Abstract
Regulated expression of AmyQ α-amylase of Bacillus amyloliquefaciens was used to examine the capacity of the protein secretion apparatus of B. subtilis. One B. subtilis cell was found to secrete maximally 10 fg of AmyQ per h. The signal peptidase SipT limits the rate of processing of the signal peptide. Another limit is set by PrsA lipoprotein. The wild-type level of PrsA was found to be 2 × 104 molecules per cell. Decreasing the cellular level of PrsA did not decrease the capacity of the protein translocation or signal peptide processing steps but dramatically affected secretion in a posttranslocational step. There was a linear correlation between the number of cellular PrsA molecules and the number of secreted AmyQ molecules over a wide range of prsA and amyQ expression levels. Significantly, even when amyQ was expressed at low levels, overproduction of PrsA enhanced its secretion. The finding is consistent with a reversible interaction between PrsA and AmyQ. The high cellular level of PrsA suggests a chaperone-like function. PrsA was also found to be essential for the viability of B. subtilis. Drastic depletion of PrsA resulted in altered cellular morphology and ultimately in cell death.
Proteins synthesized with a signal peptide are secreted from bacterial cells by the action of the protein secretion apparatus, which consists of several components involved in protein targeting, translocation, signal peptide processing, and posttranslocational folding (4, 7). Extensive studies of Escherichia coli and Bacillus subtilis have identified and characterized to a substantial extent the components of the apparatus that translocates secretory proteins across the cytoplasmic membrane. In B. subtilis, they include the SecY, SecE, and SecG proteins, which form the core of the translocation channel or translocator (30, 47) and are associated in the membrane with the SecDF protein (3). Furthermore, there are several signal peptidases (43, 45). The role of SecA ATPase on the cis side of the membrane in targeting and coupling the energy required for translocation has been well established (15, 48). Many components of the secretion apparatus are known to be under temporal control; their maximal level of expression parallels the onset of protein secretion in the early stationary growth phase (15, 43).
The stages of protein secretion that take place outside the cytoplasmic membrane are less well understood. A central feature of secretion is posttranslocational folding. The correct folding of many secreted proteins is not spontaneous but dependent on assisting folding factors. In E. coli they include protein-specific chaperones, periplasmic peptidyl-prolyl cis/trans isomerases, and enzymes (Dsb proteins) involved in the formation and rearrangement of disulfide bonds (8, 16, 19, 31, 32). The depletion of foldases such as SurA and PpiD causes misfolding stress that activates the ςE- and cpx-dependent stress response (5, 6, 31). This results in the induction of expression of the periplasmic protease and foldases (6, 37), often resulting in the degradation of misfolded and other abnormal proteins in the extracytoplasmic compartment of the cell.
In the gram-positive bacterium B. subtilis, only one protein outside the cytoplasmic membrane, PrsA, is known to be involved in protein secretion. PrsA is a lipoprotein that consists of a 33-kDa lysine-rich protein part and the N-terminal cysteine with a thiol-linked diacylglycerol anchoring the protein to the outer leaflet of the cytoplasmic membrane (21, 23, 28). The PrsA protein is crucial for efficient secretion of a number of exoproteins. In prsA mutants, the secretion and stability of some model proteins is decreased, while overproduction of PrsA enhances the secretion of exoproteins engineered to be expressed at a high level (18, 23, 28). Although the nature of the PrsA protein hints at an activity outside the cytoplasmic membrane, its mode of action and interaction with other components of the secretion apparatus and the specific steps(s) of secretion in which it is involved remain to be elucidated.
High-level expression of secretory proteins can saturate the secretion apparatus in B. subtilis. In strains overexpressing the α-amylase of B. amyloliquefaciens (AmyQ) or of B. licheniformis (AmyL), there is cell-associated accumulation of precursors with uncleaved signal peptide (pre-AmyQ and pre-AmyL), either associated with the cytoplasmic membrane or in the cytoplasm (15, 22). There were no cell-associated precursors of overexpressed levansucrase or α-amylase (AmyE) of B. subtilis, but processed, mature protein accumulated with slow release from the cell, indicating a rate-limiting step for these proteins after translocation and cleavage of the signal peptide (26). High-level expression of AmyQ also resulted in cell-associated accumulation of mature α-amylase in addition to the precursor (22). However, quantitative studies on the capacity of the bacterial protein secretion apparatus are few (2), and there are no published data on B. subtilis. Furthermore, it is not known how the translocator complex interacts with the components involved in the later stages of secretion, including posttranslocational folding; the contribution of these two stages to the overall secretion capacity remains unclear.
In this work, we first determined the maximal capacity of the wild-type secretion apparatus of B. subtilis in terms of translocation and the processing of pre-AmyQ. The PrsA protein was found to act independently of the translocator complex, its activity being confined to the cellular compartment outside the cell membrane. The rate of AmyQ secretion was dependent on the cellular level of PrsA in a manner consistent with a mechanism affecting folding of AmyQ. The necessity of PrsA for viability was established, suggesting that the role of PrsA in the folding of extracytoplasmic proteins is not restricted to exoproteins.
MATERIALS AND METHODS
Plasmids, bacterial strains, and growth conditions.
Plasmids and B. subtilis strains are listed in Tables 1 and 2, respectively. Bacteria were grown in L broth, modified 2× L broth (2% tryptone, 1% yeast extract, 1% NaCl), and Spizizen's minimal salts medium. Modified 2× L broth was supplemented with 2% starch when AmyQ secretion was studied. For plasmid maintenance, chloramphenicol (5 μg/ml), kanamycin (10 μg/ml), erythromycin (1 or 100 μg/ml), or ampicillin (100 μg/ml) was added to the culture media.
TABLE 1.
Plasmids used in this study
| Plasmid | Characteristics | Reference or source |
|---|---|---|
| pDG148 | Apr Kmr, Pspac | 42 |
| pGDL100 | Kmr, contains replication functions of the Lactococcus lactis WG2 plasmid pWVO1, encodes pre-(A13i)-β-lactamase and sipT | 43 |
| pKTH10 | Kmr, pUB110 derivative carrying the α-amylase gene (amyQ) of B. amyloliquefaciens | 35 |
| pKTH277 | Cmr Emr, pHP13 derivative carrying the prsA gene | 21 |
| pKTH1601 | Apr Tetr, Cmr, pJH101 derivative carrying the amyQ gene and a 0.9-kb fragment of the ywlG region of the B. subtilis chromosome | 20 |
| pKTH3326 | Emr Cmr, pE194 derivative carrying a prsA deletion cassette | This study |
| pKTH3327 | pDG148 derivative with the prsA gene placed under Pspac control | This study |
| pKTH3339 | pSX50 derivative for expression of xylose-inducible amyQ | This study |
| pKTH3362 | pKTH1601 derivative carrying the prsA gene | This study |
| pKTH3384 | pMUTIN4 derivative with a 0.3-kb 5′ fragment of the prsA gene | This study |
| pMUTIN4 | Apr Emr, integrable vector | 46 |
| pSX50 | Cmr, Pxyn | 14 |
TABLE 2.
B. subtilis strains used in this study
| Strain | Characteristics | Reference or source |
|---|---|---|
| IH6525 | prsA3 hisA1 trpC2 (pKTH10) | 22 |
| IH6538 | glyB133 hisA1 trpC2 | 21 |
| IH6961 | glyB133 (::pKTH1601) | S. Leskeläa |
| IH6973 | Low protease (total) sacA321 (pKTH3327) | This study |
| IH7075 | IH6973 prsA::cat (pKTH3327) | This study |
| IH7118 | IH6525(::pKTH3362) | This study |
| IH7120 | IH6538(::pKTH3362) | This study |
| IH7136 | IH6538(::pKTH1601, pKTH3327) | This study |
| IH7138 | IH6538(::pKTH3362, pKTH10) | This study |
| IH7171 | prsA3 glyB133 hisA1 | H.-L. Hyyryläinena |
| IH7162 | IH7136 prsA::cat | This study |
| IH7163 | IH7138 prsA::cat | This study |
| IH7211 | 168(::pKTH3384) | This study |
| IH7441 | IH6538(pKTH3339) | This study |
| IH7558 | IH7171(pKTH3339) | This study |
| IH7673 | IH6538(::pKTH3384, pKTH3339) | This study |
| IH7983 | IH7441(pGDL100) | This study |
| 168 | trpC2 | 1A1 in BGSCb |
National Public Health Institute, Helsinki, Finland.
