Abstract
Extracellular sulfatase-2 (Sulf-2) influences receptor–ligand binding and subsequent signaling by chemokines and growth factors, yet Sulf-2 remains unexplored in inflammatory cytokine signaling in the context of rheumatoid arthritis (RA). In the present study, we characterized Sulf-2 expression in RA and investigated its potential role in TNF-α-induced synovial inflammation using primary human RA synovial fibroblasts (RASFs). Sulf-2 expression was significantly higher in serum and synovial tissues from patients with RA and in synovium and serum from hTNFtg mice. RNA sequencing analysis of TNF-α-stimulated RASFs showed that Sulf-2 siRNA modulated ~2500 genes compared to scrambled siRNA. Ingenuity Pathway Analysis of RNA sequencing data identified Sulf-2 as a primary target in fibroblasts and macrophages in RA. Western blot, ELISA, and qRT‒PCR analyses confirmed that Sulf-2 knockdown reduced the TNF-α-induced expression of ICAM1, VCAM1, CAD11, PDPN, CCL5, CX3CL1, CXCL10, and CXCL11. Signaling studies identified the protein kinase C-delta (PKCδ) and c-Jun N-terminal kinase (JNK) pathways as key in the TNF-α-mediated induction of proteins related to cellular adhesion and invasion. Knockdown of Sulf-2 abrogated TNF-α-induced RASF proliferation. Sulf-2 knockdown with siRNA and inhibition by OKN-007 suppressed the TNF-α-induced phosphorylation of PKCδ and JNK, thereby suppressing the nuclear translocation and DNA binding activity of the transcription factors AP-1 and NF-κBp65 in human RASFs. Interestingly, Sulf-2 expression positively correlated with the expression of TNF receptor 1, and coimmunoprecipitation assays demonstrated the binding of these two proteins, suggesting they exhibit crosstalk in TNF-α signaling. This study identified a novel role of Sulf-2 in TNF-α signaling and the activation of RA synoviocytes, providing the rationale for evaluating the therapeutic targeting of Sulf-2 in preclinical models of RA.
Keywords: Sulfatase-2, TNF-α, Rheumatoid arthritis, Synovial fibroblasts, Signal transduction
Subject terms: Tumour-necrosis factors, Target identification, Mechanisms of disease
Introduction
Tumor necrosis factor-alpha (TNF-α) is a crucial proinflammatory cytokine produced in response to infection and injury that is also involved in the maintenance of tissue homeostasis [1, 2]. However, dysregulated TNF-α signaling drives the pathogenesis of autoimmune inflammatory conditions, including rheumatoid arthritis (RA) [1]. While RA joint pathogenesis is influenced by infiltrating immune cells, TNF-α-activated RA synovial fibroblasts (RASFs) are key aggressors [3]. TNF-α binding to TNFR1 initiates canonical nuclear factor-κB (NF-κB) signaling [4]. By signaling through TNFR1 and NF-κB, TNF-α induces the production of other cytokines, including IL-1β and IL-6, and chemokines involved in the recruitment and activation of leukocytes [5]. In RA, TNF-α promotes hyperplasia of synovial tissue, leading to the progressive destruction of articular cartilage and bone [6]. Anti-TNF therapies have significantly improved the management of autoimmune rheumatic diseases [7], but their long-term use carries the risk of serious infection, immunogenicity, and malignancy [8]. Thus, there is a need to identify alternative approaches for regulating TNF-α signaling to limit its role in RA pathogenesis.
The extracellular sulfatases sulfatase-1 (Sulf-1) and sulfatase-2 (Sulf-2) regulate the receptor–ligand binding of various growth factors, chemokines, and cytokines to significantly modulate downstream signaling [9, 10]. Sulf-2 activity has been shown to mobilize growth factors and chemokines, including VEGF, FGF-1, and SDF-1, from their binding sites [11]. In liver, pancreas and lung tumors, overexpression of the SULF2 gene correlates with increased tumorigenicity, and Sulf-2 has been identified as a novel therapeutic target in cancer [10]. The small-molecule Sulf-2 inhibitor OKN-007 (also known as NZY-059) [12] is currently in clinical trials with temozolomide adjuvant therapy for glioblastoma, aiming to demonstrate that the pharmacologic inhibition of Sulf-2 is safe and effective [13].
A recent review compiled existing evidence supporting the likely roles of extracellular sulfatases in inflammation [14]; however, the potential involvement of Sulf-1 and Sulf-2 in TNF-α signaling and RA remains unexplored. Given the considerable presence of extracellular sulfatases in musculoskeletal tissue, the logical hypothesis is that Sulf-2 plays a role in synovial pathogenesis in RA. Otsuki et al. detected an age-dependent increase in Sulf-1 and Sulf-2 expression in mouse cartilage and in human OA cartilage compared to normal cartilage [15]. In another study, Ratzka et al. determined that Sulf-1 and Sulf-2 are expressed in overlapping patterns during embryonic bone formation, and double knockouts of these two proteins showed skeletal abnormalities, including shorter bones and vertebral fusion [16] Similarly, Zaman et al. found higher Sulf-2 expression during both bone development and fracture healing [17]. While these studies provide data on the differential expression of extracellular sulfatases in musculoskeletal tissue disease states, our study deciphered the previously undescribed pathological relevance of Sulf-2 in TNF-α-induced signaling, activation and proliferation of RASFs, which are a major component of the invasive synovial pannus in RA.
The present study aimed to characterize the expression of sulfatases in RA patient samples and to determine their potential role in TNF-α-mediated inflammatory signaling in human RASFs.
Materials and methods
Detailed information about reagents, antibodies, mice, IHC and RNA sequencing are provided in the SI Materials and Methods.
Detection of extracellular sulfatases in human synovial tissues and serum
Deidentified synovial tissues from nondiseased deceased individuals and from RA patients who had undergone total joint replacement surgery or synovectomy were obtained from the Cooperative Human Tissue Network (Columbus, OH) or National Disease Research Interchange (Philadelphia, PA) under a protocol approved by the Washington State University Institutional Review Board (IRB) (approval no. 17249). Beyond the specification of “synovial membrane”, no description of the location within the synovium was provided by the vendors. RA patients had not been treated with biologics. Table S1 shows the demographics of the participants whose tissues or cells were studied. The IRB of Washington State University determined these experiments to be exempt from the requirement for informed consent, as deidentified human materials that were not collected specifically for the purpose of this research were utilized, and there was no mechanism to link the samples or their associated data to living individuals.
To determine gene and protein expression in human synovial tissues, total RNA was extracted from six RA and six nondiseased frozen tissues using an RNeasy mini kit (Qiagen, Hilden, Germany), and the expression of SULF1, SULF2, TNFRSF1A and TNFRSF1B was measured by quantitative real-time PCR (qRT‒PCR). Sulf-1, Sulf-2, TNFR1 and TNFR2 protein levels were determined by Western blotting.
Human serum samples were purchased from commercial vendors (Discovery Life Sciences, Newtown, PA; Innovative Research, Novi, MI; or Sanguine Biosciences, Los Angeles, CA) or generously shared by Dr. Cynthia Crowson (Mayo Clinic, Rochester, MN). Table S2 shows a demographic summary of the human serum donors. Sera from RA patients (n = 48), multiple sclerosis (MS) patients (n = 8) and nondiseased individuals (n = 50) were tested using Sulf-2 (XPEH0934) and Sulf-1 (XPEH15391) ELISAs from XpressBio (Ballenger Creek, MD).
Extracellular sulfatases in wild-type (WT) and human TNF-α-transgenic (hTNFtg) mice
Formalin-fixed, paraffin-embedded ankle sections were prepared in Dr. Edward M. Schwartz’s laboratory as previously described [18, 19]. Specimens from littermate WT C57BL/6 mice were provided as controls. The expression of Sulf-1 and Sulf-2 was determined in ankle joint sections from male WT and hTNFtg mice by fluorescence immunohistochemistry (IHC). Serum samples from male and female WT or hTNFtg mice were tested using ELISA kits for murine Sulf-1 (MBS2890096) and Sulf-2 (MBS2706314) from MyBioSource (San Diego, CA). Please see the SI Materials and Methods for details regarding the mice (Table S4) and IHC protocol.
