Summary
Ficus benghalensis and Ficus religiosa are large woody trees well known for their long lifespan, ecological and traditional significance, and medicinal properties. To understand the genomic and evolutionary aspects of these characteristics, the whole genomes of these Ficus species were sequenced using 10x Genomics linked reads and Oxford Nanopore long reads. The draft genomes of F. benghalensis and F. religiosa comprised of 392.89 Mbp and 332.97 Mbp, respectively. We established the genome-wide phylogenetic positions of the two Ficus species with respect to 50 other Angiosperm species. Comparative evolutionary analyses with other phylogenetically closer Eudicot species revealed adaptive evolution in genes involved in key cellular mechanisms associated with prolonged survival including phytohormones signaling, senescence, disease resistance, and abiotic stress tolerance, which provide genomic insights into the mechanisms conferring longevity and suggest that longevity is a multifaceted phenomenon. This study also provides clues on the existence of CAM pathway in these Ficus species.
Subject areas: Plant biology, Plant genetics, Plant evolution, Genomics
Graphical abstract

Highlights
-
•
First whole genome assemblies of Ficus benghalensis and Ficus religiosa trees
-
•
Genome-wide phylogeny with 50 other Angiosperm species
-
•
Evolution of genes in phytohormone signaling, senescence, and stress tolerance
-
•
Genomic insights into longevity-regulating mechanisms
Plant biology; Plant Genetics; Plant evolution; Genomics
Introduction
Longevity is an intriguing characteristic of exclusive plant species that show lifespans ranging from hundreds to thousands of years. It is also a key constituent of demographic storage as the long-lived plants such as Pinus, Oak maintain the ecosystem (Piovesan and Biondi, 2021; Roeder et al., 2021). Several factors have been proposed to achieve longevity in plants such as sustained growth over a long time period, resistance to biotic and abiotic stress (major drivers of mortality, particularly in larger trees), accumulation and transmission of somatic mutations, expansion of disease resistance gene families, phytohormones signaling, etc. (Bartrina et al., 2017; Munné-Bosch, 2018; Piovesan and Biondi, 2021; Plomion et al., 2018). In the Indian subcontinent, the two Ficus species, F. benghalensis and F. religiosa, are the best-known examples of trees with the longest lifespans.
Ficus is one of the most diverse genus that originated around 80–90 million years ago, and consists of ∼800 species (Harrison, 2005; Singh et al., 2011). Ficus trees (also known as fig trees) that belong to the plant family Moraceae are also known for their well-developed morphological characteristics, and religious, ecological, evolutionary, and medicinal importance. Ficus species possess several characteristics such as smaller seed size, higher growth rate, higher fecundity, etc. that make them evolutionarily flexible to grow in diverse environmental habitats (Harrison, 2005). Species from the Ficus genus occupy diverse ecological niches in different life forms such as free-living, epiphytes, semi-epiphytes, lithophytes, or rheophytes (Shi et al., 2018). Ficus trees are considered as keystone species in a tropical ecosystem as they sustain a large number of animal species with fig fruits that are produced throughout the year (Shanahan et al., 2001). Furthermore, Ficus trees (including F. benghalensis and F. religiosa) act as forest restoration agents due to their larger canopy areas that favor plant species dispersed by frugivores as well as other species (Cottee-Jones et al., 2016). The co-evolution of Ficus and their pollinator species such as fig wasps is a well-known example to show obligate mutualism (GALIL and EISIKOWITCH, 1971; Zhang et al., 2020), where Ficus trees interact with their wasp pollinators by attracting them with volatile organic compounds (VOCs) (Grison-Pigé et al., 2002) for pollination. Interestingly, not all members from this genus show longevity unlike the F. benghalensis and F. religiosa species.
F. benghalensis, also known as Indian banyan tree, is the National tree of India, and is native to tropical, sub-tropical regions in South Asia (Gopukumar and Praseetha, 2015). It is also known for its traditional religious, mythological significance in Hinduism and Buddhism, and their widespread branches signify eternal life (Gopukumar and Praseetha, 2015). F. benghalensis (a strangler fig) is a large woody tree with around 30 m height, and typically has a long life spanning several centuries. The largest banyan tree (Thimmamma Marrimanu) is located in Southern India, and is 550 years old. It starts growing as an epiphyte in early days, and in course of time it disperses thick plentiful aerial roots from the spreading branches to cover several hundred meters and grows independently (Moustafa, 2020).
F. religiosa, also known as “sacred fig”, “Bodhi tree”, “Peepal”, or “Ashwattha”, is a large hemi epiphyte, deciduous tree with a long average lifespan of 900–1,500 years (Devanesan et al., 2018; Vidyapeeth, 2019). It is also a well-known tree for its mythological and religious importance in Buddhism and Hinduism, and is known to have the longest lifespan in Hindu mythology (Sitaramam et al., 2009; Vidyapeeth, 2019). Both F. benghalensis and F. religiosa are large perennial trees with 20–30 m height, are epiphytic in young stages, and the adult trees have widespread branches (Devanesan et al., 2018; Galil, 1984). One of the key differences between the two trees is the absence of aerial roots in F. religiosa, unlike F. benghalensis. The extracts from bark, leaf, and fruit of these trees possess various bioactive compounds, e.g., flavonoid, terpenoid, alkaloid, tannin, saponin, etc. that confer a large number of pharmacological activities, e.g., anti-inflammatory, antihelminthic, antioxidant, antimicrobial, antitumor, antipyretic, antidiabetic, immunomodulatory, and wound healing properties (Devanesan et al., 2018; Gopukumar and Praseetha, 2015; Kumar Makhija et al., 2010), and can also be used as potential therapeutic agents (Chaudhary et al., 2017).
Therefore, the noteworthy long lifespans and the ecological and medicinal significance displayed by these two tropical Ficus species motivated us to perform their genome sequencing to understand the genomic basis of such traits. The whole genome of a few Ficus species such as Ficus carica, Ficus hispida, Ficus microcarpa, and Ficus erecta (Mori et al., 2017; Shirasawa et al., 2020; Zhang et al., 2020), and chloroplast genome of other Ficus species such as Ficus pumila (Huang et al., 2022; Liang et al., 2022) have been sequenced; however, the whole genomes of these two key Ficus species showing longevity were sequenced for the first time in this study. The genomes of both the species, F. benghalensis and F. religiosa, are known to be diploid (2n = 26) with an estimated genome size of 686 Mbp according to Plant DNA C-values Database (Pellicer and Leitch, 2020). We carried out the genome sequencing and analysis of F. benghalensis and F. religiosa using 10x Genomics linked reads and Oxford Nanopore long reads. We also performed transcriptome sequencing of these species that helped in comprehensive genome annotation. The phylogenetic position of the two Ficus species was resolved using other available Eudicot species, and comparative evolutionary analyses with selected Angiosperm plant species revealed that genes required for plant growth, development, and stress tolerance mechanisms are highly evolved in these species, which are perhaps responsible for the longevity of these two Ficus species.
Results
Genome and transcriptome sequencing
The estimated genome sizes of F. benghalensis and F. religiosa species were 381.9 and 337.7 Mbp, respectively, using GenomeScope v2.0 (Ranallo-Benavidez et al., 2020) (Figures S1A and S1B). A total of 49 (128.3X coverage) and 9 Gb (23.6X coverage) of genomic data were generated from F. benghalensis leaf tissue using 10x Genomics (Chromium) and Oxford Nanopore technologies, respectively. Furthermore, a total of 7.4 Gb of RNA-Seq data were also generated from the leaf tissue of this species (Tables S1 and S2). For F. religiosa species, 10x Genomics and Oxford Nanopore sequencing technologies were used to generate 48.7 (144.1X) and 7.2 Gb (21.3X) of genomic data, respectively. Furthermore, a total of 10.8 Gb of RNA-Seq data were also generated from the leaf tissue of this species (Tables S1 and S2).
Genome assembly
The heterozygosity content for F. benghalensis was estimated to be 1.78% from the k-mer analysis (Figure S1A). After scaffolding, the final genome assembly of F. benghalensis had a total size of 392.89 Mbp with 4,822 sequences and was close to the estimated genome size of 381.9 Mbp (Figure S1A). The assembly had a scaffold N50 value of 486.9 Kbp, longest scaffold size of 5.4 Mbp, and the GC-content of 34.54% (Table S3). 98.39% of the barcode-filtered linked-reads and 93.11% adapter-processed Nanopore reads could be mapped on the final genome assembly. Furthermore, the genome assembly had 96.4% complete BUSCOs and 1.7% of fragmented BUSCOs, using embryophyta_odb10 gene set (Figure S1C and Table S4).
The heterozygosity content for F. religiosa genome was estimated to be 1.62% from the k-mer analysis (Figure S1B). The final genome assembly of F. religiosa had a total size of 332.97 Mbp, which was also close to the predicted genome size of 337.7 Mbp and consisted of 6,087 sequences (longest scaffold of 4.51 Mbp), a scaffold N50 value of 553.4 Kbp, and a GC-content of 34.31% (Figure S1B and Table S3). 98.14% of the barcode-filtered 10x Genomics linked reads and 93.99% of the adapter-processed Nanopore long reads could be mapped to the final genome assembly. Furthermore, the final genome assembly had 95.6% complete BUSCOs, and 1.1% of fragmented BUSCOs, using embryophyta_odb10 gene set (Figure S1C and Table S4). Thus, the hybrid sequencing and assembly approach implemented using 10x Genomics linked reads and Oxford Nanopore long reads helped in the successful construction of F. benghalensis and F. religiosa genomes with decent scaffold N50 values and BUSCO scores. The comparatively higher N50 value of F. religiosa genome assembly could be due to the lower heterozygosity compared to F. benghalensis genome.
The estimated and assembled genome sizes of the Ficus species in this study were smaller than their genome sizes reported at Plant C-values Database, which perhaps depends on the usage of different reference standard plant species while estimating the genome size in the earlier used method (Doležel and Bartoš, 2005). Also, the genome sizes of these two Ficus species were similar to F. microcarpa genome and slightly larger than other Ficus—F. carica, F. hispida, and F. erecta genomes (Mori et al., 2017; Shirasawa et al., 2020; Zhang et al., 2020).
Sequence variation analysis in these final genome assemblies using barcode-filtered 10x Genomics reads identified 1,772,802 nucleotide positions (0.45%) as variant sites in F. benghalensis, and 1,466,212 variant sites (0.44%) in F. religiosa. The variant sites were in 3,254 and 3,859 contigs (after scaffolding) in F. benghalensis and F. religiosa, respectively. Depth distribution analysis showed the presence of major peaks and secondary peaks for both the Ficus genomes (Figures S2A–S2D, S3A, and S3B). Furthermore, the scaffolds showing read depths in the secondary peak regions (Figures S3A and S3B) could be representing redundancy in the assembly perhaps due to the presence of highly heterozygous regions (Table S5). Furthermore, the scaffolds showing zero read depth after mapping 10x Genomics and Oxford Nanopore reads could be a result of assembly errors (Table S6).
Genome annotation
Repeat-masking of the F. benghalensis genome using a de novo repeat library showed that among the repeat classes, the interspersed repeats constituted 47.08% (including 2.70% Ty1/Copia, 8.28% Gypsy/DIRS1 elements, and 32.88% unclassified) (Figure S1D and Table S7), and tandem repeats constituted 7.01% of the genome. Thus, ∼54% of the F. benghalensis genome was predicted to consist of simple and interspersed repeat regions.
Similarly, repeat-masking of F. religiosa genome assembly showed that 41.68% of the genome was constituted of interspersed repeats (including 1.55% Ty1/Copia, 4.51% Gypsy/DIRS1 elements, and 33.35% unclassified) (Figure S1D and Table S8), and 8.84% of the genome was constituted of simple repeats. Thus, ∼51% of the F. religiosa genome assembly consisted of repetitive regions (including both simple and interspersed repeats). Among the LTR retrotransposon class of repeats, Gypsy/DIRS1 elements were higher than Ty1/Copia elements in both the species (Tables S7 and S8), similar to other Ficus genomes (Shirasawa et al., 2020; Usai et al., 2020; Zhang et al., 2020). Thus, both the Ficus genomes contained lower repetitive content compared to highly repetitive larger plant genomes. Additionally, non-coding RNAs were also detected in the final genome assemblies of F. benghalensis and F. religiosa (Table 1).