BGSC, Bacillus Genetic Stock Center, Columbus, Ohio.
DNA manipulation and strain constructions.
To construct plasmid pKTH3327, in which the expression of prsA is under the control of the spac promoter (Pspac), a fragment containing the ribosome-binding site, the coding region, and the transcription termination site of the prsA gene was amplified by PCR and then cloned into the HindIII site of pDG148 (42). The PCR primers were 5′-CTCCACAAGCTTGGAATGATTAGGAGTGTT-3′ and 5′-CTCCACAAGCTTGCAGTTCTCAGCAGCATG-3′, and the template was the DNA of pKTH277 (21). The chromosomal prsA gene was made controllable by using pMUTIN4 (46). A 0.3-kb fragment of prsA containing the ribosome-binding site and part of the coding region was amplified with primers 5′-CTCCACAAGCTTGGAATGATTAGGAGTGTT-3′ and 5′-GCGGATCCCGAGGGCAGTATATTG-3′, and the obtained fragment was cloned between HindIII and BamHI sites of pMUTIN4. One plasmid with the prsA fragment, pKTH3384, was transformed into B. subtilis strain 168. The plasmid was integrated into the prsA locus by a Campbell-type mechanism, and the expected construct, which placed the prsA gene under Pspac, was confirmed by Southern blotting. To disrupt the chromosomal prsA gene, we constructed a DNA cassette containing a fragment from the 5′ flanking region of prsA, the chloramphenicol acetyltransferase gene (cat) of pC194, and a fragment from the 3′ flanking region of prsA. The fragments of the 5′ and 3′ regions were 524 and 500 bp and were 154 nucleotides upstream and 76 nucleotides downstream from the coding region of the prsA gene, respectively. Plasmid pKTH3326, which carries the cassette, was linearized with BclI and transformed into strain IH6973 by selecting for chloramphenicol resistance. One transformant, which was Cmr but Ems and shown by PCR and Southern hybridization to have the prsA gene replaced with the cat gene, was named IH7075.
Strain IH7163 was constructed as follows. The 1.7-kb EcoRI-BamHI fragment of pKTH3327 containing the Pspac-controlled prsA (Pspac-prsA) and PpenP-lacI genes were inserted between the respective sites of pKTH1601 to construct pKTH3362. pKTH3362 also carries a 0.9-kb fragment of the ywlG region of the B. subtilis chromosome (24), enabling integration of the plasmid into the chromosome by a Campbell-type mechanism (10, 11, 20). A prsA3 mutant, IH6525, was transformed with pKTH3362 for Cmr to obtain IH7118, and isopropyl-β-d-thiogalactopyranoside (IPTG)-dependent expression of Pspac-prsA and complementation of the prsA3 mutation was confirmed. Then a glyB auxotrophic strain, IH6538, was transformed with the chromosome of IH7118 to introduce the IPTG-inducible prsA gene into this strain. The strain obtained, IH7120, was made AmyQ secreting by transformation with pKTH10. Finally, the native prsA gene was disrupted by transformation with the chromosome of IH7075 (prsA::cat) and selecting for glyB+. The prsA::cat marker of IH7075 is cotransformed with the glyB marker at a high frequency (about 40%). IPTG dependency of AmyQ secretion revealed recombinants of prsA::cat, one of which was named IH7163. Strain IH7136 harbors plasmid pKTH3327 (Pspac-prsA) and one copy of the amyQ gene integrated in the chromosome (ywlG locus). To construct IH7162, the chromosomal prsA gene of IH7136 was disrupted by transformation with the DNA of IH7075 as above.
In pKTH3339, the amyQ gene was placed under Pxyn control by inserting a PCR fragment (template pKTH10) containing the ribosome-binding site, the coding region, and the transcription termination site of amyQ between ClaI and SalI sites of the expression vector pSX50 (14).
Quantitative immunoblotting.
Cells were harvested from 1 ml of culture by centrifugation and resuspended in 100 μl of protoplast buffer (20 mM potassium phosphate [pH 7.5], 15 mM MgCl2, 20% sucrose, 1 mg of lysozyme/ml). The cell suspension was incubated for 15 min at 37°C; 25 μl of sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (63 mM Tris-HCl [pH 6.5], 2% SDS, 5% β-mercaptoethanol, 9% glycerol) was added to the cell suspension and to 0.1 ml of culture medium prior to boiling for 10 min and the separation of proteins by SDS-PAGE using the Mini-Protean II electrophoresis system (Bio-Rad). Proteins were transferred to an Immobilon-P membrane (Millipore) in a Mini Trans-Blot cell (Bio-Rad) and visualized by specific antisera, horseradish peroxidase-conjugated goat anti-rabbit immunoglobulin IgG (Bio-Rad), and ECL (enhanced chemiluminescence) immunoblotting detection reagents (Amersham). The membrane was placed on Hyperfilm-βmax (Amersham), and densities of protein bands were measured quantitatively by optical scanning with Bio Image (Milligen/Biosearch).
Determination of the specific secretion rate and number of AmyQ and PrsA molecules.
In all gels subjected to quantitative immunoblotting, a series of dilutions of purified AmyQ or PrsA protein with a known amount was applied in addition to the cell samples to be analyzed. Integrated optical densities of protein bands of AmyQ and PrsA visualized by ECL immunodetection were obtained by optical scanning. Amounts of the proteins in the cell samples were calculated from a standard curve drawn with data for the standards. The specific secretion rates of AmyQ were obtained by first plotting α-amylase concentrations in the medium versus time of incubation. Tangents of the resulting curves yielded the secretion rates, which were then divided by cell densities to obtain the specific secretion rates. Cell densities were determined by microscopically counting cells with a Petroff-Hausser counting chamber. The specific secretion rates were expressed as femtograms per hour per cell. The number of AmyQ molecules in the culture medium and PrsA molecules in the cell were calculated using the molecular weights 54,820 and 30,640 for AmyQ and PrsA, respectively.
Protease accessibility of pre-AmyQ in protoplasts.
The expression of Pxyn-amyQ in IH7441(pKTH3339) and IH7558(prsA3, pKTH3339) was induced at a cell density of 100 Klett units (Klett 100) with 0.2% xylose. Cells were collected after 1 h of induction by centrifugation (2,500 × g, 10 min) and resuspended in 1 ml of protoplast buffer (20 mM potassium phosphate [pH 7.5], 15 mM MgCl2, 20% sucrose, 1 mg of lysozyme/ml) (fivefold concentrated). After incubation at 37°C for 30 min, the formed protoplasts were centrifuged at 2,500 × g for 5 min, and the pellet was resuspended in 1 ml of protoplast buffer. The resuspended protoplasts were then divided into three portions. One portion was diluted by adding only protoplast buffer (twofold-concentrated preparation). Another was similarly diluted with protoplast buffer containing 1 mg of trypsin/ml. The protoplasts in the third portion were solubilized in 2% Triton X-100 and then diluted with trypsin-containing protoplast buffer. These three protoplast preparations were incubated at 37°C for 30 min, after which trypsin inhibitor (final concentration, 1.2 mg/ml) was added. The trypsin-treated and nontreated protoplasts were then centrifuged at 5,000 × g for 2 min. The pelleted protoplasts were solubilized in SDS-PAGE sample buffer and then boiled for 10 min. The contents of AmyQ, PrsA, and GroEL (a cytoplasmic protein marker) in the samples (and in the trypsin-treated Triton X-100-solubilized sample) were analyzed by immunoblotting with specific antibodies.
