%), antigen detection (14·5%), light microscopy (11·1%), patient antibody detection (1·6%), electron microscopy (0·7%) and unknown/not recorded (22·3%). The genomic detection (PCR) method was first introduced to detect HPIV in 2004 in England and Wales, and has become the dominant method since 2007. The use of the light microscopy method showed a decreasing trend over the study period while culture and antigen detection methods are still commonly in use. The test methods were grouped into four groups: (a) antibody detection including antibody detection, antibody detection – rising titre and IgM antibody detection; (b) antigen detection including antigen detection, culture, light/electron microscopy, (c) PCR and (d) methods not recorded (unknown). The changing trend in proportion of each test method group within each year is shown in Figure 4. It shows that PCR test increased markedly since 2007 while antigen detection methods decreased continuously; antibody test methods remained at low levels across the study period, probably due to small numbers being tested using these methods.
Fig. 4.
Changing trend in testing method proportion each year in England and Wales, 1998–2013 (LabBase System).
Clinical features
Clinical features were only recorded in a small proportion of patients (7·1%) in the database during the study period. Among those where clinical feature information was available, 237/1460 (16·2%) were recorded as having respiratory illnesses (bronchiolitis, pneumonia or croup) but the majority of the available data on clinical feature were recorded as ‘Outbreak/Cluster’, 1065/1460 (72·9%).
Setting of sampling
In the analysis on settings of sampling, information was available for 54·8% of patients. Most specimens were taken from hospital inpatients (81·9%), followed by hospital outpatients (10·9%); only 2·8% were reported as from the community patients through their family doctors (2·8%).
The random-effects model for incidence rates showed that, compared to the 15–44 years age group (reference group), the HPIV IRR in the <1 year age group was 261 times higher, the 1–4 years age group was 20 times higher, the 5–14 and 45–64 years age groups were 1·8 and 1·6 times higher, respectively, while for those aged ⩾65 years there was no significant difference (Table 1). The model also indicates an overall significant increasing trend of cases reported over time, particularly in older age groups while no statistically significant increase was observed in the <1 year age group (Table 2). The model accounts for the varying number of reporting laboratories and changes in demographics in the population.
Table 1.
Incidence rate ratio of parainfluenza infection by age group, England and Wales, 1998–2013
| Age group | IRR | 95% CI | P |
|---|---|---|---|
| <1 | 260·90 | 229·10–297·20 | <0·001 |
| 1–4 | 20·02 | 17·45–22·94 | <0·001 |
| 5–14 | 1·76 | 1·48–2·07 | <0·001 |
| 15–44 | 1 (ref.) | ||
| 45–64 | 1·58 | 1·35–1·84 | <0·001 |
| ⩾65 | 1·11 | 0·92–1·33 | 0·132 |
IRR, Incidence rate ratio; CI, confidence interval.
Table 2.
Age-specific trend (annual rate of change) of parainfluenza infection by age group, England and Wales, 1998–2013
| Age group | IRR | 95% CI | P |
|---|---|---|---|
| <1 | 1·03 | 0·99–1·07 | 0·066 |
| 1–4 | 1·10 | 1·06–1·15 | <0·001 |
| 5–14 | 1·20 | 1·14–1·25 | <0·001 |
| 15–44 | 1·15 | 1·10–1·20 | <0·001 |
| 45–64 | 1·19 | 1·13–1·24 | <0·001 |
| ⩾65 | 1·27 | 1·21–1·33 | <0·001 |
IRR, Incidence rate ratio; CI, confidence interval.
Data from RDMS
The weekly positive numbers and weekly proportion positive (%) by HPIV type extracted from RDMS over the period between 2010 and 2013 are presented in Figure 5. It demonstrates that HPIV-3 was the major circulating type with regular peaks between spring and early summer each year. The peaks and patterns of numbers of positive detection and proportion positive were closely matched for HPIV-3. Both HPIV-1 and HPIV-2 mainly circulated in odd numbered years. HPIV-4 peaked every year towards the end of year between October and December. The pattern of proportion positives generally matched well with number of detections.
Fig. 5.