Table 1.
Genome annotation statistics of the two Ficus species
| Parameters | F. benghalensis | F. religiosa |
|---|---|---|
| No.a of coding genes obtained from MAKER | 29,524 | 27,544 |
| No. of coding genes with AED value < 0.5 | 27,674 (93.73%) | 26,126 (94.85%) |
| Final no. of coding genes after length-based filtering (High-confidence gene set) | 25,016 | 23,929 |
| Presence of BUSCOs in coding gene set (using embryophyta_odb10 gene set) | 86.3% complete, 8.1% fragmented | 90.1% complete, 4.2% fragmented |
| No. of rRNAs | 1,028 | 1,135 |
| No. of tRNAs (decoding standard amino acids) | 679 | 668 |
| No. of miRNAs (homology-based) | 195 | 185 |
| No. of coding genes mapped against NCBI-nr database | 24,909 | 23,638 |
| No. of coding genes mapped against Swiss-Prot database | 20,947 | 19,850 |
| No. of coding genes mapped against Pfam-A database | 20,541 | 19,437 |
No.: Number.
Prior to gene set construction, quality-filtered RNA-Seq data for F. benghalensis species were de novo assembled, which resulted in a total of 10,156 transcripts (used as a set of empirical evidence in MAKER pipeline (Campbell et al., 2014)). In case of F. religiosa species, quality-filtered RNA-Seq data from this study and from a previous study (Matasci et al., 2014) were used for de novo transcriptome assembly, which resulted in 74,514 assembled transcripts that were used for evidence-based alignments in MAKER genome annotation pipeline along with the protein evidences from other publicly available related species (see STAR Methods). A total of 25,016 (94.4% BUSCOs) and 23,929 (94.3% BUSCOs) high-confidence coding genes were predicted for F. benghalensis and F. religiosa, respectively, after AED value-based (AED values < 0.5) and length-based (≥300 bp) filtering of the MAKER-derived gene models (Table 1). In these high-confidence gene sets, 90 F. benghalensis genes (0.36%) and 262 F. religiosa genes (1.09%) could not find a match in any of the following reference databases—NCBI-nr, Swiss-Prot, and Pfam-A (Table 1).
The usage of final genome assemblies along with transcriptome and protein sequences as empirical evidence, and AED value criteria of <0.5 as quality control metric ensured the construction of the high-confidence gene set. Presence of more than 94% (complete and fragmented) BUSCO genes in these high-confidence gene sets further attested to the comprehensiveness of the coding gene sets of both the species. The number of observed high-confidence coding genes was also similar to the other Ficus species (F. hispida and F. microcarpa) (Zhang et al., 2020).
Synteny analysis
Intra-species collinear blocks were identified using MCScanX (Wang et al., 2012), which showed 23.72% collinearity (5,933 out of 25,016 genes) among F. benghalensis coding genes and 18.65% collinearity (4,463 out of 23,929 genes) among F. religiosa coding genes. Furthermore, 48.44% dispersed, 4.37% proximal, 12.93% tandem, 23.72% segmental duplicated genes, and 10.54% singleton genes were detected in F. benghalensis coding gene set. F. religiosa contained 53.21% dispersed, 3.63% proximal, 12.99% tandem, 18.65% segmental duplicated genes, and 11.52% singleton coding genes. Furthermore, 503 inter-species syntenic blocks were identified, which involved 58.33% of the total coding genes predicted in F. benghalensis and F. religiosa genomes, including 14,688 F. benghalensis syntelogs (58.71% of the predicted genes) and 13,861 F. religiosa syntelogs (57.93% of the predicted genes). The observed high percentage of duplicated genes in these genomes could have occurred due to the expansion of gene families that provide long lifespan to plants, as in the case of Oak genome (Plomion et al., 2018). Furthermore, the high percentage of inter-species collinearity between the two Ficus genomes might be a result of Ficus hybridization events caused by frequent host switching of the fig pollinators (Wang et al., 2021).
Demographic history and phylogenetic position of Ficus species
Effective population size (Ne) estimation for the two Ficus species using PSMC (Li and Durbin, 2011) analysis showed that both the species suffered one major population bottleneck in a similar time period—around 0.8 million years ago (Figure 1) in the Pleistocene epoch of the Geological timescale, which is similar to the population demographic history of other tropical forest plants (Bharatraj et al., 2020). The predicted population bottleneck events in both these Ficus species appear to be a consequence of the major glaciation events in Pleistocene (Verbitsky et al., 2018), and the environmental changes (warm and cold conditions prevailing for longer time, lack of rainfall, etc.) caused in the Middle Pleistocene transition (Clark et al., 2006; Dupont et al., 2001). PSMC analysis also showed that these Ficus species could not recover from the long-term decrease in effective population size caused by the bottleneck events.
Figure 1.
Comparative demographic histories of F. benghalensis and F. religiosa
Light green and light pink lines denote to the bootstrap values used in PSMC analysis.
Phylogenetic position of F. benghalensis was resolved in comparison with 50 other Eudicot species spanning a total of 15 plant orders, along with Zea mays as an outgroup species. A total of 469 fuzzy one-to-one orthogroups were constructed across all 52 selected species including F. benghalensis, F. religiosa, and four other Ficus species (F. hispida, F. microcarpa, F. carica, and F. erecta). A maximum likelihood-based phylogenetic tree was constructed (with no polytomy) using the concatenated and filtered protein sequence alignments of these orthogroups containing a total of 525,369 alignment positions (Figure 2).
Figure 2.
Phylogenetic position of the Ficus trees
Species phylogenetic tree of F. benghalensis and F. religiosa with respect to 49 other selected Eudicot species (spanning 15 plant orders), and Zea mays (monocot) as outgroup species. The numbers mentioned in green and red color denote to the number of expanded and contracted gene families, respectively, in the corresponding species. Divergence time between F. benghalensis and F. religiosa species was 55 MYA according to TimeTree database v5.0 (Hedges et al., 2006).
F. benghalensis and F. microcarpa were found to be the closest related species to each other and were placed in the same clade. F. religiosa was the phylogenetically closer species of F. benghalensis and F. microcarpa compared to the other Ficus species used in this study. Species from other genera of the same phylogenetic order Rosales were placed in the adjacent clades to these Ficus species, among which, Cannabis sativa belonging to the plant family Cannabaceae was the closest of Ficus species and shared the most recent common ancestor. Species belonging to Rosales plant order were closer to species from Fabales and Cucurbitales orders in comparison to other selected species, which is also supported by studies based on 18S rDNA, atpB and rbcL gene sequences (Soltis et al., 2000), and other previous reports (Lee et al., 2005).
The phylogenetic positions of the plant orders—Rosales, Fabales, and Cucurbitales—were compared and found to be comparatively closer to the plant orders Brassicales, Malpighiales, Sapindales, and Malvales. The plant orders Apiales, Asterales, Solanales, Gentianales, Lamiales, Ericales, Vitales, and Caryophyllales diverged earlier and among them, the species from the plant order Caryophyllales (Beta vulgaris) was found to diverge the earliest among all the selected Eudicot species, and hence it was the most distant Eudicot species to the two Ficus species. The relative phylogenetic positions of the plant orders analyzed in this study were also supported by previous studies (Mahajan et al., 2021; Soltis et al., 2000).
Expanded/contracted gene families
A total of 68,557 gene families were obtained from the set of coding gene sequences of the six Ficus species and 46 other plant species that were used for phylogenetic tree construction. 31,177 families were retained after the clade and size-based filtering of gene families. In F. benghalensis 1,562 families were found expanded and 2,242 families showed contraction in gene number. In F. religiosa species, 1,265 and 7,358 families were found to be expanded and contracted, respectively. The highly expanded gene families (18 families with ≥10 expanded genes) in F. benghalensis consisted of ABC transporters, cAMP signaling pathway, phenylpropanoid biosynthesis, flavonoid biosynthesis, and other KEGG pathways. Similarly, 17 highly expanded gene families in F. religiosa included KEGG pathways such as starch and sucrose metabolism, plant-pathogen interaction, longevity-regulating pathway, ABC transporter, phenylpropanoid biosynthesis, protein processing in ER, and others. It is to be noted that the expanded gene families in these Ficus species were involved in disease resistance functions in plants.
Among the annotated gene families that showed high contraction (≥10 contracted genes) in F. benghalensis, 28 gene families were involved in tryptophan metabolism, plant hormone signal transduction, and other KEGG pathways, and 31 gene families in F. religiosa were involved in phenylpropanoid biosynthesis, flavonoid biosynthesis, amino sugar and nucleotide sugar metabolism, and other KEGG pathways. It was noted that among the list of highly expanded gene families observed in F. benghalensis, three families showed contraction or no expansion/contraction in F. religiosa species (Table S9), which perhaps hints toward their role in species-specific evolution in the two Ficus species.
Genes with signatures of adaptive evolution
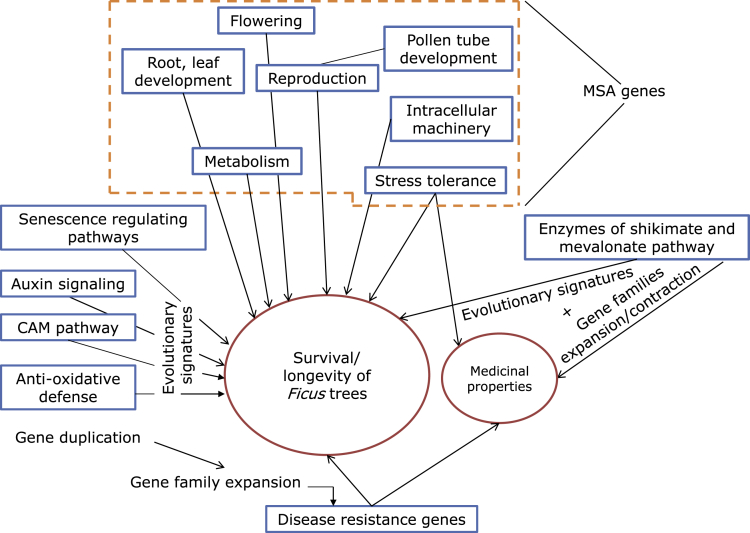
A total of 7,038 orthogroups identified across the 20 species including F. benghalensis, F. religiosa, and four other Ficus species (F. hispida, F. microcarpa, F. carica, and F. erecta) (see STAR Methods) were used for the evolutionary analyses to identify the Ficus species-specific genes showing signatures of adaptive evolution. Among the F. benghalensis species-specific genes, 596 genes had unique amino acid substitutions compared to the other plant species. Of which, substitutions in 415 of these genes were identified to have a functional impact. Also, 85 genes showed positive selection (p values <0.05), and 14 genes showed higher rates of evolution, i.e., higher branch length distance values compared to the other species. Additionally, 205 genes containing positively selected codon sites were found with >95% probability, which is a site-specific evolutionary signature. A total of 17 genes were identified as the genes with multiple signs of adaptive evolution (MSA) (see STAR Methods), and were associated with functions that are essential for long-time survival of plants, e.g., root development, cellular machinery, metabolism, etc. (Data S1 and Figure 3).
Figure 3.
Evolutionary significant biological processes responsible for Ficus longevity
Functional categories of genes involved in providing long lifespan of Ficus trees. See also Tables S21 and S22.
Similarly, in F. religiosa species, a total of 62 genes were positively selected (p values <0.05), and 14 genes showed higher rate of nucleotide divergence. Among the 751 genes that showed unique amino acid substitution, 536 genes that contained substitutions with predicted functional impacts were considered for further analysis. Also, 246 genes contained positively selected codon sites (>95% probability). Thus, a total of 19 genes were identified as MSA genes and were found to be involved in functions essential for providing longevity in plants such as root development, reproduction, metabolism, etc. (Data S2 and Figure 3). The distribution of genes with signatures of adaptive evolution in various KEGG pathways, COG categories, and GO categories is mentioned in Tables S10–S22.
