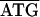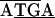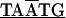Abstract
Psyllids, like aphids, feed on plant phloem sap and are obligately associated with prokaryotic endosymbionts acquired through vertical transmission from an ancestral infection. We have sequenced 37 kb of DNA of the genome of Carsonella ruddii, the endosymbiont of psyllids, and found that it has a number of unusual properties revealing a more extreme case of degeneration than was previously reported from studies of eubacterial genomes, including that of the aphid endosymbiont Buchnera aphidicola. Among the unusual properties are an exceptionally low guanine-plus-cytosine content (19.9%), almost complete absence of intergenic spaces, operon fusion, and lack of the usual promoter sequences upstream of 16S rDNA. These features suggest the synthesis of long mRNAs and translational coupling. The most extreme instances of base compositional bias occur in the genes encoding proteins that have less highly conserved amino acid sequences; the guanine-plus-cytosine content of some protein-coding sequences is as low as 10%. The shift in base composition has a large effect on proteins: in polypeptides of C. ruddii, half of the residues consist of five amino acids with codons low in guanine plus cytosine. Furthermore, the proteins of C. ruddii are reduced in size, with an average of about 9% fewer amino acids than in homologous proteins of related bacteria. These observations suggest that the C. ruddii genome is not subject to constraints that limit the evolution of other known eubacteria.
The insect suborder Sternorrhyncha (Hemiptera), which includes psyllids, also contains aphids, mealybugs, and whiteflies, all of which feed primarily or exclusively on plant phloem sap (5). All of these insects contain primary endosymbionts corresponding to different bacterial clades (11, 12, 18; P. Baumann, N. A. Moran, and L. Baumann, http://link.springer.de/link /service/books/10125/). The most extensively studied endosymbionts are those from aphids (3, 10; Baumann et al., website). The clade constituting these endosymbionts has been given the designation Buchnera aphidicola, and phylogenies based on several molecules indicate that the closest free-living relatives are members of the Enterobacteriaceae. Previously we have sequenced about 130 kb of DNA from B. aphidicola of the aphid Schizaphis graminum (Baumann et al., website), and recently the complete sequence of the 640-kb genome of the endosymbiont of the aphid Acyrthosiphon pisum has been determined (24). These genetic studies indicate that in B. aphidicola there has been a major reduction in the gene content with a retention of the genes necessary for a variety of housekeeping functions as well as the synthesis of essential amino acids and riboflavin (3, 24; Baumann et al., website). The biosynthetic functions of B. aphidicola are also supported by numerous nutritional studies (10; Baumann et al., website).
Psyllids, or “jumping plant lice” (Hemiptera: Psyllidae), differ from aphids in many aspects of life history and biogeography but are similar in that they feed on phloem sap (5, 14) and possess maternally transmitted endosymbionts (6, 11, 26; Baumann et al., website). Phloem sap of most plants is deficient in essential amino acids (23), suggesting that B. aphidicola and the endosymbionts of psyllids have the same functions related to host nutrition. The primary endosymbionts of psyllids, designated Carsonella ruddii, constitute a unique lineage within the γ3 subdivision of the Proteobacteria (11, 26, 28). They are located within host cells called bacteriocytes, where they are enclosed by host-derived membrane vesicles; the multicellular structure containing the bacteriocytes is called a bacteriome (6, 7, 11, 31). In a recent study of 32 psyllid species, the phylogenetic tree derived from the 16S-23S rDNA of C. ruddii agreed with the tree derived from a host gene, a result consistent with a single infection of a psyllid ancestor and subsequent vertical transmission (cospeciation) of endosymbionts and hosts (28). Some psyllid species also contain morphologically diverse secondary (S) endosymbionts (6, 11, 26); molecular phylogenetic analyses indicate that S-endosymbionts result from multiple infections of hosts and possible horizontal transmission among them (29). Since C. ruddii appears to be present in all psyllids and the S-endosymbionts are absent in about one-third of the species (28), C. ruddii alone must be able to fulfill all of the necessary functions of the endosymbiotic association.
The 16S and 23S rDNAs of C. ruddii have G+C contents lower than those of any other known bacteria (11, 26, 28). In addition, the 3′ end of C. ruddii 16S rDNA lacks a sequence complementary to the ribosomal binding site (RBS) of mRNA (Shine-Dalgarno sequence) (28). These observations suggested that the genome of C. ruddii may also have unusual properties and led to the present study, in which we describe the results of a sequence analysis of 37 kb of C. ruddii DNA.
MATERIALS AND METHODS
General methods.
Standard molecular biology methods were used in this study (2, 22). Additional methods have been described in our past publications and include the isolation of total psyllid DNA, restriction enzyme and Southern blot analyses, and cloning into λZAP (Strategene, La Jolla, Calif.) (28; Baumann et al., website). The nucleotide sequence of C. ruddii DNA was determined at the University of Arizona (Tucson) LMSE sequencing facility. Besides T3 and T7 primers, custom-made oligonucleotide primers were designed for sequencing. For sequence determination of some DNA fragments, a double-stranded DNA nested deletion kit (Pharmacia, Picataway, N.J.) was used.
Most of the sequence data involved the psyllids Pachypsylla venusta and P. celtidis; additional psyllids used were Acizzia uncatoides, Bactericera cockerelli, Ctenarytaina eucalypti, C. longicauda, C. spatulata, Calophya schini, Heteropsylla cubana, and Trioza eugenia. P. venusta and P. celtidis contained only C. ruddii and lacked the S-endosymbiont (28).
General approach.