Weekly positive number of detection and weekly proportion positive (%) by human parainfluenza virus (HPIV) type from the Respiratory DataMart System in England, 2010–2013.
DISCUSSION
This report describes the epidemiological and clinical characteristics of parainfluenza infections based on laboratory surveillance data in England and Wales in the period 1998 and 2013. First, the main epidemiological features of parainfluenza virus infection in this study period have remained generally similar in terms of sex, serotype and seasonal pattern as reported in the previous study between 1975 and 1997 [22]. Second, there has been an age shift observed, with a decline in the proportion of the total positive reports in the <1-year-olds, with a concomitant increase in all other age groups, especially those aged ⩾65 years. Third, there has been an increase in HPIV-4 infections in recent years, although HPIV-3 remains the major type of infection (68·3%) with peaks occurring between March and June each year.
In many respects the epidemiology in this current analysis is similar to previous studies [2, 9, 13, 22, 26–32], with HPIV-3 being the most common type of HPIV infection with regular annual peaks between spring and summer while other types of HPIV mainly occur between autumn and winter, with the most vulnerable population for HPIV infection remaining in young children aged <5 years with a slight male dominance. However, comparing our results with those in an earlier US report [2], we found that although the seasonality of HPIV-3 infections was similar, our data found varied biennial peaks for HPIV-1 infections, mainly in odd-numbered years between 2003 and 2013 but the biennial peaks were not clearly presented between 1998 and 2002 while the US data showed clear and consistent biennial peaks in odd-numbered years for HPIV-1. The biennial peaks for HPIV-2 infections were also clearly present in our data in odd-numbered years in the autumn as in the US data. The pattern of biennial peaks in HPIV-1 was also observed in other US studies [3, 7, 9, 33, 34], whereas HPIV-2 was seen in even-numbered years in other studies [27, 34] in Japan and the United States. The reasons for these geographical differences in circulation of HPIV types is unclear, but highlights the importance of ensuring local studies are undertaken to fully understand the epidemiology.
We found evidence of a long-term shift in the age distribution over the study period suggesting a relative decline for the most commonly affected population of HPIV infection in the <1-year-olds while the proportions for all the other age groups increased, especially in the elderly population, although the absolute number of infections continued to increase across all age groups to varying degrees (Fig. 3). This means that although the <1 year age group remains the most commonly affected population with HPIV infection, it is becoming more common to detect HPIV infections in older age groups, with the ⩾65 years age group having the fastest increase in detections in recent years. Such a shift in the age distribution has not been reported by other studies to the best of our knowledge. The decrease in infants aged <1 year might be due to, in our speculation, less chance of exposure in recent years (less sibling exposure) or more maternal historic infection due to passive antibody protection but many other factors might also play a role in this phenomenon. Our LabBase database only collects positive test results from routine laboratory tests. The increased detection of HPIV infection in older age groups could also be due to a range of factors, such as more laboratories testing for HPIV; more HPIV circulating or HPIV infection becoming more severe. The increased availability of multiplex respiratory virus testing kits in laboratories has made the HPIV test more readily available for any suspected cases, and so it is likely that the test no longer focused on young children. Indeed previous studies suggest that HPIV infection in adults is often mild [2, 13] and the highest HPIV hospitalization rate is in children aged <6 months [8, 12]. Therefore the more likely explanations for the increase in older age groups might be the increased availability of multiplex test kits and more laboratory testing since the 2009 influenza pandemic.
Our data showed a significant increase in HPIV-4 detection in recent years with annual peaks between autumn and winter (Figs 1 and 3). This increasing trend has, to the best of our knowledge, not been reported previously in the UK. Specific HPIV-4 testing is often not available in diagnostic laboratories hence the data are too sparse to reveal any pattern [2, 33]. However, a recent 4-year study showed that in China HPIV-4 infection is not a rare event and it is even slightly more common than HPIV-2 infection [26]. Moreover, outbreaks caused by HPIV-4 had been reported previously [35]. It is worth noting that even before the start of the 2009 pandemic in the UK, parainfluenza virus had already shown varied degrees of increased activities since autumn 2007 and activity levels have remained higher since then (Figs 1 and 4). This increased pattern was also seen for other respiratory viruses such as rhinovirus and adenovirus (data not shown). The reasons for the increase are not completely clear but are likely related to the increased testing and reporting ability of laboratories and increased availability of multiplex respiratory virus testing kits in laboratories. Clinicians are required to make decisions around which patients to test, for which pathogens, based upon patients’ presenting symptoms and clinical needs. However, the testing of patients when they present with symptoms of URTI, LRTI, bronchitis or croup is most commonly undertaken for not one, but a set of common respiratory viruses including HPIV based upon the particular multiple panel in that laboratory.