Culture and treatment of human RASFs
Primary human RASFs were isolated from deidentified synovial tissues as previously described [20]. RASFs were cultured in RPMI 1640 with 2 mM L-glutamine and 10% heat-inactivated fetal bovine serum in a humidified incubator at 37 °C with 5% CO2. Experiments were performed using RASFs between passages 4 and 10. Human RASFs were grown to 80% confluence and serum-starved overnight prior to stimulation.
Signaling pathways related to the TNF-α-mediated induction of adhesion/migration molecules were evaluated by pretreating RASFs with chemical inhibitors for 2 h, followed by stimulation with TNF-α (20 ng/ml) for 24 h. The inhibitors included 200 μM PDTC (NF-κB), 10 μM SP600125 (JNK), 10 μM SB203980 (p38), 10 μM PD98059 (ERK1/2), 25 μM TC-ASK10 (ASK1), 10 μM AG490 (JAK2/3, STAT3), 10 μM rottlerin (PKCδ), and 20 μM LY294002 (PI3K, Akt). Confirmatory testing with the specific PKCδ inhibitor NP627 was performed by pretreating RASF2 of 2 h with 5, 10 or 20 nM NP627, followed by stimulation with TNF-α (20 ng/ml) for 24 h. To test the effects of a small-molecule Sulf-2 inhibitor on TNF-α-induced proteins, RASFs were pretreated with OKN-007 (0–1000 µM) for 12 h, followed by TNF-α (20 ng/ml) stimulation for 24 h. Nonstimulated (NS) and TNF-α-stimulated RASFs without inhibitor served as controls. Adhesion molecules were measured by Western blotting, and chemokines in conditioned media were quantified by ELISA.
Experiments investigating the release of soluble Sulf-2 from TNF-α-stimulated RASFs
To compare the levels of soluble Sulf-2 released from NS and TNF-α-stimulated primary human RASFs in culture, RASFs from four patients were serum-starved overnight, followed by the addition of fresh serum-free medium. RASFs treated with TNF-α (20 ng/ml) were compared to NS controls.
To determine the effects of a broad-spectrum matrix metalloproteinase (MMP) inhibitor on the solubilization of Sulf-2, RASFs from three patients were serum-starved overnight and then exposed to fresh serum-free medium. Cells were exposed to the following treatments for 72 h: control (NS), GM6001 (20 µM; MMP inhibitor), TNF-α (20 ng/ml), or GM6001 + TNF-α.
To ascertain the effect of signaling inhibitors on the TNF-α-induced release of soluble Sulf-2, RASFs from three patients were serum-starved overnight and then exposed to fresh serum-free medium. Cells were exposed to the following treatments for 72 h: control (NS), TNF-α (20 ng/ml), NP627 (20 nM; PKCδi) + TNF-α, PDTC (200 µM; NF-κBi) + TNF-α, and SP600125 (10 µM JNKi) + TNF-α. Soluble Sulf-2 levels in the conditioned medium were quantitated by a Sulf-2 ELISA kit (XPEH0934) from XpressBio (Ballenger Creek, MD).
Transient transfection of siRNA
Human RASFs were transfected with small interfering RNA (siRNA) using Lipofectamine® 2000 (Invitrogen, Waltham, MA) to deliver 130 pmol siRNA targeting Sulf-1, Sulf-2, or scrambled sequence (negative control, NC) in a 6-well format. After 48 h, RASFs were serum-starved overnight and then treated with TNF-α (20 ng/ml). TNF-α-stimulated RASFs transfected with NC siRNA served as a positive control; NS RASFs transfected with NC siRNA served as a negative control. After TNF-α stimulation for 8 h, total RNA was isolated, and qRT‒PCR was performed to determine the effects on target gene expression. Cells stimulated with TNF-α for 30 min were evaluated by Western blotting of phosphorylated MAPK and PKCδ intermediates and used to prepare cytoplasmic and nuclear extracts. Cells stimulated with TNF-α for 24 h were used to evaluate the siRNA-mediated changes in TNF-α-induced proteins.
RNA library preparation and sequencing
Details of the treatment, RNA preparation and sequencing are provided in the SI Materials and Methods. Expression values for each target gene were determined as reads per million (RPM) mapped reads for each sample as described previously [21].
Bioinformatics analysis of differentially expressed genes (DEGs)
Statistical analysis was performed using R version 4.1.0, and differential gene expression analysis was conducted with the edgeR package version 3.34.0 [22]. Genes with low expression were filtered out using the filterByExpr() function with the minimum count needed for a portion of the samples set to 10. Normalization was performed with the calcNormFactors() function using a trimmed mean of M-values approach [23]. Counts were fit to a negative binomial linear model, and statistical testing was performed using a paired design between treatment groups. p values were adjusted using the Benjamin–Hochberg correction, and significant differentially expressed genes (DEGs) were defined as those with a false discovery rate (FDR) <0.05.
Volcano plots of the top 2500 significant DEGs in RASFs when comparing NS/NCsi vs. TNF/NCsi and TNF/NCsi vs. TNF/Sulf-2si were generated using GraphPad Prism (GraphPad Software Corporation, San Diego, CA). Preliminary gene ontology (GO) analysis was conducted using Ingenuity Pathway Analysis (IPA®; Qiagen, Hilden, Germany). We performed detailed GO-based identification of enriched biological processes, interaction clusters and transcription factors using Metascape [24]. Heatmaps were created using www.heatmapper.ca [25].
Quantitative reverse transcription PCR analyses
Total RNA was reverse transcribed using the SuperScript™ First Strand Synthesis kit (Life Technologies, Carlsbad, CA). Relative mRNA expression was determined by qRT‒PCR using SYBR® Green PCR master mix (Life Technologies) and quantitated by the ΔΔCt method with GAPDH as the endogenous control. Supplementary Table S3 shows the custom primer sequences.
Chemokine quantification by ELISA
ENA-78/CXCL5, RANTES/CCL5, fractalkine/CX3CL1, IP-10/CXCL10, and I-TAC/CXCL11 were quantified in conditioned media using DuoSet ELISA kits (R&D Systems, Minneapolis, MN).
Western blotting
Whole-cell lysates were prepared, and protein was quantitated using a DC Protein Assay (Bio-Rad, Hercules, CA). Western blot analysis of the cell lysates (30 µg) was performed as described in our earlier study [21]. Sulf-2 is translated as a 125 kD preproprotein, from which two subunits are proteolytically processed and bound by disulfide bonds to form the mature heterodimer [26]. To measure cellular Sulf-2 levels for the determination of siRNA knockdown or changes in response to TNF-α, densitometry was performed on the 125 kD Sulf-2 proprotein band. The band corresponding to the 75 kD subunit of mature heterodimeric Sulf-2 was found to bind TNFR1 in the coimmunoprecipitation assay. Densitometry was performed using Image Lab 6.0 software to measure expression values (adjusted volume intensity), and data are presented as the mean ± standard error of the mean (SEM).
Cell proliferation assay
A CyQUANT® Direct Cell Proliferation Assay Kit (C35011) was used to evaluate the effect of Sulf-2 siRNA on the TNF-α-induced proliferation of RASFs. After 8 h of transfection with Sulf-2 or NC siRNA, 1 × 104 cells in 100 μl of complete medium were seeded in 96-well plates. After 48 h, the cells were serum-starved for 2 h and then treated with 20 ng/ml TNF-α for 24 h. Next, 100 μl of CyQUANT GR dye/lysis buffer (prepared per the manufacturer’s instructions) was added to each sample well, and the plates were incubated at 37 °C for 60 min. The fluorescence was measured at 485/535 nm.