In our previous studies on Buchnera aphidicola (endosymbiont of aphids), we developed oligonucleotide primers based on conserved protein sequences, amplified the DNAs by PCR, cloned the fragments into pBluescript (Stratagene), and subsequently determined their nucleotide sequence. These DNA fragments were used as probes for restriction enzyme and Southern blot analyses of endosymbiont DNA. Based on these results, appropriate restriction enzyme-digested DNA fragments were cloned into λZAP (Stratagene) and sequenced (3). Initially this approach was suitable for the cloning of a 4.9-kb DNA fragment from C. ruddii-P. venusta which contained atp genes (Fig. 1B). Subsequent attempts to clone a portion of the 16S rDNA and the region upstream, as well as a portion of the 23S rDNA and the region downstream (Fig. 1C), using these methods failed due to instability of the recombinants or insertion of Escherichia coli sequences. Consequently, we used two approaches to generate sets of overlapping 1.5- to 3.8-kb fragments. In the first approach, we selected proteins that are highly conserved and tend to be clustered in single regulatory units (Fig. 1) (21). Synthetic oligonucleotide primers were designed based on conserved amino acid sequences of homologous proteins. These primers also contained, at the 5′ ends, sequences for restriction enzymes. Following amplification by PCR, the DNA was digested with the appropriate restriction enzymes and cloned into pBluescript (Stratagene) or pWSK130 (a low-copy vector) (33), and the nucleotide sequence was determined. The sequences of the oligonucleotide primers and their positions on the sequenced DNA fragments will be made available on request. In some cases we were not able to extend the sequence using this approach and recourse was made to unidirectional PCR.
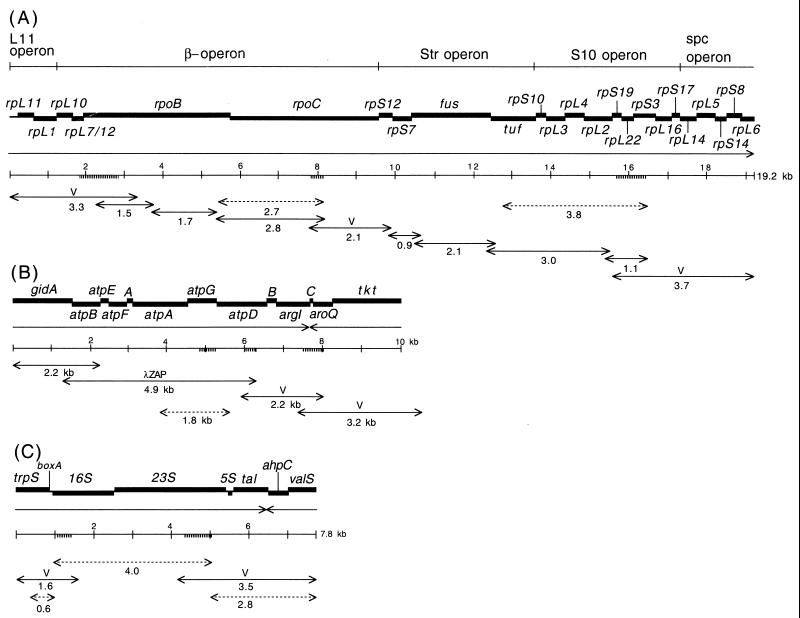
FIG. 1.
Genetic maps of DNA fragments from C. ruddii. (A) The 19.2-kb DNA fragment of C. ruddii-P. venusta; (B) the 10.0-kb DNA fragment of C. ruddii-P. venusta; (C) the 7.8-kb DNA fragment of C. ruddii-P. celtidis. Thick lines, structural genes; thick striped lines, positions of probes used for restriction enzyme and Southern blot analyses to obtain fragments for Vectorette II unidirectional PCR; single-headed arrows, direction of transcription; double-headed arrows with V on top, fragments obtained with Vectorette II unidirectional PCR; double-headed arrows, overlapping DNA fragments obtained by PCR; double-headed dashed arrows, DNA fragments also obtained from endosymbionts of other psyllid species.
Unidirectional PCR.
We used the Vectorette II system (Sigma-Genosys, St. Louis, Mo.) and the instructions provided by the manufacturer. From the nucleotide sequence of endosymbiont DNA fragments, we obtained a useable restriction enzyme site (close to the end which was to be extended) and the sequence for two oligonucleotide primers from which a probe was amplified by PCR. Restriction enzyme and Southern blot analyses were performed using this probe, the restriction enzyme that digests the site in the known sequence, and additional restriction enzymes. In most cases 1.6- to 3.5-kb DNA fragments were selected. The endosymbiont-psyllid DNA was digested with a combination of two restriction enzymes, and the appropriate-sized fragment was eluted from agarose gels. The ends were filled in with the Klenow fragment (2), and the DNA fragments were blunt-end ligated to phosphorylated BamHI linkers (New England Biolabs, Beverly, Mass.) and subsequently digested with BamHI. Following removal of the linkers, the fragments were ligated to BamHI-compatible Vectorette II DNA. Using an oligonucleotide primer derived from the known sequence, which also contained a restriction enzyme site(s), and a Vectorette II primer, the DNA was amplified by PCR, digested with the appropriate restriction enzymes, and cloned into either pBluescript or pWSK130. The primers used for unidirectional PCR are available upon request.
PCR conditions.
PCR amplification was performed in a 50-μl volume containing 50 ng of total psyllid DNA, 5 mM MgCl2, 0.2 mM deoxynucleoside triphosphate mix, 1 μM primers, and 2 U of Bio-X-ACT DNA polymerase in Optibuffer (Bioline, London, United Kingdom). PCR products larger than 1 kb were amplified by 30 cycles consisting of a 30-s denaturation at 94°C, a 2-min annealing at 48 to 50°C, and a 5-min elongation at 70°C. For PCR products smaller than 1 kb, the annealing and elongation times were reduced to 30 s and 1 min, respectively.
Analysis of the DNA.
We used GeneJockey II (Biosoft, Ferguson, Mo.) to identify open reading frames (ORFs) and Blast searches (National Center for Biotechnology Information, Bethesda, Md.) for proteins with amino acid sequence similarity. Alignment of amino acids was performed using Gap (Genetics Computer Group, Madison, Wis.). In comparative studies, sequences of B. aphidicola, Richettsia prowazekii, and Escherichia coli were also included (accession numbers AF000398, AJ235269, and U00096, respectively).
Nucleotide sequence accession numbers.
