Abstract
rsmBEcc specifies a nontranslatable RNA regulator that controls exoprotein production and pathogenicity in soft rot-causing Erwinia carotovora subsp. carotovora. This effect of rsmBEcc RNA is mediated mostly by neutralizing the function of RsmAEcc, an RNA-binding protein of E. carotovora subsp. carotovora, which acts as a global negative regulator. To determine the occurrence of functional homologs of rsmBEcc in non-soft-rot-causing Erwinia species, we cloned the rsmB genes of E. amylovora (rsmBEa) and E. herbicola pv. gypsophilae (rsmBEhg). We show that rsmBEa in E. amylovora positively regulates extracellular polysaccharide (EPS) production, motility, and pathogenicity. In E. herbicola pv. gypsophilae, rsmBEhg elevates the levels of transcripts of a cytokinin (etz) gene and stimulates the production of EPS and yellow pigment as well as motility. RsmAEa and RsmAEhg have more than 93% identity to RsmAEcc and, like the latter, function as negative regulators by affecting the transcript stability of the target gene. The rsmB genes reverse the negative effects of RsmAEa, RsmAEhg, and RsmAEcc, but the extent of reversal is highest with homologous combinations of rsm genes. These observations and findings that rsmBEa and rsmBEhg RNA bind RsmAEcc indicate that the rsmB effect is channeled via RsmA. Additional support for this conclusion comes from the observation that the rsmB genes are much more effective as positive regulators in a RsmA+ strain of E. carotovora subsp. carotovora than in its RsmA− derivative. E. herbicola pv. gypsophilae produces a 290-base rsmB transcript that is not subject to processing. By contrast, E. amylovora produces 430- and 300-base rsmB transcripts, the latter presumably derived by processing of the primary transcript as previously noted with the transcripts of rsmBEcc. Southern blot hybridizations revealed the presence of rsmB homologs in E. carotovora, E. chrysanthemi, E. amylovora, E. herbicola, E. stewartii and E. rhapontici, as well as in other enterobacteria such as Escherichia coli, Salmonella enterica serovar Typhimurium, Serratia marcescens, Shigella flexneri, Enterobacter aerogenes, Klebsiella pneumoniae, Yersinia enterocolitica, and Y. pseudotuberculosis. A comparison of rsmB sequences from several of these enterobacterial species revealed a highly conserved 34-mer region which is predicted to play a role in positive regulation by rsmB RNA.
In many host-pathogen systems, disease development requires coordinate expression of sets of genes in response to various signals and environmental cues (9, 13). Regulation of these genes, like that of the housekeeping genes, is subject to both transcriptional and posttranscriptional control. We have determined that posttranscriptional regulation mediated by the RsmA-rsmB pair is the most critical factor in soft-rot-causing Erwinia. RsmA is a small RNA binding protein, which acts by reducing the half-life of mRNA species (21, 33). rsmB specifies an untranslated regulatory RNA (21) and neutralizes the effect of RsmA. RsmA and rsmB RNA control many phenotypes in soft-rot-causing Erwinia, including the production of pectate lyase, polygalacturonase, cellulase, protease, harpin, motility, flagellum formation, antibiotic, pigment, elicitation of the HR (hypersensitive reaction), and pathogenicity (10, 26). Erwinia carotovora subsp. carotovora strain 71 produces two rsmB RNA species: a primary RNA of 479 bases which is processed to yield a 259-base RNA, designated rsmB' RNA (21). The rsmB' RNA has the regulatory functions attributed to rsmB. Romeo and associates have identified a 360-base csrB RNA in Escherichia coli which is functionally very similar to rsmB RNA, except that there is no evidence that csrB RNA is processed (33).
Preliminary trials with the cloned E. carotovora subsp. carotovora genes revealed transdominant regulatory effects of rsmA and rsmB genes in non-soft-rot-causing Erwinia species such as E. amylovora, the representative of the Amylovora group (31), and E. stewartii and E. herbicola pv. gypsophilae, the representatives of the Herbicola group (31) (now members of the genus Pantoea [14]). Furthermore, homologs of rsmA exist in these bacteria and in all other Erwinia species tested (10). These observations prompted the hypothesis that the regulatory pair controls the production of pathogenicity factors in these non-soft-rot-causing Erwinia species as well. We should note that pathogenicity of the Amylovora and Herbicola (Pantoea) groups of bacteria is determined not by extracellular enzymes, as in soft-rot-causing Erwinia, but by factors such as extracellular polysaccharide (EPS) or growth hormones (4, 23). Since rsmB homologs of these bacteria have not been examined, it was of interest to compare the structural and functional characteristics of rsmB genes of the three groups of bacteria. We report here (i) cloning of rsmA and rsmB genes of E. amylovora (rsmAEa and rsmBEa) and E. herbicola pv. gypsophilae (rsmAEhg and rsmBEhg), (ii) nucleotide sequence or deduced amino acid sequence homologies of these genes, (iii) effects of rsmA and rsmB in homologous and heterologous bacterial species, and (iv) reversal of the RsmA effect by rsmB. We also show that E. amylovora strain E9 produces two rsmB RNA species whereas E. herbicola pv. gypsophilae possesses a single rsmB transcript species. Our findings and the physical evidence for the occurrence of RsmA homologs (10) suggest that the RsmA-rsmB regulatory system has been conserved in enterobacterial species. Moreover, we have identified a region of rsmB RNA which has been conserved in enterobacterial species.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. The strains carrying drug markers were maintained on Luria-Bertani (LB) agar containing appropriate antibiotics. The wild-type strains were maintained on LB agar. The composition of LB medium, minimal salts medium, KB medium, nutrient gelatin (NG) agar, and polygalacturonate-yeast extract agar (PYA) have been described previously (2, 8). When required, antibiotics were supplemented as follows (in micrograms per milliliter): ampicillin, 100; kanamycin, 50; spectinomycin, 50; tetracycline, 10. Media were solidified by the addition of 1.5% (wt/vol) agar.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relative characteristics | Reference or source |
|---|---|---|
| Strains | ||
| E. carotovora subsp. carotovora | ||
| Ecc71 | Wild type | 39 |
| AC5071 | RsmA of Ecc71 | 25 |
| E. amylovora E9 | Wild type | 32 |
| E. herbicola pv. gypsophilae PD713 | Wild type | 17 |
| E. carotovora subsp. atroseptica Eca12 | Wild type | 39 |
| E. carotovora subsp. betavasculorum Ecb11129 | Wild type | J. E. Loper |
| E. chrysanthemi EC16 | Wild type | 7 |
| E. rhapontici Er1 | Wild type | 24 |
| E. herbicola EH105 | Wild type | 10 |
| E. stewartii Es1 | Wild type | 10 |
| S. enterica serovar Typhimurium LT2 | Wild type | 10 |
| Serratia marcescens Sm1 | Wild type | 10 |
| Yersinia pseudotuberculosis Yp1 | Wild type | 7 |
| Yersinia enterocolitica 8081V | Wild type | S. A. Munnich |
| Shigella flexneri Sf1 | Wild type | 10 |
| Enterobacter aerogenes Ena1 | Wild type | 10 |
| Klebsiella pneumoniae KP1 | Wild type | 10 |
| E. coli | ||
| DH5α | Φ80lacZ M15 (lacZYA-argF)U169 hsdR17 recA1 endA1 thi-1 | Gibco BRL |
| K-12 | Wild type | 37 |
| Plasmids | ||
| pCL1920 | Spcr Smr | 16 |
| pCL1921 | Spcr Smr | 16 |
| pRK415 | Tcr | 15 |
| pBluescript SK(+) | Apr | Stratagene |
| pSF6 | Mob+ Spcr Smr | 36 |
| pMBL-R.73 | Apr, 0.73-kb EcoRI fragment containing pre-etz plus etz in pBluescript SK(+) | 17 |
| pAKC781 | Apr, peh-1+ | 19 |
| pAKC783 | Apr, pel-1+ | 19 |
| pAKC891 | Spcr, 3.0-kb EcoRI-SacI fragment containing rsmAEhg in pCL1921 | This work |
| pAKC892 | Tcr, 3.0-kb EcoRI-HindIII fragment of pAKC891 containing rsmAEhg in pRK415 | This work |
| pAKC120 | Spcr, pSF6 cosmid containing rsmAEa | This work |
| pAKC893 | Spcr, 4.5-kb HindIII fragment containing rsmAEa in pCL1920 | This work |
| pAKC894 | Tcr, 4.5-kb HindIII fragment containing rsmAEa in pRK415 | This work |
| pAKC878 | Tcr, 9.0-kb EcoRI fragment containing rsmAEcc in pRK415 | 21 |
| pAKC679 | AepH+ (rsmBEcc), Spcr | 28 |
| pAKC1004 | Spcr, plac-rsmB′Ecc, nt +220 to +540 of rsmBEcc DNA in pCL1920 | 21 |
| pAKC1042 | Spcr, 1.7-kb HincII fragment containing rsmBEhg in pCL1920 | This work |
| pAKC1043 | Spcr, 1.7-kb BamHI fragment containing rsmBEa in pCL1920 | This work |
| pAKC1044 | Apr, nt-62 to +193 of rsmBEa DNA in pBluescript SK(+) | This work |
| pAKC1045 | Apr, nt +1 to +455 of rsmBEa DNA in pBluescript SK(+) | This work |
| pAKC1046 | Apr, at +1 to +310 rsmBEhg of DNA in pBluescript SK(+) | This work |
| PAKC1049 | Spcr, plac-rsmBEcc, nt +1 to +540 of rsmBEcc DNA in pCL1920 | This work |
| pAKC1061 | Spcr, plac-rsmBEhg, nt +26 to +761 of rsmBEhg DNA in pCL1920 | This work |
| pAKC1062 | Spcr, plac-rsmBEa, nt +1 to +455 of rsmBEa DNA in pCL1920 | This work |
| pAKC1063 | Spcr, plac-rsmB′Ea, nt +135 to +455 of rsmBEa DNA in pCL1920 | This work |
Extracellular enzyme assays.