Clinical features were only available in a very small proportion of patients (7·1%) in our data during the study period. The majority of the available recordings on clinical features were ‘Outbreak/Cluster’ (72·9%), where it is not possible to further clarify what kind of outbreaks/clusters they might be or other underlying conditions or diseases the patients might have. Previous outbreak reports of HPIV-3 infections [18, 36] have shown that HPIV can be introduced from the community to vulnerable hospitalized patients, e.g. adult bone marrow transplant unit or haematopoietic stem cell transplant patients in adult haematology unit, resulting in nosocomial transmission with severe complications and even death. This highlights the importance of strong respiratory virus surveillance and of the rapid implementation of standard respiratory infection control and prevention measures particularly for these high-risk patients and the staff responsible for them to minimize the risk of onward transmission.
There are several limitations of the PHE LabBase database. Firstly the database only contains positive testing results. However, since the 2009 influenza pandemic, a new laboratory surveillance system, the RDMS, for influenza and other respiratory viruses has been established in England [25], which collects both positive and negative test results. This means that the proportion of positive samples (positivity) can be calculated. Clear annual seasonality of HPIV infection was shown in the RDMS using both indicators of the proportion positive and the number of positive detections. The patterns of proportion positive and number of positive detections were reassuringly generally matched well except the positivity for HPIV-1 and HPIV-4 which were peaked earlier than the actual positive numbers during the 2011/2012 season (Fig. 5). We believe that positivity from RDMS is a more appropriate indicator or proxy for infection rate reflecting infection intensity or incidence rate in the population than the actual absolute positive numbers which would be influenced by the total number of samples tested as more testing would result in more positive results. Secondly minimal clinical and outcome information is held within both the LabBase and RDMS databases, which prevented us from giving full description or inferences on other clinical characteristics and impact of HPIV infections. Because all samples from both primary and secondary healthcare settings were included in both of the LabBase and RDMS databases, although more samples come from secondary healthcare settings due to clinical needs and based on our earlier work [25], we do not believe that this difference would have biased our results too much on the overall national epidemiological characteristics of the HPIV infections.
The most important limitation for our study using passive, laboratory-based surveillance data may be the changes of testing methods and testing patterns across the study years. The PCR method has become a more common method reported into LabBase since 2007, which could affect whether infants, young children, and older adults are tested more often or not, and certainly will have impact on the relative proportions of detections across the age spectrum over time. Therefore cautions are needed when explaining the long-term trend of the pathogen detections. Changes in medical care seeking behaviour may also affect laboratory-based surveillance systems in countries where medical care is not provided free of charge. However, in the UK people are provided with medical care free of charge at the point of care under the NHS. Therefore the changes in medical care-seeking behaviour may not have much significant impact to our results.
In summary, our results show strong evidence that it is important to ensure a strengthened national laboratory surveillance system is set up to monitor the changes for commonly circulated respiratory viruses to provide insights into seasonality, temporal trends along with other epidemiological characteristics regarding age, sex, serotypes as well as clinical features of HPIV infections. The proportion of HPIV infection in infants aged <1 year has dropped significantly over the study period, whereas HPIV infections in other age groups are increasing. Consistent annual seasonality of HPIV-3 and biennial seasonality for HPIV-1 and HPIV-2 are also important feature of HPIV infection. The more commonly detected infections with HPIV-4 need careful and continuous monitoring. Therefore, based on this study, the clinical and public health implications are: (a) when dealing with patients presenting with respiratory symptoms, be aware of possible parainfluenza infection as a potential differential diagnosis. HPIV-3 is the main serotype circulating during the spring-summer period while other serotypes (HPIV-1, HPIV-2, HPIV-4) circulate at low levels during the autumn-winter period; (b) although HPIV infection is mainly observed in the <5-year-olds, older people in the ⩾65 years age group are also commonly affected; (c) no vaccine or antiviral medicines with proven benefit for HPIV are available at present. Therefore the main medical management would be supportive, and prevention and control measures relying on standard respiratory hygiene measures. In conclusion, ongoing surveillance will be invaluable in monitoring the temporal trends, estimating disease burden and providing differential diagnosis between influenza infections and other infections, and evaluating impact of any future development on treatment and prevention measures.