Transwell invasion assay
Invasion through a Matrigel layer was measured to compare the invasiveness of TNF-α-stimulated RASFs transfected with scrambled control (NC) siRNA or Sulf-2 siRNA. Cell invasion was evaluated using the Transwell method (Corning, Corning, NY, USA). After 8 h of transfection with Sulf-2 or NC siRNA, 1 × 105 cells in 100 μl of complete medium were seeded directly into the wells of Transwell® chambers (8.0 µm Transparent PET Membrane, Corning, Cat. No. 353097) coated with Matrigel™ (Corning, Cat. No. CB-40234). The lower chamber was filled with 600 μl of complete growth medium (RPMI-1640 with 10% FBS). The cells were incubated at 37 °C and 5% CO2 for a total of 48 h of transfection. The cells were starved in serum-free RPMI for 2 h. After starvation, the cells were treated with TNF-α (20 ng/ml) for 48 h or left unstimulated (controls). Noninvading cells on the upper membrane were removed with a cotton swab. Invading cells on the lower surface of the membrane were stained using 0.1% crystal violet and counted in three random fields using ImageJ software, and the mean was calculated.
Coimmunoprecipitation assay
RASFs from three patients were grown to 80% confluency in 150 mm dishes and then serum-starved overnight. For cells from each patient, one plate was not stimulated (NS), and one plate was stimulated with TNF-α (20 ng/ml) for 30 min. Cells were washed twice with ice-cold PBS, followed by collection and lysis in 500 μl of RIPA buffer. Lysates were then utilized for immunoprecipitation assays as described in the SI Materials and Methods.
Preparation of nuclear extracts and DNA binding assay
RASFs were transfected with NC or Sulf-2 siRNA for 48 h and then treated with TNF-α (20 ng/ml) for 30 min. NS RASFs transfected with NC siRNA served as a control. Nuclear fractions were prepared as described previously [27] and evaluated for phosphorylated signaling kinase expression using Western blotting. Five micrograms of total protein per sample was tested in DNA binding activity assays for NF-κBp65 (Cayman Chemicals, Ann Arbor, MI) and c-Jun (Ray Biotech, Peachtree Corners, GA) according to the manufacturers’ instructions.
Statistical analysis
The results are expressed as the mean ± SEM. The Pearson correlation was calculated in GraphPad Prism (GraphPad Software Corporation, San Diego, CA) to investigate the coexpression of SULF1 or SULF2 with TNFRSF1A. Comparisons of two groups were performed using two-tailed Student’s t test. Multigroup comparisons were performed using one-way analysis of variance followed by Dunnett’s post hoc test to compare treatments to a control value. p < 0.05 was the threshold for statistical significance.
Results
The expression of Sulf-2 is significantly increased in RA patient synovial tissues and serum and is induced by TNF-α stimulation of human RASFs
In the published literature, extracellular sulfatases have remained largely uncharacterized in the context of inflammation and RA. We quantified the gene expression of SULF1 and SULF2 in homogenized normal synovial tissues (NLSTs) and RA synovial tissues (RASTs) by qRT–PCR (Fig. 1A). RASTs were derived from patients with at least 2 years of clinically established RA who were not treated with biologics. Sulf-1 mRNA levels were 1.9-fold higher (p < 0.05) and Sulf-2 mRNA levels were 3.0-fold higher (p < 0.001) in RASTs than in NLSTs. We also compared the gene and protein expression of TNFRSF1A/TNFR1 and TNFRSF1B/TNFR2 in NLSTs and RASTs (Fig. S1A). Interestingly, Pearson correlation analysis of the qRT‒PCR results from the 12 synovial tissues tested showed a positive linear association between the expression of SULF2 and TNFRSF1A (r = 0.67, p < 0.05) (Fig. 1A). No association was found for SULF1 and TNFRSF1A (data not shown).
Fig. 1.
Sulfatase-2 expression is significantly higher in human RA synovial tissues and serum, and SULF2 expression is transiently induced by TNF-α stimulation of RASFs in vitro. A qRT‒PCR analysis revealed significantly higher SULF1 and SULF2 mRNA levels in human RASTs than in NLSTs. Pearson correlation analysis revealed positive coexpression of TNFRSF1A and SULF2. B RASTs have significantly elevated Sulf-2 protein levels compared to NLSTs, as determined by Western blotting followed by densitometry analysis. C Quantitative ELISAs revealed significantly higher soluble Sulf-2 levels in serum from RA patients than in that from normal controls and MS patients. Soluble Sulf-1 was not detected. D Primary human RA synovial fibroblasts stimulated with TNF-α (20 ng/ml) for 0–48 h in vitro showed a transient increase in Sulf-1 and Sulf-2 protein levels in cell lysates. E In a separate experiment, soluble Sulf-2 levels in conditioned medium were quantitated by ELISA. Data are presented as the mean ± SEM. *p < 0.05; **p < 0.01; ***p < 0.001
Protein levels of the extracellular sulfatases were also significantly elevated in RA patient synovial tissues. Densitometric analysis of the Western blots showed ~1.5-fold higher Sulf-1 protein levels (p = 0.06) and five-fold higher Sulf-2 protein levels in RASTs than in NLSTs (p < 0.01) (Fig. 1B).
Sulf-1 and Sulf-2 can be released in enzymatically active soluble forms from cell membranes into the conditioned medium of cultured cells and in human bodily fluids in certain disease states [10]. We tested human serum samples from healthy donors and RA and MS patients for soluble Sulf-1 and Sulf-2. Osteoarthritis patient serum samples were not utilized as controls since elevated expression of Sulf-1 and Sulf-2 has been reported in OA cartilage, which could have introduced a confounding factor in our study [15]. MS patient sera were included for comparison, as MS is an autoimmune disease without joint synovial pathogenesis. Sulf-1 was undetectable in normal and RA serum samples (data not shown). In contrast, soluble Sulf-2 levels were significantly higher (p < 0.001) in sera of RA patients (31.0 ± 4 ng/ml) than in those of normal controls (13.6 ± 1.4 ng/ml) or patients with MS (17.0 ± 1.0 ng/ml) (Fig. 1C). Sulf-2 levels were slightly higher in males than females, although this difference was not statistically significant.
In cultured human RASFs, we observed a transient and time-dependent increase in Sulf-1 and Sulf-2 upon TNF-α stimulation (Figs. 1D and S1B). The observed increase in Sulf-1 was detected in cell lysates up to 24–48 h after the initiation of TNF-α treatment. We detected a rapid, slight increase in Sulf-2 protein levels as early as 2–6 h after the initiation of TNF-α stimulation, but these levels decreased to those in the NS group by 12–24 h. In a separate experiment, we detected an increase in soluble Sulf-2 in the conditioned medium after TNF-α stimulation for 72 h (Fig. 1D).
A broad-spectrum MMP inhibitor was associated with a 20% average reduction in soluble Sulf-2 release from TNF-α-stimulated RASFs compared to TNF-α alone (n = 3), although the difference did not reach statistical significance (Fig. S1C). We investigated whether TNF-α regulates Sulf-2 expression via the same key signaling pathways identified in our experiments. We addressed this question by measuring Sulf-2 levels in RASF lysates and soluble Sulf-2 levels in conditioned medium of RASFs from three patients at 6 and 72 h, respectively, after exposure to TNF-α alone or to TNF-α and key signaling inhibitors. Signaling inhibitors had no effect on Sulf-2 levels in RASF lysates (data not shown). ELISA analysis of the conditioned medium confirmed the ~1.5-fold increase in soluble Sulf-2 from TNF-α-stimulated RASFs, as we observed in the earlier experiments shown in Fig. 1E. NP627 (20 nM), a PKCδ inhibitor, had no effect. Interestingly, the NF-κB inhibitor PDTC (200 µM) and the JNK inhibitor SP600125 were associated with an additional 1.5-fold increase and 3-fold increase, respectively, in the release of soluble Sulf-2 over TNF-α stimulation alone (p < 0.05 and p < 0.001, respectively). These data are included in Fig. S1D within the Supplementary Materials. While we do not have a clear explanation for this observation, one hypothesis is that TNF-α-induced proteases, MMPs, and disintegrin and metalloproteinase domain-containing protein 10 (ADAM-10) or ADAM-17 may be involved in the process independent of these signaling proteins.