Evolutionary analyses of the two Ficus species provide clues for longevity
Plant growth and development
In F. benghalensis, the MSA genes were mainly involved in root growth, pollen tube and seed development, leaf formation, cell wall synthesis, metabolism, and other developmental processes (Table S23). The MSA genes of F. religiosa were mainly associated with root cell elongation, cell proliferation, seed and pollen tube growth, lateral organ development, controlling flowering time, metabolism, intracellular transport, etc. (Table S24). Functions of these MSA genes in both the Ficus species were obtained from previous experimental studies (Hinckley et al., 2019; Kinoshita et al., 2010; Liu et al., 2003; Madison et al., 2015; Muñoz-Bertomeu et al., 2010; Schiøtt et al., 2004).
Using the protein-protein interaction data of Arabidopsis genes orthologous to Ficus genes present in STRING database, five out of 17 F. benghalensis MSA genes were found to interact with at least one other MSA gene based on protein co-expression evidence. AT5G41040 and GAPDH, GAPDH and ACA9, and AT5G24690 and AT1G80410 showed protein-protein interaction in F. benghalensis. In F. religiosa also, five out of 19 MSA genes showed protein-protein interaction with at least one other MSA gene based on experimental or co-expression evidence present in the STRING database. HAC1 and GAPDH (experimental evidence), GAPDH and TGA9 (co-expression evidence), and AT1G05010 and AT2G39210 (co-expression evidence) showed protein-protein interaction. These genes in both the species were involved in plant organ development, phytohormones signaling, and stress responses in plants (Hinckley et al., 2019; Houben and Van de Poel, 2019; X. Li et al., 2019b; Murmu et al., 2010; Schiøtt et al., 2004).
CAM pathway is an adaptation to photosynthetic machinery that is majorly observed in plants in arid conditions or epiphytes or hemiepiphytes (Shameer et al., 2018; Zotz, 2004), and plants that exhibit both C3 and CAM pathways of photosynthesis show leaf longevity (Lüttge, 2010). Among the six major enzymes (PEPCK, PPDK, ME, MDH, PEPC, and CA) involved in dark and light phases of CAM pathway (Shameer et al., 2018), CA showed unique amino acid substitution with functional impact in F. benghalensis. PEPC and PPDK regulatory protein contained unique substitutions with functional impact, and CA showed both unique substitution and positively selected codon site in F. religiosa.
Auxin is a phytohormone involved in plant root growth and lateral root formation (Fukui and Hayashi, 2018). Among the two major enzymes (TAA1 and YUCCA) responsible for auxin biosynthesis (Fukui and Hayashi, 2018), TAA1 showed unique substitution with functional impact in F. benghalensis. Among the genes important for auxin signaling (Fukui and Hayashi, 2018), GH3, AUX1, and ABCB had unique substitution with functional impact in F. benghalensis (Figure 4A). In F. religiosa species, ARF, GH3, and DAO showed unique substitution with functional impact (Figure 4B).
Figure 4.
Evolutionary signatures in auxin signaling and plant senescence regulatory pathways
(A) Adaptive evolution of auxin biosynthesis and signaling pathway in F. benghalensis.
(B) Adaptive evolution of auxin biosynthesis and signaling pathway in F. religiosa.
(C) Adaptive evolution of genes involved in regulating senescence in F. benghalensis.
(D) Adaptive evolution of genes involved in regulating senescence in F. religiosa.
Plant senescence or aging process is regulated by various phytohormones signaling and has a proposed role in increased lifespan or longevity (Chen et al., 2017; Khan et al., 2014). The genes containing unique substitutions in F. benghalensis include AHK3 that regulates longevity (Bartrina et al., 2017), and AHP, MYC2, RGL1, LOX2, and Invertase (Figure 4C). In case of F. religiosa, EIN2, ARF2, and RGL1 had uniquely substituted amino acid positions, and SAG, WRKY53, and WRKY70 showed positively selected codon site (Figure 4D).
Gene family expansion/contraction analysis revealed that the highly expanded gene families of both the species were involved in disease resistance functions in plants. Notably, one of these families in both the Ficus species was CC-NBS-LRR class gene family, which is one of the best-characterized disease resistance families (R-proteins) in plants (Knepper and Day, 2010). The gene structure of the longest gene identified from this family revealed presence of three exons and two introns in F. benghalensis, two exons and one intron in F. religiosa, and the presence of nucleotide-binding (NB) and leucine-rich repeat (LRR) domains that is the characteristic feature of this gene family (Arora et al., 2019) (Figures 5A and 5B). Gene expression analysis of CC-NBS-LRR gene family showed that five out of 22, and seven out of 23 coding genes present in this family showed TPM (Transcripts Per Million) value > 1 in F. benghalensis and F. religiosa, respectively (Figures 5D and 5E) (see STAR Methods). Also, in F. benghalensis species, one of the considerably expanded gene families was xyloglucan endotransglucosylase/hydrolase, which aids in canker disease resistance in plants (Q. Li et al., 2019a; Shirasawa et al., 2020). The longest gene from this family in F. benghalensis consisted of three introns and four exons (Figure 5C), and six out of 31 coding genes showed TPM value > 1 in this family (Figure 5F). Gene number expansion in the considerably expanded gene families of both the Ficus species occurred due to the various types of gene duplications as obtained from MCScanX analysis.
Figure 5.
Structure and expression of genes from the disease resistance gene families
(A) Gene structure of the longest gene from the expanded CC-NBS-LRR gene family (R-gene family) in F. benghalensis.
(B) Gene structure of the longest gene from the expanded CC-NBS-LRR gene family in F. religiosa.
(C) Gene structure of the longest gene from the expanded xyloglucan endotransglucosylase/hydrolase (canker resistance) gene family in F. benghalensis.
(D) Expression level (TPM values) of the genes included in CC-NBS-LRR gene family in F. benghalensis.
(E) Expression level (TPM values) of the genes included in CC-NBS-LRR gene family in F. religiosa (data not shown for TPM value > 100).
(F) Expression level (TPM values) of the genes included in xyloglucan endotransglucosylase/hydrolase (canker resistance) gene family in F. benghalensis.
Plant stress tolerance
In F. benghalensis species, 15 out of 17 MSA genes were also associated with tolerance against environmental stress responses in plants. The genes were mainly involved in tolerance against drought, oxidative stress, osmotic stress, and other abiotic and biotic stress responses against pathogens (Afzal et al., 2016; Almaghrabi et al., 2019; Ghosh et al., 2022; Singh et al., 2018; Wi et al., 2020) (Table S23). In F. religiosa, 17 out of 19 MSA genes were known associated with stress tolerance activities of plants such as osmotic stress, oxidative stress, other ABA-mediated abiotic stress, and phytohormones signaling-based pathogen defense responses (Kachroo et al., 2003; Li et al., 2019b; Sharma et al., 2018; Yuan et al., 2018) (Table S24).
Plants protect themselves against the toxic reactive oxygen species (ROS)-mediated cellular damage caused by biotic and abiotic stress responses by means of their antioxidative mechanisms (Pandey et al., 2015). In F. religiosa, three enzymes (APX, MDHAR, and DHAR) involved in Ascorbate-Glutathione (AsA-GSH) pathway (a crucial pathway in ROS homeostasis (Pandey et al., 2015)) showed unique substitution with functional impact, and Glutathione S-transferase was positively selected. In F. benghalensis, the MDHAR enzyme showed unique substitution, CAT was positively selected, and Glutathione S-transferase had positively selected codon site. Furthermore, genes involved in glutamate signaling and metabolism that are required for oxidative stress tolerance, senescence, and development in plants showed unique substitution with functional impact in these Ficus species (GLR3.3 in F. benghalensis, and 5-oxoprolinase in F. religiosa) (Qiu et al., 2020).
Pollination in Ficus and floral longevity
Efficient pollination has been related to floral longevity and reproductive ecology of plants (Castro et al., 2008). For pollination in Ficus, shikimate and mevalonate pathways are the two important pathways that produce VOCs (volatile organic compounds) present in floral scents of Ficus trees, which are responsible for fig-wasp (fig pollinator) interactions that help to maintain obligate mutualism between Ficus trees and wasps (Grison-Pigé et al., 2002; Zhang et al., 2020). All seven enzymes involved in shikimate pathway and mevalonate pathway were present in the high-confidence coding gene set of F. religiosa and F. benghalensis. In F. religiosa, AACT (involved in mevalonate pathway) showed positively selected codon site, and mevalonate kinase (involved in mevalonate pathway) showed unique amino acid substitution with functional impact compared to their orthologs in other 15 species. Furthermore, mevalonate pathway is also the precursor to terpenoid backbone biosynthesis. GGPPS that is involved in the downstream terpenoid backbone biosynthesis pathway also showed unique substitution with functional impact. In F. benghalensis, EPSPS (involved in shikimate pathway) and GGPPS (involved in terpenoid biosynthesis pathway) showed unique amino acid substitution with functional impact, and HMGR (involved in mevalonate pathway) showed positively selected codon site.
Enzymes involved in mevalonate and shikimate pathway were also analyzed for gene family expansion/contraction. One gene family of mevalonate pathway (AACT) was contracted in F. benghalensis, whereas two families (MK and AACT) were contracted in F. religiosa. For the gene families containing genes involved in shikimate pathway, one expanded (shikimate kinase) gene family was found in F. benghalensis, whereas one contracted family (DAHP synthase) was found in F. religiosa. Gene expansion in shikimate kinase gene family of F. benghalensis was accounted by gene duplication events as found in the gene duplication analysis in this study.
Evolutionary analyses excluding the other Ficus species showed a greater number of genes with adaptive evolution
Analyses of genes showing evolutionary signatures excluding the four other Ficus species, i.e., across 16 Eudicot species (see STAR Methods) showed presence of 45 and 55 MSA genes in F. benghalensis and F. religiosa, respectively. In F. benghalensis, the MSA genes were mainly involved in root gravitropism response, root thigmomorphogenesis, root elongation, pollen tube development, auxin transport, reproductive growth, flower development, intracellular protein transport, stress tolerance, and other developmental processes. The MSA genes of F. religiosa were mainly associated with root cell elongation, leaf development, cell proliferation, phytohormones signaling, reproductive growth, controlling flowering time, formation of cellular components, stress tolerance, etc. (Tables S25 and S26).
Discussion
In this study, we have performed the first whole genome sequencing of the two dicot Ficus tree species—F. benghalensis and F. religiosa, belonging to Moraceae family, and successfully constructed their draft genomes using a hybrid sequencing and assembly approach. The comprehensive genome-wide phylogeny constructed here included both the Ficus species, and consisted of a total of 52 species covering 15 phylogenetic plant orders, which will serve as a valuable reference for future phylogenetic studies. In the comparative evolutionary analyses, the inclusion of closely related plant species (according to the above phylogenetic tree) helped in precise identification of genes with evolutionary signatures in F. benghalensis and F. religiosa species. Also, the inclusion of other plant species with long lifespan, including Actinidia chinensis, Coffea canephora, Olea europaea, Pistacia vera, Populus trichocarpa, and Vitis vinifera, in the comparative analysis helped in the identification of adaptively evolved genes, which could have played a significant role in longevity of these two Ficus species. Furthermore, the inclusion of coding genes from other Ficus species, both short-lived and long-lived, helped to identify plausible species-specific Ficus genes responsible for the morphological characteristics and evolutionary adaptation in these two plant species. Evolutionary analyses including the other Ficus species (F. hispida, F. microcarpa, F. carica, and F. erecta) showed a lesser number of genes having adaptive evolution compared to the analyses excluding the other Ficus species. This eliminates the possibility of finding genes that are also under selection in the short-lived Ficus species, and thus adds to the significance of these results.