The following are the GenBank accession numbers (in parentheses) for the sequences of the fragments obtained in this study: C. ruddii-P. venusta 19.2 kb (Fig. 1A), AF274444; C. ruddii-C. eucalypti 3.8 kb (Fig. 1A), AF250389; C. ruddii-P. celtidis 3.8 kb (Fig. 1A), AF250390; C. ruddii-P. venusta 10.0 kb (Fig. 1B), AF291051; and C. ruddii-P. celtidis 7.8 kb (Fig. 1C), AF211141. In addition, sequences of rRNA operons (complete or partial) for the following endosymbionts were deposited: C. ruddii-P. venusta, AF211143; C. ruddii-C. eucalypti, AF211133; C. ruddii-A. uncatoides, AF211124; C. ruddii-C. longicauda, AF211134; C. ruddii-C. spatulata, AF211135; C. ruddii-B. cockerelli, AF211126; C. ruddii-C. schini, AF211132; C. ruddii-H. cubana, AF211138; and C. ruddii-T. eugenia, AF211151.
RESULTS
General properties of the C. ruddii DNA.
The three DNA fragments of C. ruddii DNA were 19,209, 10,049, and 7,806 bp in length (total, 37,057 bp) (Fig. 1A, B, and C, respectively). Searches in databases (September 2000) identified 38 ORFs as corresponding to known genes; 37 were represented in the E. coli genome, and 1 (aroQ) was found in Haemophilus influenzae. The total G+C content of the DNA fragments was 19.9 mol%. When the genes coding for rRNA (which have a higher G+C content) were excluded, the G+C content was 18.0 mol%. All genes on the longest fragment (Fig. 1A) are transcribed in one direction. The genes on the other fragments (Fig. 1B and C) are transcribed in both directions.
General properties of the ORFs.
A list of the C. ruddii genes together with the product designations, G+C contents, and percent amino acid identities to E. coli homologs is presented in Table 1. The highest G+C contents are in the 16S and 23S rDNA (35.6 and 33.1%, respectively). The range of G+C contents of protein-coding genes is 9.9 (atpF) to 28.3 (tuf) mol%. A plot of the percent amino acid identity between the C. ruddii and E. coli proteins against the mol% G+C content of the C. ruddii genes (Fig. 2) indicates a correlation between the conservation of the amino acid sequence of a protein and the G+C content of the C. ruddii gene encoding the protein. Less highly conserved protein sequences are encoded by genes with lower G+C contents.
TABLE 1.
Genes found C. ruddiia
| Gene | Protein | G+C content (mol%) | % Amino acid identityb |
|---|---|---|---|
| Pentose phosphate nonoxidative branch | |||
| talc | Transaldolase | 16.4 | 36.1 |
| tkt | Transketolase | 17.6 | 33.2 |
| ATP proton motive force | |||
| atpA | ATP synthase, α subunit | 24.3 | 51.6 |
| atpB | ATP synthase, subunit a | 12.8 | 31.4 |
| atpD | ATP synthase, β subunit | 25.8 | 68.7 |
| atpE | ATP synthase, subunit c | 23.9 | 52.0 |
| atpF | ATP synthase, subunit b | 9.9 | 22.1 |
| atpG | ATP synthase, γ subunit | 13.2 | 33.3 |
| Amino acid biosynthesis | |||
| Glutamate family | |||
| argI | Ornithine transcarbamylase | 13.1 | 28.7 |
| Aromatic amino acid family | |||
| aroQ | 3-Dehydroquinate dehydratase | 14.6 | 33.8d |
| rRNA | |||
| rrf | 5S rRNA | 26.8 | 50.0 |
| rrl | 23S rRNA | 33.1 | 68.7 |
| rrs | 16S rRNA | 35.6 | 73.2 |
| Ribosomal proteins | |||
| rpS3 | Ribosomal protein S3 (rpsC) | 17.5 | 29.5 |
| rpS7 | Ribosomal protein S7 (rpsG) | 18.5 | 30.8 |
| rpS8 | Ribosomal protein S8 (rpsH) | 13.4 | 21.7 |
| rpS10 | Ribosomal protein S10 (rpsJ) | 12.8 | 27.6 |
| rpS12 | Ribosomal protein S12 (rpsL) | 25.1 | 63.3 |
| rpS14 | Ribosomal protein S14 (rpsN) | 14.2 | 31.6 |
| rpS17 | Ribosomal protein S17 (rpsQ) | 14.7 | 35.4 |
| rpS19 | Ribosomal protein S19 (rpsS) | 20.6 | 45.5 |
| rpL1 | Ribosomal protein L1 (rplA) | 11.3 | 23.8 |
| rpL2 | Ribosomal protein L2 (rplB) | 25.4 | 44.6 |
| rpL3 | Ribosomal protein L3 (rplC) | 20.2 | 40.8 |
| rpL4 | Ribosomal protein L4 (rplD) | 14.8 | 29.0 |
| rpL5 | Ribosomal protein L5 (rplE) | 15.0 | 31.5 |
| rpL6e | Ribosomal protein L6 (rplF) | 14.6 | 19.8 |
| rpL7/12 | Ribosomal protein L7/12 (rplL) | 17.0 | 31.3 |
| rpL10 | Ribosomal protein L10 (rplJ) | 11.1 | 13.3 |
| rpL11 | Ribosomal protein L11 (rplK) | 17.5 | 30.7 |
| rpL14 | Ribosomal protein L14 (rplN) | 21.1 | 54.9 |
| rpL16 | Ribosomal protein L16 (rplP) | 21.8 | 40.6 |
| rpL22 | Ribosomal protein L22 (rplV) | 11.4 | 25.0 |
| Amino acid tRNA synthetases | |||
| trpSc,e | Tryptophanyl-tRNA synthetase | 12.2 | 20.3 |
| valSc,e | Valyl-tRNA synthetase | 10.6 | 24.8 |
| DNA replication | |||
| gidAe | Cell division protein | 17.8 | 38.3 |
| Protein translation | |||
| fus | Elongation factor G | 22.2 | 46.9 |
| tuf | Elongation factor Tu | 28.3 | 71.6 |
| RNA synthesis | |||
| rpoB | RNA polymerase, β subunit | 17.1 | 34.3 |
| rpoC | RNA polymerase, β′ subunit | 18.9 | 36.8 |
| Detoxification | |||
| ahpCc | Alkyl hydroxyperoxide reductase | 17.2 | 32.8 |
| Unidentified | |||
| ORF-A | 8.0 | ||
| ORF-B | 11.1 | ||
| ORF-C | 9.7 |
Unless otherwise indicated, all genes are from C. ruddii-P. venusta.