α-Amylase assay.
A sample of culture medium was appropriately diluted and pipetted into 1 ml of α-amylase assay buffer (50 mM morpholineethanesulfonic acid [MES; pH 6.0], 50 mM NaCl, 0.1 mM CaCl2), one-fourth of a Phadebas tablet (Pharmacia) was dispersed in the buffer, and the mixture was incubated for 1 h at 37°C. The reaction was stopped by adding 30 μl of 10 M NaOH. The mixture was then filtered through Whatman no. 1 filter paper. The intensity of the blue color in the filtrate released from the Phadebas tablet by the action of α-amylase was measured at 616 to 624 nm, using 800 to 804 nm as a reference wavelength range. Enzyme concentrations (in micrograms per milliliter) were calculated from a standard curve using purified α-amylase from B. amyloliquefaciens (Sigma).
Microscopy of cells depleted of PrsA.
IH7211 was grown in modified 2× L broth supplemented with 1 mM IPTG to a cell density of Klett 100. Cells were harvested and stored in 10% glycerol at −70°C. For microscopy of cells with low levels of PrsA, the frozen bacteria were thawed, washed, and used to inoculate 10 ml of the above medium (1/2,000 dilution) containing different concentrations of IPTG. The cultures were incubated with shaking for 7 h, and then a drop was spread on a slide, air dried, and fixed by heating. Gram staining of the cells was done by the Hucker method as described in reference 9. Cells were photographed through an Axiophot photomicroscope (Zeiss).
RESULTS
Capacity of the secretion apparatus of B. subtilis.
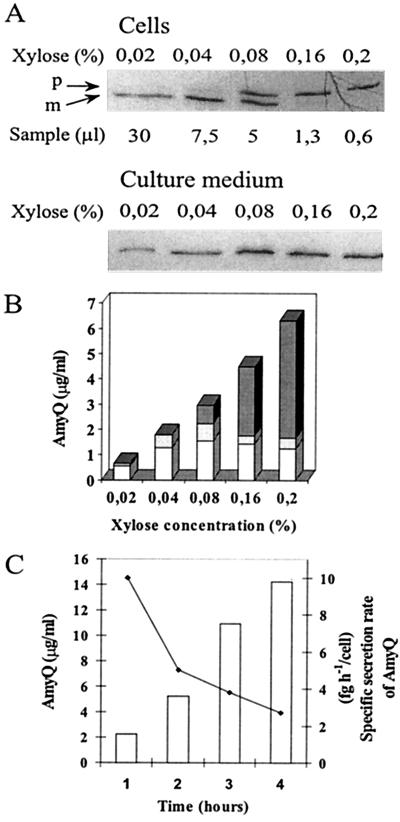
Expression of a secretory protein at a high level causes saturation of the protein secretion apparatus (27). By using a xylose-inducible expression system, we determined the threshold of saturation of the secretion apparatus for AmyQ α-amylase. In strain IH7441, amyQ has been placed under Pxyn control in plasmid pKTH3339. Pxyn-amyQ was induced in rich medium at the exponential growth phase. Steady-state levels of secreted AmyQ, cell-associated pre-AmyQ, and mature AmyQ were measured by quantitative immunoblotting. The amount of AmyQ in the culture medium increased in a xylose concentration-dependent manner but only up to 0.04%, at which point it leveled off (Fig. 1A and B). However, the total amount of AmyQ increased up to the highest xylose concentration (0.2%); high xylose concentrations caused concentration-dependent accumulation of pre-AmyQ in the cells (Fig. 1A and B). At 0.2% xylose, the precursor constituted approximately 74% of the total amount of AmyQ synthesized (Fig. 1B), while at 0.04% there was hardly any pre-AmyQ despite a peak level of secretion (Fig. 1A and B). The results indicate a clearcut threshold level of AmyQ synthesis above which the secretion apparatus is saturated.
FIG. 1.
Saturation of the secretion apparatus of B. subtilis by overexpressing AmyQ. (A) IH7441 (Pxyn-amyQ) was grown in modified 2× L broth up to a cell density of Klett 100, at which point amyQ gene expression was induced by adding xylose (concentrations were as indicated). After 1 h of induction, cell and medium fractions were separated and AmyQ in the fractions was analyzed by ECL immunoblotting. Cell samples corresponding to the indicated volumes of culture and medium samples corresponding to 10 μl of culture were immunoblotted. Precursor (p) and mature (m) forms of AmyQ are indicated by arrows. (B) The protein bands in panel A were quantified by optical scanning. For quantitation of the cell-associated mature AmyQ in cultures containing 0.16 or 0.2% xylose, another immunoblot with higher sample volumes was used. Columns show the content of secreted (white), cell-associated mature (light grey), and precursor (dark grey) forms of AmyQ in the cultures. (C) Accumulation of AmyQ in the culture medium of IH7441 (bars) induced with 0.04% xylose and specific secretion rate (line). The horizontal axis shows time after Klett 100.
We determined the specific secretion rate of AmyQ in the exponential growth phase under conditions in which the secretion apparatus was already saturated but no precursor had yet accumulated. At a cell density of Klett 100, 0.04% xylose was added and samples were taken at hourly intervals for measurements of α-amylase activity and determination of cell densities by microscopy. The specific secretion rate was 10 fg h−1/cell 1 h after the addition of xylose and decreased to 2.5 fg h−1/cell during the following 3 h. The concentration of AmyQ in the culture medium increased from 2 to 14 μg/ml during the same time period (Fig. 1C).
PrsA deficiency does not affect the cell-associated accumulation of precursors of a secretory protein.
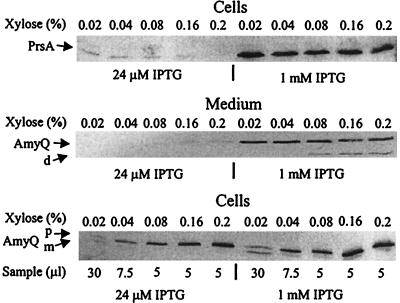
Our previous studies showed that PrsA is involved in a late stage of protein secretion (18, 21, 23, 28) but did not pinpoint the stage in the context of translocation. We have now addressed this by studying the nature and location of precursors of a secretory protein accumulating in the cell when PrsA is depleted. The model protein was again the α-amylase expressed by Pxyn-amyQ (pKTH3339). To control the expression of prsA, it was placed under the Pspac control by integrating the plasmid pMUTIN into the chromosome. The strain containing Pspac-prsA and pKTH3339 was designated IH7673. The amounts of PrsA and the cell-associated and secreted AmyQ were assayed by immunoblotting. The growth rates of this strain were similar at IPTG concentrations of between 24 μM and 1 mM (see below) and thus independent of the level of PrsA protein expressed in this range of induction.
Maximal induction of the Pspac-prsA gene was achieved in the presence of 1 mM IPTG. At this inducer concentration, an abundant band of PrsA protein was detected by immunoblotting; the PrsA level was close to that in the wild-type cells (data not shown), and was not affected by the expression level of Pxyn-amyQ (from 0.02 to 0.2% xylose [Fig. 2, top right]). Similar amounts of secreted AmyQ were found in the culture medium at inducer levels of 0.02% and higher (Fig. 2, middle), indicating that under these growth conditions the secretion apparatus was already saturated at the lowest amyQ expression level studied. Again pre-AmyQ clearly accumulated in the cells in the same xylose-dependent manner (Fig. 2, bottom) as observed in prsA+ cells (Fig. 1). A small amount of pre-AmyQ and a putative degradation product were also detected in culture media that were supplemented with high concentrations of the inducer (0.08 to 0.2% xylose), presumably due to secretion stress and cell lysis (Fig. 2, middle). Thus, consistent with the results obtained when PrsA was expressed from the native gene, there was a block in the secretion and expression level-dependent accumulation of pre-AmyQ.