Shikimate and mevalonate pathways are responsible for efficient pollination event that might influence floral longevity as well as reproductive success of the Ficus species, as in the case of Ficus altissima (Castro et al., 2008; Peng et al., 2010). The presence of evolutionary signatures in the enzymes as well as gene family expansion/contraction suggested adaptive evolution of these pathways in these Ficus species. Evolution of shikimate pathway, which is also responsible for the production of pollinator attracting VOCs (Zhang et al., 2020), suggested presence of fig-wasp obligate mutualism in these plant species. In addition, the adaptively evolved genes related to flowering also might influence Ficus pollination as well as fig-wasp mutualism and co-evolution in both these species. Similarly, the evolution of mevalonate pathway in F. religiosa that regulates plant developmental processes by aiding metabolism, phytohormones biosynthesis, isoprenoid biosynthesis, etc. (Kasahara et al., 2002; Venkateshwaran et al., 2015), indicates the adaptive evolution of plant growth and development in this species. In addition, the adaptive evolution of shikimate and mevalonate pathways, which are also the precursors for production of phenolic and terpenoid classes of secondary metabolites (Francenia Santos-Sánchez et al., 2019; Wang et al., 2019), explains the presence of medicinal properties in these plants. To support this, evolutionary signature was also observed in another enzyme involved in terpenoid backbone biosynthesis pathway in both the species. These evolutionary signatures suggest the evolutionary advantage of both these species since many of these terpenoids have a potent defensive element against pests and pathogens that is a genetically controlled trait common among long-lived species (Kitajima et al., 2018; Piovesan and Biondi, 2021; Villard et al., 2019).
Plant longevity is influenced by several factors such as phytohormones signaling, senescence regulating pathways, reproductive growth, a wide range of stress tolerance genes, reactive oxygen species, gene family expansion and gene duplication of disease-resistant genes, etc. (Bartrina et al., 2017; Plomion et al., 2018; Wang et al., 2020). One of the key findings of this study is the identification of signatures of adaptive evolution in genes that are associated with providing longevity in both the species. Particularly, the genes related to sustained growth and development—plant root development, flowering, reproductive growth, and metabolism—showed multiple signs of adaptation. The study also indicated the presence of CAM pathway of photosynthesis in F. benghalensis and F. religiosa, which is a characteristic of epiphyte or hemiepiphyte plants to conserve water storage, when they survive as epiphytes (Martin et al., 2005). Previous studies have shown that plants that use both C3 and CAM pathways of photosynthesis exhibit long-lived leaves, thus the evolutionary signatures of CAM pathway in F. benghalensis and F. religiosa perhaps also contribute to leaf longevity (Lüttge, 2010).
Notably, the genes involved in auxin signaling and plant senescence also showed signatures of adaptive evolution. The phytohormone auxin plays a major role in plant root development that aids in the growth and development of plants (Saini et al., 2013; Zhang et al., 2020). The signatures of adaptive evolution in genes involved in auxin synthesis and signaling mechanisms could explain the presence of well-developed aerial roots in F. benghalensis and provide support for the growth and survival of large Ficus trees against environmental challenges. Auxin-responsive gene families, e.g., ARF and GH3 are also known to be involved in plant disease resistance mechanisms that confer longevity in plants (Ghanashyam and Jain, 2009). Furthermore, the presence of evolutionary signatures in genes of both the species involved in phytohormone signaling pathways that function to regulate plant developmental senescence and aging processes also provides clues for the long lifespan of these Ficus species.
In addition, the expansion of selected plant disease resistance gene families through gene duplication events in the course of evolution also confers longevity in these species (Plomion et al., 2018). Furthermore, we also noted high expression levels of genes included in these distinct disease resistance gene families (Figures 5D–5F). Thus, it is tempting to speculate the collective role of gene duplication and subsequent expansion of plant disease resistance gene families, along with gene expression regulation in achieving longer lifespan (Plomion et al., 2018).
Notably, 88% and 89% of the MSA genes were also associated with tolerance against biotic and abiotic stress responses in F. benghalensis and F. religiosa, respectively, which further assist in plant survival against environmental challenges. The presence of diverse defense or stress tolerance-related genes is also an important factor in providing long-time survival of plants (Tobias and Guest, 2014). Stress response mechanisms such as JA and SA-signaling pathways also regulate secondary metabolism (Isah, 2019), which further associates with the medicinal properties of these trees (Chakraborty et al., 2021).
Thus, it is apparent from the comparative analyses that the two Ficus species show a high level of similarity in genomic characteristics and follow similar evolutionary adaptations of functional pathways that aid in their growth and survival strategies to achieve longevity. It can be speculated that to survive in diverse and challenging environmental conditions, the genes that help to sustain plant growth and development through processes such as root cell growth and proliferation, phytohormones signaling, CAM pathway, plant senescence regulating pathways, plant metabolism, cell wall and plasma membrane component biosynthesis, intracellular machinery, antioxidative defense, etc. have evolved to provide a longer lifespan in plant species such as F. benghalensis and F. religiosa, and make them the keystone plants in tropical and sub-tropical ecosystems (Harrison, 2005; Shanahan et al., 2001). The presence of adaptively evolved genes of both Ficus species (Tables S11–S13) and highly expanded gene families of F. religiosa in longevity-regulating pathways (KEGG pathway) also supports the long lifespan of these two Ficus species. In addition, identification of adaptive evolutionary signatures in the plant senescence regulating pathways, and expansion and high expression of the disease resistance gene families might also have a key role in the observed long lifespan of these Ficus species. Taken together, the availability of whole genome sequences of these two Ficus species and evolutionary insights into the mechanisms related to longevity and medicinal properties in these species will serve as a valuable resource for further genomic, ecological, and evolutionary studies on Ficus genus.
Limitations of the study
Ficus genus comprises more than 800 species and the availability of genomic data from more Ficus species will help in improving the genomic assemblies and comparative analysis. Furthermore, the experimental validation of the role of adaptively evolved genes in longevity will help in supporting our hypothesis that longevity is a multifaceted phenomenon.
STAR★Methods
Key resources table
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the Lead Contact, Vineet K. Sharma (vineetks@iiserb.ac.in).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
This study does not include experimental model or subjects.
Method details
DNA-RNA extraction and sequencing
The leaves were collected from a single F. benghalensis tree and a single F. religiosa tree located in Bhopal, India (23.2280252°N 77.2088987°E). The leaves were cleaned and processed for DNA and RNA extraction. The DNA extraction was carried out using Carlson lysis buffer by following Oxford Nanopore suggested protocol using Genomic tip 20 of Blood and cell culture kit (Qiagen, CA, USA) with few modifications. The quality check and quantification of DNA were performed on Nanodrop 8000 spectrophotometer (Thermo Fisher Scientific, USA) and Qubit 2.0 fluorometer (Invitrogen, Thermo Fisher Scientific, USA), respectively. The species were identified as F. benghalensis and F. religiosa by amplifying, sequencing and performing Nucleotide BLAST (Basic local Alignment Search Tool) of two marker genes i.e., Internal Transcribed Spacer (ITS) and Maturase K (MatK). The genomic library for linked reads was prepared on Chromium Controller instrument (10x Genomics, CA) and sequenced on Illumina Novaseq 6000 platform to produce 150 bp paired-end reads. The long read genomic library was prepared using SQK-LSK-109 kit followed by sequencing on MinION Mk1B instrument (Oxford Nanopore Technologies, UK). The total RNA was extracted using TRIzol reagent (Invitrogen, Thermofisher Scientific, USA). The transcriptomic library was prepared using Illumina TruSeq Stranded Total RNA library preparation kit with Ribo-Zero workflow and 150 bp paired-end reads were generated on Illumina Novaseq 6000 instrument (Illumina Inc., CA, USA).
Genome assembly
Genome sizes of the two Ficus species were predicted using 10x Genomics linked reads. Barcode sequences were extracted from the raw reads using available proc10xG set of python scripts (https://github.com/ucdavis-bioinformatics/proc10xG), and these pre-processed linked reads were used to estimate the genome size. Genomic heterozygosity was also estimated for the two Ficus species in order to assess the complexity of the genomes. Jellyfish v2.2.10 (Marçais and Kingsford, 2011) was used to generate k-mer count histograms using the barcode-filtered paired-end reads, and GenomeScope v2.0 (Ranallo-Benavidez et al., 2020) was used to estimate the genome sizes, and to calculate the heterozygosity profiles.
The de novo genome assembly of both the Ficus species was performed following the same approach. Oxford Nanopore raw data was basecalled using Guppy v3.2.1 (Oxford Nanopore Technologies) and adapter removal of the raw reads was performed using Porechop v0.2.3 (Oxford Nanopore Technologies). Genome assembly using these pre-processed reads was performed using wtdbg v2.0.0 (Ruan and Li, 2020) with default settings, SMARTdenovo (https://github.com/ruanjue/smartdenovo) with minimum read length of 0, Canu v2.0 (Koren et al., 2017) with default settings (after reads correction using Canu v2.0) and Flye v2.4.2 (Kolmogorov et al., 2019) with default settings. Quality statistics of all these assemblies were calculated using Quast v5.0.2 (Gurevich et al., 2013), and based on the assembled genome size and assembly statistics, the assembly derived from Flye v2.4.2 was selected for further analysis. 10x Genomics linked reads, which were processed using Longranger basic v2.2.0 (https://support.10xgenomics.com/genome-exome/software/pipelines/latest/installation), were used to polish this assembly three times using Pilon v1.23 (Walker et al., 2014). RNA-Seq data was quality-filtered using Trimmomatic v0.38 (Bolger et al., 2014) with the parameters - “LEADING:20 TRAILING:20 SLIDINGWINDOW:4:20 MINLEN:60”, and the filtered paired-end data was used for scaffolding using AGOUTI v0.3.3 (Zhang et al., 2016) to improve the assembly contiguity. The barcode-processed linked reads and pre-processed Nanopore long reads (Canu v2.0-corrected reads) were also used for further scaffolding of this assembly using ARCS v1.1.1 (Yeo et al., 2018) and LINKS v1.8.6 (Warren et al., 2015) with default settings. Contigs with length of ≥1,000 bp (after scaffolding) were extracted (assembly v0.1).
The complete 10x Genomics data of 327.41 million barcoded linked reads (116.67x raw coverage) and 324.53 million reads (125.43X raw coverage) were used for de novo assembly of F. benghalensis and F. religiosa genome, respectively, using Supernova v2.1.1 (Weisenfeld et al., 2017). The mis-assembly regions were corrected using Tigmint v1.2.1 (Jackman et al., 2018) with the barcode-processed linked reads resulted from Longranger basic v2.2.0. Quality-filtered RNA-Seq paired-end reads were used to improve the contiguity of this assembly using AGOUTI v0.3.3 (Zhang et al., 2016). Barcode-processed linked reads and error-corrected Nanopore long reads were used for further scaffolding using ARCS v1.1.1 (Yeo et al., 2018) (default settings) and LINKS v1.8.6 (Warren et al., 2015), respectively. Contigs with length of ≥1,000 bp (after scaffolding) were retained (assembly v0.2).
For each of the two species, assembly v0.1 and assembly v0.2 were merged using Quickmerge v0.3 (Chakraborty et al., 2016), which joins the gaps between the sequences in one assembly by using sequences from the other assembly, and thus improve the assembly contiguity (assembly v1). Further, scaffolds from assembly v0.2 were searched against the scaffolds from assembly v1 using BLASTN, and the unique sequences that did not match even with stringent parameters (sequence identity 10%, query coverage 10%, e-value 10−9) were directly added to assembly v1 (Jaiswal et al., 2021; Schmidt et al., 2020). In this process, 910 and 1,976 unique contigs with comparatively shorter lengths (mean sequence length = 39.1 kb and 11.6 kb) were added to the assemblies v1 of F. benghalensis and F. religiosa genomes, respectively. Gap-closing of these assemblies (assembly v2) was performed in three iterations using LR_Gapcloser (Xu et al., 2018) with error-corrected Oxford Nanopore long reads. Finally, barcode-processed 10x Genomics linked reads were used to fix any further local mis-assemblies, small indels, and base errors using Pilon v1.23 (Walker et al., 2014) to generate the final F. benghalensis and F. religiosa genome assemblies (assembly v3). Completeness of these final genome assemblies was checked using embryophyta_odb10 single-copy orthologs dataset from BUSCO v5.2.1 (Simão et al., 2015). Also, to check the comprehensiveness of the final genome assembly, barcode-filtered 10x Genomics linked reads and adapter-processed Nanopore long reads were mapped to the final genome assemblies using BWA-MEM v0.7.17 (Li, 2013), and the mapping percentage was calculated using SAMtools v1.9 “flagstat” (Li et al., 2009). The read alignment results were further analyzed using Qualimap v2.2.2 (Okonechnikov et al., 2016) to calculate the depth distribution for all base positions in the genomes. The detailed information related to genome assembly and processing steps are mentioned in Figure S4.