Expression of Sulf-1 and Sulf-2 is elevated in TNF-transgenic mice
We determined the expression of Sulf-1 and Sulf-2 in tissue sections and serum samples from a recently concluded study on WT C57BL/6 mice and hTNFtg mice [18]. Fluorescence IHC showed qualitatively higher Sulf-1 and Sulf-2 levels in the inflamed joint tissues of hTNFtg mice than in those of WT mice (Fig. 2A). ELISA analysis of mouse serum samples revealed elevated expression of Sulf-1 and Sulf-2 in hTNFtg mice (Fig. 2B); the average Sulf-1 levels were 2.75-fold higher in the sera of hTNFtg mice than in those of WT mice (p < 0.05), with the highest concentration in female hTNFtg mice. The average serum Sulf-2 levels in males and females together were higher in hTNFtg mice than in WT mice, although this difference was not statistically significant. Soluble Sulf-2 levels were two-fold higher in female hTNFtg mice than in female WT mice (p < 0.05).
Fig. 2.
Extracellular sulfatase levels are elevated in inflamed joint tissues and serum of a human TNF-transgenic mouse model of RA. Extracellular sulfatase protein levels were compared in human TNF-transgenic mice of the 3647 line (hTNFtg) and littermate wild-type (WT) C57BL/6 mice. A Murine Sulf-1 and Sulf-2 proteins were at qualitatively higher levels in ankle sections of hTNFtg mice as determined by fluorescence IHC. Images are shown with isotype-matched controls on the same tissue and slide. H&E images illustrate synovial hyperplasia and leukocyte infiltration, which are characteristic of hTNFtg mice. Bone (B) and synovial tissues (S) are labeled. B Murine Sulf-1 and Sulf-2 proteins in serum samples from hTNFtg and littermate WT mice were quantitated by ELISA. Data are presented as the mean ± SEM. *p < 0.05
RNA-seq shows global gene expression changes induced by TNF-α in human RASFs
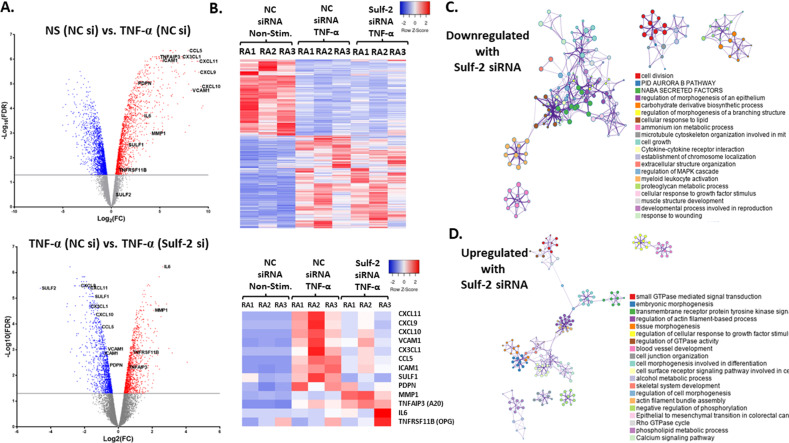
Despite the importance of TNF-α and fibroblasts in RA synovial hyperplasia and joint destruction, there is a lack of information or RNA-seq data documenting the DEGs in response to TNF-α in human RASFs. TNF-α significantly modulated the expression of 3634 out of 20,803 genes on our RNA-seq array at a threshold of FDR < 0.05. A volcano plot comparing TNF-α/NC siRNA and NS/NC siRNA (Fig. 3A; upper panel) showed the dramatic induction of inflammatory genes. Several TNF-α-induced genes labeled on the volcano plot have known roles in RA pathogenesis, including the chemokine genes CCL5, CX3CL1, CXCL11, CXCL9, and CXCL10; the adhesion protein genes ICAM1 and VCAM1; the migration-associated gene PDPN; and the TNFAIP3 gene, which encodes A20, a negative regulator of NF-κB inflammatory signaling. The top 2500 DEGs ranked by statistical significance are shown in the heatmap in Fig. 3B.
Fig. 3.
RNA-seq array reveals that the TNF-α-mediated induction of many inflammatory genes in human RASFs in vitro is dependent on Sulf-2. A (Upper volcano plot): Gene expression profiles of TNF-α-stimulated and NS RASFs are compared in a volcano plot. Genes of interest in this study are labeled, including TNF-α-induced inflammatory genes that were highly significant (up) and highly upregulated (right). (Lower volcano plot): TNF-α-stimulated RASFs with or without Sulf-2 are compared in a volcano plot. For GO analysis, DEGs were identified by |log2FC| > 1 and FDR < 0.05. B The upper heatmap shows the relative gene expression of the top 236 DEGs by FDR in the comparison between unstimulated and TNF-α-stimulated RASFs. Modulation of TNF-α-induced gene expression can be seen in RASFs with Sulf-2 knockdown. The lower heatmap shows the relative expression of selected gene targets that are relevant to RA pathogenesis. Blue represents lower expression, and red represents higher expression. C–D Diagrams depict networks of enriched GO terms for the 414 downregulated genes (panel C) and 315 upregulated genes (panel D) in TNF-α-stimulated RASFs with Sulf-2 knockdown in vitro; node size represents the number of input genes included in a specific term, colors indicate a shared cluster ID, and line thickness represents the GO similarity score
Separate GO analysis was performed for DEGs induced or suppressed by ≥2-fold by TNF-α treatment in RASFs. TNF-α upregulated 1127 genes in RASFs (≥2-fold, FDR < 0.05). GO analysis revealed closely related clusters involved in inflammation and immunity, including “cytokine signaling in the immune system, response to virus, and response to interferon-gamma” (Fig. S2A, B), and the top enriched transcriptional networks (Fig. S2C). TNF-α downregulated the expression of 1038 genes in RASFs (≥2-fold FDR < 0.05). GO analysis revealed diverse and loosely interconnected processes (Fig. S2D, E), including “small GTPase-mediated signal transduction, embryonic morphogenesis, transmembrane receptor protein tyrosine kinase signaling” (Fig. S2E), and a downregulated transcriptional network regulated by TP53 (Fig. S2F).
RNA-seq shows the impact of Sulf-2 on TNF-α-induced inflammatory activation in RASFs
We used a loss-of-function approach and RNA-seq analysis to assess the role of Sulf-2 in TNF-α-stimulated RASFs. The qRT‒PCR results demonstrated successful knockdown of Sulf-2 mRNA, with >90% reduction in the Sulf-2 siRNA group compared to the NC siRNA group (p < 0.05) (Fig. S2G). In TNF-α-stimulated RASFs, the absence of Sulf-2 significantly modulated 2502 genes (FDR < 0.05) compared to the NC siRNA condition. The volcano plot (Fig. 3A; lower panel) shows a striking reduction in many inflammatory genes compared to TNF-α alone. Notably, while the transcript levels of the TNF-α-induced chemokines CCL5, CX3CL1, CXCL11, CXCL9, CXCL10 and adhesion molecules ICAM1, VCAM1 and PDPN decreased in the absence of Sulf-2, those of the protective factors TNFAIP3 (anti-inflammatory A20) and TNFRSF11B (bone-protective osteoprotegerin) increased beyond the level induced by TNF-α alone. MMP1 and IL6 gene expression levels also further increased after TNF-α stimulation in the context of Sulf-2 knockdown. The changes in gene expression of 236 DEGs are shown in the heatmap in Fig. 3B.