ACKNOWLEDGEMENTS
We are very grateful to laboratories for their continuous support in submitting their test data to LabBase (now called SGSS) and RDMS systems.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.CDC. National Center for Infectious Disease – The Respiratory and Enteric Viruses Branch (http://www.cdc.gov/ncidod/dvrd/revb/index.htm). 2007.
- 2.Fry AM, et al. Seasonal trends of human parainfluenza viral infections: United States, 1990–2004. Clinical Infectious Diseases 2006; 43: 1016–1022. [DOI] [PubMed] [Google Scholar]
- 3.Hall CB. Respiratory syncytial virus and parainfluenza virus. New England Journal of Medicine 2001; 344: 1917–1928. [DOI] [PubMed] [Google Scholar]
- 4.Schmidt AC, et al. Progress in the development of human parainfluenza virus vaccines. Expert Review of Respiratory Medicine 2011; 5: 515–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Glezen WP, et al. Parainfluenza virus type 3: seasonality and risk of infection and reinfection in young children. Journal of Infectious Diseases 1984; 150: 851–857. [DOI] [PubMed] [Google Scholar]
- 6.Henrickson KJ. Parainfluenza viruses. Clinical Microbiology Reviews 2003; 16: 242–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Counihan ME, et al. Human parainfluenza virus-associated hospitalizations among children less than five years of age in the United States. Pediatric Infectious Disease Journal 2001; 20: 646–653. [DOI] [PubMed] [Google Scholar]
- 8.Iwane MK, et al. Population-based surveillance for hospitalizations associated with respiratory syncytial virus, influenza virus, and parainfluenza viruses among young children. Pediatrics 2004; 113: 1758–1764. [DOI] [PubMed] [Google Scholar]
- 9.Reed G, et al. Epidemiology and clinical impact of parainfluenza virus infections in otherwise healthy infants and young children <5 years old. Journal of Infectious Diseases 1997; 175: 807–813. [DOI] [PubMed] [Google Scholar]
- 10.Senchi K. Development of oligomannose-coated liposome-based nasal vaccine against human parainfluenza virus type 3. Frontiers in Microbiology 2013; 4: 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Muller-Pebody B, et al. Contribution of RSV to bronchiolitis and pneumonia-associated hospitalizations in English children, April 1995-March 1998. Epidemiology and Infection 2002; 129: 99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinberg GA, et al. Parainfluenza virus infection of young children: estimates of the population-based burden of hospitalization. Journal of Pediatrics 2009; 154: 694–699. [DOI] [PubMed] [Google Scholar]
- 13.Liu WK, et al. Epidemiology and clinical presentation of the four human parainfluenza virus types. BMC Infectious Diseases 2013; 13: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marx A, et al. Parainfluenza virus infection among adults hospitalized for lower respiratory tract infection. Clinical Infectious Diseases 1999; 29: 134–140. [DOI] [PubMed] [Google Scholar]
- 15.Glasgow KW, Tamblyn SE, Blair G. A respiratory outbreak due to parainfluenza virus type 3 in a home for the aged – Ontario. Canada Communicable Disease Report 1995; 21: 57–61. [PubMed] [Google Scholar]
- 16.Karron RA, et al. Molecular epidemiology of a parainfluenza type 3 virus outbreak on a pediatric ward. Journal of Infectious Diseases 1993; 167: 1441–1445. [DOI] [PubMed] [Google Scholar]
- 17.Wendt CH, et al. Parainfluenza virus respiratory infection after bone marrow transplantation. New England Journal of Medicine 1992; 326: 921–926. [DOI] [PubMed] [Google Scholar]
- 18.Zambon MC, et al. Molecular epidemiology of two consecutive outbreaks of parainfluenza 3 in a bone marrow transplant unit. Journal of Clinical Microbiology 1998; 36: 2289–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention. Parainfluenza outbreaks in extended-care facilities – United States. Morbidity and Mortality Weekly Report 1978; 27: 475. [Google Scholar]
- 20.Centre PHLSDS. Parainfluenza infections in the elderly 1976–82. British Medical Journal (Clinical Research Edition) 1983; 287: 1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohenthal U, et al. Parainfluenza virus type 3 infections in a hematology unit. Bone Marrow Transplantation 2001; 27: 295–300. [DOI] [PubMed] [Google Scholar]
- 22.Laurichesse H, et al. Epidemiological features of parainfluenza virus infections: laboratory surveillance in England and Wales, 1975–1997. European Journal of Epidemiology 1999; 15: 475–484. [DOI] [PubMed] [Google Scholar]
- 23.RCGP Research & Surveillance Centre. Weekly Returns Service, Annual Report 2011.