Sequence variation in the final Ficus genome assemblies was analyzed by mapping the barcode-filtered 10x Genomics linked reads to their respective whole genome assemblies using BWA-MEM v0.7.17 (Li, 2013), and SAMtools v1.9 (Li et al., 2009). Variant calling was performed using BCFtools v1.9 “mpileup” (Li, 2011) with the quality-filtration criteria - sequencing depth ≥30, quality of variant sites ≥30, and mapping quality ≥50.
Genome annotation and gene set construction
Genome annotation for F. benghalensis and F. religiosa species was performed using their corresponding final genome assemblies generated in the previous step. For repeat annotation, de novo repeat libraries were constructed for these genomes using RepeatModeler v2.0.1 (Flynn et al., 2020), which were further clustered to remove redundant sequences using CD-HIT-EST v4.8.1 (Li and Godzik, 2006) with sequence identity of 90%, and seed size of 8 bp. These resultant repeat libraries were used to soft-mask the two Ficus genomes using RepeatMasker v4.1.0 (http://www.repeatmasker.org). Additionally, simple repeat sequences were also predicted in the final genome assemblies using Tandem Repeats Finder (TRF) v4.09 (Benson, 1999).
The repeat-masked genomes of F. benghalensis and F. religiosa were used for gene set construction. MAKER v2.31.10 (Cantarel et al., 2008), the genome annotation pipeline was used to construct the coding gene sets using ab initio and evidence alignment-based approaches. Quality-filtered RNA-Seq reads (using Trimmomatic v0.38) of F. benghalensis from this study, and quality-filtered transcriptome reads of F. religiosa from this study and previous report (Matasci et al., 2014) were used to construct de novo transcriptome assemblies using Trinity v2.9.1 (Haas et al., 2013) with default parameters. The resultant transcriptome assemblies, and the protein sequences of phylogenetically closer species - Cannabis sativa, Malus domestica, Prunus avium, and Rosa chinensis that belong to the same plant order Rosales (obtained from Ensembl Plants release 48) were used as EST and protein evidence for gene set construction of the two respective Ficus species. For ab initio gene finding, AUGUSTUS v3.2.3 (Stanke et al., 2006) was used, and for empirical evidence alignments, BLAST (Altschul et al., 1990) and Exonerate v2.2.0 (https://github.com/nathanweeks/exonerate) were used. The gene models were filtered based on their Annotation Edit Distance (AED) values and length-based criteria. Only the genes with AED values < 0.5, and length (≥300 bp) were selected for the final gene set construction of F. benghalensis and F. religiosa, and termed high-confidence gene sets (Zhang et al., 2019). Completeness of these gene sets was assessed using BUSCO v5.2.1 (Simão et al., 2015) with embryophyta_odb10 dataset of single-copy orthologous genes.
Further, de novo prediction of tRNA and rRNA genes was performed using tRNAscan-SE v2.0.7 (Chan et al., 2021), and Barrnap v0.9 (https://github.com/tseemann/barrnap). Homology-based identification of miRNA gene sequences was performed using BLASTN (sequence identity 80%, e-value 10−3) against miRBase database (Griffiths-Jones et al., 2008).
PSMC analysis
Demographic history was constructed for F. benghalensis and F. religiosa species for comparative study using Pair-wise Sequentially Markovian Coalescent (PSMC) (Li and Durbin, 2011) with the whole genome sequences of F. benghalensis and F. religiosa constructed in this study. The barcode-filtered 10x Genomics linked reads of the two species, which were mapped to their respective whole genome assemblies using BWA-MEM (Li, 2013), were used to construct the consensus diploid sequences using SAMtools v1.9 “mpileup” (Li et al., 2009) and BCFtools v1.9 (Li, 2011) in a manner that the minimum read depth is a third of the average read depth, and the maximum read depth is set to the twice of the average read depth. The consensus sequences were filtered for sites with quality score <20. PSMC analysis was performed using the parameters “N30 -t5 -r5 -p4+25∗2 + 4+6” with 100 bootstrap values. Generation time of 7 years, and a mutation rate per site per generation of 1.75 × 10−8 was considered for this analysis, which was used for another Ficus species F. carica in a previous study (Bharatraj et al., 2020).
Phylogenetic analysis
Protein sequences of all 45 Eudicot species available on Ensembl (Bolser et al., 2016) plants release 48 were used to resolve the phylogenetic position of F. benghalensis and F. religiosa. The selected Eudicot species were – A. chinensis, Arabidopsis halleri, Arabidopsis lyrata, Arabidopsis thaliana, Arabis alpina, B. vulgaris, Brassica napus, Brassica oleracea, Brassica rapa, Camelina sativa, C. sativa female, Capsicum annuum, Citrullus lanatus, Citrus clementina, C. canephora, Corchorus capsularis, Cucumis melo, Cucumis sativus, Cynara cardunculus, Daucus carota, Glycine max, Gossypium raimondii, Helianthus annuus, Ipomoea triloba, Lupinus angustifolius, M. domestica Golden, Manihot esculenta, Medicago truncatula, Nicotiana attenuata, O. europaea var. sylvestris, Phaseolus vulgaris, P. vera, P. trichocarpa, P. avium, Prunus persica, Prunus dulcis, R. chinensis, Solanum lycopersicum, Solanum tuberosum, Theobroma cacao Belizian Criollo B97-61/B2, T. cacao Matina 1–6, Trifolium pratense, Vigna angularis, Vigna radiata, and V. vinifera. Along with these species, data from other available Ficus species - F. hispida (Zhang et al., 2020), F. microcarpa (Zhang et al., 2020), F. carica (Mori et al., 2017), and F. erecta (Shirasawa et al., 2020) were also used for phylogenetic analysis. Z. mays, which is a monocot plant, was used as an outgroup species, and protein sequences of Z. mays were also obtained from Ensembl plants release 48.
Proteome files of all species were used to extract the longest isoform for each protein. The resultant protein sequences were used to construct the orthologous gene sets using OrthoFinder v2.4.1 (Emms and Kelly, 2019). From these orthogroups, the fuzzy one-to-one orthogroups that contained proteins from all 52 species were extracted using KinFin v1.0 (Laetsch and Blaxter, 2017). For further analysis, these fuzzy one-to-one orthologous gene sets were processed to extract and consider only the longest protein sequence for each species. Each of these orthogroups was individually aligned using MAFFT v7.467 (Katoh and Standley, 2013). These multiple sequence alignments were filtered to remove the empty sites and were concatenated using BeforePhylo v0.9.0 (https://github.com/qiyunzhu/BeforePhylo). RAxML v8.2.12 (Stamatakis, 2014), which uses rapid hill climbing algorithm, was utilized to construct the maximum likelihood-based species phylogenetic tree with the concatenated protein sequence alignment. ‘PROTGAMMAAUTO’ amino acid substitution model, and a bootstrap value of 100 were used for phylogenetic tree construction.
Evolution of gene families
CAFÉ v4.2.1 (De Bie et al., 2006) was used to estimate the evolution of gene families across the above selected 52 species (Z. mays as an outgroup species) using the proteome files containing the longest isoform for each protein, and the species phylogenetic tree generated in the previous step. Prior to the analysis, the species phylogenetic tree (with Z. mays as an outgroup species) was converted to ultrametric tree based on the calibration point of 120 million years between F. benghalensis and B. vulgaris obtained from TimeTree database v5.0 (Hedges et al., 2006). All-versus-All BLASTP search for the protein sequences of all 52 species was performed, which was followed by clustering using MCL v14.137 (Van Dongen and Abreu-Goodger, 2012), and removal of gene families that contained genes in <2 species of the specified clades and ≥100 gene copies for at least one species. These filtered gene families, and the ultrametric species tree were utilized to analyze the expansion or contraction of gene families using two-lambda (λ) model, where λ is a random birth-death parameter. In this analysis, the species from Rosales order were given the same λ-value, while the other species were given another λ-value. The considerably expanded (with ≥10 expanded genes) and contracted (with ≥10 contracted genes) gene families for F. benghalensis and F. religiosa were extracted and compared.
Further, gene family expansion/contraction for the enzymes involved in mevalonate and shikimate pathways that play important role in Ficus species (Zhang et al., 2020) was analyzed using CAFÉ results. EC numbers of these enzymes were used to extract their sequence (specific to A. thaliana, a model Eudicot plant) from Swiss-Prot database (Bairoch and Apweiler, 2000), and the sequences were queried against the proteome of F. benghalensis and F. religiosa using BLASTP (e-value of 10−5) to identify the enzymes in these two Ficus species. Gene families of these enzymes were identified from CAFÉ results, and the expansion/contraction of these families was identified in two Ficus species compared to their immediate ancestor phylogenetic node.
Synteny analysis
Collinear blocks between F. benghalensis and F. religiosa species (inter-species), and intra-species syntenic blocks for these two species were identified using the All-versus-All BLASTP (e-value of 10−5) homology alignments of the respective protein sequences and the GFF annotations. Inter-species and intra-species synteny analyses were performed using MCScanX (Wang et al., 2012), ten genes were required to detect a collinear block (Bi et al., 2019). Also, the duplicated genes in each of the Ficus species were detected and classified into various gene duplication categories (proximal, tandem, segmental, or dispersed) using the same parameter.
Identification of evolutionary signatures
For identifying the genes with evolutionary signatures, all six Ficus species with available genomes from Rosales plant order, and only the Eudicot species from other phylogenetically closer plant orders of these Ficus species (one species from each order) were considered to avoid erroneous results and to analyze adaptive evolution of these two Ficus species across dicot plant orders. The selected 14 Eudicot species other than the six Ficus species were - A. thaliana (order Brassicales), A. chinensis (order Ericales), C. canephora (order Gentianales), C. melo (order Cucurbitales), D. carota (order Apiales), G. max (order Fabales), G. raimondii (order Malvales), H. annuus (order Asterales), O. europaea var. sylvestris (order Lamiales), P. vera (order Sapindales), P. trichocarpa (order Malpighiales), R. chinensis (order Rosales), S. tuberosum (order Solanales), and V. vinifera (order Vitales). Proteome files of these 20 species were used to construct the orthologous gene sets using OrthoFinder v2.4.1 (Emms and Kelly, 2019). Orthogroups containing protein sequences from all 20 species were identified, and only the longest protein sequence for each species was extracted from these orthogroups.
Genes with unique amino acid substitution
Each of the orthogroups across 20 species constructed in the previous step was aligned using MAFFT v7.467 (Katoh and Standley, 2013). In the multiple sequence alignments, F. benghalensis genes showing amino acid positions that were identical in other species but different in this species were considered as genes with unique amino acid substitutions in F. benghalensis. Similarly, F. religiosa genes that showed different amino acids in the sequence alignments compared to the other species were also considered as the F. religiosa genes with unique substitutions. Any gap in these alignments and ten sites around the gap were not considered in this analysis. The genes showing unique amino acid substitutions were extracted, and impact of these unique amino acid substitutions on the proteins function was predicted using Sorting Intolerant From Tolerant (SIFT) (Ng and Henikoff, 2003) with UniProt reference database (Bateman et al., 2015).