FIG. 2.
Decreased levels of PrsA protein affect only a late stage of secretion. Saturation of the secretion apparatus by overexpressed AmyQ in cells with a low level of PrsA was determined. IH7673 (Pspac-prsA Pxyn-amyQ) was grown in modified 2× L broth supplemented either with 24 μM or 1 mM IPTG. At a cell density of Klett 100, expression of Pxyn-amyQ was induced as described in the legend to Fig. 1. Cell and medium fractions were prepared, and PrsA and AmyQ content in the fractions was analyzed by ECL. The analyzed samples were from 40 μl (PrsA; top) or 7.5 μl (secreted AmyQ; middle) of culture, or as indicated (cell-associated AmyQ; bottom). Precursor (p) and mature (m) forms and degradation product (d) are indicated by arrows.
When expression of the Pspac-prsA gene was induced with 24 μM IPTG, a low level of PrsA protein was detectable in an immunoblot assay. Again, the protein level was hardly dependent on the expression of Pxyn-amyQ (Fig. 2A, top left). Although the growth rate was not affected, the cells formed long filaments (see below). Even at this low level of PrsA, the amount and pattern of cell-associated AmyQ proteins were quite similar to those detected in the cells containing a high level of PrsA (Fig. 2, bottom). In particular, ratios of cell-associated pre-AmyQ to processed AmyQ were similar in both cases; the drastic accumulation of pre-AmyQ began at the level of AmyQ expression obtained at 0.04% xylose. However, there was strikingly little accumulation of AmyQ in culture medium when the PrsA level was low (Fig. 2, middle).
The accumulated pre-AmyQ is primarily exposed on the outer surface of protoplasts.
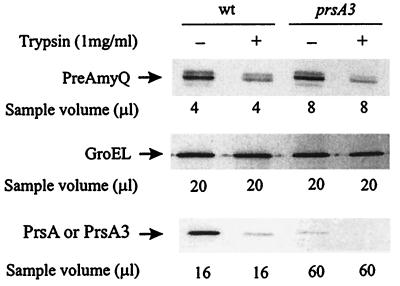
To identify the rate-limiting step in the secretion of overexpressed AmyQ, we used protease accessibility in protoplasts to determine the cellular location of the accumulated precursor. We also compared the protease accessibility of pre-AmyQ in protoplasts of a prsA mutant (prsA3 [23]) and its wild-type parent to investigate whether PrsA deficiency increases the proportion of intracellular pre-AmyQ, although it did not affect the pre-AmyQ/AmyQ ratio or the threshold of saturation. The prsA3 mutation causes the change of Asp268 to Asn268 near the C terminus of PrsA.
IH7441(pKTH3339) and IH7558(prsA3, pKTH3339) were grown in modified 2× L broth, and Pxyn-amyQ of pKTH3339 was induced at the cell density of Klett 100 with 0.2% xylose to express pre-AmyQ at a level sufficient to saturate the secretion apparatus. After 1 h of induction, cells were harvested and protoplasts were prepared (see Materials and Methods). The protoplasts were either not treated or treated with trypsin (1 mg/ml) or with trypsin and Triton X-100 (2%), followed by measurement of the levels of pre-AmyQ, GroEL, and PrsA by immunoblotting. The level of GroEL, a trypsin-sensitive cytoplasmic protein, was determined to monitor protoplast lysis during trypsin treatment. The presence of inverted membrane vesicles in the protoplast preparations was estimated by the degree of degradation of PrsA, which is a lipoprotein and thus is located on the outer surface of the cell membrane.
GroEL was almost completely protected from degradation in both strains, indicating that the protoplasts were intact, as shown in Fig. 3. The protoplast preparation of wild-type cells was also nearly devoid of inverted membrane vesicles, as indicated by almost complete degradation of PrsA (Fig. 3). The level of PrsA3 protein in the prsA3 mutant is less than 10% of that in the wild type (Fig. 3) (23), due to proteolytic degradation during posttranslocational folding by some unknown membrane- or cell wall-associated protease(s). Interestingly, approximately 70% of the pre-AmyQ was degraded after trypsin treatment of protoplasts of both strains (Fig. 3). No traces of pre-AmyQ or GroEL were detected when protoplasts were lysed with Triton X-100 (not shown). These findings suggest that the major rate-limiting step is in a posttranslocational phase of secretion and that PrsA deficiency does not increase the level of intracellular pre-AmyQ.
FIG. 3.
Accessibility of pre-AmyQ in protoplasts to external trypsin. IH7441(pKTH3339) and IH7558(prsA3, pKTH3339) were induced with 0.2% xylose to express Pxyn-amyQ at a level that saturates the secretion apparatus. Protoplasts were prepared under osmotic protection and treated with trypsin. The level of pre-AmyQ was analyzed by quantitative immunoblotting. The levels of GroEL and PrsA were also analyzed to assess the degree of protoplast lysis and the presence of inverted membrane vesicles, respectively. The analyzed samples correspond to the indicated volumes of original cultures. wt, wild type.
The rate of signal peptide processing limits the secretion of overexpressed AmyQ.
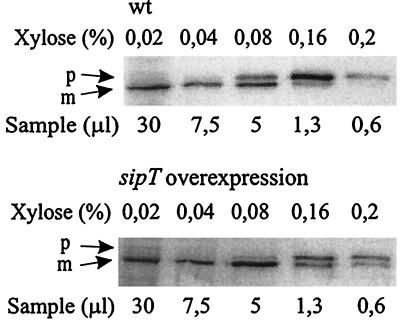
The above result suggested that a posttranslocational step is rate limiting in the secretion of overexpressed AmyQ. To test if this step is the processing of the signal peptide, we overexpressed sipT, encoding the main signal peptidase responsible for the processing of pre-AmyQ (44), in IH7983(pGDL100, pKTH3339) and carried out the saturation analysis as for Fig. 1.
The results (Fig. 4) showed that in IH7983, the threshold of saturation of the secretion apparatus clearly was at a higher expression level of Pxyn-amyQ (0.16% xylose) than in the strain with a wild-type level of SipT (0.08% xylose). At the highest expression levels (0.16 and 0.2% xylose), the proportion of mature AmyQ in cells of the SipT overproducer was significantly higher than in those of the wild type. Thus, SipT indeed is one rate-limiting factor in the secretion of AmyQ.
FIG. 4.
Effect of sipT overexpression on the threshold of saturation of the secretion apparatus. The wild-type (wt) strain IH7441(pKTH3339) and its derivative strain harboring plasmid pGDL100 with the sipT gene were cultivated as described in the legend to Fig. 1A, and the expression of Pxyn-amyQ was induced with different concentrations of xylose as indicated. The levels of cellular pre-AmyQ and AmyQ were analyzed by immunoblotting. Precursor (p) and mature (m) forms of AmyQ are indicated by arrows.
Stoichiometric requirement of PrsA protein for extracellular accumulation of overexpressed α-amylase.
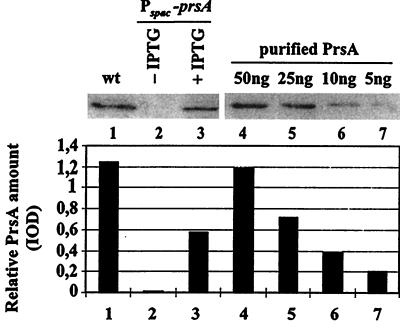
To relate the rate of α-amylase secretion to the number of PrsA molecules in the cell, we constructed strain IH7163. It secretes constitutively α-amylase at a level saturating the secretion machinery, and its prsA has been placed under the Pspac control (see Table 1 and Materials and Methods). IH7163 was cultivated in the presence of different concentrations of IPTG (0 to 1,000 μM); samples were withdrawn in late exponential growth phase (3 h after the culture had reached the turbidity of Klett 100). This growth phase was chosen in order to obtain high concentrations of AmyQ in the medium while avoiding the accumulation of extracellular proteases (22). The cell density was determined by a direct cell count. Quantitation of PrsA and secreted AmyQ was performed by immunoblotting. PrsA protein was barely detectable in uninduced cultures (Fig. 5), whereas full induction of the Pspac-prsA gene (100 μM IPTG) resulted in a more than 10-fold enhancement of the PrsA level (Fig. 5). This was about 40% of the level observed in the wild-type cultures (Fig. 5), indicating that Pspac is a weaker promoter than the prsA gene's own promoter.