Unless otherwise indicated, all comparisons are to E. coli proteins.
Genes from C. ruddii-P. celtidis.
Comparison to H. influenzae protein.
Partial sequence.
FIG. 2.
Comparisons of the percent amino acid (AA) sequence identity (ID) of homologous proteins of C. ruddii and E. coli with the percent G+C content of the C. ruddii genes.
Amino acid composition and codon usage.
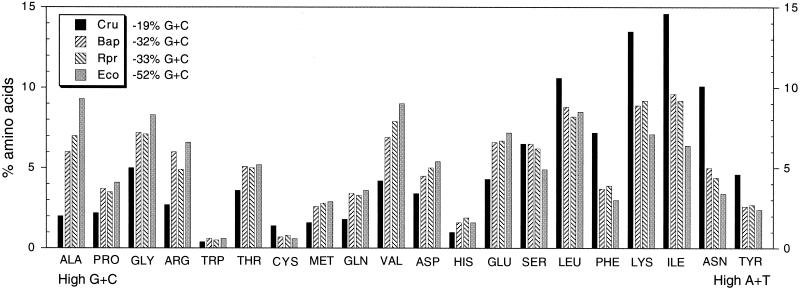
In C. ruddii, the reduction in G+C content has had a major effect on polypeptide sequences, causing an increased frequency of amino acids that utilize high-A+T-containing codons. Although such a shift occurs in the endosymbiont B. aphidicola and the intracellular pathogen R. prowazekii, it is most drastic in C. ruddii, in which biased base composition shows a more extreme effect on polypeptide sequences than in any previously studied bacterium (Fig. 3). Within the 32 polypeptide sequences inferred for C. ruddii that have homologs in B. aphidicola, R. prowazekii, and E. coli, 50% of the residues consist of five amino acids for which the corresponding codons have maximum A and T content (phenylalanine, lysine, isoleucine, asparagine, and tyrosine). In comparison, these amino acids comprise only 29.3% of the amino acids of B. aphidicola, 29.4% of the amino acids of R. prowazekii, and 22.3% of the amino acids of E. coli, for the same genes. This shift also affects the charge of the proteins. The calculated isoelectric points of the combined proteins of C. ruddii, B. aphidicola, R. prowazekii, and E. coli were 10.1, 9.9, 9.6, and 9.2, respectively.
FIG. 3.
Amino acid compositions and the A+T and G+C content of codons of homologous proteins of C. ruddii (Cru), B. aphidicola (Bap), R. prowazekii (Rpr), and E. coli (Eco). The G+C contents are for the total coding regions compared. The genes compared are listed in Fig. 1A and include gidA, atpB, atpE, atpF, atpA, atpG, and atpD (Fig. 1B) and trpS (Fig. 1C).
Intergenic spaces.
An inspection of the putative coding regions of C. ruddii DNA indicates a major reduction or elimination of intergenic spaces. A summary of the nucleotide sequences found between 34 adjacent genes, which are transcribed in the same direction, is presented in Table 2. In 85% of the adjacent genes there are overlapping regions between the stop codons for the upstream genes and the initiating codons for the downstream genes. The most frequent overlap arrangement (44%) is ATGA, in which the last 3 nucleotides (nt) are used as a stop codon for the upstream protein and the first 3 nt are used as the initiating methionine for the downstream protein. In 35% of the cases, the overlapping regions are longer. Even in the 15% of the adjacent gene pairs that contain an intergenic space, it is short (7 nt or less). The elimination of intergenic spaces also results in operon fusion. In E. coli and many other organisms, the genes in Fig. 1A are part of five different transcription units (L11 operon, β operon, Str operon, S10 operon, and spc operon) (13, 16). In C. ruddii, these operons appear to be part of a single transcription unit.
TABLE 2.
Nucleotide sequences between adjacent genesa
Based on comparisons of 34 adjacent genes that are transcribed in the same direction.
Overline, initiating codon; underline, stop codon.
−, nucleotide overlap between start and stop codons; +, space between start and stop codons.
The 3.8-kb tuf-rpS3 DNA fragment designated by dashed double-headed arrows in Fig. 1A was also sequenced from C. ruddii of two additional psyllid species. Comparisons indicate that there is both conservation and variation in some of the protein initiation and termination arrangements. In C. ruddii-P. venusta, C. ruddii-P. celtidis, and C. ruddii-C. eucalypti, the junctions between rpS10-rpL3, rpL3-rpL4, and rpL4-rpL2 were the same (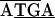 ; the initiating codon is overlined, and the stop codon is underlined). There were differences in the remaining junctions. Between tuf-rpS10 of C. ruddii-P. venusta and C. ruddii-P. celtidis there was
; the initiating codon is overlined, and the stop codon is underlined). There were differences in the remaining junctions. Between tuf-rpS10 of C. ruddii-P. venusta and C. ruddii-P. celtidis there was 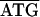 ATTAA, while in C. ruddii-C. eucalypti the sequence was
ATTAA, while in C. ruddii-C. eucalypti the sequence was  . Between rpL2-rpS19 of C. ruddii-P. venusta and C. ruddii-P. celtidis the sequence was
. Between rpL2-rpS19 of C. ruddii-P. venusta and C. ruddii-P. celtidis the sequence was 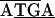 , while in C. ruddii-C. eucalypti the sequence was