Genes with higher nucleotide divergence
The protein sequence alignments of the orthogroups across 20 species generated in the previous step were used to construct individual maximum likelihood-based phylogenetic tree for each orthogroup using RAxML v8.2.12 (Stamatakis, 2014) with ‘PROTGAMMAAUTO’ amino acid substitution model and 100 bootstrap values. Root-to-tip branch length distances were estimated for genes from all selected species in these phylogenetic trees using “adephylo” (Jombart and Dray, 2010) package in R. F. benghalensis and F. religiosa specific genes with relatively higher branch length distance values compared to the other species were extracted separately as the genes with higher rate of evolution.
Positively selected genes
The nucleotide sequences of all the orthologous gene sets constructed across 20 species were individually aligned using MAFFT v7.467 (Katoh and Standley, 2013). Further, RAxML v8.2.12 (Stamatakis, 2014) was used to construct the species phylogenetic tree for these selected 20 species in a similar way as mentioned in the earlier steps (see Phylogenetic analysis step). This species phylogenetic tree and the nucleotide alignments for the orthogroups in PHYLIP format were used to separately detect the F. benghalensis and F. religiosa genes with positive selection using a branch-site model utilized by “codeml” program available in PAML v4.9a (Yang, 2007). Likelihood-ratio tests were performed from the obtained log likelihood values, and genes qualifying against the null model having FDR-corrected p values of <0.05 (obtained from chi-square analysis) were termed positively selected genes. Additionally, Bayes Empirical Bayes (BEB) analysis was carried out to identify the codon sites of F. benghalensis and F. religiosa genes that showed positive selection with >95% probability for the foreground lineage.
The genes that showed more than one of these above mentioned evolutionary signatures, namely - positive selection, higher nucleotide divergence rate, and unique substitution with functional impact, were extracted and considered as the genes with multiple signatures of adaptive evolution (MSA genes) (Jaiswal et al., 2018; Mittal et al., 2019).
The analysis to identify the MSA genes was also performed excluding the other four Ficus species (F. hispida, F. microcarpa, F. carica and F. erecta), across 16 species, to compare the MSA genes with those obtained including other short-lived and long-lived Ficus species.
Functional annotation
The complete high-confidence gene sets of F. benghalensis and F. religiosa were annotated against NCBI Non-Redundant (nr) database using BLASTP (e-value of 10−5), Swiss-Prot (Bairoch and Apweiler, 2000) database using BLASTP (e-value of 10−5), and Pfam-A v32.0 (Bateman, 2004) database using HMMER v3.3 (Finn et al., 2011) (e-value of 10−5). The species-specific gene sets with evolutionary signatures were assigned to KEGG Orthology (KO) categories and KEGG pathways using KAAS v2.1 genome annotation server (Moriya et al., 2007). These genes were also annotated to different COG categories using eggNOG-mapper v2 (Huerta-Cepas et al., 2017). Gene Ontology (GO) categories were also assigned to these genes using the over-representation analysis in WebGeStalt web server (Liao et al., 2019). Further, functional annotations of these gene sets were screened, and curated manually. The MSA genes were also checked for protein-protein interaction data in STRING v11.0 database (Szklarczyk et al., 2017) using information from previously reported studies, and the interacting genes were analyzed. Enzymes involved in characteristic functional pathways were searched for their presence in the gene sets showing evolutionary signatures, and in the expanded/contracted gene families.
Disease-resistance gene families
Gene structures for the notable disease-resistance gene families in the Ficus species (identified from CAFÉ analysis) were constructed by mapping the longest coding gene of the corresponding gene families on the final genome assemblies of the two Ficus species using Exonerate v2.2.0 (https://github.com/nathanweeks/exonerate) with 90% match and 90% quality threshold. Protein domain searching in the corresponding amino acid sequences was performed using Motif Scan (https://myhits.sib.swiss/cgi-bin/motif_scan). Gene expression level (in terms of TPM value) of all the identified coding genes present in these respective gene families was estimated by mapping the quality-filtered paired-end RNA-Seq reads generated in this study on the coding gene sets of both the Ficus species using Kallisto v0.46.0 (Bray et al., 2016).
Quantification and statistical analysis
Statistical tests in the positive selection analysis were performed using PAML v4.9a (Yang, 2007). p values mentioned in the Gene Ontology over-representation analysis were calculated using WebGeStalt web server (Liao et al., 2019). Gene expression values (in terms of TPM values) of the Ficus coding genes were quantified using Kallisto v0.46.0 (Bray et al., 2016).
Additional resources
This study does not report additional resources.
Acknowledgments
A.C., S.M., and M.S.B. thank Council of Scientific and Industrial Research (CSIR) for fellowship. The authors also thank the NGS facility at IISER Bhopal and the intramural research funds provided by IISER Bhopal.
Author contributions
V.K.S. conceived and coordinated the project. S.M. performed DNA-RNA extraction, prepared samples for sequencing, performed Nanopore sequencing, and species identification assays. A.C. and V.K.S. designed computational framework of the study. A.C. performed all the computational analyses presented in the study, functional annotation of gene sets, and constructed all the figures. A.C., M.S.B., and V.K.S. analyzed the data and interpreted the results. A.C., S.M., and V.K.S. wrote the manuscript. All the authors have read and approved the final version of the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: October 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.105100.
Supplemental information
Data and code availability
-
•
The raw reads obtained from genome and transcriptome sequencing of both F. benghalensis (BioProject accession – PRJNA759132, BioSample accession – SAMN21156846, SRA accessions – SRR15676288, SRR15676289, SRR15676290), and F. religiosa (BioProject accession – PRJNA759116, BioSample accession – SAMN21156584, SRA accessions – SRR15676280, SRR15676281, SRR15676282) species have been deposited in NCBI SRA database.
-
•
This study does not report original code.
-
•
We have provided all detailed information to reanalyze this study in STAR Methods to the best of our knowledge. Any additional information is available from the lead contact upon request.
References
- Afzal Z., Howton T.C., Sun Y., Mukhtar M.S. The roles of aquaporins in plant stress responses. J. Dev. Biol. 2016;4:E9. doi: 10.3390/jdb4010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almaghrabi B., Ali M.A., Zahoor A., Shah K.H., Bohlmann H. Arabidopsis thionin-like genes are involved in resistance against the beet-cyst nematode (Heterodera schachtii) Plant Physiol. Biochem. 2019;140:55–67. doi: 10.1016/j.plaphy.2019.05.005. [DOI] [PubMed] [Google Scholar]
- Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arora S., Steuernagel B., Gaurav K., Chandramohan S., Long Y., Matny O., Johnson R., Enk J., Periyannan S., Singh N., et al. Resistance gene cloning from a wild crop relative by sequence capture and association genetics. Nat. Biotechnol. 2019;37:139–143. doi: 10.1038/s41587-018-0007-9. [DOI] [PubMed] [Google Scholar]
- Bairoch A., Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartrina I., Jensen H., Novák O., Strnad M., Werner T., Schmülling T. Gain-of-Function mutants of the cytokinin receptors AHK2 and AHK3 regulate plant organ size, flowering time and plant longevity. Plant Physiol. 2017;173:1783–1797. doi: 10.1104/PP.16.01903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Coin L., Durbin R., Finn R.D., Hollich V., Griffiths-Jones S., Khanna A., Marshall M., Moxon S., Sonnhammer E.L.L., et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., UniProt Consortium. Martin M.J., O’Donovan C., Magrane M., Apweiler R., Alpi E., Antunes R., Arganiska J., Bely B., et al. UniProt: a hub for protein information. Nucleic Acids Res. 2015;43:D204–D212. doi: 10.1093/nar/gku989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharatraj A., Sharad S., Raghavendra S., Satish B.N., Kushalappa G., Vijay N. The genome sequence of Mesua ferrea and comparative demographic histories of forest trees. Gene. 2020;769:145214. doi: 10.1016/j.gene.2020.145214. [DOI] [PubMed] [Google Scholar]
- Bi Q., Zhao Y., Du W., Lu Y., Gui L., Zheng Z., Yu H., Cui Y., Liu Z., Cui T., et al. Pseudomolecule-level assembly of the Chinese oil tree yellowhorn (Xanthoceras sorbifolium) genome. GigaScience. 2019;8:giz070. doi: 10.1093/gigascience/giz070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger A.M., Lohse M., Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolser D., Staines D.M., Pritchard E., Kersey P. Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomics data. Methods Mol. Biol. 2016 doi: 10.1007/978-1-4939-3167-5_6. [DOI] [PubMed] [Google Scholar]
- Bray N.L., Pimentel H., Melsted P., Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- Campbell M.S., Holt C., Moore B., Yandell M. Genome annotation and curation using MAKER and MAKER-P. Curr. Protoc. Bioinformatics. 2014;48:4.11.1–4.11.39. doi: 10.1002/0471250953.bi0411s48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B.L., Korf I., Robb S.M.C., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M. MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 2008;18:188–196. doi: 10.1101/gr.6743907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro S., Silveira P., Navarro L. Effect of pollination on floral longevity and costs of delaying fertilization in the out-crossing Polygala vayredae Costa (Polygalaceae) Ann. Bot. 2008;102:1043–1048. doi: 10.1093/aob/mcn184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty A., Mahajan S., Jaiswal S.K., Sharma V.K. Genome sequencing of turmeric provides evolutionary insights into its medicinal properties. Commun. Biol. 2021;4:1193. doi: 10.1038/s42003-021-02720-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty M., Baldwin-Brown J.G., Long A.D., Emerson J.J. Contiguous and accurate de novo assembly of metazoan genomes with modest long read coverage. Nucleic Acids Res. 2016;44:e147. doi: 10.1093/nar/gkw654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan P.P., Lin B.Y., Mak A.J., Lowe T.M. TRNAscan-SE 2.0: improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021;49:9077–9096. doi: 10.1093/nar/gkab688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhary A., Yadav B.S., Singh S., Maurya P.K., Mishra A., Srivastva S., Varadwaj P.K., Singh N.K., Mani A. Docking-based screening of Ficus religiosa phytochemicals as inhibitors of human histamine H2 receptor. Pharmacogn. Mag. 2017;13:S706–S714. doi: 10.4103/pm.pm_49_17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Xiang S., Chen Y., Li D., Yu D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol. Plant. 2017;10:1174–1189. doi: 10.1016/j.molp.2017.07.008. [DOI] [PubMed] [Google Scholar]
- Clark P.U., Archer D., Pollard D., Blum J.D., Rial J.A., Brovkin V., Mix A.C., Pisias N.G., Roy M. The middle Pleistocene transition: characteristics, mechanisms, and implications for long-term changes in atmospheric pCO2. Quat. Sci. Rev. 2006;25:3150–3184. doi: 10.1016/j.quascirev.2006.07.008. [DOI] [Google Scholar]
- Cottee-Jones H.E.W., Bajpai O., Chaudhary L.B., Whittaker R.J. The importance of Ficus (Moraceae) trees for tropical forest restoration. Biotropica. 2016;48:413–419. doi: 10.1111/btp.12304. [DOI] [Google Scholar]