Preliminary GO comparisons using IPA® showed that the absence of Sulf-2 modulated genes in the canonical pathway “role of macrophages, fibroblasts, and endothelial cells in rheumatoid arthritis” (p = 7.26E−8) (Fig. S2H). Separate GO analysis was conducted for DEGs that were upregulated or downregulated ≥2-fold by TNF-α/Sulf-2 siRNA compared to TNF-α/NC siRNA. All RASFs in these comparisons were stimulated with TNF-α, and the only variable was the absence of Sulf-2. The expression of 414 genes was suppressed (≥2-fold, FDR < 0.05) by Sulf-2 knockdown; these genes included TNF-α-induced inflammatory genes with important roles in RA pathogenesis. The GO-enriched biological processes included cell division, PID Aurora B pathway (cytokinesis in mitosis), NABA secreted factors (glycoproteins, collagens, and proteoglycans), cell growth, cytokine‒cytokine receptor interaction, regulation of MAPK cascade, and myeloid leukocyte activation (Figs. 3C and S2I).The top transcription factors that regulate these genes included Kruppel-like factor 5 (which regulates cell proliferation), inflammatory interferon regulatory factor 1 (IRF1), NF-κB p65 (RELA) and p50 (NFκB1) (Fig. S2J). The expression of 315 genes increased (≥2-fold, FDR < 0.05). GO analysis identified the enriched biological processes, which included vasculature development and response to peptide (Figs. 3D and S2K). The enriched transcriptional networks are shown in Fig. S2L. The RNA-seq array findings were validated by qRT‒PCR analysis of additional human RASFs; this validation confirmed the significant decreases in the TNF-α-induced levels of CCL5, CX3CL1, CXCL11, CXCL10, CAD11, ICAM1, VCAM1 and PDPN mRNA after Sulf-2 knockdown compared to TNF-α/NC (Fig. S2M, p < 0.05 for all). Taken together, these results suggest that Sulf-2 plays an important but previously undescribed role in TNF-α inflammatory signaling.
Knockdown of Sulf-2 inhibits the expression of TNF-α-induced adhesion proteins and chemokines
Next, we confirmed our RNA-seq and qRT‒PCR findings at the protein level. Significant time-dependent increases in cadherin-11, ICAM-1, VCAM-1, and podoplanin in TNF-α-treated RASFs were confirmed by Western blotting (Fig. S3A). We knocked down Sulf-2 and Sulf-1 before stimulating RASFs with TNF-α for 24 h and found that Sulf-2 and Sulf-1 protein levels were reduced by >60% (p < 0.05) (Fig. 4A). Compared to TNF-α/NC siRNA, TNF-α/ Sulf-2 siRNA led to a 52% reduction in ICAM-1 (p < 0.05), a 40% reduction in VCAM-1 (p < 0.05), and a 39% reduction in Cadherin-11 (N.S.) expression, while no significant reductions were observed with TNF-α/Sulf-1 siRNA (Fig. 4A).
Fig. 4.
Knockdown of Sulf-2 or pretreatment of human RASFs with the Sulf-2 inhibitor OKN-007 inhibits the TNF-α-mediated induction of adhesion proteins and chemokines. Human RASFs were transfected with scrambled (NC), Sulf-2 or Sulf-1 siRNA (130 pmol) for 48 h, followed by serum starvation and stimulation with TNF-α (20 ng/ml) for 24 h. Knockdown of Sulf-2 significantly inhibited the TNF-α-mediated induction of ICAM-1 and VCAM-1 (A) and of IP-10/CXCL10, and I-TAC/CXCL11 (B). Pretreatment of RASFs for 12 h with the Sulf-2 inhibitor OKN-007 (0–1 mM) followed by TNF-α (20 ng/ml) stimulation for 24 h resulted in the dose-dependent inhibition of the TNF-α-mediated induction of ICAM-1 and VCAM-1 (C) and RANTES/CCL5, ENA-78/CXCL5, IP-10/CXCL10 and I-TAC/CXCL11 (D)
CXCL10 and CXCL11 were among the top TNF-α-induced genes in RASFs identified in the RNA-seq array, and Sulf-2 knockdown significantly impeded their induction. The qRT‒PCR results confirmed that the absence of Sulf-2 decreased the TNF-α-mediated induction of both chemokine genes by greater than 60%. To verify this finding at the protein level and to compare the roles of Sulf-2 and Sulf-1, both sulfatases were knocked down by siRNA, and the TNF-α-mediated induction of CXCL10/IP-10 and CXCL11/I-TAC was determined by analyzing RASF conditioned media (Fig. 4B). The results differed between Sulf-1 knockdown and Sulf-2 knockdown; Sulf-1 siRNA evoked a significant 67% reduction in CXCL10/IP-10 (p < 0.01) but a nonsignificant decrease in CXCL11/I-TAC, whereas Sulf-2 siRNA resulted in a 74% reduction in CXCL10/IP-10 (p < 0.001) and a 58% reduction in CXCL11/I-TAC (p < 0.05).
The Sulf-2 inhibitor OKN-007 abrogates TNF-α-induced adhesion protein expression and chemokine production
OKN-007 has been reported to inhibit Sulf-2 activity [12]. OKN-007 alone at a dose of 0–1000 µM did not significantly affect RASF viability, but in combination with TNF-α, it modestly reduced RASF viability at 500 and 1000 µM (p < 0.05) (Fig. S3B). We evaluated the inhibition of Sulf-2 by OKN-007 in TNF-α-stimulated RASFs to support our findings with siRNA-mediated Sulf-2 knockdown. OKN-007 (500 and 1000 µM) decreased TNF-α-induced ICAM-1 expression by 20% and 36% and VCAM-1 expression by 23% and 35%, respectively (p < 0.05) (Fig. 4C), and inhibited CXCL5/ENA-78 (66–75%), CCL5/RANTES (~65%), CXCL10/IP-10 (80–95%), and CXCL11/I-TAC (43–65%) (p < 0.05) production in human RASFs (Fig. 4D).
Sulf-2 mediates TNF-α signaling primarily via the PKCδ and JNK signaling pathways
We investigated the signaling pathways through which TNF-α induces adhesion molecules in RASFs. Human RASFs were preincubated for 2 h with a panel of signaling inhibitors, followed by stimulation with TNF-α for 24 h. The protein levels of adhesion molecules were measured by Western blotting (Fig. 5A). For cadherin-11, a significant reduction of ~50% was observed with NF-κB, JNK, ASK1, JAK2, and PI3K inhibitors (p < 0.05), and a 67% reduction was observed with the PKCδ inhibitor (p < 0.01). For ICAM-1 and VCAM-1, inhibition of PKCδ led to 94% and 97% reductions, respectively; TNF-α-induced VCAM-1 levels were also reduced to ~50% by the ASK1, JAK2, and PI3K inhibitors (p < 0.01). Podoplanin levels were reduced by 40% in the presence of a JNK inhibitor and by 60% in the presence of rottlerin, an inhibitor of PKCδ (p < 0.05). The PKCδ pathway appeared to be the most significant common pathway, so we confirmed these results using the newly developed small-molecule PKCδ inhibitor NP627 (Fig. 5B). NP627 (20 nM) reduced the TNF-α-induced expression of cadherin-11 by 43% (p < 0.05), VCAM-1 by 52% (p < 0.01) and ICAM-1 by 53% (p < 0.05). NP627 did not exhibit cytotoxicity at this concentration in an MTT assay (Fig. S4). These results indicate that PKCδ and JNK are critical pathways in the TNF-α-mediated induction of adhesion molecules in RASFs. Furthermore, silencing of Sulf-2 reduced the TNF-α-induced phosphorylation of PKCδ by 52% and of JNK by 36% (p < 0.05) (Fig. 6A), with no change in the phosphorylation of p38 or ERK, suggesting that Sulf-2 regulates the TNF-α-mediated expression of adhesion and migration molecules in RASFs through the PKCδ and JNK pathways.
Fig. 5.
TNF-α induces adhesion proteins in human RASFs via the PKCδ (JNK and NF-κB) signaling pathway. Knockdown of Sulf-2 inhibits the TNF-α-induced phosphorylation of PKCδ and JNK. A Human RASFs were preincubated for 2 h with a panel of signaling inhibitors, followed by stimulation with TNF-α for 24 h. Western blots were probed for ICAM-1, VCAM-1, cadherin-11 and podoplanin. Inhibitors of NF-κB, JNK and, most prominently, PKCδ led to significant reductions in TNF-α-induced adhesion proteins, revealing the importance of these pathways. B The central role of PKCδ was confirmed by the dose-dependent decrease in ICAM-1, VCAM-1 and cadherin-11 levels in RASFs after treatment with the PKCδI-specific small-molecule inhibitor NP627
Fig. 6.