, while in C. ruddii-C. eucalypti the sequence was 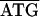 TCTAG. Between rpS19-rpL22 of C. ruddii-P. venusta and C. ruddii-P. celtidis the sequence was
TCTAG. Between rpS19-rpL22 of C. ruddii-P. venusta and C. ruddii-P. celtidis the sequence was 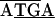 , while in C. ruddii-C. eucalypti the sequence was
, while in C. ruddii-C. eucalypti the sequence was  TTA6TAA. Between rpL22-rpS3 of C. ruddii-P. venusta and C. ruddii-C. eucalypti the sequence was
TTA6TAA. Between rpL22-rpS3 of C. ruddii-P. venusta and C. ruddii-C. eucalypti the sequence was 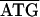 GGA8TTAA, while in C. ruddii-P. celtidis the sequence was
GGA8TTAA, while in C. ruddii-P. celtidis the sequence was 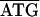 GGTAA. Similarly, the sequence of a 2.8-kb 23S-valS C. ruddii DNA fragment (Fig. 1C) was determined for C. ruddii of three additional psyllid species. In C. ruddii-P. celtidis, C. ruddii-C. eucalypti, C. ruddii-A. uncatoides, and C. ruddii-C. longicauda there was an overlapping region between the initiating ATG of aphC and the stop codon of valS, consisting of 23, 14, 8, and 14 nt, respectively. Previously it was found that the rpoB-rpoC junction (Fig. 1A, 2.7-kb fragment) of 13 strains of C. ruddii involved the sequence
GGTAA. Similarly, the sequence of a 2.8-kb 23S-valS C. ruddii DNA fragment (Fig. 1C) was determined for C. ruddii of three additional psyllid species. In C. ruddii-P. celtidis, C. ruddii-C. eucalypti, C. ruddii-A. uncatoides, and C. ruddii-C. longicauda there was an overlapping region between the initiating ATG of aphC and the stop codon of valS, consisting of 23, 14, 8, and 14 nt, respectively. Previously it was found that the rpoB-rpoC junction (Fig. 1A, 2.7-kb fragment) of 13 strains of C. ruddii involved the sequence 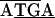 while in the related species C. ruddii-H. cubana and C. ruddii-H. texana, the sequence in the junctions was TAAAAA
while in the related species C. ruddii-H. cubana and C. ruddii-H. texana, the sequence in the junctions was TAAAAA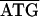 (30). Similarly, in a previous study of the atpAGD region (Fig. 1B, 1.8-kb fragment) from 31 strains of C. ruddii, it was found that in all cases the junction region was
(30). Similarly, in a previous study of the atpAGD region (Fig. 1B, 1.8-kb fragment) from 31 strains of C. ruddii, it was found that in all cases the junction region was 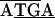 (30).
(30).
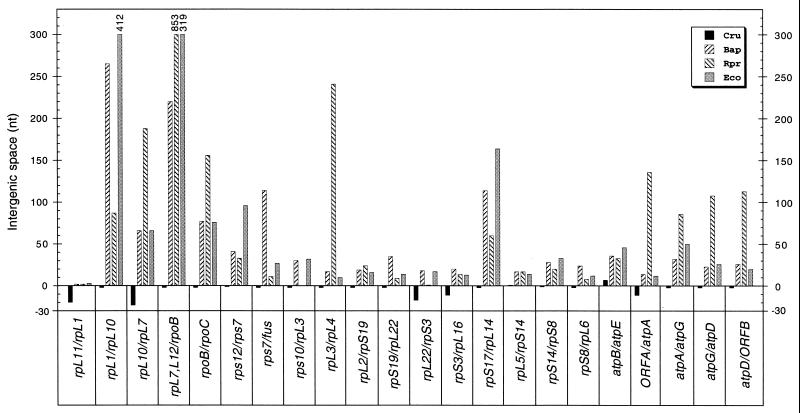
For 22 pairs of genes transcribed in the same direction, the flanking genes are the same in C. ruddii as in B. aphidicola, R. prowazekii, and E. coli, allowing direct comparison of the homologous intergenic space. For every one of these 22 cases, the space between genes in C. ruddii is absent or smaller than in any of these other three organisms (Fig. 4). This indicates that C. ruddii differs from all of these organisms in the elimination or reduction of intergenic spaces throughout the genome.
FIG. 4.
Comparison of the nucleotide length of the intergenic spaces between the same adjacent gene pairs (transcribed in the same direction) of C. ruddii (Cru), B. aphidicola (Bap), R. prowazekii (Rpr), and E. coli (Eco). ORFA and ORFB of Cru are found in the positions corresponding to atpH and atpC, respectively of B. aphidicola, R. prowazekii, and E. coli. Numbers on top of three bars indicate the lengths (in nucleotides) of the intergenic spaces.
The same tendency to minimize sequence length is evident in the two cases in which adjacent genes are transcribed in opposite directions, with end points flanking each other. In argI-ORFC (Fig. 1B) there is an overlap of 26 nt involving the sequence TCAN18TAG, in which the last triplet is the stop codon for argI and the complement of the first triplet is the stop codon of the putative ORFC. In the ahpC-tal region (Fig. 1C) of C. ruddii-P. celtidis, C. ruddii-P. venusta, C. ruddii-C. eucalypti, C. ruddii-C. longicauda, and C. ruddii-C. spatulata, translation termination involves the sequence TTATAA, in which the last triplet is the stop codon for tal while the complement of the first triplet is the stop codon of ahpC. In C. ruddii-A. uncatoides, termination involves the shorter sequence TTAA, in which the last 3 nt are the stop codon for tal while the complement of the first 3 nt (two of which overlap with the stop codon for tal) are the stop codon for ahpC.
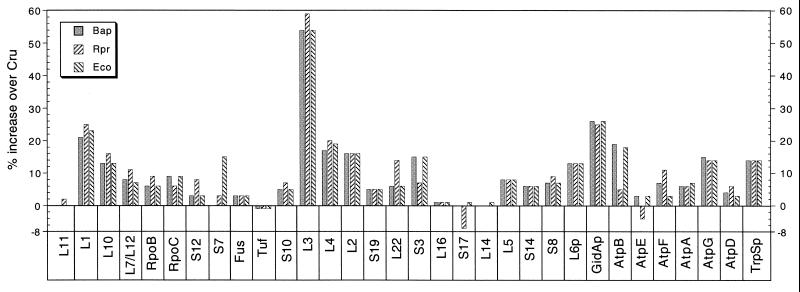
Protein size.