- De Bie T., Cristianini N., Demuth J.P., Hahn M.W. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22:1269–1271. doi: 10.1093/bioinformatics/btl097. [DOI] [PubMed] [Google Scholar]
- Devanesan E.B., Vijaya Anand A., Kumar P.S., Vinayagamoorthy P., Basavaraju P. Phytochemistry and pharmacology of ficus religiosa. Syst. Rev. Pharm. 2018;9:45–48. doi: 10.5530/srp.2018.1.9. [DOI] [Google Scholar]
- Doležel J., Bartoš J. Plant DNA flow cytometry and estimation of nuclear genome size. Ann. Bot. 2005;95:99–110. doi: 10.1093/aob/mci005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont L.M., Donner B., Schneider R., Wefer G. Mid-Pleistocene environmental change in tropical Africa began as early as 1.05 Ma. Geology. 2001;29:195–198. doi: 10.1130/0091-7613(2001)029<0195:MPECIT>2.0.CO;2. [DOI] [Google Scholar]
- Emms D.M., Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn R.D., Clements J., Eddy S.R. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn J.M., Hubley R., Goubert C., Rosen J., Clark A.G., Feschotte C., Smit A.F. RepeatModeler2 for automated genomic discovery of transposable element families. Proc. Natl. Acad. Sci. USA. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francenia Santos-Sánchez N., Salas-Coronado R., Hernández-Carlos B., Villanueva-Cañongo C. Plant Physiological Aspects of Phenolic Compounds. 2019. Shikimic acid pathway in biosynthesis of phenolic compounds. [DOI] [Google Scholar]
- Fukui K., Hayashi K.I. Manipulation and sensing of auxin metabolism, transport and signaling. Plant Cell Physiol. 2018;59:1500–1510. doi: 10.1093/PCP/PCY076. [DOI] [PubMed] [Google Scholar]
- Galil J. Ficus religiosa L.–the tree-splitter. Bot. J. Linn. Soc. 1984;88:185–203. doi: 10.1111/j.1095-8339.1984.tb01570.x. [DOI] [Google Scholar]
- GALIL J., EISIKOWITCH D. Studies on mutualistic symbiosis between syconia and sycophilous wasps in monoecious figs. New Phytol. 1971;70:773–787. doi: 10.1111/j.1469-8137.1971.tb02578.x. [DOI] [Google Scholar]
- Ghanashyam C., Jain M. Role of auxin-responsive genes in biotic stress responses. Plant Signal. Behav. 2009;4:846–848. doi: 10.4161/psb.4.9.9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh S., Bheri M., Bisht D., Pandey G.K. Calcium signaling and transport machinery: potential for development of stress tolerance in plants. Curr. Plant Biol. 2022;29:100235. doi: 10.1016/j.cpb.2022.100235. [DOI] [Google Scholar]
- Gopukumar S.T., Praseetha P.K. Ficus benghalensis Linn – the sacred Indian medicinal tree with potent pharmacological remedies. Int. J. Pharm. Sci. Rev. Res. 2015;32:223–227. [Google Scholar]
- Griffiths-Jones S., Saini H.K., Van Dongen S., Enright A.J. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grison-Pigé L., Bessière J.M., Hossaert-McKey M. Specific attraction of fig-pollinating wasps: role of volatile compounds released by tropical figs. J. Chem. Ecol. 2002;28:283–295. doi: 10.1023/A:1017930023741. [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas B.J., Papanicolaou A., Yassour M., Grabherr M., Blood P.D., Bowden J., Couger M.B., Eccles D., Li B., Lieber M., et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R.D. Figs and the diversity of tropical rainforests. Bioscience. 2005;55:1053–1064. doi: 10.1641/0006-3568(2005)055[1053:FATDOT]2.0.CO;2. [DOI] [Google Scholar]
- Hedges S.B., Dudley J., Kumar S. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22:2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- Hinckley W.E., Keymanesh K., Cordova J.A., Brusslan J.A. The HAC1 histone acetyltransferase promotes leaf senescence and regulates the expression of ERF022. Plant Direct. 2019;3:e00159. doi: 10.1002/pld3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houben M., Van de Poel B. 1-aminocyclopropane-1-carboxylic acid oxidase (ACO): the enzyme that makes the plant hormone ethylene. Front. Plant Sci. 2019;10:695. doi: 10.3389/fpls.2019.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Li J., Yang Z., An W., Xie C., Liu S., Zheng X. Comprehensive analysis of complete chloroplast genome and phylogenetic aspects of ten Ficus species. BMC Plant Biol. 2022;22:253–315. doi: 10.1186/S12870-022-03643-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., Von Mering C., Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isah T. Stress and defense responses in plant secondary metabolites production. Biol. Res. 2019;52:39. doi: 10.1186/s40659-019-0246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman S.D., Coombe L., Chu J., Warren R.L., Vandervalk B.P., Yeo S., Xue Z., Mohamadi H., Bohlmann J., Jones S.J.M., Birol I. Tigmint: correcting assembly errors using linked reads from large molecules. BMC Bioinf. 2018;19:393. doi: 10.1186/s12859-018-2425-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S.K., Gupta A., Saxena R., Prasoodanan V.P.K., Sharma A.K., Mittal P., Roy A., Shafer A.B.A., Vijay N., Sharma V.K. Genome sequence of peacock reveals the peculiar case of a glittering bird. Front. Genet. 2018;9:392. doi: 10.3389/fgene.2018.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiswal S.K., Mahajan S., Chakraborty A., Kumar S., Sharma V.K. The genome sequence of Aloe vera reveals adaptive evolution of drought tolerance mechanisms. iScience. 2021;24:102079. doi: 10.1016/j.isci.2021.102079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jombart T., Balloux F., Dray S. Adephylo: exploratory analyses for the phylogenetic comparative method. Bioinformatics. 2010;26:1907–1909. doi: 10.1093/bioinformatics/btq292. [DOI] [PubMed] [Google Scholar]
- Kachroo A., Lapchyk L., Fukushige H., Hildebrand D., Klessig D., Kachroo P. Plastidial fatty acid signaling modulates salicylic acid- and jasmonic acid-mediated defense pathways in the Arabidopsis ssi2 mutant. Plant Cell. 2003;15:2952–2965. doi: 10.1105/tpc.017301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara H., Hanada A., Kuzuyama T., Takagi M., Kamiya Y., Yamaguchi S. Contribution of the mevalonate and methylerythritol phosphate pathways to the biosynthesis of Gibberellins in Arabidopsis. J. Biol. Chem. 2002;277:45188–45194. doi: 10.1074/jbc.M208659200. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M., Rozhon W., Poppenberger B. E-mail experimental section/mini-review the role of hormones in the aging of plants-A mini-review. Gerontology. 2014;60:49–55. doi: 10.1159/000354334. [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development. 2010;137:3911–3920. doi: 10.1242/dev.048199. [DOI] [PubMed] [Google Scholar]
- Kitajima S., Aoki W., Shibata D., Nakajima D., Sakurai N., Yazaki K., Munakata R., Taira T., Kobayashi M., Aburaya S., et al. Comparative multi-omics analysis reveals diverse latex-based defense strategies against pests among latex-producing organs of the fig tree (Ficus carica) Planta. 2018;247:1423–1438. doi: 10.1007/S00425-018-2880-3/FIGURES/8. [DOI] [PubMed] [Google Scholar]
- Knepper C., Day B. From perception to activation: the molecular-genetic and biochemical landscape of disease resistance signaling in plants. Arabidopsis Book. 2010;8:e012. doi: 10.1199/TAB.0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolmogorov M., Yuan J., Lin Y., Pevzner P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- Koren S., Walenz B.P., Berlin K., Miller J.R., Bergman N.H., Phillippy A.M. Canu: scalable and accurate long-read assembly via adaptive κ-mer weighting and repeat separation. Genome Res. 2017;27:722–736. doi: 10.1101/gr.215087.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar Makhija I., Sharma I.P., Khamar D. Phytochemistry and Pharmacological properties of Ficus religiosa: an overview. Ann. Biol. Res. 2010;1:171–180. [Google Scholar]
- Laetsch D.R., Blaxter M.L. KinFin: software for taxon-aware analysis of clustered protein sequences. G3. 2017;7:3349–3357. doi: 10.1534/g3.117.300233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.Y., Baum S.F., Oh S.H., Jiang C.Z., Chen J.C., Bowman J.L. Recruitment of CRABS CLAW to promote nectary development within the eudicot clade. Development. 2005;132:5021–5032. doi: 10.1242/dev.02067. [DOI] [PubMed] [Google Scholar]
- Li H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv. 2013 doi: 10.48550/arXiv.1303.3997. Preprint at. [DOI] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Hu A., Dou W., Qi J., Long Q., Zou X., Lei T., Yao L., He Y., Chen S. Systematic analysis and functional validation of Citrus XTH genes reveal the role of Csxth04 in Citrus bacterial canker resistance and tolerance. Front. Plant Sci. 2019;10:1109. doi: 10.3389/FPLS.2019.01109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Li X., Wei W., Li F., Zhang L., Deng X., Liu Y., Yang S. The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for abiotic stress response in wheat. Int. J. Mol. Sci. 2019;20:E1104. doi: 10.3390/ijms20051104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D.Y., Lv H., Liao J. The complete chloroplast genome of Ficus pumila, a functional plant in East Asia. Mitochondrial DNA B Resour. 2022;7:326–327. doi: 10.1080/23802359.2022.2031328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Xu Z.H., Luo D., Xue H.W. Roles of OsCKI1, a rice casein kinase I, in root development and plant hormone sensitivity. Plant J. 2003;36:189–202. doi: 10.1046/j.1365-313X.2003.01866.x. [DOI] [PubMed] [Google Scholar]
- Lüttge U. Ability of crassulacean acid metabolism plants to overcome interacting stresses in tropical environments. AoB Plants. 2010;2010:plq005. doi: 10.1093/aobpla/plq005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison S.L., Buchanan M.L., Glass J.D., McClain T.F., Park E., Nebenführ A. Class XI myosins move specific organelles in pollen tubes and are required for normal fertility and pollen tube growth in Arabidopsis. Plant Physiol. 2015;169:1946–1960. doi: 10.1104/pp.15.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahajan S., Chakraborty A., Sil T., Sharma V.K. Genome sequencing and assembly of Tinospora cordifolia (Giloy) plant. bioRxiv. 2021 doi: 10.1101/2021.08.02.454741. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marçais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S.L., Davis R., Protti P., Lin T., Lin S., Martin C.E. The occurrence of crassulacean acid metabolism in epiphytic ferns, with an emphasis on the vittariaceae. Int. J. Plant Sci. 2005;166:623–630. doi: 10.1086/430334. [DOI] [Google Scholar]
- Matasci N., Hung L.H., Yan Z., Carpenter E.J., Wickett N.J., Mirarab S., Nguyen N., Warnow T., Ayyampalayam S., Barker M., et al. Data access for the 1,000 Plants (1KP) project. GigaScience. 2014;3:17. doi: 10.1186/2047-217X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittal P., Jaiswal S.K., Vijay N., Saxena R., Sharma V.K. Comparative analysis of corrected tiger genome provides clues to its neuronal evolution. Sci. Rep. 2019;9:18459. doi: 10.1038/s41598-019-54838-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K., Shirasawa K., Nogata H., Hirata C., Tashiro K., Habu T., Kim S., Himeno S., Kuhara S., Ikegami H. Identification of RAN1 orthologue associated with sex determination through whole genome sequencing analysis in fig (Ficus carica L.) Sci. Rep. 2017;7:41124. doi: 10.1038/srep41124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y., Itoh M., Okuda S., Yoshizawa A.C., Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:W182–W185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A. Modern Fruit Industry. 2020. A threatened introduced species ( Ficus benghalensis L.) in ismailia, Egypt. [DOI] [Google Scholar]
- Munné-Bosch S. Limits to tree growth and longevity. Trends Plant Sci. 2018;23:985–993. doi: 10.1016/j.tplants.2018.08.001. [DOI] [PubMed] [Google Scholar]