Growth conditions, preparation of culture supernatant, quantitative assay conditions for pectate lyase (Pel) and semiquantitative assay conditions for pectate lyase (Pel), polygalacturonase (Peh), cellulase (Cel), and protease (Prt) have been previously described (5, 29).
EPS production and motility assays.
Cultures of E. herbicola pv. gypsophilae or E. amylovora were patched or streaked on minimal salts medium plus sucrose (1%, wt/vol) and spectinomycin for EPS detection and stab inoculated into KB soft agar (0.4%, wt/vol) plus spectinomycin with a needle for examination of motility. Bacteria were incubated at 28°C for 24 h. Motility and EPS production of the bacteria were visually examined.
Pathogenicity assays on apple shoots.
Pathogenicity assays on apple shoots were carried out essentially as previously described (26), cell suspensions (30 μl containing 2 × 108 CFU) of E. amylovora strain E9 carrying the rsmAEa+ plasmid, rsmBEa+ plasmid, or the cloning vector, pCL1920, were applied to the cut surface of each petiole. Inoculated plants were incubated at 28°C with a 14-h light/10-h dark regime until disease symptoms appeared.
DNA techniques.
Standard procedures were used in the isolation of plasmid and chromosomal DNAs, transformation, restriction endonuclease digests, gel electrophoresis, and DNA ligation (34). Southern hybridizations were carried out as previously described (10). Restriction and modifying enzymes were obtained from Promega Biotec (Madison, Wis.). The Prime-a-Gene DNA labeling system of Promega Biotec was used for labeling DNA probes. PCR was performed as described by Liu et al. (21). By using the degenerate primers, DB1 (5′-YMADGGACACCTCCAGG-3′) and DB2 (5′-WCTGYRCYCCCGGTTCG-3′) (Y = C or T; M = A or C; D = A, G, or T; W = A or T; R = A or G), in the rsmB sequence, we amplified the rsmB DNA in the following bacterial species: Pseudomonas fluorescens, Serratia marcescens, Shigella flexneri, Enterobactor arogenes, Salmonella enterica serovar Typhimurium, and Klebsiella pneumoniae (Table 1).
The plac-rsmB transcription fusions were constructed as follows. The 0.7-kb HincII-EcoRV fragment from pAKC1042 was cloned into the SmaI site of pCL1920 to produce pAKC1061, which contains plac-rsmBEhg. The 473-bp DNA fragment was amplified by PCR using the KpnI-tagged primer AGGGTACCGTTGCGAAGGAACAGCATG and HindIII-tagged primer AGAAGCTTAAAGGGGGCACTGTATAAACA and template DNA pAKC1043. After digestion with endonucleases, the DNA fragment was cloned into pCL1920 to produce pAKC1062 which carries plac-rsmBEa. Similarly, pAKC1063, which contains plac-rsmB'Ea was constructed by cloning the 326-bp DNA fragment into pCL1920. The DNA fragment was amplified by PCR using the KpnI-tagged primer AGGGTACCTCTCCAGGATGGAGAAACG and the HindIII-tagged primer AGAAGCTTAAAGGGGGCACTGTATAAACA and template DNA pAKC1043 and digested with endonucleases KpnI and HindIII. To construct pAKC1049 containing plac-rsmBEcc the KpnI-tagged primer AGGGTACCAAGTTAGTAACCGGTTACA and P7 primer GCAAGCTTCTTCACAACGTGGCGCTACAT (21) and template DNA pAKC679 were used for PCR. The 558-bp PCR product was digested with HindIII and KpnI and cloned into pCL1920.
RNA preparation and Northern hybridization.
Bacteria were grown in 20 ml of LB or minimal salts medium supplemented with sucrose (0.5%, wt/vol) and appropriate antibiotics at 28°C to a turbidity of ca. 200 Klett units. Total RNA was then extracted by the method of Aiba et al. (1). The procedure used for Northern blot analysis was previously described (6, 22).
A 730-bp EcoRI fragment containing pre-etz and etz from pMBL-R.73 (17) was used as a probe. A 695-bp HincII-EcoRV fragment from pAKC1042 was used as the rsmBEhg probe. A 310-bp PstI-BglII fragment from pAKC1043 was used as the rsmBEa probe. A 314-bp EcoRV-KpnI fragment from pAKC783 was used as the pel-1 probe (19). A 743-bp HindIII fragment from pAKC781 was used as the peh-1 probe (19).
Primer extension analysis and RNase protection assay.
Primer extension was performed as specified by the manufacturer (Promega Biotec). An aliquot (10 pmol) of primer EhgB1 (5′-TGCTCAATCCTGAGCGATCCTG-3′) (nucleotides [nt] +103 to +125) or EaP6 (5′-CTTCATCCTGAAGCCTGTCCCTG-3′) (nt +214 to +236) was end labeled with T4 polynucleotide kinase and [γ-32P]ATP. A 20-μg portion of total RNA from E. herbicola pv. gypsophilae or E. amylovora and 100 fmol of 32P-labeled primer in 11 μl of primer extension buffer were incubated at 58°C for 20 min and cooled for 10 min at room temperature for annealing. Reverse transcription reaction was carried out with avian myeloblastosis virus reverse transcriptase at 42°C for 30 min.
The RNase protection assay was carried out as described by Liu et al. (20). The 272-bp rsmBEa DNA fragment (corresponding to at −62 to +194) amplified from pAKC1043 by PCR using BamHI-tagged primer EaP13 (5′-AGGGATCCAATAGCCTAAATAGCCGCTC-3′) and XbaI-tagged primer EaP15 (5′-AGTCTAGAATCCTGTTATCATCCATGAACTGCCG-3′) was cloned into pBluescript SK(+) to produce pAKC1044. XbaI-digested pAKC1044 was used as the template for in vitro transcription. The in vitro-synthesized RNA probes were labeled with [γ-32P]UTP by using T7 polymerase as specified by the manufacturer instructions (Promega Biotec). The DNA template was then removed from the RNA probes by DNase treatment. A 20-μg RNA sample from E. amylovora strain E9 and 105 cpm of RNA probe were incubated in 30 μl of hybridization buffer (80% formamide, 40 mM PIPES [pH 6.7], 400 mM NaCl, 1 mM EDTA) overnight at 45°C and then digested with 300 μl of RNase solution (10 mM Tris [pH 7.5], 5 mM EDTA, 300 mM NaCl, 40 μg of RNase A per ml, 2 μg of RNase T1 per ml) at 30°C for 1 h.
RNA stability assays.
Cultures were grown at 28°C in minimal salts medium plus sucrose (0.5%, wt/vol) and spectinomycin to a turbidity of ca. 160 Klett units, and rifampicin was added to a final concentration of 200 μg/ml. Aliquots (10 ml) were collected at 0, 2.5, 5, 7.5, 10, and 15 min in tubes containing 5 ml of diethylpyrocarbonate-treated ice-cold water. Total RNA was extracted by the method of Aiba et al. (1), and Northern blot analysis was performed by the procedures described by Chatterjee et al. (6) and Liu et al. (22). After being washed, the blots were exposed to X-ray film. The Metamorph imaging system (Universal Imaging Corp.) was used for the densitometric analysis of the autoradiograms. All experiments were performed three times or more, and the results were reproducible.
RNA mobility shift assays.