In Fig. 5 the sizes of 32 C. ruddii proteins are compared with those of homologous proteins of B. aphidicola, R. prowazekii, and E. coli. In 29 of the 32 cases, the C. ruddii proteins were shorter than any of the homologs. The numbers of amino acids in proteins of B. aphidicola, R. prowazekii, and E. coli averaged 9.2, 9.1, and 9.5%, respectively, greater than those in the C. ruddii proteins. The major size difference appears in ribosomal protein L3, which has a substantial deletion at the N terminus. We have also cloned and sequenced the 3.8 kb tuf-rpS3 fragment from C. ruddii-P. celtidis and C. ruddii- C. eucalypti (Fig. 1A). A comparison of the lengths of the proteins in C. ruddii from these three psyllid species indicated that they are identical or nearly identical in size. The number of amino acids is listed in parentheses following the protein designation in the order C. ruddii-P. venusta, C. ruddii-P. celtidis, and C. ruddii-C. eucalypti (where the size is constant only one value is given): Tuf, partial 312, 312, 310; RpS10, 98; RpL3, 136; RpL4, 172; RpL2, 235, 235, 234; RpS19, 88; RpL22, 104, 101, 105; RpS3, partial 148, 148, 150.
FIG. 5.
Comparison of the percent increase in the amino acid content of homologous proteins that are present in B. aphidicola (Bap), R. prowazekii (Rpr), and E. coli (Eco) over that in C. ruddii (Cru).
rRNA operon.
Sequence comparison of the DNA region upstream of rRNA operons of B. aphidicola indicated that a sequence resembling the −35, −10 promoter region is conserved and is followed by downstream conserved sequences resembling boxA and boxC (3; Baumann et al., website). In addition, inverted repeats resembling rho-independent terminators following the rRNA operons were found. We have used a similar comparative approach to look for sequences conserved upstream of 16S rDNA which might correspond to putative promoter regions. The results are presented in Fig. 6A and indicate conservation of a sequence resembling boxA; no sequences resembling the −35, −10 promoter region were found upstream of the putative boxA, in the last 200 nt of trpS. Similarly, there is no sequence conservation following 5S rDNA (Fig. 6B) and no significant inverted repeats resembling rho-independent terminators (25). These results suggest that the 16S-23S-5S rRNA genes are part of a larger transcription unit that includes upstream and downstream genes (Fig. 1C).
FIG. 6.
Comparisons of the intergenic sequence upstream of 16S (A) and downstream of 5S (B) in C. ruddii from different psyllids. Ba, B. aphidicola; Ec, E. coli. Nucleotides in bold letters indicate conserved sequences that are putative ends or beginnings of genes.
DISCUSSION
General properties.
C. ruddii has a unique combination of properties distinguishing it from other known bacteria. These properties consist of (i) extremely low G+C content (18% for the protein-coding regions); (ii) elimination or reduction of intergenic spaces, sometimes resulting in fusion of operons; (iii) absence of the complement of an RBS at the 3′ end of 16S rDNA (28) and an RBS preceding any structural genes; (iv) a reduction in protein size; (v) absence of conserved sequences corresponding to the −35, −10 promoter region preceding 16S rDNA; and (vi) absence of inverted repeats characteristic of rho-independent terminators following 5S rDNA. The absence of intergenic spaces and the complement of the RBS at the 3′ end of 16S rDNA suggest that long polycistronic mRNAs are made and translational coupling occurs during protein synthesis (25). The organization of the rRNA genes is highly conserved in bacteria, and they are generally arranged as a single transcription unit with recognizable, conserved −35, −10 promoter regions and a rho-independent terminator(s) (19). The absence of these sequences in C. ruddii suggests that these genes are part of a larger transcription unit. Outside of the rRNA genes, the only conserved sequence that was found preceded 16S rDNA (Fig. 6A) and had some resemblance to boxA, which is involved in antitermination (4, 27). The absence of a sequence complementary to the RBS at the 3′ end of 16S-like rRNA has also been noted in animal mitochondria (35).
The elimination or reduction of intergenic spaces and the reduction in the protein sizes indicate a history of mutational pressure and/or selection favoring reduced sequence length. One interpretation of the tendency toward elimination of intergenic spacers and reduction in gene length in C. ruddii is that mutations are biased in favor of deletions relative to insertions and that selection to oppose these deletions is often weak or ineffective (due to genetic drift). An analysis of mutational patterns in pseudogenes of Rickettsia species indicated a mutational bias favoring deletions (1). It is possible that C. ruddii experiences similar mutational bias but relaxed selection for conservation of function (or less efficient selection due to increased genetic drift) results in even greater shrinkage. Alternatively, selection might have favored the reduction of sequence length for some unknown reason. However, the length reduction occurs both in spacers (Fig. 4) and in protein-coding sequences (Fig. 5), which are generally subject to different kinds of selection. The bias toward A+T in DNA sequences is most readily explained as the result of mutational pressure. The pattern in Fig. 2 suggests that this mutational pressure is opposed by selection for conservation of function, with the result that genes encoding proteins which are poorly conserved have a higher A+T content. Conservative selection opposing the mutational bias appears to be weaker or less efficient than in other bacteria, with the result that C. ruddii has the most extreme bias toward A+T known for bacteria (15). A similarly low G+C content has been found in the genome of the eukaryote Plasmodium (20).
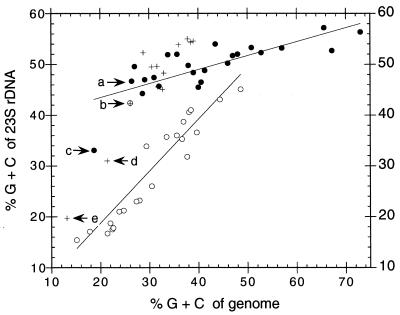
The unique combination of properties found in C. ruddii is also illustrated by a comparison of the G+C contents of homologous rRNA subunits of bacteria, plastids, and mitochondria with the G+C contents of their genomes (or representative C. ruddii genomic fragments). Since the results for small (16S and 16S-like) and large (23S and 23S-like) subunits of rDNA are very similar, only the data for 23S rDNA are presented (Fig. 7). Two different patterns are discerned, one for bacteria and plastids and one for and mitochondria. The distinctness of the two patterns suggests the existence of different sets of constraints on allowable evolutionary change, although the reasons for these constraints are not understood. In Fig. 7, arrow (a) designates B. aphidicola, which is at the low end of the bacterial G+C content (the range for bacteria is 27 to 73 mol%). The remaining arrows designate entities which do not fit within the bacterial or mitochondrial pattern. Arrow b corresponds to the mitochondrion of the protozoan Reclinomonas americana, which is unique in that it retains many bacterial attributes such as the presence of −35, −10 promoter sequences and a mitochondrion-encoded α2ββ′ς RNA polymerase (17). Arrow c designates C. ruddii, which is distinct from the bacterial pattern and tends toward mitochondria. Arrows d and e designate highly unusual, extrachromosomal DNA molecules which are found within membrane-bounded structures in cells of the Apicomplexa (genera Plasmodium and Toxoplasma) (34). These structures appear to be remnants of plastids which have lost their photosynthetic function and are involved in fatty acid biosynthesis (32). Their position is distinct from plastids and tends toward mitochondria.