- Muñoz-Bertomeu J., Cascales-Miñana B., Irles-Segura A., Mateu I., Nunes-Nesi A., Fernie A.R., Segura J., Ros R. The plastidial glyceraldehyde-3-phosphate dehydrogenase is critical for viable pollen development in Arabidopsis. Plant Physiol. 2010 doi: 10.1104/pp.109.150458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J., Bush M.J., de Long C., Li S., Xu M., Khan M., Malcolmson C., Fobert P.R., Zachgo S., Hepworth S.R. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiol. 2010;154:1492–1504. doi: 10.1104/pp.110.159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng P.C., Henikoff S. SIFT: predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okonechnikov K., Conesa A., García-Alcalde F. Qualimap 2: advanced multi-sample quality control for high-throughput sequencing data. Bioinformatics. 2016;32:292–294. doi: 10.1093/bioinformatics/btv566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Singh J., Achary V.M.M., Reddy M.K. Redox homeostasis via gene families of ascorbate-glutathione pathway. Front. Environ. Sci. 2015;3:1–14. doi: 10.3389/fenvs.2015.00025. [DOI] [Google Scholar]
- Pellicer J., Leitch I.J. The Plant DNA C-values database (release 7.1): an updated online repository of plant genome size data for comparative studies. New Phytol. 2020;226:301–305. doi: 10.1111/nph.16261. [DOI] [PubMed] [Google Scholar]
- Peng Y.Q., Compton S.G., Yang D.R. The reproductive success of Ficus altissima and its pollinator in a strongly seasonal environment: Xishuangbanna, Southwestern China. Plant Ecol. 2010;209:227–236. doi: 10.1007/S11258-009-9690-4/FIGURES/4. [DOI] [Google Scholar]
- Piovesan G., Biondi F. On tree longevity. New Phytol. 2021;231:1318–1337. doi: 10.1111/NPH.17148. [DOI] [PubMed] [Google Scholar]
- Plomion C., Aury J.-M., Amselem J., Leroy T., Murat F., Duplessis S., Faye S., Francillonne N., Labadie K., Le Provost G., et al. Oak genome reveals facets of long lifespan. Nat. Plants. 2018;4:440–452. doi: 10.1038/s41477-018-0172-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X.-M., Sun Y.-Y., Ye X.-Y., Li Z.-G. Signaling role of glutamate in plants. Front. Plant Sci. 2019;10:1743. doi: 10.3389/FPLS.2019.01743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranallo-Benavidez T.R., Jaron K.S., Schatz M.C. GenomeScope 2.0 and Smudgeplot for reference-free profiling of polyploid genomes. Nat. Commun. 2020;11:1432. doi: 10.1038/s41467-020-14998-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeder A., Schweingruber F.H., Ebeling A., Eisenhauer N., Fischer M., Roscher C. Plant diversity effects on plant longevity and their relationships to population stability in experimental grasslands. J. Ecol. 2021;109:2566–2579. doi: 10.1111/1365-2745.13661. [DOI] [Google Scholar]
- Ruan J., Li H. Fast and accurate long-read assembly with wtdbg2. Nat. Methods. 2020;17:155–158. doi: 10.1038/s41592-019-0669-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S., Sharma I., Kaur N., Pati P.K. Auxin: a master regulator in plant root development. Plant Cell Rep. 2013;32:741–757. doi: 10.1007/S00299-013-1430-5. [DOI] [PubMed] [Google Scholar]
- Schiøtt M., Romanowsky S.M., Bækgaard L., Jakobsen M.K., Palmgren M.G., Harper J.F. A plant plasma membrane Ca2+ pump is required for normal pollen tube growth and fertilization. Proc. Natl. Acad. Sci. USA. 2004;101:9502–9507. doi: 10.1073/pnas.0401542101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt H., Hellmann S.L., Waldvogel A.M., Feldmeyer B., Hankeln T., Pfenninger M. A high-quality genome assembly from short and long reads for the non-biting midge chironomus riparius (diptera) G3. 2020;10:1151–1157. doi: 10.1534/G3.119.400710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shameer S., Baghalian K., Cheung C.Y.M., Ratcliffe R.G., Sweetlove L.J. Computational analysis of the productivity potential of CAM. Nat. Plants. 2018;4:165–171. doi: 10.1038/s41477-018-0112-2. [DOI] [PubMed] [Google Scholar]
- Shanahan M., So S., Compton S.G., Corlett R. Fig-eating by vertebrate frugivores: a global review. Biol. Rev. Camb. Philos. Soc. 2001;76:529–572. doi: 10.1017/S1464793101005760. [DOI] [PubMed] [Google Scholar]
- Sharma A., Sharma D., Verma S.K. In silico study of Iron, zinc and copper binding proteins of Pseudomonas syringae pv. lapsa: emphasis on secreted metalloproteins. Front. Microbiol. 2018;9:1838. doi: 10.3389/fmicb.2018.01838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Mon A.M., Fu Y., Zhang Y., Wang C., Yang X., Wang Y. The genus Ficus (Moraceae) used in diet: its plant diversity, distribution, traditional uses and ethnopharmacological importance. J. Ethnopharmacol. 2018;226:185–196. doi: 10.1016/j.jep.2018.07.027. [DOI] [PubMed] [Google Scholar]
- Shirasawa K., Yakushiji H., Nishimura R., Morita T., Jikumaru S., Ikegami H., Toyoda A., Hirakawa H., Isobe S. The Ficus erecta genome aids Ceratocystis canker resistance breeding in common fig (F. carica) Plant J. 2020;102:1313–1322. doi: 10.1111/tpj.14703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- Singh D., Singh B., Goel R.K. Traditional uses, phytochemistry and pharmacology of Ficus religiosa: a review. J. Ethnopharmacol. 2011;134:565–583. doi: 10.1016/j.jep.2011.01.046. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Sung T.-Y., Chung T.-Y., Lin S.-Y., Lin S.-C., Liao J.-C., Hsieh W.-Y., Hsieh M.-H. ACR11 modulates levels of reactive oxygen species and salicylic acid-associated defense response in Arabidopsis. Sci. Rep. 2018;8:11851. doi: 10.1038/S41598-018-30304-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitaramam V., Jog S.R., Tetali P. Ecology of Ficus religiosa accounts for its association with religion. Curr. Sci. 2009;97 [Google Scholar]
- Soltis D., Soltis P.S., Chase M.W., Mort M.E., Albach D.C., Zanis M., Savolainen V., Hahn W.H., Hoot S.B., Fay M.F., et al. Angiosperm phylogeny inferred from 18S rDNA, rbcL, and atpB sequences. Bot. J. Linn. Soc. 2000;133:381–461. doi: 10.1006/bojl.2000.0380. [DOI] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B. AUGUSTUS: a b initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34:W435–W439. doi: 10.1093/nar/gkl200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P., et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias P.A., Guest D.I. Tree immunity: growing old without antibodies. Trends Plant Sci. 2014;19:367–370. doi: 10.1016/J.TPLANTS.2014.01.011. [DOI] [PubMed] [Google Scholar]
- Usai G., Mascagni F., Giordani T., Vangelisti A., Bosi E., Zuccolo A., Ceccarelli M., King R., Hassani-Pak K., Zambrano L.S., et al. Epigenetic patterns within the haplotype phased fig (Ficus carica L.) genome. Plant J. 2020;102:600–614. doi: 10.1111/TPJ.14635. [DOI] [PubMed] [Google Scholar]
- Van Dongen S., Abreu-Goodger C. Using MCL to extract clusters from networks. Methods Mol. Biol. 2012;804:281–295. doi: 10.1007/978-1-61779-361-5_15. [DOI] [PubMed] [Google Scholar]
- Venkateshwaran M., Jayaraman D., Chabaud M., Genre A., Balloon A.J., Maeda J., Forshey K., Den Os D., Kwiecien N.W., Coon J.J., et al. A role for the mevalonate pathway in early plant symbiotic signaling. Proc. Natl. Acad. Sci. USA. 2015;112:9781–9786. doi: 10.1073/pnas.1413762112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbitsky M.Y., Crucifix M., Volobuev D.M. A theory of Pleistocene glacial rhythmicity. Earth Syst. Dyn. 2018;9:1025–1043. doi: 10.5194/esd-9-1025-2018. [DOI] [Google Scholar]
- Vidyapeeth J. Vol. 6. 2019. pp. 901–906. (A Review on Ficus Religiosa (Sacred Fig)). [Google Scholar]
- Villard C., Larbat R., Munakata R., Hehn A. Defence mechanisms of Ficus: pyramiding strategies to cope with pests and pathogens. Planta. 2019;249:617–633. doi: 10.1007/S00425-019-03098-2. [DOI] [PubMed] [Google Scholar]
- Walker B.J., Abeel T., Shea T., Priest M., Abouelliel A., Sakthikumar S., Cuomo C.A., Zeng Q., Wortman J., Young S.K., Earl A.M. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9:e112963. doi: 10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G., Zhang X., Herre E.A., McKey D., Machado C.A., Yu W.-B., Cannon C.H., Arnold M.L., Pereira R.A.S., Ming R., et al. Genomic evidence of prevalent hybridization throughout the evolutionary history of the fig-wasp pollination mutualism. Nat. Commun. 2021;12:718–814. doi: 10.1038/s41467-021-20957-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Quan S., Xiao H. Towards efficient terpenoid biosynthesis: manipulating IPP and DMAPP supply. Bioresour. Bioprocess. 2019;6:6. doi: 10.1186/s40643-019-0242-z. [DOI] [Google Scholar]
- Wang Y., Kumaishi K., Suzuki T., Ichihashi Y., Yamaguchi N., Shirakawa M., Ito T. Morphological and physiological framework underlying plant longevity in Arabidopsis thaliana. Front. Plant Sci. 2020;11:1693. doi: 10.3389/FPLS.2020.600726/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Tang H., Debarry J.D., Tan X., Li J., Wang X., Lee T.H., Jin H., Marler B., Guo H., et al. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012;40:e49. doi: 10.1093/nar/gkr1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R.L., Yang C., Vandervalk B.P., Behsaz B., Lagman A., Jones S.J.M., Birol I. LINKS: scalable, alignment-free scaffolding of draft genomes with long reads. GigaScience. 2015;4:35. doi: 10.1186/s13742-015-0076-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisenfeld N.I., Kumar V., Shah P., Church D.M., Jaffe D.B. Direct determination of diploid genome sequences. Genome Res. 2017;27:757–767. doi: 10.1101/gr.214874.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wi J., Na Y., Yang E., Lee J.H., Jeong W.J., Choi D.W. Arabidopsis AtMPV17, a homolog of mice MPV17, enhances osmotic stress tolerance. Physiol. Mol. Biol. Plants. 2020;26:1341–1348. doi: 10.1007/s12298-020-00834-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G.C., Xu T.J., Zhu R., Zhang Y., Li S.Q., Wang H.W., Li J.T. LR-Gapcloser: a tiling path-based gap closer that uses long reads to complete genome assembly. GigaScience. 2018;8 doi: 10.1093/gigascience/giy157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Paml 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yeo S., Coombe L., Warren R.L., Chu J., Birol I. ARCS: scaffolding genome drafts with linked reads. Bioinformatics. 2018;34:725–731. doi: 10.1093/bioinformatics/btx675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan N., Yuan S., Li Z., Zhou M., Wu P., Hu Q., Mendu V., Wang L., Luo H. STRESS INDUCED FACTOR 2, a leucine-rich repeat kinase regulates basal plant pathogen defense. Plant Physiol. 2018;176:3062–3080. doi: 10.1104/pp.17.01266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Hu J., Han X., Li J., Gao Y., Richards C.M., Zhang C., Tian Y., Liu G., Gul H., et al. A high-quality apple genome assembly reveals the association of a retrotransposon and red fruit colour. Nat. Commun. 2019;10:1494. doi: 10.1038/s41467-019-09518-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.V., Zhuo L., Hahn M.W. AGOUTI: improving genome assembly and annotation using transcriptome data. GigaScience. 2016;5:31. doi: 10.1186/s13742-016-0136-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Wang G., Zhang S., Chen S., Wang Y., Wen P., Ma X., Shi Y., Qi R., Yang Y., et al. Genomes of the banyan tree and pollinator wasp provide insights into fig-wasp coevolution. Cell. 2020;183:875–889.e17. doi: 10.1016/j.cell.2020.09.043. [DOI] [PubMed] [Google Scholar]
- Zotz G. How prevalent is crassulacean acid metabolism among vascular epiphytes? Oecologia. 2004;138:184–192. doi: 10.1007/s00442-003-1418-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The raw reads obtained from genome and transcriptome sequencing of both F. benghalensis (BioProject accession – PRJNA759132, BioSample accession – SAMN21156846, SRA accessions – SRR15676288, SRR15676289, SRR15676290), and F. religiosa (BioProject accession – PRJNA759116, BioSample accession – SAMN21156584, SRA accessions – SRR15676280, SRR15676281, SRR15676282) species have been deposited in NCBI SRA database.
-
•
This study does not report original code.
-
•
We have provided all detailed information to reanalyze this study in STAR Methods to the best of our knowledge. Any additional information is available from the lead contact upon request.