Using primers KpnI-tagged EaP10 (5′-AGGGTACCGTTGCGAAGGAACAGCATG-3′) and HindIII-tagged EaP12 (5′-AGAAGCTTAAAGGGGGCACTGTATAAACA-3′) or KpnI-tagged EhgB11 (5′-AGGGTACCACTGCAGGAGGCTCAGGAA-3′) and HindIII-tagged EhgB12 (5′-AGAAGCTTAAAGGGAGCACTGTATAAACA-3′), the PCR-amplified DNA fragment corresponding to nt +1 to +455 of rsmBEa or nt +1 to +310 of rsmBEhg was cloned in pBluescript SK(+) to produce pAKC1045 and pAKC1046. The rsmBEa and rsmBEhg RNA probes were synthesized in vitro from the HindIII-digested pAKC1045 and pAKC1046 DNAs by T7 RNA polymerase in the presence of [α-32P]UTP. The RNA-protein interaction was assayed in 20 μl of binding buffer (10 mM Tris-acetate [pH 7.5], 10 mM MgCl2, 50 mM NaCl, 50 mM KCl, 10 mM dithiothreitol, 5% [wt/vol] glycerol) containing 4000 cpm of labeled RNA (0.1 ng), with or without purified His6-tagged RsmAEcc and a 50-fold excess of unlabeled RNAs or yeast tRNA (Gibco BRL). After incubation for 30 min at room temperature, the reaction mixtures were loaded on a prerun 5% (wt/vol) polyacrylamide gel containing 5% (wt/vol) glycerol at 4°C. Electrophoresis was continued in 0.5× Tris-borate-EDTA (TBE) running buffer for another 4 h at 4°C. The gel was dried and exposed to X-ray film.
RESULTS AND DISCUSSION
As stated above, physical evidence has established the presence of rsmA homologs in various enterobacterial species (10). Furthermore, several studies have revealed that rsmA-rsmB and csrA-csrB work together to modulate gene expression in E. carotovora subsp. carotovora and Escherichia coli, respectively (18, 21). However, prior to this work, there was no evidence for RsmA-plus-rsmB-mediated gene regulation beyond these bacterial species. To alleviate this deficiency, we cloned and characterized rsmB genes from two Erwinia species that do not cause soft rot disease, i.e., E. amylovora and E. herbicola pv. gypsophylae, and obtained physical evidence for the occurrence of rsmB homologs in Erwinia and other enterobacterial species.
Cloning of rsmB from E. herbicola pv. gypsophilae strain PD713 and E. amylovora strain E9.
To clone the rsmB genes, genomic libraries of E. herbicola pv. gypsophilae strain PD713 and E. amylovora strain E9 were transferred by triparental matings into E. carotovora subsp. carotovora strain Ecc71 or strain AC5071 carrying the E9 rsmA+ plasmid, pAKC120 (Table 1). Transconjugants were screened for protease production on nutrient gelatin agar medium, and colonies showing higher protease activity were tested for their levels of pectinase and cellulase activities. The clones showing higher levels of all these enzymes were presumed to carry rsmB+ plasmids. In this manner, we obtained several E9 and PD713 clones which produced elevated levels of enzymatic activities. Plasmid DNAs isolated from these colonies were subsequently analyzed by Southern blot hybridization under low-stringency conditions using the transcribed region of rsmBEcc as the probe. One E9 clone and two clones from PD713 hybridized with the probe. By a subcloning and functional assay, the rsmBEhg gene was localized in a 1.7-kb HincII DNA fragment and the rsmBEa gene was localized in a 1.7-kb BamHI DNA fragment.
Characterization of the rsmB genes of E. amylovora and E. herbicola pv. gypsophilae.
To gain a better understanding of the structure and function of the rsmB genes of E. amylovora and E. herbicola pv. gypsophilae, we determined the nucleotide sequences of the 1.7-kb BamHI fragment and the 1.7-kb HincII fragment that contain the E. amylovora rsmB gene (rsmBEa) and the E. herbicola pv. gypsophilae rsmB gene (rsmBEhg), respectively. A homology search revealed that the nucleotide sequences of rsmBEa and rsmBEhg have high similarities to those of rsmBEcc of E. carotovora subsp. carotovora (21) and csrB of Escherichia coli (see below and Fig. 1A). Like csrB (18) and rsmBEcc (21), the rsmBEa and rsmBEhg genes contain no apparent open reading frames, suggesting that they encode RNA regulators rather than protein products.
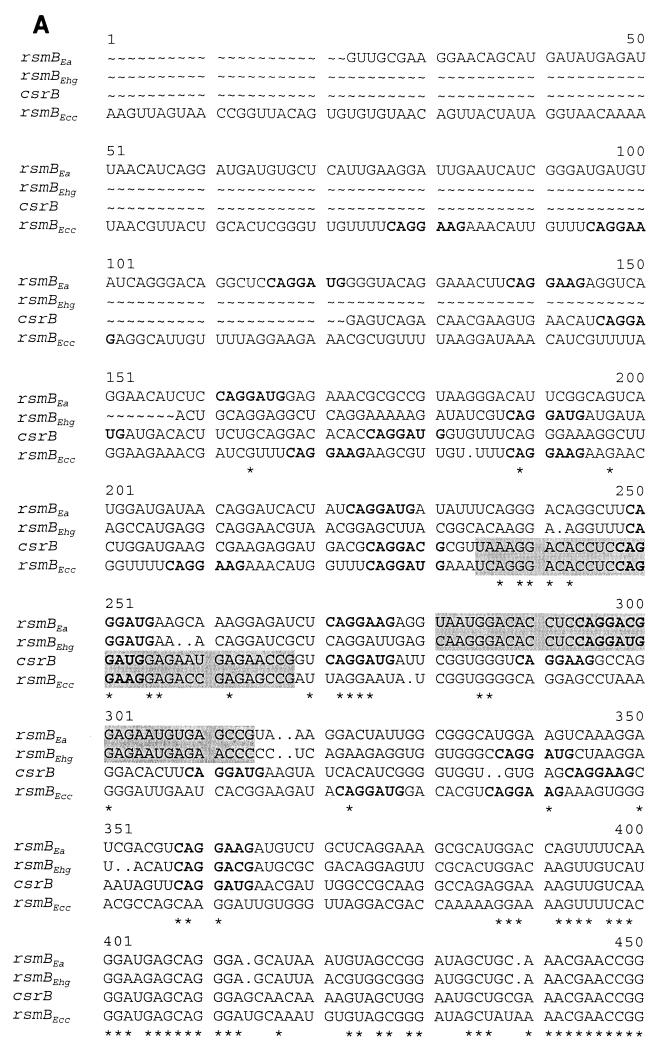
FIG. 1.
(A) Alignment of the ribonucleotide sequences of rsmB genes from E. amylovora strain E9 (Ea), E. herbicola pv. gypsophilae strain PD713 (Ehg), E. carotovora subsp. carotovora strain Ecc71(Ecc), and csrB from Escherichia coli. Asterisks indicate identical amino acids, and dots indicate conserved substitutions. Arrows indicate the processed start sites of rsmBEa RNA and rsmBEcc RNA. The 34-mer consensus sequences in rsmB are shown against shaded backgrounds. The 7-base repeats are in boldface type. (B) Alignment of the deduced amino acid sequence of RsmA from E. herbicola pv. gypsophilae strain PD713, E. amylovora strain E9, E. carotovora subsp. carotovora strain Ecc71, and CsrA from Escherichia coli.
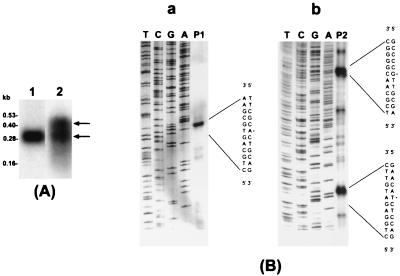
To characterize rsmB RNAs, we performed Northern blot analyses. As shown in Fig. 2A, the E. herbicola pv. gypsophilae probe hybridized to a 290-base transcript. By contrast, with E9 total RNA, we detected two bands of about 300 and 430 bases. To further characterize the transcripts, we performed primer extension analysis (Fig. 2B) and RNase protection assays (data not shown). Using appropriate oligonucleotide primers, we detected that the 5′ end of rsmBEhg was positioned at a single adenosine residue and that two 5′ ends of the rsmBEa transcripts, separated by 133 nt, were localized at the guanosine and thymidine residues, respectively (Fig. 2B). Analysis of the 3′ sequence revealed that each rsmB RNA species contains a strong stem-loop structure (Fig. 1A), which can function as rho-independent transcription terminators as defined for rsmBEcc (21). Therefore, the sizes of rsmB RNA species, stretching from the 5′ ends identified by primer extension or RNase protection assays to their 3′ rho-independent terminators, are 310 nt for rsmBEhg and 317 and 451 nt for the rsmBEa RNA species. These observations confirm that E. amylovora produces two rsmB transcripts, the shorter one spanning the 3′ end and the larger one extending further upstream of the shorter one (Fig. 2). We do not know the mechanism underlying the production of these two rsmB RNA species by E. amylovora. However, extrapolating from the findings with rsmBEcc (21), we consider it most likely that the 451-nt transcript represents the primary transcript whereas the 317-nt transcript is the processed product of the primary transcript. Since we have not detected sequences resembling typical enterobacterial promoters upstream of the 5′ end of the 317-base transcript, we consider it unlikely that the 451- and the 317-nt transcripts result from initiation of transcription from different start sites.
FIG. 2.