FIG. 5.
Quantitation of expression of Pspac-prsA. Whole-cell samples were prepared from late-exponential-phase cultures of IH7163 (Pspac-prsA) grown in the presence or absence of 100 μM IPTG and of IH6538, an isogenic wild-type (wt) strain. Samples corresponding to 30 μl of the cultures were subjected to SDS-PAGE (12% gel), and PrsA was analyzed by ECL immunoblotting with anti-PrsA serum. PrsA was quantitated by optical scanning using purified PrsA as a standard. The lower panel shows the integrated optical densities (IOD) of the bands in the upper panel.
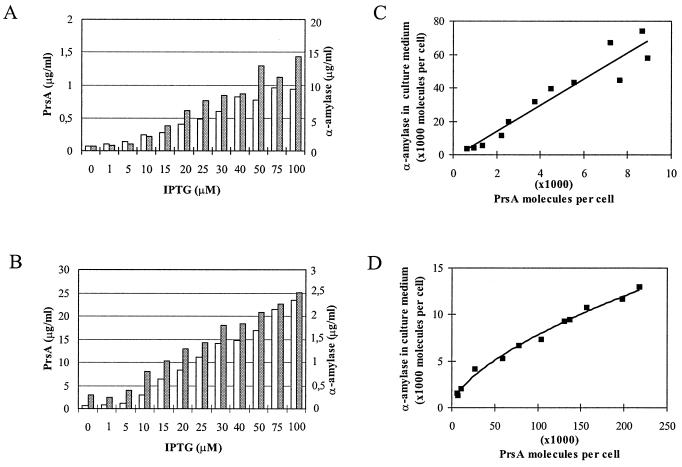
In fully induced cultures, there was about 10 μg of secreted α-amylase per ml (Fig. 6A), which is comparable to the level found in cultures with a massive cell-associated accumulation of pre-AmyQ (Fig. 1C). At intermediate concentrations of IPTG, the amylase level in the medium was dependent on the inducer concentration up to 50 μM (Fig. 6A). Under these conditions, there was a linear correlation between the number of PrsA molecules in the cell and the number of AmyQ molecules in the culture medium; a twofold increase in the number of PrsA molecules caused nearly a twofold increase in the number of secreted AmyQ molecules (Fig. 6C).
FIG. 6.
Dependence of AmyQ secretion on the content of PrsA protein in the cell. Strains IH7163(::pKTH3362, pKTH10) (A and C) and IH7162(::pKTH1601, pKTH3327) (B and D) were grown to late exponential phase in the presence of various concentrations of IPTG. The cellular level of PrsA (white bars) was determined as described in the legend to Fig. 5. Secreted α-amylase (grey bars) was determined enzymatically from the culture medium. Values represent the means of triplicate determinations. Numbers of PrsA and AmyQ molecules (C and D) per cell were calculated using the data in panels A and B, and cell densities of the cultures were determined by counting with a Petroff-Hausser counting chamber.
Molar excess of PrsA stimulates the secretion of α-amylase even when expressed at a low level.
The observations described above show that AmyQ secretion is stoichiometrically dependent on PrsA when the cell expresses AmyQ at a high level. To examine whether such dependence was simply due to the limiting threshold level of PrsA, we also studied the secretion of α-amylase in the reverse situation. We constructed strain IH7162, which overexpresses PrsA from the Pspac-prsA gene in plasmid pKTH3327 and expresses AmyQ at a low level from a single copy of amyQ integrated in the chromosome.
In the culture maximally induced with IPTG (100 μM), the level of PrsA protein was about 10-fold higher than in the wild type (data not shown) and thus about 20-fold higher than in IH7163, in which there is only one copy of the Pspac-prsA gene (Fig. 5 and 6A). Although expression of the chromosomal amyQ gene was far below the level required to cause saturation of the secretion apparatus in the experiment illustrated in Fig. 1C (Fig. 6B; compare also Fig. 6A and B), an almost linear dependence of AmyQ secretion on the level of PrsA was seen up to 2.2 × 105 PrsA molecules per cell (Fig. 6D). Furthermore, there was no indication that the AmyQ accumulation was leveling off even at highest amounts of PrsA obtainable under the experimental conditions used.
PrsA is essential for viability of B. subtilis.
Our previous attempts to disrupt the prsA gene by Campbell-type integration were unsuccessful, suggesting that this gene is essential for the viability of Bacillus cells (23). In this study, we endeavored to demonstrate directly that the functional prsA gene is indispensable. Since the uninduced level of expression from Pspac on pMUTIN4 (46) is very low, this plasmid can be used to examine whether or not a gene is essential for growth.
The strain containing Pspac-prsA (IH7211) was grown on L plates supplemented with 1 mM IPTG. Bacteria from one colony were suspended in 1 ml of sterile water, and the viable count was determined by plating a series of diluted samples onto plates with or without the inducer. Cells that were induced to express the Pspac-prsA gene formed colonies that appeared to be identical to those of the wild type; the viable count was 3 × 107/ml (Table 3). In the absence of inducer, however, the viable count was only 8 × 103/ml, and the colonies were heterogeneous in size; most were very small. The particular colonial morphology was maintained in subcultures. Bacteria that were able to grow on the noninducing plates appear to contain spontaneous suppressor mutations, which allow some, albeit impaired, growth, even in the absence of PrsA (17).
TABLE 3.
Dependence of growth of B. subtilis on expression of prsA
| Dilution | Colonies/platea
|
|
|---|---|---|
| +IPTG | −IPTGb | |
| 10−1 | Confluent | 27 |
| 10−2 | Confluent | 8 |
| 10−3 | 2 × 103 | 1 |
| 10−4 | 1.7 × 102 | 0 |
| 10−5 | 27 | 0 |
| 10−6 | 3 | 0 |
One colony of IH7211 (Pspac-prsA) from an L plate supplemented with 1 mM IPTG was suspended in 1 ml of water and diluted in water as indicated; 100-μl samples were spread on L plates (no induction of Pspac-prsA) or L plates supplemented with 1 mM IPTG (Pspac-prsA induced). Plates were incubated for 20 h at 37°C, and raised colonies were counted.
Colonies on these plates displayed heterogeneous morphology.
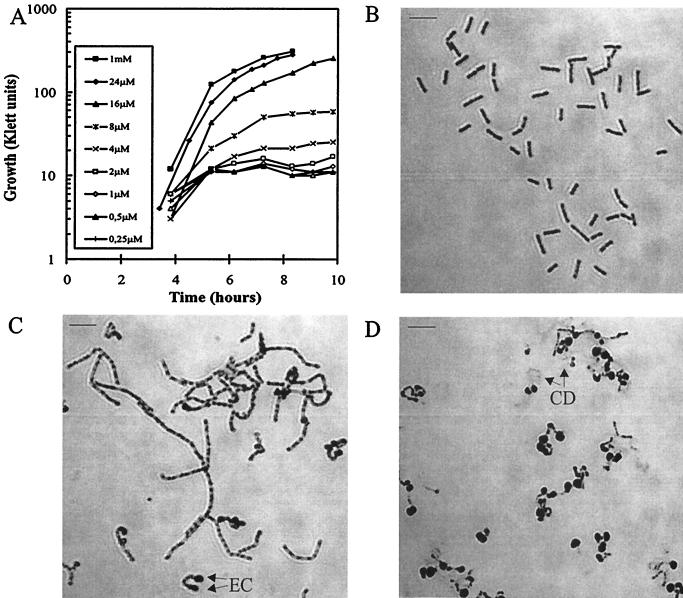
To determine the lowest level of PrsA protein able to maintain normal growth, we cultivated IH7211 in L broth supplemented with different concentrations of IPTG and examined growth and PrsA content in the culture with the lowest IPTG concentration that supported normal growth. IPTG concentrations of 16 μM or lower did not induce prsA expression sufficient for normal growth (Fig. 7A); at 2 μM or lower, practically no growth was observed. However, growth was not impaired in the presence of 24 μM IPTG (Fig. 7A).