FIG. 7.
Comparison of the G+C content of 23S and 23S-like rDNA with the G+C content of the genome. ●, bacteria; ○, mitochondria; +, plastids; arrow a, B. aphidicola; arrow b, Reclinomonas americana; arrow c, Cr-Pve; arrow d, Toxoplasma gondii; arrow e, Plasmodium falciparum. Data compiled in January 2000 from GenBank and data in reference 15.
Gene content, gene order, and physiology.
The list of detected genes presented in Table 1 indicates that C. ruddii encodes proteins and RNAs which have housekeeping functions. This includes rRNAs, ribosomal proteins, two subunits of RNA polymerase, elongation factors, and tRNA synthases. In addition, we detected many of the components of the ATP synthase. In E. coli and other organisms, the genes presented in Fig. 1A are part of five different operons (13, 16). In C. ruddii, these operons appear to be fused. The order of the genes is identical to that in E. coli (16), and the sole difference is the absence of three genes. C. ruddii lacks rpL23, rpL29, and rpL24, which in E. coli and many other organisms are found following rpL4, rpL16, and rpL14, respectively (Fig. 1A) (13, 16). In B. aphidicola and many other organisms, the gene order of the ATP synthase is atpBEFHAGDC (9). In C. ruddii, part of this order is maintained (Fig. 1B). However, we have not been able to detect atpH or atpC. ORFA and ORFB are found in the locations where these genes are expected, but they show no significant similarity to atpH or atpC. The presence of genes encoding ATP synthase suggests that C. ruddii may be able to generate ATP from the proton motive force. Genes for transaldolase (Fig. 1B) and transketolase (Fig. 1C) were detected, suggesting a functional nonoxidative pentose phosphate cycle. In addition, genes encoding proteins involved in the synthesis of amino acids of the glutamate and aspartate family were found. These results suggest that C. ruddii may be able to synthesize essential amino acids for the psyllid host as in the case of B. aphidicola, the endosymbiont of aphids (10, 24; Baumann et al., website).
Comparison with B. aphidicola.
Both C. ruddii and B. aphidicola are associated with insect hosts that feed on plant phloem sap and thus have similar nutritional needs. These endosymbiotic associations result from infections by two different bacterial ancestors, both from the γ division of the proteobacteria (11, 18, 28; Baumann et al., website). It is likely that C. ruddii retains genes for essential amino acid pathways, as in B. aphidicola. Some of the same genomewide features appear in both species and can be interpreted as convergences; these include increased A+T content (18), accelerated sequence evolution (18, 26, 28), and shortened proteins (8). Although a slight reduction in protein length has been noted previously for B. aphidicola relative to E. coli (8), the reduction is much more severe in C. ruddii, as is evident in Fig. 5. B. aphidicola has intergenic spaces similar to those found in other bacteria (Fig. 4), and the 3′-end of its 16S rDNA has the complement of the RBS (3, 24; Baumann et al., website). The G+C contents of both the B. aphidicola genome and the B. aphidicola rRNA are comparable to those of other bacteria at the lower end of the known scale (Fig. 7). In C. ruddii, the modifications are considerably more drastic, resulting in an even lower G+C content of the DNA and almost complete elimination of intergenic spaces. B. aphidicola and some other bacteria with low G+C contents have very small genomes, having lost most of the genes found in ancestors (1, 24); future studies determining the C. ruddii genome size will be of interest.
ACKNOWLEDGMENTS
This material is based on work supported by National Science Foundation awards MCB-9807145 (to P.B.) and DEB-9978518 (to N.A.M. and P.B.) and the University of California Experiment Station (to P.B.).