(A) Northern blot analysis of rsmB transcripts of E. herbicola pv. gypsophilae strain PD713 (lane 1) and E. amylovora strain E9 (lane 2). The positions of two rsmB transcripts of E9 are indicated by arrows. (B) Primer extension analysis of the 5′ ends of rsmB transcripts from E. herbicola pv. gypsophilae strain PD713 (a) and E. amylovora strain E9 (b). The portions of sequences pertinent to the 5′ ends are shown. The A residue in lane P1 is identified as the 5′ end of rsmB mRNA of PD713; the G and T residues in lane P2 are indicated as the two 5′ ends of rsmB mRNA of E9.
To determine the minimal size of rsmB required for its biological function, we constructed several plac-rsmB plasmids. In these constructs, the rsmB DNA fragments that do not carry the promoter regions were subcloned into the pCL1920 vector. Similar to the positive control pAKC1049 and pAKC1004 plasmids, which contain plac-rsmBEcc and plac-rsmB'Ecc, respectively, all of these constructs were able to not only stimulate extracellular Pel production in E. carotovora subsp. carotovora strain 71 and its RsmA-deficient derivative AC5071 but also reverse the negative effect of RsmA on extracellular Pel production (see Tables 2 and 3). These data allow several conclusions; (i) the rsmBEa and rsmBEhg RNA species are biologically active and are responsible for the activation of extracellular enzyme production; (ii) these RNA species specifically act against the negative effect of RsmA; and (iii) the putative processed rsmB'Ea RNA species is still active, although it is less active than the primary rsmBEa RNA.
TABLE 2.
Reversal of negative effects of rsmA on extracellular Pel production by rsmB in RsmA− strain AC5071 of E. carotovora subsp. carotovora
| Bacterial constructa | Relevant genotype | Pel activityb (10−2) | Relative activity |
|---|---|---|---|
| AC5071/pAKC878 + pCL1920 | rsmAEcc + vector | 0.32 ± 0.01 | 1.0 |
| + pAKC1049 | rsmAEcc + plac-rsmBEcc | 14.4 ± 1.19 | 45.0 |
| + pAKC1004 | rsmAEcc + plac-rsmB′Ecc | 2.46 ± 0.06 | 7.6 |
| + pAKC1061 | rsmAEcc + plac-rsmBEhg | 2.38 ± 0.013 | 7.4 |
| + pAKC1062 | rsmAEcc + plac-rsmBEa | 1.88 ± 0.07 | 5.8 |
| + pAKC1063 | rsmAEcc + plac-rsmB′Ea | 1.63 ± 0.04 | 5.0 |
| AC5071/pAKC892 + pCL1920 | rsmAEhg + vector | 8.63 ± 0.45 | 1.0 |
| + pAKC1049 | rsmAEhg + plac-rsmBEcc | 14.8 ± 0.4 | 1.7 |
| + pAKC1004 | rsmAEhg + plac-rsmB′Ecc | 11.03 ± 0.25 | 1.2 |
| + pAKC1061 | rsmAEhg + plac-rsmBEhg | 196.33 ± 6.6 | 22.7 |
| + pAKC1043 | rsmAEhg + plac-rsmBEa | 110.83 ± 10.9 | 12.7 |
| + pAKC679 | rsmAEhg + plac-rsmB′Ea | 21.47 ± 0.37 | 2.5 |
| AC5071/pAKC894 + pCL1920 | rsmAEa + vector | 12.67 ± 0.57 | 1.0 |
| + pAKC1049 | rsmAEa + plac-rsmBEcc | 18.3 ± 1.4 | 1.5 |
| + pAKC1004 | rsmAEa + plac-rsmB′Ecc | 16.06 ± 0.4 | 1.2 |
| + pAKC1061 | rsmAEa + plac-rsmBEhg | 70.7 ± 4.6 | 5.6 |
| + pAKC1043 | rsmAEa + plac-rsmBEa | 152.17 ± 7.5 | 12 |
| + pAKC679 | rsmAEa + plac-rsmB′Ea | 99.42 ± 4.8 | 7.8 |
Strains were grown at 28°C in minimal salts medium plus sucrose (0.5%, wt/vol) to an absorbance at 600 nm of 2.3, and the culture supernatants were assayed for enzymatic activities.
Pel activity is expressed as units per absorbance unit of culture at 600 nm.
TABLE 3.
Multiple-copy effects of rsmB on extracellular Pel production in E. carotovora subsp. carotovora strains Ecc71 (RsmA+) and AC5071 (RsmA−)
| Bacterial constructa | Relevant phenotype | Pel activityb (10−2) | Relative activity |
|---|---|---|---|
| Ecc71/pCL1920 | RsmA+/vector | 6.6 ± 0.6 | 1.0 |
| Ecc71/pAKC1049 | RsmA+/plac-rsmBEcc | 532 ± 6.5 | 80.6 |
| Ecc71/pAKC1004 | RsmA+/plac-rsmB′Ecc | 149.2 ± 4.2 | 22.6 |
| Ecc71/pAKC1061 | RsmA+/plac-rsmBEhg | 64.3 ± 1.6 | 9.7 |
| Ecc71/pAKC1062 | RsmA+/plac-rsmB′Ea | 41.9 ± 1.5 | 6.3 |
| Ecc71/pAKC1063 | RsmA+/plac-rsmB′Ea | 17.9 ± 0.2 | 2.7 |
| AC5071/pCL1920 | RsmA−/vector | 212.2 ± 4.3 | 1.0 |
| AC5071/pAKC1049 | RsmA−/plac-rsmBEcc | 903.6 ± 3.5 | 4.2 |
| AC5071/pAKC1004 | RsmA−/plac-rsmB′Ecc | 764 ± 12 | 4.0 |
| AC5071/pAKC1061 | RsmA−/plac-rsmBEhg | 502.6 ± 11 | 2.4 |
| AC5071/pAKC1062 | RsmA−/plac-rsmBEa | 725.6 ± 6.8 | 3.4 |
| AC5071/pAKC1063 | RsmA−/plac-rsmB′Ea | 562 ± 5.5 | 2.7 |
Strains were grown at 28°C in minimal salts medium plus sucrose (0.5%, wt/vol) and spectinomycin to an absorbance at 600 nm of 2.3, and the culture supernatants were assayed for enzymatic activities.
Pel activity is expressed as units per absorbance unit of culture at 600 nm.
Our findings reported here and elsewhere (21) reveal two classes of rsmB genes. In one class, the larger primary transcript presumably is processed to yield a smaller and relatively stable RNA species. The other class comprises the rsmB genes that yield a small and stable RNA species not subject to processing. Moreover, based on our findings with strain Ecc71 rsmB transcripts (21), we postulate that the primary transcript binds more RsmA molecules than the processed rsmB RNA does, thereby more effectively lowering the pool of free RsmA. In fact, the analysis of primary and secondary structures of RNA species (data not shown) clearly shows the availability of more putative RsmA binding sites in the primary transcript than in the processed RNA.
Analysis of rsmB and csrB RNA sequences.
A database search revealed that rsmBEa and rsmBEhg RNA sequences are homologs of rsmBEcc of E. carotovora subsp. carotovora (21) and csrB of Escherichia coli (18). Alignment of the ribonucleotide sequences of rsmB from E. carotovora subsp. carotovora, E. herbicola pv. gypsophilae, and E. amylovora, as well as csrB, is shown in Fig. 1A. In addition, phylogram analysis shows that the rsmB RNAs of E. amylovora and E. herbicola pv. gypsophilae fall into a subgroup, suggesting that rsmBEa and rsmBEhg may be evolutionarily and functionally closer. It also was evident that the rsmBEa and rsmBEhg RNAs are genetically closer to the csrB RNA than to the rsmBEcc RNA (data not shown). A noteworthy feature is that the homologies among these RNAs are higher at their 3′ ends than at the 5′ ends. While the last 100-base sequence of rsmBEcc is critical for RNA stability, this sequence is not involved in regulating gene expression (21; Y. Liu and A. K. Chatterjee, unpublished data). Therefore, we propose that the 100-base sequence at the 3′ end, the most highly conserved sequence in the rsmB and csrB RNAs, plays roles in the transcription termination and RNA stability due to its extremely stable stem-loop structure. Moreover, this RNA region does not possess the putative sequences that can be bound by RsmA and CsrA proteins (see below).