FIG. 7.
Morphological changes in cells depleted of PrsA. (A) Growth of IH7211 (Pspac-prsA) in the presence of different concentrations of IPTG. Photomicrographs show filament formation and cell shape of IH7211 grown in the presence of 32 μM IPTG (B), 16 μM IPTG (C), or 8 μM IPTG (D). Cells were Gram stained for microscopic examination. EC and CD denote enlarged cell and cell debris, respectively. The scale bars represent 8 μm.
The morphological changes in cells with low levels of PrsA were also studied. When growth was only slightly inhibited (16 μM IPTG), cells formed long filaments (compare Fig. 7B and C) with swelling at the ends of some of the filaments. The growth abnormalities observed at 8 μM IPTG were more severe, the cells becoming enlarged and spherical (Fig. 7D). Such cells appeared to be quite fragile, as indicated by the cell debris seen in Fig. 7D.
DISCUSSION
The objective of this investigation was to characterize the relationship of the PrsA protein with the translocase complex, its mode of action, its interaction with secreted proteins, and its role in cell viability.
We showed that PrsA is a highly abundant protein in B. subtilis. As measured by an immunoblot assay, PrsA protein is present at approximately 2 × 104 molecules per wild-type cell, a level clearly higher than that of most other membrane proteins. Estimates of the levels of different membrane components of the preprotein translocase (Sec) in E. coli have given values from fewer than 30 to 900 molecules per cell (33, 36). Similar quantitation has not been carried out with the corresponding Sec components of B. subtilis, but if their cellular levels are even approximately similar to those in E. coli, PrsA protein is indeed in high molar excess compared to the translocase complex. It thus seems unlikely that the PrsA protein is associated firmly and in a stoichiometric way with the translocase complex. The enhanced secretion in strains overproducing PrsA at up to 2 × 105 molecules per cell also argues against a stoichiometric relationship.
The high number of PrsA molecules, its lipoprotein nature, and possible free lateral movement in the membrane may ensure that enough PrsA molecules are available to interact with translocated secreted protein molecules even without specific association with the translocase complex.
What could be the mode of action of PrsA? Several lines of consideration point to a role in the posttranslocational folding of secreted proteins. First, PrsA shows high sequence homology with the parvulin (PpiC) family of peptidyl-prolyl cis/trans isomerases (39, 41). One of them, SurA, is a major periplasmic foldase of E. coli assisting in the assembly of outer membrane proteins. Second, there is sequence homology in the whole length of PrsA with the lactococcal PrtM lipoprotein, a dedicated folding factor of the PrtP exoprotease (12, 13, 21). Third, there is extensive cell-associated degradation of some secreted model proteins when the level of functional PrsA is low (18, 28), consistent with the depletion of a folding factor. However, the mode of action of PrsA as well as the steps of the secretion pathway where it interacts with the process of secretion are not known.
Experiments with PrsA depletion now show that the PrsA protein is not involved in translocation or in the processing of secreted proteins. We could demonstrate the saturation of the secretion apparatus in terms of the cell-associated accumulation of pre-AmyQ when a threshold level of expression was increased. Neither the threshold level nor the pattern of cell-associated AmyQ (ratio of pre-AmyQ to processed AmyQ) was affected by the depletion of PrsA. This result is consistent with a similar finding that a defect in the PrsA protein does not disturb the rate of processing of the signal peptide of pre-AmyQ as analyzed by pulse-chase labeling (28). Moreover, we have also now demonstrated that PrsA deficiency does not increase the proportion of intracellular pre-AmyQ. However, there was a decreased amount of AmyQ secreted into the culture medium. This would be consistent with the involvement of the PrsA protein in a posttranslocational stage of secretion and with the degradation of secreted proteins after their translocation when PrsA is deficient. Depletion of the PrsA protein also showed that the secretion of AmyQ was linearly dependent on the cellular level of PrsA under conditions where AmyQ was expressed at a level high enough to saturate the secretion apparatus and the number of PrsA molecules in the cell was below that of the wild type (between 500 and 8,000 molecules per cell). This finding is consistent with our previous studies of prsA point mutants and strains that overexpress PrsA, pointing to PrsA as a bottleneck for the secretion of highly expressed exoproteins (23). Strikingly, however, there was also a dependence of AmyQ secretion on the level of PrsA protein when AmyQ was expressed at a low level. This stimulation was not linear, but even at the highest achievable level of PrsA (2.2 × 105 molecules per cell), secretion did not level off. These findings suggest a mode of action for PrsA that involves reversible association and disassociation of PrsA with α-amylase, compatible both with chaperone and foldase-like modes of activity.
The dependence of AmyQ secretion on PrsA, even when there was clearly a huge (about 1,000-fold) excess of the number of PrsA molecules over those of translocase complexes could also mean that only a fraction of PrsA molecules are readily available at the site where posttranslocational folding takes place. Thus, the number of PrsA molecules adjacent to the translocase could be limiting in spite of an apparent large overall excess. Alternatively, such an excess may be advantageous for the cell under some growth conditions, as has been demonstrated in the case of the DivIB cell division protein: much higher levels of DivIB are needed for normal growth at 47°C than at 30°C (40). Our findings also suggest that overexpression of PrsA is beneficial not only in biotechnical applications in which a secreted protein saturates the secretion apparatus (23) but also at lower levels of expression. This might be of special importance for the production of valuable heterologous proteins when the prime goal is to minimize degradation and ensure correct folding of the product. To determine the secretion capacity of B. subtilis, we studied the secretion of α-amylase expressed from an inducible promoter. In a cell with wild-type levels of all components of the secretion apparatus, the secretion of α-amylase into the culture medium was limited to 10 fg h−1/cell. At this secretion level, one cell secreted about 30 AmyQ molecules per s. Estimating that there are approximately 300 translocases in one cell, a new AmyQ molecule is released from the secretion pathway every 10 s, in perfect agreement with the estimated translocation rate of individual precursor proteins in E. coli (2). However, we found that in B. subtilis translocation is not a rate-limiting step in the secretion of AmyQ. Most of the pre-AmyQ which accumulated in the cell when overexpressed was at least partly translocated, as indicated by its accessibility in protoplasts to degradation by external trypsin.
Our results showed that there are in B. subtilis at least two factors the amount of which limits the secretion of overexpressed AmyQ, PrsA and SipT. The availability of SipT limits the rate of the signal peptide cleavage. The overexpression of sipT, however, does not enhance the accumulation of AmyQ in the growth medium (data not shown), although it enhances the processing. The availability of PrsA most probably limits the secretion rate to 10 fg h−1/cell, since concentrations of PrsA below or above the wild-type level can significantly restrict or enhance, respectively, AmyQ secretion, most probably by affecting posttranslocational folding (above results and reference 23).
To examine the effects of decreased levels of PrsA protein on B. subtilis viability, we constructed a controllable prsA gene. Immunoblotting showed that when the level of PrsA protein in the cells was decreased to below 1% (about 200 molecules per cell) of that in the wild type, cells stopped dividing and their morphology changed strikingly. About 200 molecules per cell still supported normal growth, but at this PrsA level, the cells formed long filaments. When the PrsA level was decreased further, cells became distorted (enlarged and spherical) and finally lysed. The morphological changes correlated well with drastically decreased plating efficiency of the strain on media devoid of IPTG.