Knockdown of Sulf-2 inhibits the TNF-α-induced phosphorylation of PKCδ and JNK and TNF-α-induced RASF proliferation and invasion. A siRNA-mediated knockdown of Sulf-2 in human RASFs significantly inhibited the TNF-α-induced phosphorylation of PKCδ and JNK. Data are presented as the mean ± SEM. *p < 0.05. B Knockdown of Sulf-2 inhibited the TNF-α-induced proliferation of RASFs to almost basal levels. C The Transwell invasion assay showed a trend in decreased (25%) TNF-α-induced RASF invasion in the presence of Sulf-2 siRNA (p < 0.07)
Knockdown of Sulf-2 inhibits TNF-α-induced RASF proliferation and invasion
The results of the proliferation assay (Fig. 6B) showed a two-fold increase in RASF proliferation in response to 24 h of TNF-α stimulation, and this increase was markedly inhibited by Sulf-2 siRNA, almost to basal levels. In addition, the Transwell invasion assay results showed a trend toward a reduction (25%) in TNF-α-induced RASF invasion upon exposure to Sulf-2 siRNA (p < 0.07) (Fig. 6C).
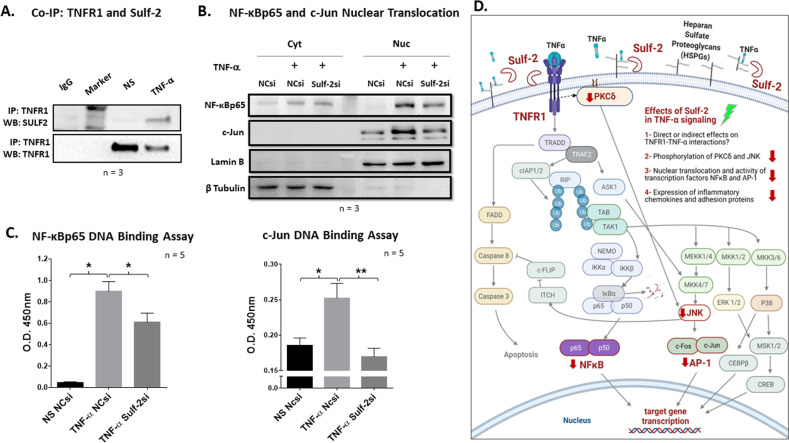
Immunoprecipitation assay reveals Sulf-2 and TNF receptor 1 binding
The results of our coimmunoprecipitation assay showed that Sulf-2 associates with TNFR1 in TNF-α-stimulated RASFs but not in unstimulated controls (Fig. 7A). TNF-α was previously shown to induce the release of its own receptor TNFR1 in the full 55 kD form, and soluble TNFR1 was reported to exert anti-inflammatory effects by acting as a sink for soluble TNF-α [28–31]. In accordance with previous reports, our immunoprecipitation assay showed a decrease in TNFR1 within RASF lysates after only 30 min of TNF-α stimulation. We confirmed the solubilization of TNFR1 by Western blot analysis of concentrated supernatants of TNF-α-stimulated RASFs from three patients (Fig. S1E).
Fig. 7.
Sulf-2 binds TNF receptor 1, and knockdown of Sulf-2 reduces the TNF-α-induced nuclear translocation and DNA binding activity of inflammatory transcription factors. A Immunoprecipitation of TNF receptor 1 revealed binding with the 75 kD subunit of the mature Sulf-2 heterodimeric protein in TNF-α-stimulated human RASFs. B Knockdown of Sulf-2 in RASFs inhibited the TNF-α-induced nuclear translocation of c-Jun and NF-κBp65. C Knockdown of Sulf-2 significantly reduced the TNF-α-induced DNA binding activity of NF-κBp65 and c-Jun. Data are presented as the mean ± SEM. *p < 0.05; **p < 0.01. D Schematic diagram illustrating the effects of Sulf-2 knockdown on TNF-α inflammatory signaling in human RASFs
Sulf-2 knockdown inhibits the TNF-α-induced nuclear translocation and DNA binding activity of inflammatory transcription factors
Cytoplasmic and nuclear fractionation of RASFs stimulated for 30 min with TNF-α with or without Sulf-2 revealed that Sulf-2 knockdown inhibited the nuclear translocation of the activated transcription factors NF-κBp65 and c-Jun (Fig. 7B). To quantify the potential transcriptional repression of inflammatory factors, we performed specific DNA binding assays for NF-κB and c-Jun. The absence of Sulf-2 reduced NF-κBp65 and c-Jun DNA binding activity by 32% (p < 0.05) and 33% (p < 0.01), respectively (Fig. 7C), suggesting that Sulf-2 contributes to TNF-α signaling through NF-κB and AP-1. A schematic diagram provides an understanding of the signaling pathways altered by Sulf-2 knockdown (Fig. 6C).
Discussion
This is the first study to characterize the expression and potential roles of extracellular sulfatases in TNF-α signaling and RA synovial pathogenesis. We found that elevated levels of Sulf-2 in the serum and synovial tissues strongly correlated with the presence of RA in patients and hTNFtg mice compared to nondiseased counterparts. Our RNA-seq array dataset identified the global transcriptional effects of TNF-α stimulation of human RASFs in vitro and demonstrated the significant impact of Sulf-2 knockdown on these effects. Bioinformatics analysis of DEGs and GO enrichment analysis revealed that Sulf-2 is important for the TNF-α-mediated induction of adhesion proteins and chemokines in RASFs. Signaling studies in RASFs revealed that Sulf-2 knockdown inhibited the TNF-α-induced activation of the PKCδ and JNK pathways, as well as the nuclear translocation and DNA binding activity of inflammatory transcription factors. Our co-IP experiment provided mechanistic insights by demonstrating the binding of TNFR1 receptor and Sulf-2 in TNF-α-stimulated RASFs. Knockdown of Sulf-2 abrogated TNF-α-induced RASF proliferation, suggesting that inhibition of Sulf-2 could mitigate synovial hyperplasia in RA. Together, our results provide the rationale for interventional studies to test the therapeutic potential of inhibiting Sulf-2 in preclinical models of RA.
TNF inhibitors have improved the therapeutic management of autoimmune diseases and beyond [8, 32]. However, the current limitations of anti-TNF therapy highlight the need for alternate approaches that limit the inflammatory effects of TNF-α without full systemic inhibition. Expression and ligation of TNFR1 is critical for inflammatory signaling. The importance of TNFR1 is illustrated by the fact that over 90 mutations in the TNFRSF1A gene, which encodes TNFR1, cause excessive receptor activation, defective receptor shedding and aberrantly elevated NF-κBp65 or c-Rel transcription factor activity in TNF receptor-associated periodic syndrome [33]. In a previous study, pathway network analysis of human synovial tissues identified TNFRSF1A as a seed gene with disrupted functional connectivity in expression specific to RA [34]. Pearson correlation analysis of our qRT‒PCR results in normal and RASTs revealed a significant positive association between TNFRSF1A and SULF2, suggesting shared transcriptional regulation and/or mutual involvement in RA pathogenesis. The pathology in the hTNFtg mouse model is mediated through TNFR1 because human TNF-α shows species-specific binding to murine TNFR1 and not TNFR2 [19]. Our observation of enhanced expression of Sulf-1 and Sulf-2 in the synovium and serum of hTNFtg mice compared to WT mice supports our findings in human RASTs and serum.