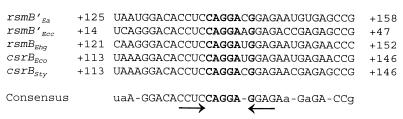
Liu et al. (18) proposed that CsrA binds csrB RNA, most probably to the 7-base repeats in the RNA molecules. The repeats contain the consensus sequence 5′-CAGGA(U/C)G-3′. Repeats carrying the identical consensus sequence also have been found within the rsmBEcc RNA as well as in rsmBEa RNA and rsmBEhg RNA (Fig. 1A), and there is evidence to suggest that these can be bound in vitro by purified RsmAEcc protein (18, 21). However, whether these sequences represent the specific recognition site for CsrA and RsmAEcc proteins is not known. Also, the significance of the sequences flanking the 7-base core sequence is unknown. A comparison of the rsmB and csrB RNA sequences reveals a 34-mer consensus sequence in each RNA species (Fig. 3). The 34-mer sequence has the following specific characteristics in addition to the presence of the 5′-CAGGA(U/C)G-3′ sequence in the middle. (i) The 34-mer sequence is much longer than the previously determined 7-base 5′-CAGGA(U/C)G-3′ sequence. (ii) In each rsmB RNA species, only one copy of this 34-mer sequence exists, whereas additional 5′-CAGGA(U/C)G-3′ sequences are present as multiple modules in every RNA species, i.e., five copies in rsmBEhg, eight copies in rsmBEa, and nine copies in rsmBEcc and csrB. (iii) In instances where there is evidence for RNA processing, the 34-mer consensus sequence occurs in the rsmB′ RNA species, the 3′-end processed product of the primary transcripts. In Ecc71, the rsmB′ RNA, rather than the 5′ region, is responsible for the regulatory role of rsmB RNA, i.e., the activation of exoprotein production (21). (iv) The 34-mer consensus sequence is predicted to carry a conserved secondary structure consisting of a 4-nt stem and a 5-nt [5′-AGGAA(U/C)-3′] loop (Fig. 3). Based on the specific features of the 34-mer consensus sequence, we propose this motif as the signature sequence for the growing rsmB/csrB regulatory RNA family. Indeed, a database search using the 34-mer consensus sequence revealed a single motif in the putative rsmB/csrB homolog of S. enterica serovar Typhimurium (accession number, gi 5730336) (Fig. 3 csrBSty). Furthermore, to test the proposal that the 34-mer motif is present in every rsmB homolog, degenerate primers were designed to be complementary to the nucleotide sequences within the 34-mer motif and to the 3′ terminator region. PCR analysis revealed amplification of rsmB DNA fragments from Serratia marcescens, Shigella flexneri, and Enterobacter aerogenes; moreover, nucleotide sequence analysis confirmed that each PCR product carried one 34-mer motif (W. L. Ma and A. K. Chatterjee, unpublished data). These structural characteristics suggest that the 34-mer sequence represents an ancient motif arising prior to the evolutionary separation of the genera tested and that this motif is important for the basic biological function of the rsmB/csrB RNA regulator.
FIG. 3.
Alignment of the 34-mer consensus sequences in the rsmB RNA species of E. amylovora strain E9 (Ea), E. carotovora subsp. carotovora strain Ecc71 (Ecc), E. herbicola pv. gypsophilae strain PD713 (Ehg), and csrB RNA of Escherichia coli (Eco), and S. enterica serovar Typhimurium (Sty). Positions refer to the nucleotide sequences relative to the 5′ of rsmB′ of E. amylovora and E. carotovora subsp. carotovora and 5′ of rsmB of E. herbicola pv. gypsophilae, Escherichia coli, and S. enterica serovar Typhimurium. Inverted arrows indicate the 4-nt stems of the stem-loop structure. The identical nucleotides of the 7-base repeats are in boldface type.
Effects of rsmBEhg in E. herbicola pv. gypsophilae and E. carotovora subsp. carotovora.
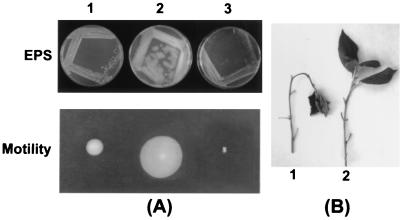
To determine the effects of multiple copies of rsmBEhg, pAKC1042 containing the rsmB gene or the vector pCL1920 was transformed into E. herbicola pv. gypsophilae strain PD713 and E. carotovora subsp. carotovora strain Ecc71. The E. herbicola pv. gypsophilae constructs were tested for extracellular polysaccharide production and motility. As shown in Fig. 4A, columns 1 and 2, multiple copies of rsmBEhg activate EPS production and stimulate swarming motility in E. herbicola pv. gypsophilae strain PD713. Previous studies have shown that rsmBEcc activates pathogenicity factor production (21, 28). To ascertain if rsmBEhg would affect pathogenicity factors, such as phytohormones, we performed Northern blot analysis of the cytokinin (etz) genes in the E. herbicola pv. gypsophilae constructs. Total RNA samples of PD713 carrying pCL1920 or its rsmBEhg+ derivative were hybridized with pre-etz plus etz (17) as the probe. The results show that two RNA bands of 1.4 and 1.0 kb hybridized with the probe, consistent with the previously reported pre-etz plus etz RNA profile of PD713 (17). Also, the levels of pre-etz and etz transcripts were higher with multiple copies of rsmBEhg than with the vector control (Fig. 4B).
FIG. 4.
Effects of multiple copies of rsmBEhg and rsmAEhg in E. herbicola pv. gypsophilae strain PD713. (A) EPS production and motility of E. herbicola pv. gypsophilae strain PD713 carrying pCL1920 (cloning vector, column 1), pAKC1042 (rsmBEhg+, column 2), or pAKC891 (RsmAEhg+, column 3). (B) Northern blot analysis showing multiple-copy effects of rsmAEhg and rsmBEhg on etz (cytokinin gene) transcripts in E. herbicola pv. gypsophilae strain PD713. Lanes: 1, PD713/pCL1920; 2, PD713/pAKC891; 3, PD713/pAKC1042. Each lane contained 15 μg of total RNA.
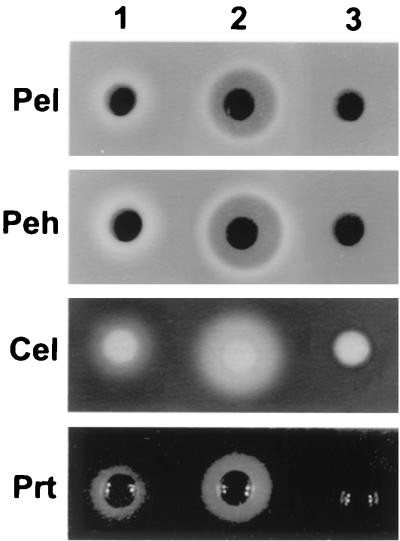
E. carotovora subsp. carotovora strain Ecc71 carrying the rsmBEhg+ plasmid or the cloning vector was tested for extracellular enzyme production. The bacteria were grown in minimal salts medium supplemented with sucrose (0.5%, wt/vol) and spectinomycin, and the culture supernatants were assayed for Pel, polygalacturonase (Peh), protease (Prt) and cellulase (Cel) activities. Figure 5 (columns 1 and 2) shows that Ecc71 carrying the rsmBEhg+ plasmid produced higher levels of Pel, Peh, Prt, and Cel than did Ecc71 carrying the cloning vector pCL1920. The effects of rsmBEhg in both homologous and heterologous systems indicate that rsmBEhg is functionally similar to rsmBEcc (21) (Table 3).
FIG. 5.
Effects of multiple copies of rsmBEhg and rsmAEhg on extracellular enzyme production in E. carotovora subsp. carotovora strain Ecc71. Ecc71 carrying the cloning vector pCL1920 (column 1), pAKC1042 (rsmBEhg+, column 2), or pAKC891 (RsmAEhg+, column 3) were grown in minimal salts medium plus sucrose (0.5%, wt/vol) and spectinomycin to a turbidity of ca. 200 Klett units, and the culture supernatant was used for an agarose plate assay of Pel, Peh, Prt, and Cel activities. Each well contained 20 μl of culture supernatant.
Effects of rsmBEa in E. amylovora and E. carotovora subsp. carotovora.
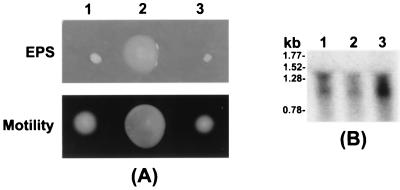
In studies similar to that with rsmBEhg described above, we determined the effects of multiple copies of rsmBEa gene in E. amylovora and E. carotovora subsp. carotovora. The rsmBEa+ plasmid, pAKC1043, or the cloning vector, pCL1920, was transformed into E. amylovora strain E9 and E. carotovora subsp. carotovora strain Ecc71. The E. amylovora constructs were tested for EPS production, motility, and pathogenicity on apple shoots. As shown in Fig. 6A (columns 1 and 2), E. amylovora E9 carrying multiple copies of rsmBEa produced copious amounts of EPS compared to E9 carrying the vector. The bacteria carrying pAKC1043 also were more motile on semisolid agar medium. Furthermore, compared to E9 carrying pCL1920, E9 carrying the rsmBEa+ plasmid wilted apple shoots in a shorter time (data not shown). Like the rsmBEhg gene, rsmBEa activated the production of Pel, Peh, Cel, and Prt in E. carotovora subsp. carotovora strain Ecc71 (data not shown).