REFERENCES
- 1.Andersson J O, Andersson S G. Genome degradation is an ongoing process in Rickettsia. Mol Biol Evol. 1999;16:1178–1191. doi: 10.1093/oxfordjournals.molbev.a026208. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 2000. [Google Scholar]
- 3.Baumann P, Baumann L, Lai C Y, Rouhbakhsh D, Moran N A, Clark M A. Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 4.Berg K L, Squires C, Squires C L. Ribosomal RNA operon anti-termination. Function of leader and spacer region boxB-boxA sequences and their conservation in diverse micro-organisms. J Mol Biol. 1989;209:345–358. doi: 10.1016/0022-2836(89)90002-8. [DOI] [PubMed] [Google Scholar]
- 5.Borror D J, Triplehorn C A, Johnson N F. An introduction to the study of insects. Fort Worth, Tex: The W. B. Saunders Co.; 1989. pp. 335–345. [Google Scholar]
- 6.Buchner P. Endosymbiosis of animals with plant microorganisms. New York, N.Y: Interscience; 1965. pp. 210–332. [Google Scholar]
- 7.Chang K P, Musgrave A J. Histochemistry and ultrastructure of the mycetome and its “symbiotes” in the pear psylla, Psylla pyricola Foerster (Homoptera) Tissue Cell. 1969;1:597–606. doi: 10.1016/s0040-8166(69)80034-0. [DOI] [PubMed] [Google Scholar]
- 8.Charles H, Mouchiroud D, Lobry J, Goncalves I, Rahbe Y. Gene size reduction in the bacterial aphid endosymbiont, Buchnera. Mol Biol Evol. 1999;16:1820–1822. doi: 10.1093/oxfordjournals.molbev.a026096. [DOI] [PubMed] [Google Scholar]
- 9.Clark M A, Baumann P. The (F1F0) ATP synthase of Buchnera aphidicola (endosymbiont of aphids): genetic analysis of the putative ATP operon. Curr Microbiol. 1997;35:84–89. doi: 10.1007/s002849900217. [DOI] [PubMed] [Google Scholar]
- 10.Douglas A E. Nutritional interactions in insect-microbial symbioses: aphids and their symbiotic bacteria Buchnera. Annu Rev Entomol. 1998;43:17–37. doi: 10.1146/annurev.ento.43.1.17. [DOI] [PubMed] [Google Scholar]
- 11.Fukatsu T, Nikoh N. Two intracellular symbiotic bacteria from the mulberry psyllid Anomoneura mori (Insecta, Homoptera) Appl Environ Microbiol. 1998;64:3599–3606. doi: 10.1128/aem.64.10.3599-3606.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukatsu T, Nikoh N. Endosymbiotic microbiota of the bamboo pseudococcid Antonina crawii (Insecta, Homoptera) Appl Environ Microbiol. 2000;66:643–650. doi: 10.1128/aem.66.2.643-650.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hansmann S, Martin W. Phylogeny of 33 ribosomal and six other proteins encoded in an ancient gene cluster that is conserved across prokaryotic genomes: influence of excluding poorly alignable sites from analysis. Int J Syst Evol Microbiol. 2000;50:1655–1663. doi: 10.1099/00207713-50-4-1655. [DOI] [PubMed] [Google Scholar]
- 14.Hodkinson I D. The biology of the Psylloidea (Homoptera): a review. Bull Entomol Res. 1974;64:325–339. [Google Scholar]
- 15.Holt J G, editor. Bergey's manual of systematic bacteriology. I to IV. Baltimore, Md: The Williams & Wilkins Co.; 1984–1989. [Google Scholar]
- 16.Keener J, Nomura M. Regulation of ribosome synthesis. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 1417–1431. [Google Scholar]
- 17.Lang B F, Burger G, O'Kelly C J, Cedergren R, Golding G B, Lemieux C, Sankoff D, Turmel M, Gray M W. An ancestral mitochondrial DNA resembling a eubacterial genome in miniature. Nature. 1997;387:493–497. doi: 10.1038/387493a0. [DOI] [PubMed] [Google Scholar]
- 18.Moran N A, Telang A. Bacteriocyte-associated symbionts of insects: a variety of insect groups harbor ancient prokaryotic endosymbionts. Bioscience. 1998;48:295–304. [Google Scholar]
- 19.Munson M A, Baumann L, Baumann P. Buchnera aphidicola (a prokaryotic endosymbiont of aphids) contains a putative 16S rRNA operon unlinked to the 23S rRNA-encoding gene: sequence determination, and promoter and terminator analysis. Gene. 1993;137:171–178. doi: 10.1016/0378-1119(93)90003-l. [DOI] [PubMed] [Google Scholar]
- 20.Musto H, Romero H, Zavala A, Jabbari K, Bernardi G. Synonymous codon choices in the extremely GC-poor genome of Plasmodium falciparum: compositional constraints and translational selection. J Mol Evol. 1999;49:27–35. doi: 10.1007/pl00006531. [DOI] [PubMed] [Google Scholar]
- 21.Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Sandström J, Moran N. How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl. 1999;91:203–210. [Google Scholar]
- 24.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Mutualism as revealed at the genomic level: the whole genome sequence of Buchnera sp. APS, an endocellula bacterial symbiont of aphids. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 25.Snyder L, Champness W. Molecular genetics of bacteria. Washington, D.C.: ASM Press; 1997. [Google Scholar]
- 26.Spaulding A W, von Dohlen C D. Phylogenetic characterization and molecular evolution of bacterial endosymbionts in psyllids (Hemiptera: Sternorrhyncha) Mol Biol Evol. 1998;15:1506–1513. doi: 10.1093/oxfordjournals.molbev.a025878. [DOI] [PubMed] [Google Scholar]
- 27.Squires C L, Greenblatt J, Li J, Condon C, Squires C L. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci USA. 1993;90:970–974. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thao M L, Moran N A, Abbot P, Brennan E B, Burckhardt D H, Baumann P. Cospeciation of psyllids and their prokaryotic endosymbionts. Appl Environ Microbiol. 2000;66:2898–2905. doi: 10.1128/aem.66.7.2898-2905.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thao M L, Clark M A, Baumann L, Brennan E B, Moran N A, Baumann P. Secondary endosymbionts of psyllids have been acquired multiple times. Curr Microbiol. 2000;41:300–304. doi: 10.1007/s002840010138. [DOI] [PubMed] [Google Scholar]
- 30.Thao, M. L., M. A. Clark, D. H. Burckhardt, N. A. Moran, and P. Baumann. Phylogenetic analysis of vertically transmitted psyllid endosymbionts (Carsonella ruddii) based on atpAGD and rpoBC; comparisons with 16S–23S rDNA-derived phylogeny. Curr. Microbiol., in press. [DOI] [PubMed]
- 31.Waku Y, Endo Y. Ultrastructure and life cycle of the symbionts in a Homopteran insect, Anomoneura mori Schwartz (Psyllidae) Appl Entomol Zool. 1987;22:630–637. [Google Scholar]
- 32.Waller R F, Keeling P K, Donald R G K, Striepen B, Handman E, Lang-Unnasch N, Cowman A F, Besra G S, Roos D S, McFadden G I. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci USA. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang R F, Kushner S R. Construction of versatile low-copy-number vectors for cloning, sequencing and gene expression in Escherichia coli. Gene. 1991;100:195–199. [PubMed] [Google Scholar]
- 34.Wilson R J M, Williamson D H. Extrachromosomal DNA in the Apicomplexa. Microbiol Mol Biol Rev. 1997;61:1–16. doi: 10.1128/mmbr.61.1.1-16.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolstenholme D R. Animal mitochondrial DNA: structure and evolution. Int Rev Cytol. 1992;141:173–214. doi: 10.1016/s0074-7696(08)62066-5. [DOI] [PubMed] [Google Scholar]