Having noted a positive correlation between the gene expression of SULF2 and TNFRSF1A, we hypothesized that an interaction between the encoded proteins may be the potential mechanism by which Sulf-2 promotes TNF-α signaling. We performed a coimmunoprecipitation assay, which showed that Sulf-2 associates with TNFR1 in TNF-α-stimulated RASFs but not in unstimulated controls. This is the first time a noncanonical cell surface partner has been identified for TNFR1, and this result warrants further in-depth mutational and computational analysis to identify potential interaction sites between these two extracellular proteins and their relevance to TNF-α signaling in RASFs and potentially other cell types.
Sulf-2 can stabilize TNFR1 on the cell membrane by participating in coreceptor assembly with TNFR1 and heparan sulfate proteoglycans (HSPGs). Furthermore, by binding to soluble TNFR1, Sulf-2 could potentially interfere with the anti-inflammatory effects of the soluble receptor. Further studies are warranted to investigate the possible regulatory or functional association between Sulf-2 and TNFR1 in inflammation.
Through their enzymatic activity, Sulf-1 and Sulf-2 cleave 6-O-sulfate groups from heparan sulfate (HS) chains on HSPGs on the cell surface [9, 35]. Regions of dense sulfation on the 6th carbon of glucosamine in HS are known binding sites for inflammatory chemokines, including platelet factor-4/CXCL4, RANTES/CCL5, IL-8/CXCL8 and stromal cell-derived factor 1/CXCL12 [14]. Sulf-2 is known to promote Wnt signaling and inhibit fibroblast growth factor-2 signaling [10]. However, the specific effects of extracellular sulfatase activity on the signaling of many growth factors, chemokines and cytokines have not been elucidated. Treatment of primary human umbilical vein endothelial cells with TNF-α for 24 h led to decreased 6-O-sulfation of membrane HSPGs [36], suggesting an increase in Sulf-1/Sulf-2 activity. Extracellular sulfatases respond to inflammatory cytokines in certain contexts. Increased Sulf-1 expression was observed in human lung fibroblast cells stimulated with TNF-α [37]. Similarly, in vitro stimulation with transforming growth factor β induced the expression of Sulf-2 in renal epithelial cells [38] and of both Sulf-1 and Sulf-2 in lung fibroblasts [39]. In a recent study, Kim et al. reported increased Sulf-1 and Sulf-2 expression in CD68+ macrophages in response to inflammation [40]. Our findings build on this foundation by providing molecular insights into the role of Sulf-2 in RA pathogenesis.
Although Sulf-2 has not been studied in arthritis, it is being investigated as a cancer drug target [10, 12]. In head and neck squamous cell carcinoma (HNSCC), elevated mRNA and protein levels of Sulf-2 correlate with tumor progression and poor overall survival [41]. Flowers et al. reported significant overexpression of Sulf-2 in 57% of HNSCC tumors and in the saliva of patients compared to healthy controls, leading to the proposal of Sulf-2 as a biomarker [42]. Although no link with Sulf-2 has been reported, aberrantly elevated NF-κB activation is a well-established feature and pathogenic mechanism in HNSCC [43]. It would be intriguing to determine if these independent observations stem from increased TNF-α signaling due to the overexpression of Sulf-2.
TNF inhibitors have radically changed the treatment of patients with refractory RA, but they are expensive, immunosuppressive, and sometimes unsuitable for long-term use. Existing RA therapies are directed at B cells, T cells, and cytokines. No existing medication for RA targets RASFs despite a body of molecular and clinical evidence for their active role in RA pathogenesis [3]. Our results suggest that targeting Sulf-2 as an adjunct therapeutic approach to limit TNF-α-induced synovial hyperplasia and tissue destruction may represent a unique opportunity to suppress synovial inflammation (but not systemic host defenses) and improve conventional RA therapy.
Supplementary information
Acknowledgements
The authors thank the Cooperative Human Tissue Network and National Disease Research Interchange for providing human synovial tissue specimens.
Author contributions
RJS, AKS, and SA designed this study. RJS, AKS, JV, SUH, and HMK performed the experiments. RJS, AKS, JV, SUH, SAK, and BSK analyzed the data. RJS wrote the manuscript with participation from SA. DAF and CSC provided human RASFs and serum samples, respectively, as well as clinical guidance and manuscript feedback. SA provided support for the study.
Funding
This study was supported by the NIH/NIAMS F31 Fellowship AR-076204–01 (RJS), Rheumatology Research Foundation Graduate Student Preceptorship Award (RJS/SA), and NIH/NIAMS R01 Grant AR-072615 (SA). Research by SUH and JV was made possible through the WSU College of Pharmacy and Pharmaceutical Sciences Honors Research Program.
Competing interests
The authors declare no competing interests.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00913-x.
References
- 1.Puimège L, Libert C, Van, Hauwermeiren F. Regulation and dysregulation of tumor necrosis factor receptor-1. Cytokine Growth Factor Rev. 2014;25:285–300. doi: 10.1016/j.cytogfr.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 3.Bartok B, Firestein GS. Fibroblast-like synoviocytes: key effector cells in rheumatoid arthritis. Immunol Rev. 2010;233:233–55. doi: 10.1111/j.0105-2896.2009.00859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lowenthal JW, Ballard DW, Bohnlein E, Greene WC. Tumor necrosis factor alpha induces proteins that bind specifically to kappa B-like enhancer elements and regulate interleukin 2 receptor alpha-chain gene expression in primary human T lymphocytes. Proc Natl Acad Sci USA. 1989;86:2331–5. doi: 10.1073/pnas.86.7.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roach DR, Bean AG, Demangel C, France MP, Briscoe H, Britton WJ. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J Immunol. 2002;168:4620–7. doi: 10.4049/jimmunol.168.9.4620. [DOI] [PubMed] [Google Scholar]
- 6.Asif Amin M, Fox DA, Ruth JH. Synovial cellular and molecular markers in rheumatoid arthritis. Semin Immunopathol. 2017;39:385–93. doi: 10.1007/s00281-017-0631-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldmann M, Maini RN. Anti-TNF alpha therapy of rheumatoid arthritis: what have we learned? Annu Rev Immunol. 2001;19:163–96. doi: 10.1146/annurev.immunol.19.1.163. [DOI] [PubMed] [Google Scholar]
- 8.Gerriets V, Bansal P, Goyal A, Khaddour K. Tumor necrosis factor inhibitors, in StatPearls. Treasure Island (FL): StatPearls; 2021. [PubMed]
- 9.El Masri R, Seffouh A, Lortat-Jacob H, Vives RR. The “in and out” of glucosamine 6- O-sulfation: the 6th sense of heparan sulfate. Glycoconj J. 2017;34:285–98. doi: 10.1007/s10719-016-9736-5. [DOI] [PubMed] [Google Scholar]