FIG. 6.
Effects of multiple copies of rsmBEa and rsmAEa in E. amylovora strain E9. (A) EPS production and motility of E. amylovora strain E9 carrying pCL1920 (cloning vector, column 1), pAKC1043 (rsmBEa+, column 2), or pAKC893 (RsmAEa+, column 3). (B) Pathogenicity of E. amylovora strain E9 carrying the cloning vector pSF6 (B1) and the rsmAEa+ plasmid pAKC120 (B2). E. amylovora strain E9 carrying the vector pSF6 caused the apple shoots to bent down and wilt, whereas E9 carrying the rsmAEa+ plasmid pAKC120 was nonpathogenic in apple shoots.
Neutralization of RsmA by rsmB.
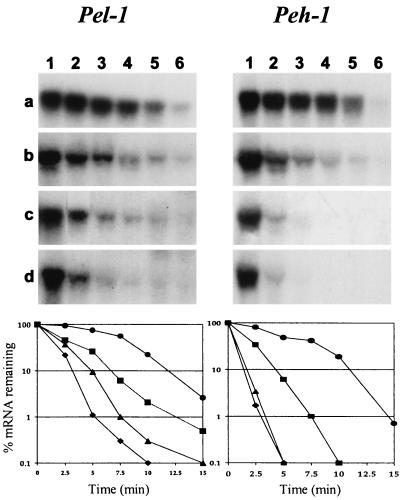
Since rsmBEcc neutralizes the effect of RsmA in E. carotovora subsp. carotovora (21) and csrB neutralizes the effect of CsrA in Escherichia coli (18), it was of interest to determine if the rsmB genes of E. amylovora and E. herbicola pv. gypsophilae also antagonize their own RsmA-like factors. For this propose, we cloned and sequenced the rsmA genes from E. amylovora strain E9 and E. herbicola pv. gypsophilae strain PD713. Analysis of the nucleotide sequences of the two rsmA genes revealed that 183-bp open reading frames encode the putative RsmAEhg and RsmAEa proteins, each consisting of 61 amino acid residues. A homology search disclosed that RsmAEhg and RsmAEa have 93 and 95% identity to RsmAEcc, respectively (Fig. 1B). In E. carotovora subsp. carotovora strain Ecc71, the RsmAEhg+ plasmid pAKC891 repressed extracellular enzyme (Pel, Peh, Cel, and Prt) production as determined by agarose plate assays (Fig. 5, column 3, shows the suppressive effect of rsmAEhg). In addition, in E. herbicola pv. gypsophilae strain PD713, the RsmAEhg+ plasmid pAKC891 repressed EPS production, motility, and the levels of cytokinin transcripts (Fig. 4A, columns 1 and 3, and Fig. 4B). Similarly, RsmAEa+ plasmid pAKC893 suppressed motility on semisolid medium and suppressed EPS production (Fig. 6A, columns 1 and 3) in E. amylovora strain E9. Moreover, E. amylovora strain E9 carrying rsmAEa cosmid pAKC120 was nonpathogenic in apple shoots (Fig. 6B). Since RsmAEhg and RsmAEa promote pel-1 and peh-1 mRNA degradation in E. carotovora subsp. carotovora RsmA strain AC5071 (Fig. 7), it is most likely that, like RsmAEcc and CsrA, these RsmA species also affect mRNA stability and consequently the cognate phenotypes.
FIG. 7.
mRNA stability of pel-1 and peh-1 in E. carotovora subsp. carotovora strain AC5071 (RsmA−) carrying pCL1920 (cloning vector, row a), pAKC891 (RsmAEhg+, row b), pAKC893 (RsmAEa+, row c), or pAKC880 (RsmAEcc+, row d). Rifampin (200 μg/ml) was added to the cultures at a turbidity of ca.160 Klett units, and RNA was extracted at 0 min (lane 1), 2.5 min (lane 2), 5 min (lane 3), 7.5 min (lane 4), 10 min (lane 5), and 15 min (lane 6). Northern hybridization was performed at 65°C with [α-P32]dATP-labeled pel-1 and peh-1 probes. Each lane in row a contained 5 μg of total RNA, and each lane in rows b, c, and d contained 30 μg of total RNA. The blots were exposed to X-ray film for 24 h and analyzed by densitometric scanning. The densitometric results (percentage of remaining mRNA) were plotted against time after rifampin treatment. The circles, squares, triangles, and diamonds represent AC5071 carrying pCL1920, pAKC891, pAKC893, or pAKC880, respectively.
The cloning of both the rsmA and rsmB genes of E. amylovora strain E9 and E. herbicola pv. gypsophilae strain PD713 and the availability of a well-characterized RsmA− strain of E. carotovora subsp. carotovora (AC5071) made it possible for us to test the interaction of these genes. Each of the AC5071 derivatives carrying rsmAEcc, rsmAEa or rsmAEhg was transformed with pAKC1004 (plac-rsmB′Ecc), pAKC1049 (plac-rsmBEcc), pAKC1062 (plac-rsmBEa), pAKC1063 (plac-rsmB′Ea), pAKC1061 (plac-rsmBEhg), or vector pCL1920. The bacterial constructs were assayed for extracellular Pel activity. The data in Table 2 show that the three rsmB genes could reverse the repressive effects of all the rsmA species; i.e., the genes are functional in heterologous systems. However, the efficiency of reversal of rsmB varies depending on the source of the rsmA species. While rsmBEcc RNA was most effective in reversing the effect of RsmAEcc, it was least effective in neutralizing the effects of RsmAEa or RsmAEhg. On the other hand, each of these RsmA species was more effectively neutralized by the cognate rsmB RNA species. These observations imply a degree of specificity in RsmA-rsmB interaction. In this context, it is perhaps significant that rsmBEa and rsmBEhg RNA species belong to the same subgroup whereas rsmBEcc RNA belongs to another subgroup genetically distant from rsmBEa and rsmBEhg RNAs. These observations raise the possibility that structural differences among the three rsmB RNA species could account for the differential effects of rsmB RNAs on RsmA species. Table 2 also shows that in the RsmAEcc− background, the suppressive effect on Pel production by the three rsmA species varied: rsmAEcc was most suppressive, followed by rsmAEa and then rsmAEhg. However, genetic variation among the three RsmA proteins is minor, with alterations mostly occurring within the last 6 amino acid residues within their C-terminal regions (Fig. 1B). It remains to be determined if the variations in RsmA proteins affect the binding specificity of rsmB RNAs. However, we prefer the view that the differences among these rsmB RNA species probably determine the efficiency of interactions with RsmA species and the consequent extracellular Pel production. In addition, these differences in RNA may affect the RsmA-independent regulatory pathway(s) through which rsmB acts to regulate gene expression (21).
To ascertain that the rsmB effects were mediated via RsmA, we compared the effects of plasmids carrying the rsmB genes in RsmA+ and RsmA− strains of E. carotovora subsp. carotovora. The data (Table 3) show that rsmBEa and rsmBEhg better stimulated enzyme production in RsmA+ bacteria than in the RsmA− strain. In RsmA− bacteria, the degrees of stimulation of Pel production were three- and twofold, which contrasts with six- and ninefold stimulation of Pel production in RsmA+ bacteria by the rsmBEa and rsmBEhg genes. Generally similar effects were seen with rsmBEcc, although the degree of stimulation in RsmA+ bacteria was much higher with this gene (80-fold stimulation) than with the rsmB genes from E. amylovora (6-fold stimulation) or E. herbicola pv. gypsophilae (9-fold stimulation). These differences notwithstanding, the data allow the conclusion that rsmB activates gene expression by neutralizing the RsmA effect. However, these results also indicate that rsmB RNA species play an additional regulatory role in the activation of extracellular enzyme production, which is RsmA independent.
rsmB RNAs bind RsmAEcc.
Previous studies with Escherichia coli and E. carotovora subsp. carotovora have shown that RsmA/CsrA binds rsmB RNA/csrB RNA (18, 21). Subsequent studies with E. carotovora subsp. carotovora have established that most of the regulatory effect of rsmB is channeled via RsmA. In light of the effects of rsmBEa and rsmBEhg on RsmAEcc, we considered it important to determine if RsmAEcc binds these rsmB RNA species. The results of gel mobility shift assays (Fig. 8) show that the purified RsmAEcc protein binds each of those two rsmB RNA species. The RsmAEcc-rsmB binding was prevented by the addition of excess unlabeled RNA to the reaction mixture (Fig. 8, lanes 3 and 7). In addition, in vitro studies suggest specificity in the binding of RsmAEcc and these rsmB RNA species, since yeast tRNA had no effect on this binding (lanes 4 and 8).
FIG. 8.