The morphological changes in cells resulting from PrsA depletion closely resemble those observed when the synthesis of cell wall polymers is deficient. There is both filamental growth and cell lysis in mutants deficient in the synthetic pathway of peptidoglycan (1) or if synthesis is inhibited by β-lactamases (34, 38). Abnormal spherical cells have been observed when the synthesis of wall teichoic acids (polyglycerophosphate) is impaired (25, 29). The synthesis of this cell wall polymer is also essential for cell growth (29). This may indicate a role of PrsA in the folding or activity of enzymes in the synthetic pathway of cell wall polymers or their interaction with their substrates.
Notably, there were also stable revertants that formed colonies of various morphologies when PrsA was depleted; most colonies were pinpoint in size. This raises also the interesting possibility that some of the revertants have been caused by mutations in genes other than prsA. Indeed, we have observed that the growth inhibition caused by PrsA depletion can be partially suppressed by targeted mutations in genes affecting the charge of anionic cell wall polymers (17).
PrsA is the only known extracytoplasmic folding factor in B. subtilis. The B. subtilis genome sequence has revealed another putative member of the parvulin family, YacD, with similarity to PrsA. This protein, however, is nonessential for viability and secretion (O. Tunnela, unpublished result). The absolute requirement for PrsA raises the question of whether PrsA homoloques of pathogenic gram-positive bacteria could be used as new targets for antibiotic design, especially considering its extracytoplasmic location.
Our results, although suggesting interactions between PrsA and secreted proteins, cannot rule out completely the possibility of other effects, such as those mediated by the inhibition of unbalanced and harmful activity of cell wall or membrane-associated proteases by PrsA. To fully understand the function of PrsA, it will be important to develop methods for an in vitro assay of PrsA activity, to examine the structure-function relationships of PrsA, and to identify its putative interactions with proteins of the cytoplasmic membrane or the cell wall and with components of the peptidoglycan-teichoic acid matrix.
ACKNOWLEDGMENTS
We thank H. Tjalsma for plasmid pGDL100. We also thank B. K. Zimmerman for critical reading of the manuscript and correction of English.
M.V. was supported by a fellowship from the Graduate School in Microbiology (University of Helsinki). This work was supported in part by the Academy of Finland (grant 40905) and Biotech grant Bio-CT96-0097 from the Commission of the European Union (CEU).
REFERENCES
- 1.Archibald A R, Hancock I C, Harwood C R. Cell wall structure, synthesis and turnover. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria. Washington, D.C.: American Society for Microbiology; 1993. pp. 381–410. [Google Scholar]
- 2.Bassilana M, Wickner W. Purified Escherichia coli preprotein translocase catalyzes multiple cycles of precursor protein translocation. Biochemistry. 1993;32:2626–2630. doi: 10.1021/bi00061a021. [DOI] [PubMed] [Google Scholar]
- 3.Bolhuis A, Broekhuizen C P, Sorokin A, van Roosmalen M L, Venema G, Bron S, Quax W J, van Dijl J M. SecDF of Bacillus subtilis, a molecular Siamese twin required for the efficient secretion of proteins. J Biol Chem. 1998;273:21217–21224. doi: 10.1074/jbc.273.33.21217. [DOI] [PubMed] [Google Scholar]
- 4.Bron S, Bolhuis A, Tjalsma H, Holsappel S, Venema G, van Dijl J M. Protein secretion and possible roles for multiple signal peptidases for precursor processing in Bacilli. J Biotechnol. 1998;64:3–13. doi: 10.1016/s0168-1656(98)00099-6. [DOI] [PubMed] [Google Scholar]
- 5.Connolly L, De Las Penas A, Alba B M, Gross C A. The response to extracytoplasmic stress in Escherichia coli is controlled by partially overlapping pathways. Genes Dev. 1997;11:2012–2021. doi: 10.1101/gad.11.15.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Danese P N, Silhavy T J. The sigma-E and the Cpx signal transduction system control the synthesis of periplasmic protein-folding enzymes in Escherichia coli. Genes Dev. 1997;11:1183–1193. doi: 10.1101/gad.11.9.1183. [DOI] [PubMed] [Google Scholar]
- 7.Danese P N, Silhavy T J. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 8.Dartigalongue C, Raina S. A new heat-shock gene, ppiD, encodes a peptidyl-prolyl isomerase required for folding of outer membrane proteins in Escherichia coli. EMBO J. 1998;17:3968–3980. doi: 10.1093/emboj/17.14.3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doetsch R N. Determinative methods of light microscopy. In: Gerhardt P, Murray R G E, Costilow R N, Nester E W, Wood W A, Krieg N R, Phillips G B, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 21–33. [Google Scholar]
- 10.Ferrari F A, Nguyen A, Lang D, Hoch J A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983;154:15131515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujiwara S, Tsubokura N, Kurusu Y, Minami K, Kobayashi Y. Heat-inducible translational coupling in Bacillus subtilis. Nucleic Acids Res. 1990;18:739–744. doi: 10.1093/nar/18.4.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haandrikman A J, Kok J, Laan H, Soemitro S, Ledeboer A M, Konings W N, Venema G. Identification of a gene required for maturation of an extracellular lactococcal serine proteinase. J Bacteriol. 1989;171:2789–2794. doi: 10.1128/jb.171.5.2789-2794.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haandrikman A J, Kok J, Venema G. Lactococcal proteinase maturation protein PrtM is a lipoprotein. J Bacteriol. 1991;173:4517–4525. doi: 10.1128/jb.173.14.4517-4525.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hastrup S, Jacobs M F. Lethal phenotype conferred by xylose-induced overproduction of Apr-LacZ fusion protein. In: Zukowski M M, Ganesan A T, Hoch J A, editors. Genetics and biotechnology of bacilli. San Diego, Calif: Academic Press; 1990. p. 3341. [Google Scholar]
- 15.Herbort M, Klein M, Manting E H, Driessen A J M, Freudl R. Temporal expression of the Bacillus subtilis secA gene, encoding a central component of the preprotein translocase. J Bacteriol. 1999;181:493–500. doi: 10.1128/jb.181.2.493-500.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hultgren S J, Jacob-Dubuisson F, Jones C H, Branden C I. PapD and superfamily of periplasmic immunoglobulin-like pilus chaperones. Adv Protein Chem. 1993;44:99–123. doi: 10.1016/s0065-3233(08)60565-3. [DOI] [PubMed] [Google Scholar]
- 17.Hyyryläinen H-L, Vitikainen M, Thwaite J, Wu H Y, Sarvas M, Harwood C R, Kontinen V P, Stephenson K. d-Alanine substitution of teichoic acids as a modulator of protein folding and stability at the cytoplasmic membrane/cell wall interface of Bacillus subtilis. J Biol Chem. 2000;275:26696–26703. doi: 10.1074/jbc.M003804200. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs M, Anderssen J B, Kontinen V P, Sarvas M. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequence. Mol Microbiol. 1993;8:957–966. doi: 10.1111/j.1365-2958.1993.tb01640.x. [DOI] [PubMed] [Google Scholar]