- 10.Rosen SD, Lemjabbar-Alaoui H. Sulf-2: an extracellular modulator of cell signaling and a cancer target candidate. Expert Opin Ther Targets. 2010;14:935–49. doi: 10.1517/14728222.2010.504718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Uchimura K, Morimoto-Tomita M, Bistrup A, Li J, Lyon M, Gallagher J, et al. HSulf- 2, an extracellular endoglucosamine-6-sulfatase, selectively mobilizes heparinbound growth factors and chemokines: effects on VEGF, FGF-1, and SDF-1. BMC Biochem. 2006;7:2. doi: 10.1186/1471-2091-7-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zheng X, Gai X, Han S, Moser CD, Hu C, Shire AM, et al. The human sulfatase 2inhibitor 2,4-disulfonylphenyl-tert-butylnitrone (OKN-007) has an antitumor effect in hepatocellular carcinoma mediated via suppression of TGFB1/SMAD2 and Hedgehog/GLI1 signaling. Genes Chromosomes Cancer. 2013;52:225–36. doi: 10.1002/gcc.22022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Towner RA, Hocker J, Smith N, Saunders D, Battiste J, Hanas J. OKN-007 Alters Protein Expression Profiles in High-Grade Gliomas: Mass Spectral Analysis of Blood Sera. Brain Sci. 2022;12:100. doi: 10.3390/brainsci12010100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.El Masri R, Crétinon Y, Gout E, Vivès RR. HS and inflammation: a potential playground for the Sulfs? Front Immunol. 2020;11:570. doi: 10.3389/fimmu.2020.00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otsuki S, Taniguchi N, Grogan SP, D'Lima D, Kinoshita M, Lotz M. Expression of novel extracellular sulfatases Sulf-1 and Sulf-2 in normal and osteoarthritic articular cartilage. Arthritis Res Ther. 2008;10:R61. doi: 10.1186/ar2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ratzka A, Kalus I, Moser M, Dierks T, Mundlos S, Vortkamp A. Redundant function of the heparan sulfate 6-O-endosulfatases Sulf1 and Sulf2 during skeletal development. Dev Dyn. 2008;237:339–53. doi: 10.1002/dvdy.21423. [DOI] [PubMed] [Google Scholar]
- 17.Zaman G, Staines KA, Farquharson C, Newton PT, Dudhia J, Chenu C, et al. Expression of Sulf1 and Sulf2 in cartilage, bone and endochondral fracture healing. Histochem Cell Biol. 2016;145:67–79. doi: 10.1007/s00418-015-1365-8. [DOI] [PubMed] [Google Scholar]
- 18.Bell RD, Wu EK, Rudmann CA, Forney M, Kaiser CRW, Wood RW, et al. Selective sexual dimorphisms in musculoskeletal and cardiopulmonary pathologic manifestations and mortality incidence in the tumor necrosis factor-transgenic mouse model of rheumatoid arthritis. Arthritis Rheumatol. 2019;71:1512–23. doi: 10.1002/art.40903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, Schwarz EM. The TNF-alpha transgenic mouse model of inflammatory arthritis. Springer Semin Immunopathol. 2003;25:19–33. doi: 10.1007/s00281-003-0125-3. [DOI] [PubMed] [Google Scholar]
- 20.Tsai C, Diaz LA, Jr., Singer NG, Li LL, Kirsch AH, Mitra R, et al. Responsiveness of human T lymphocytes to bacterial superantigens presented by cultured rheumatoid arthritis synoviocytes. Arthritis Rheum. 1996;39:125–36. doi: 10.1002/art.1780390117. [DOI] [PubMed] [Google Scholar]
- 21.Haque M, Singh AK, Ouseph MM, Ahmed S. Regulation of synovial inflammation and tissue destruction by guanylate binding protein 5 in synovial fibroblasts from patients with rheumatoid arthritis and rats with adjuvant-induced arthritis. Arthritis Rheumatol. 2021;73:943–54. doi: 10.1002/art.41611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robinson MD, Oshlack A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010;11:R25. doi: 10.1186/gb-2010-11-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systemslevel datasets. Nat Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, et al. Heatmapper: web-enabled heat mapping for all. Nucleic Acids Res. 2016;44:W147–53. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singer MS, Phillips JJ, Lemjabbar-Alaoui H, Wang YQ, Wu J, Goldman R, et al. SULF2, a heparan sulfate endosulfatase, is present in the blood of healthy individuals and increases in cirrhosis. Clin Chim Acta. 2015;440:72–8. doi: 10.1016/j.cca.2014.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed S, Pakozdi A, Koch AE. Regulation of interleukin-1beta-induced chemokine production and matrix metalloproteinase 2 activation by epigallocatechin-3- gallate in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2006;54:2393–401. doi: 10.1002/art.22023. [DOI] [PubMed] [Google Scholar]
- 28.Redl H, Schlag G, Adolf GR, Natmessnig B, Davies J. Tumor necrosis factor (TNF)- dependent shedding of the p55 TNF receptor in a baboon model of bacteremia. Infect Immun. 1995;63:297–300. doi: 10.1128/iai.63.1.297-300.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lantz M, Malik S, Slevin ML, Olsson I. Infusion of tumor necrosis factor (TNF) causes an increase in circulating TNF-binding protein in humans. Cytokine. 1990;2:402–6. doi: 10.1016/1043-4666(90)90048-x. [DOI] [PubMed] [Google Scholar]
- 30.Hawari FI, Rouhani FN, Cui X, Yu ZX, Buckley C, Kaler M, et al. Release of fulllength 55-kDa TNF receptor 1 in exosome-like vesicles: a mechanism for generation of soluble cytokine receptors. Proc Natl Acad Sci USA. 2004;101:1297–302. doi: 10.1073/pnas.0307981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bradley JR. TNF-mediated inflammatory disease. J Pathol. 2008;214:149–60. doi: 10.1002/path.2287. [DOI] [PubMed] [Google Scholar]
- 32.Mercogliano MF, Bruni S, Mauro F, Elizalde PV, Schillaci R. Harnessing tumor necrosis factor alpha to achieve effective cancer immunotherapy. Cancers. 2021;13:564. doi: 10.3390/cancers13030564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner MD, Nedjai B, Hurst T, Pennington DJ. Cytokines and chemokines: at the crossroads of cell signalling and inflammatory disease. Biochim Biophys Acta. 2014;1843:2563–82. doi: 10.1016/j.bbamcr.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 34.Sabir JSM, El Omri A, Banaganapalli B, Al-Shaeri MA, Alkenani NA, Sabir MJ, et al. Dissecting the role of NF-κb protein family and its regulators in rheumatoid arthritis using weighted gene co-expression network. Front Genet. 2019;10:1163. doi: 10.3389/fgene.2019.01163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lanzi C, Zaffaroni N, Cassinelli G. Targeting heparan sulfate proteoglycans and their modifying enzymes to enhance anticancer chemotherapy efficacy and overcome drug resistance. Curr Med Chem. 2017;24:2860–86. doi: 10.2174/0929867324666170216114248. [DOI] [PubMed] [Google Scholar]
- 36.Reine TM, Kusche-Gullberg M, Feta A, Jenssen T, Kolset SO. Heparan sulfate expression is affected by inflammatory stimuli in primary human endothelial cells. Glycoconj J. 2012;29:67–76. doi: 10.1007/s10719-011-9365-y. [DOI] [PubMed] [Google Scholar]
- 37.Sikora AS, Hellec C, Carpentier M, Martinez P, Delos M, Denys A, et al. Tumournecrosis factor-α induces heparan sulfate 6-O-endosulfatase 1 (Sulf-1) expression in fibroblasts. Int J Biochem Cell Biol. 2016;80:57–65. doi: 10.1016/j.biocel.2016.09.021. [DOI] [PubMed] [Google Scholar]
- 38.Alhasan AA, Spielhofer J, Kusche-Gullberg M, Kirby JA, Ali S. Role of 6-Osulfated heparan sulfate in chronic renal fibrosis. J Biol Chem. 2014;289:20295–306. doi: 10.1074/jbc.M114.554691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yue X, Li X, Nguyen HT, Chin DR, Sullivan DE, Lasky JA. Transforming growth factor-beta1 induces heparan sulfate 6-O-endosulfatase 1 expression in vitro and in vivo. J Biol Chem. 2008;283:20397–407. doi: 10.1074/jbc.M802850200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim HJ, Kim HS, Hong YH. Sulfatase 1 and sulfatase 2 as novel regulators of macrophage antigen presentation and phagocytosis. Yeungnam Univ J Med. 2021;38:326–36. doi: 10.12701/yujm.2021.01025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Y, Ahn J, Raghunathan R, Kallakury BV, Davidson B, Kennedy ZB, et al. Expression of the extracellular sulfatase SULF2 affects survival of head and neck squamous cell carcinoma patients. Front Oncol. 2020;10:582827. doi: 10.3389/fonc.2020.582827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Flowers SA, Zhou X, Wu J, Wang Y, Makambi K, Kallakury BV, et al. Expression of the extracellular sulfatase SULF2 is associated with squamous cell carcinoma of the head and neck. Oncotarget. 2016;7:43177–87. doi: 10.18632/oncotarget.9506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen CT, Ricker JL, Chen Z, Van Waes C. Role of activated nuclear factor-kappaB in the pathogenesis and therapy of squamous cell carcinoma of the head and neck. Head Neck. 2007;29:959–71. doi: 10.1002/hed.20615. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.