RNA mobility shift assay for binding of RsmAEcc to the rsmB RNAs. rsmB RNA probes of E. herbicola pv. gypsophilae and E. amylovora were synthesized from pAKC1046 and pAKC1045 in vitro by T7 RNA polymerase in the presence of [α-32P]-UTP. Labeled RNAs (0.1 ng, 4,000 cpm) were incubated without RsmA, with 5.0 ng of affinity-purified RsmA, with 5.0 ng of affinity-purified RsmA in the presence of a 50-fold excess of unlabeled probes, or with 5.0 ng of affinity-purified RsmA in the presence of a 50-fold excess of yeast tRNA. Lanes: 1, labeled rsmBEa RNA; 2, labeled rsmBEa RNA plus RsmAEcc; 3, labeled rsmBEa RNA plus RsmAEcc plus 50-fold unlabeled rsmBEa RNA; 4, labeled rsmBEa RNA plus RsmAEcc plus 50-fold yeast tRNA; 5, labeled rsmBEhg RNA; 6, labeled rsmBEhg RNA plus RsmAEcc; 7, labeled rsmBEhg RNA plus RsmAEcc plus 50-fold unlabeled rsmBEhg RNA; 8, labeled rsmBEhg RNA plus RsmAEcc plus 50-fold yeast tRNA.
Occurrence of rsmB homologs in Erwinia species and other enterobacteria.
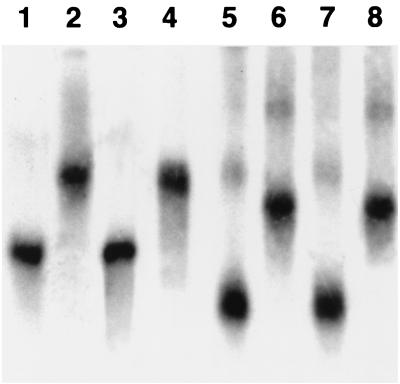
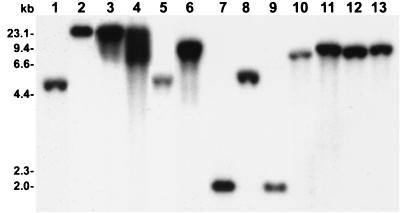
It has been established that rsmA occurs in enterobacteria and P. aeruginosa (38) and perhaps even in other bacteria such as Bacillus subtilis and Haemophilus influenzae (12, 35). Our previous findings (28) have revealed that rsmBEcc (formerly aepH) occurs in E. carotovora subsp. carotovora and E. carotovora subsp. atroseptica strains. To examine if an rsmB homolog occurs in other Erwinia and enterobacterial species, we conducted Southern hybridizations with the rsmB probe from E. herbicola pv. gypsophilae strain PD713. The data (Fig. 9) show that rsmBEhg hybridized to all Erwinia and other enterobacterial species tested, indicating the presence of rsmB homologs in these bacteria. To further strengthen this physical evidence, we used E. coli csrB as the probe. The same size bands that hybridized with the rsmBEa probe also hybridized with the csrB probe (data not shown). These data demonstrate that rsmB sequences have been conserved in these bacteria. However, the differences in the sizes of the hybridizing fragments suggest that sequences of the rsmB-like genes and the DNA flanking them may have diverged.
FIG. 9.
Southern hybridization of EcoRI-digested chromosomal DNAs of Erwinia and other enterobacterial strains with rsmB of E. herbicola pv. gypsophilae strain PD713. Lanes: 1, E. chrysanthemi strain Ec16; 2, E. amylovora strain E9; 3, E. herbicola strain EH105; 4, E. herbicola pv. gypsophilae strain PD713; 5, E. rhapontici strain Erl; 6, E. stewartii strain DC283; 7, Escherichia coli strain K-12; 8, S. enterica serovar Typhimurium strain LT2; 9, Serratia marcescens strain Sm1; 10, Yersinia pseudotuberculosis strain Yp1; 11, Shigella flexneri strain Sf1; 12, Enterobacter aerogenes strain Ena1; 13, Klebsiella pneumoniae strain Kp1. Southern hybridization was performed at 65°C. The blot was washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) for 15 min at room temperature followed by 30 min in 2× SSC–0.1% sodium dodecyl sulfate at 65°C. A 500-bp HincII-EcoRV fragment from pAKC1042 was used as the rsmBEhg probe.
In conclusion, we have characterized the rsmB genes cloned from E. amylovora and E. herbicola pv. gypsophilae. These genes have high levels of genetic, structural, and functional homology among themselves and to that of E. carotovora subsp. carotovora. The suppression of EPS production and pathogenicity of E. amylovora by RsmAEa, the inhibition of expression of a cytokinin gene in E. herbicola pv. gypsophilae by RsmAEhg, and the reversal of the negative effects of the RsmA species by the rsmB genes in E. carotovora subsp. carotovora demonstrate that the rsmA-rsmB pairs play important regulatory roles in these bacteria. The fully conserved KH motifs in putative products of these rsmA genes strongly suggest that RsmAEa and RsmAEhg, like RsmAEcc, bind RNA species. Indeed, studies with the pel and peh genes suggest that RsmAEhg and RsmAEa affect mRNA stability, most probably by binding and promoting mRNA decay. rsmBEhg and rsmBEa counteract the RsmA effects, probably by reducing the pool of free RsmA due to the formation of a biologically inactive RsmA-rsmB ribonucleoprotein complex. The occurrence of rsmB homologs in all enterobacterial species included in this study strongly suggests that many more bacteria employ this regulatory system to modulate their gene expression. This is clearly supported by the pleiotropic effect of RsmA of P. aeruginosa and P. fluorescens (3). Our work has also raised several other issues that await clarification. For example, what is the teleological significance of the processing of rsmB transcripts in some bacteria but not in others? Does RsmA binding prime RNAs for degradation by nucleases? Does RsmA act in conjunction with other factors which are responsible for the decay of specific transcripts? Since the RsmA-rsmB pair controls many factors important in bacterial ecology and in the production of useful metabolites, we expect that these and other issues will be resolved in the near future.
ACKNOWLEDGMENTS
Our work was supported by the National Science Foundation (grant MCB-9728505) and the Food for the 21st Century program of the University of Missouri.
We thank S. Manulis for E. herbicola pv. gypsophilae strains and the etz plasmid, and we thank Jeanne Erickson and Judy D. Wall for reviewing the manuscript.
Footnotes
We affectionately dedicate this paper to the memory of Robert N. Goodman, whose insight and numerous contributions have led to a better understanding of the biology of these and other plant-pathogenic bacteria.
Journal series 13,022 of the Missouri Agriculture Experiment Station.