- 19.Jones C H, Danese P N, Pinkner J S, Silhavy T J, Hultgren S J. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 1997;16:6394–6406. doi: 10.1093/emboj/16.21.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kallio P, Palva A, Palva I. Enhancement of α-amylase production by integration and amplifying the α-amylase gene of Bacillus amyloliquefaciens in the genome of Bacillus subtilis. Appl Microbiol Biotechnol. 1987;27:64–71. [Google Scholar]
- 21.Kontinen V P, Saris P, Sarvas M. A gene (prsA) of Bacillus subtilis involved in a novel, late stage of protein export. Mol Microbiol. 1991;5:1273–1283. doi: 10.1111/j.1365-2958.1991.tb01901.x. [DOI] [PubMed] [Google Scholar]
- 22.Kontinen V P, Sarvas M. Mutants of Bacillus subtilis defective in protein export. J Gen Microbiol. 1988;134:2333–2344. doi: 10.1099/00221287-134-8-2333. [DOI] [PubMed] [Google Scholar]
- 23.Kontinen V P, Sarvas M. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol Microbiol. 1993;8:727–737. doi: 10.1111/j.1365-2958.1993.tb01616.x. [DOI] [PubMed] [Google Scholar]
- 24.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 25.Lazarevic V, Karamata D. The tagGH operon of Bacillus subtilis 168 encodes a two-component ABC transporter involved in the metabolism of two wall teichoic acids. Mol Microbiol. 1995;16:345–355. doi: 10.1111/j.1365-2958.1995.tb02306.x. [DOI] [PubMed] [Google Scholar]
- 26.Leloup L, Haddaoui E A, Chambert R. Characterization of the rate-limiting step of the secretion of Bacillus subtilis α-amylase overproduced during the exponential phase of growth. Microbiology. 1997;143:3295–3303. doi: 10.1099/00221287-143-10-3295. [DOI] [PubMed] [Google Scholar]
- 27.Leskelä S, Wahlström E, Hyyryläinen H-L, Jacobs M, Palva A, Sarvas M, Kontinen V P. Ecs, an ABC transporter of Bacillus subtilis: dual signal transduction functions affecting expression of secreted proteins, as well as their secretion. Mol Microbiol. 1999;31:533–543. doi: 10.1046/j.1365-2958.1999.01194.x. [DOI] [PubMed] [Google Scholar]
- 28.Leskelä S, Wahlström E, Kontinen V P, Sarvas M. Lipid modification of prelipoproteins is dispensable for growth but essential for efficient protein secretion in Bacillus subtilis: characterization of the lgt gene. Mol Microbiol. 1999;31:1075–1085. doi: 10.1046/j.1365-2958.1999.01247.x. [DOI] [PubMed] [Google Scholar]
- 29.Mauel C, Young M, Margot P, Karamata D. The essential nature of teichoic acids in Bacillus subtilis as revealed by insertional mutagenesis. Mol Gen Genet. 1989;215:388–394. doi: 10.1007/BF00427034. [DOI] [PubMed] [Google Scholar]
- 30.Meyer T H, Menétrét J-F, Breitling R, Miller K R, Akey C W, Rapoport T A. The bacterial SecY/E translocation complex forms channel-like structures similar to those of the eukaryotic Sec61p complex. J Mol Biol. 1999;258:1789–1800. doi: 10.1006/jmbi.1998.2413. [DOI] [PubMed] [Google Scholar]
- 31.Missiakas D, Betton J M, Raina S. New components of protein folding in extracytoplasmic compartments of Escherichia coli SurA, FkpA and Skp/OmpH. Mol Microbiol. 1996;21:871–874. doi: 10.1046/j.1365-2958.1996.561412.x. [DOI] [PubMed] [Google Scholar]
- 32.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizushima S, Tokuda H, Matsuyama S I. Molecular characterization of Sec proteins comprising the protein secretory machinery of Escherichia coli. In: Neupert W, Lill R, editors. Membrane biogenesis and protein targeting. Amsterdam, The Netherlands: Elsevier Science Publishers; 1992. p. 2132. [Google Scholar]
- 34.Murray T, Popham D L, Setlow P. Bacillus subtilis cells lacking penicillin-binding protein 1 require increased levels of divalent cations for growth. J Bacteriol. 1998;180:4555–4563. doi: 10.1128/jb.180.17.4555-4563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palva I. Molecular cloning of α-amylase gene from Bacillus amyloliquefaciens and its expression in Bacillus subtilis. Gene. 1982;19:81–87. doi: 10.1016/0378-1119(82)90191-3. [DOI] [PubMed] [Google Scholar]
- 36.Pogliano K J, Beckwith J. Genetic and molecular characterization of the Escherichia coli secD operon and its products. J Bacteriol. 1994;176:804–814. doi: 10.1128/jb.176.3.804-814.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pogliano J, Lynch A S, Belin D, Lin E C C, Beckwith J. Regulation of Escherichia coli cell envelope proteins involved in protein folding and degradation by the Cpx two-component system. Genes Dev. 1997;11:1169–1182. doi: 10.1101/gad.11.9.1169. [DOI] [PubMed] [Google Scholar]
- 38.Popham D L, Setlow P. Phenotypes of Bacillus subtilis mutants lacking multiple class a high-molecular-weight penicillin-binding proteins. J Bacteriol. 1996;178:2079–2085. doi: 10.1128/jb.178.7.2079-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahfeld J-U, Rucknagel K P, Schelbert B, Ludvig B, Hacker J, Mann K, Fischer G. Confirmation of the existence of a third family among peptidyl-prolyl cis/trans isomerases. Amino acid sequence and recombinant production of parvulin. FEBS Lett. 1994;352:180–184. doi: 10.1016/0014-5793(94)00932-5. [DOI] [PubMed] [Google Scholar]
- 40.Rowland S L, Katis V L, Partridge S R, Wake R G. DivIB, FtsZ and cell division in Bacillus subtilis. Mol Microbiol. 1997;23:295–302. doi: 10.1046/j.1365-2958.1997.2141580.x. [DOI] [PubMed] [Google Scholar]
- 41.Rudd K E, Sofia H J, Koonin E V, Plunkett III G, Lazar S, Rouviere P E. A new family of peptidyl-prolyl isomerases. Trends Biochem Sci. 1995;20:12–14. doi: 10.1016/s0968-0004(00)88940-9. [DOI] [PubMed] [Google Scholar]
- 42.Stragier P, Bonamy C, Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988;52:697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- 43.Tjalsma H, Noback M A, Bron S, Venema G, Yamane K, van Dijl J M. Bacillus subtilis contains four closely related type I signal peptidases with overlapping substrate specificities. J Biol Chem. 1997;272:25983–25992. doi: 10.1074/jbc.272.41.25983. [DOI] [PubMed] [Google Scholar]
- 44.Tjalsma H, Bolhuis A, van Roosmalen M L, Wiegert T, Schumann W, Broekhuizen C P, Quax W J, Venema G, Bron S, van Dijl J M. Functional analysis of the secretory precursor processing machinery of Bacillus subtilis: identification of a eubacterial homolog of archaeal and eukaryotic signal peptidases. Genes Dev. 1998;12:2318–2331. doi: 10.1101/gad.12.15.2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tjalsma H, Kontinen V P, Pragai Z, Wu H, Meima R, Venema G, Bron S, Sarvas M, van Dijl J M. The role of lipoprotein processing by signal peptidase II in the Gram-positive eubacterium Bacillus subtilis. Signal peptidase II is required for the efficient secretion of α-amylase, a non-lipoprotein. J Biol Chem. 1999;274:1698–1707. doi: 10.1074/jbc.274.3.1698. [DOI] [PubMed] [Google Scholar]
- 46.Vagner V, Dervyn E, Ehrlich S D. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology. 1998;144:3097–3104. doi: 10.1099/00221287-144-11-3097. [DOI] [PubMed] [Google Scholar]
- 47.van Wely K H M, Swaving J, Broekhuizen C P, Rose M, Quax W J, Driessen A J. Functional identification of the product of the Bacillus subtilis yvaL gene as a SecG homologue. J Bacteriol. 1999;181:1786–1792. doi: 10.1128/jb.181.6.1786-1792.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.van der Wolk J, Klose M, Breukink E, Demel R A, de Kruijff B, Freudl R, Driessen A J M. Characterization of a Bacillus subtilis SecA mutant protein deficient in translocation ATPase and release from the membrane. Mol Microbiol. 1993;8:31–42. doi: 10.1111/j.1365-2958.1993.tb01200.x. [DOI] [PubMed] [Google Scholar]