- 24.Grant AD, Eke B. Application of information technology to the laboratory reporting of communicable disease in England and Wales. Communicable Disease Report. CDR Review 1993; 3: R75–R78. [PubMed] [Google Scholar]
- 25.Zhao H, et al. A new laboratory-based surveillance system (Respiratory DataMart System) for influenza and other respiratory viruses in England: results and experience from 2009 to 2012. Eurosurveillance 2014; 19(3). [DOI] [PubMed] [Google Scholar]
- 26.Shi W, et al. Prevalence of human parainfluenza virus in patients with acute respiratory tract infections in Beijing, 2011–2014. Influenza and Other Respiratory Viruses 2015; 9: 305–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mizuta K, et al. Epidemiology of parainfluenza virus types 1, 2 and 3 infections based on virus isolation between 2002 and 2011 in Yamagata, Japan. Microbiology and Immunology 2012; 56: 855–858. [DOI] [PubMed] [Google Scholar]
- 28.De Silva LM, Cloonan MJ. Brief report: parainfluenza virus type 3 infections: findings in Sydney and some observations on variations in seasonality world-wide. Journal of Medical Virology 1991; 35: 19–21. [DOI] [PubMed] [Google Scholar]
- 29.Easton AJ, Eglin RP. Epidemiology of parainfluenza virus type 3 in England and Wales over a ten-year period. Epidemiology and Infection 1989; 102: 531–535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Glezen P, Denny FW. Epidemiology of acute lower respiratory disease in children. New England Journal of Medicine 1973; 288: 498–505. [DOI] [PubMed] [Google Scholar]
- 31.Glezen WP, et al. Epidemiologic patterns of acute lower respiratory disease of children in a pediatric practice. Journal of Paediatrics 1971; 78: 397–406. [DOI] [PubMed] [Google Scholar]
- 32.Marx A, et al. Pediatric hospitalizations for croup (laryngotracheobronchitis): biennial increases associated with human parainfluenza virus 1 epidemics. Journal of Infectious Diseases 1997; 176: 1423–1427. [DOI] [PubMed] [Google Scholar]
- 33.Abedi GR, et al. Estimates of parainfluenza virus-associated hospitalizations and cost among children aged less than 5 years in the United States, 1998–2010. Journal of the Pediatric Infectious Diseases Society. Published online: 2 June 2014. doi: 10.1093/jpids/piu047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Knott AM, Long CE, Hall CB. Parainfluenza viral infections in pediatric outpatients: seasonal patterns and clinical characteristics. Pediatric Infectious Diseases Journal 1994; 13: 269–273. [DOI] [PubMed] [Google Scholar]
- 35.Lau SK, et al. Human parainfluenza virus 4 outbreak and the role of diagnostic tests. Journal of Clinical Microbiology 2005; 43: 4515–4521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jalal H, et al. Molecular investigations of an outbreak of parainfluenza virus type 3 and respiratory syncytial virus infections in a hematology unit. Journal of Clinical Microbiology 2007; 45: 1690–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]