REFERENCES
- 1.Aiba H, Adhya S, de Crombrugghe B. Evidence for two functional gal promoters in intact Escherichia coli. J Biol Chem. 1981;256:111905–11910. [PubMed] [Google Scholar]
- 2.Barras F, Thurn K K, Chatterjee A K. Resolution of four pectate lyase structural genes of Erweinia chrysanthemi (EC16) and characterization of the enzymes produced in Escherichia coli. Mol Gen Genet. 1987;209:319–325. doi: 10.1007/BF00329660. [DOI] [PubMed] [Google Scholar]
- 3.Blumer C, Heeb S, Pessi G, Haas D. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc Natl Acad Aci USA. 1999;96:14073–14078. doi: 10.1073/pnas.96.24.14073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bugert P, Geider K. Molecular analysis of the ams operon required for exopolysaccharide synthesis of Erwinia amylovora. Mol Microbiol. 1995;15:917–933. doi: 10.1111/j.1365-2958.1995.tb02361.x. [DOI] [PubMed] [Google Scholar]
- 5.Chatterjee A, Liu Y, Dumenyo C K, Chatterjee A K. Inactivation of rsmA leads to overproduction of extracellular pectinases, cellulases, and proteases in Erwinia carotovora subsp. carotovora in the absence of the starvation/cell density-sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Appl Environ Microbiol. 1995;61:1959–1967. doi: 10.1128/aem.61.5.1959-1967.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee A, McEvoy J L, Chambost J P, Blasco F, Chatterjee A K. Nucleotide sequence and molecular characterization of pnlA, the structural gene for damage-inducible pectin lyase of Erwinia carotovora subsp. carotovora 71. J Bacteriol. 1991;173:1765–1769. doi: 10.1128/jb.173.5.1765-1769.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chatterjee A K, Buchanan G E, Behrens M K, Starr M P. Synthesis and excretion of polygalacturonic acid trans-eliminase in Erwinia, Yersinia, and Klebsiella species. Can J Microbiol. 1979;25:94–102. doi: 10.1139/m79-014. [DOI] [PubMed] [Google Scholar]
- 8.Chatterjee A K, Thurn K K, Tyrell D J. Isolation and characterization of Tn5 insertion mutants of Erwinia chrysanthemi that are deficient in polygalacturonate catabolic enzymes oligogalacturonate lyase and 3-deoxy-d-glycero-2,5-hexodiulosonate dehydrogenase. J Bacteriol. 1985;162:708–714. doi: 10.1128/jb.162.2.708-714.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cotter P A, Miller J F. In vivo and ex vivo regulation of bacterial virulence gene expression. Curr Opin Microbiol. 1998;1:17–26. doi: 10.1016/s1369-5274(98)80138-0. [DOI] [PubMed] [Google Scholar]
- 10.Cui Y, Chatterjee A, Liu Y, Dumenyo C K, Chatterjee A K. Identification of a global repressor gene, rsmA, of Erwinia carotovora subsp. carotovora that controls extracellular enzymes, N-(3-oxohexanoyl)-l-homoserine lactone, and pathogenicity in soft-rotting Erwinia spp. J Bacteriol. 1995;177:5108–5115. doi: 10.1128/jb.177.17.5108-5115.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui Y, Mukherjee A, Dumenyo C K, Liu Y, Chatterjee A K. rsmC of the soft-rotting bacterium Erwinia carotovora subsp. carotovora negatively controls extracellular enzyme and harpinEcc production and virulence by modulating levels of regulatory RNA (rsmB) and RNA-binding protein (RsmA) J Bacteriol. 1999;181:6042–6052. doi: 10.1128/jb.181.19.6042-6052.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, Fitzhugh W, Fields C A, Gocayne J D, Scott J D, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 13.Grant M, Mansfield J. Early events in host-pathogen interactions. Curr Opin Plant Biol. 1999;2:312–319. doi: 10.1016/S1369-5266(99)80055-7. [DOI] [PubMed] [Google Scholar]
- 14.Hauben L, Moore E R B, Vauterin L, Steenackers M, Mergaert J, Verdonck L, Swings J. Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst Appl Microbiol. 1998;21:384–397. doi: 10.1016/S0723-2020(98)80048-9. [DOI] [PubMed] [Google Scholar]
- 15.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 16.Lerner C G, Inouye M. Low copy number pasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white inset screening capability. Gene. 1990;18:4631. doi: 10.1093/nar/18.15.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichter A, Barash I, Valinsky L, Manulis S. The genes involved in cytokinin biosynthesis in Erwinia herbicola pv. gypsophilae: characterization and role in gall formation. J Bacteriol. 1995;177:4457–4465. doi: 10.1128/jb.177.15.4457-4465.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu M Y, Gui G, Wei B, Preston III J F, Oakford L, Yueksel U, Giedroc D P, Romeo T. The RNA molecule CsrB binds to the global regulatory protein CsrA and antagonizes its activity in Escherichia coli. J Biol Chem. 1997;272:17502–17510. doi: 10.1074/jbc.272.28.17502. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Chatterjee A, Chatterjee A K. Nucleotide sequence and expression of a novel pectate lyase gene (pel-3) and a closely linked endopolygalacturonase gene (peh-l) of Erwinia carotovora subsp. carotovora 71. Appl Environ Microbiol. 1994;60:2545–2552. doi: 10.1128/aem.60.7.2545-2552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu Y, Chatterjee A, Chatterjee A K. Nucleotide sequence, organization and expression of rdgA and rdgB genes that regulate pectin lyase production in the plant pathogenic bacterium Erwinia carotovora subsp. carotovora in response to DNA-damaging agents. Mol Microbiol. 1994;14:999–1010. doi: 10.1111/j.1365-2958.1994.tb01334.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu Y, Cui Y, Mukherjee A, Chatterjee A K. Characterization of a novel RNA regulator of Erwinia carotovora ssp. carotovora that controls production of extracellular enzymes and secondary metabolites. Mol Microbiol. 1998;29:219–234. doi: 10.1046/j.1365-2958.1998.00924.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Murata H, Chatterjee A, Chatterjee A K. Characterization of a novel regulatory gene aepA that controls extracellular enzyme production in the phytopathogenic bacterium Erwinia carotovora ssp carotovora. Mol Plant-Microbe Interact. 1993;6:299–308. doi: 10.1094/mpmi-6-299. [DOI] [PubMed] [Google Scholar]
- 23.Manulis S, Havivchesner A, Brandl M T, Lindow S E, Barash I. Differential involvement of indole-3-acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae. Mol Plant-Microbe Interact. 1998;11:634–642. doi: 10.1094/MPMI.1998.11.7.634. [DOI] [PubMed] [Google Scholar]
- 24.McEvoy J L, Murata H, Chatterjee A K. Molecular cloning and characterization of an Erwinia carotovora subsp. carotovora pectin lyase gene that responds to DNA-damaging agents. J Bacteriol. 1990;172:3284–3289. doi: 10.1128/jb.172.6.3284-3289.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukherjee A, Cui Y, Liu Y, Chatterjee A K. Molecular characterization and expression of the Erwinia carotovora hrpNEcc gene, which encodes an elicitor of the hypersensitive reaction. Mol Plant-Microbe Interact. 1997;10:462–71. doi: 10.1094/MPMI.1997.10.4.462. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee A, Cui Y, Liu Y, Dumenyo C K, Chatterjee A K. Global regulation in Erwinia species by Erwinia carotovora rsmA, a homologue of Escherichia coli csrA: repression of secondary metabolites, pathogenicity and hypersensitive reaction. Microbiology. 1996;142:427–434. doi: 10.1099/13500872-142-2-427. [DOI] [PubMed] [Google Scholar]
- 27.Mukherjee A, Cui Y, Ma W, Liu Y, Chatterjee A K. hexA of Erwinia carotovora ssp. carotovora strain Ecc71 negatively regulates production of RpoS and rsmB RNA, a global regulator of extracellular proteins, plant virulence and the quorum sensing signal, N-(3-oxohexanoyl)-l-homoserine lactone. Environ Microbiol. 2000;2:203–215. doi: 10.1046/j.1462-2920.2000.00093.x. [DOI] [PubMed] [Google Scholar]
- 28.Murata H, Chatterjee A, Liu Y, Chatterjee A K. Regulation of the production of extracellular pectinase, cellulase, and protease in the soft rot bacterium Erwinia carotovora subsp. carotovora: evidence that aepH of E. carotovora subsp. carotovora 71 activates gene expression in E. carotovora subsp. carotovora, E. carotovora subsp. atroseptica, and Escherichia coli. Appl Environ Microbiol. 1994;60:3150–3159. doi: 10.1128/aem.60.9.3150-3159.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murata H, McEvoy J L, Chatterjee A, Collmer A, Chatterjee A K. Molecular cloning of an aepA gene that activates production of extracellular pectolytic, cellulolytic, and proteolytic enzymes in Erwinia carotovora subsp. carotovora. Mol Plant-Microbe Interact. 1991;4:239–246. [Google Scholar]
- 30.Musco G, Stier G, Joseph C, Morelli M A C, Nilges M, Gibson T J, Pastore A. Three-dimensional structure and stability of the KH domain—molecular insights into the fragile X syndrome. Cell. 1996;85:237–245. doi: 10.1016/s0092-8674(00)81100-9. [DOI] [PubMed] [Google Scholar]
- 31.Perombelon M C M. The genus Erwinia. In: Balows A, Trüper H G, Dworkin M, Schleifer K-H, editors. The prokaryotes: a handbook on the biology of bacteria. Ecophysiology, isolation, identification, applications. New York, N.Y: Springer-Verlag; 1992. pp. 2899–2921. [Google Scholar]
- 32.Politis D, Goodman R N. Fine structure of extracellular polysaccharide of Erwinia amylovora. Appl Environ Microbiol. 1980;40:596–607. doi: 10.1128/aem.40.3.596-607.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Romeo T. Global regulation by the small RNA-binding protein CsrA and the non-coding RNA molecule CsrB. Mol Microbiol. 1998;29:1321–1330. doi: 10.1046/j.1365-2958.1998.01021.x. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Habor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Seldo B, Lazarevic V, Mauel C, Karamata D. Sequence of the 305 degrees-307 degrees region of the Bacillus subtilis chromosome. Microbiology. 1996;142:3079–3088. doi: 10.1099/13500872-142-11-3079. [DOI] [PubMed] [Google Scholar]
- 36.Selvaraj G, Fong Y C, Iyer V N. A portable DNA sequence carrying the cohesive site (cos) of the bacteriophage lambda and the mob (mobilization) region of the broad-host-range plasmid RK2: a module for the construction of new cosmids. Gene. 1984;32:235–241. doi: 10.1016/0378-1119(84)90051-9. [DOI] [PubMed] [Google Scholar]
- 37.Thurn K K, Chatterjee A K. Isolation and protein composition of the outer membrane of Erwinia amylovora. Curr Microbiol. 1982;7:87–92. [Google Scholar]
- 38.White D, Hart M E, Romeo T. Phylogenetic distribution of the global regulatory gene csrA among eubacteria. Gene. 1996;182:221–223. doi: 10.1016/s0378-1119(96)00547-1. [DOI] [PubMed] [Google Scholar]
- 39.Zink R T, Kemble R J, Chatterjee A K. Transposon Tn5 mutagenesis in Erwinia carotovora subsp. carotovora and E. carotovora subsp. atroseptica. J Bacteriol. 1984;157:809–814. doi: 10.1128/jb.157.3.809-814.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]