Graphical abstract
Keywords: Angiogenesis, Single-cell multi-omics, Functional enrichment, Biological networks, Unsupervised and supervised data fusion, Gene prioritization
Abstract
Multi-omics technologies are being increasingly utilized in angiogenesis research. Yet, computational methods have not been widely used for angiogenic target discovery and prioritization in this field, partly because (wet-lab) vascular biologists are insufficiently familiar with computational biology tools and the opportunities they may offer. With this review, written for vascular biologists who lack expertise in computational methods, we aspire to break boundaries between both fields and to illustrate the potential of these tools for future angiogenic target discovery. We provide a comprehensive survey of currently available computational approaches that may be useful in prioritizing candidate genes, predicting associated mechanisms, and identifying their specificity to endothelial cell subtypes. We specifically highlight tools that use flexible, machine learning frameworks for large-scale data integration and gene prioritization. For each purpose-oriented category of tools, we describe underlying conceptual principles, highlight interesting applications and discuss limitations. Finally, we will discuss challenges and recommend some guidelines which can help to optimize the process of accurate target discovery.
1. Introduction
Angiogenesis has broad pathophysiological implications in promoting disorders like cancer, ischemia, inflammation, infection and immune responses [1]. Some disorders are characterized by abnormal, excessive angiogenesis, whereas others are typified by sparse angiogenesis with vessel regression. Angiogenic therapeutic strategies aim to normalize and restore blood vessels, thereby regulating tissue oxygenation and nutrient supply. Depending upon the disorder, most of the available therapies focus on either blocking growth factor signaling pathways (e. g. vascular endothelial growth factor (VEGF) signaling), thereby blocking angiogenesis (anti-angiogenic therapy (AAT)), or delivering components, mostly growth factors, to promote angiogenesis (pro-angiogenic therapy) [2]. In both types of therapies, inappropriate tuning of VEGF levels can lead to an increase in leaky or regressed blood vessels, as opposed to the anticipated normalization. Even in metastatic tumors where anti-angiogenic therapeutics have been widely tested, anti-VEGF targeted therapies show a large variability in response across tumor types and are often characterized by resistance and insufficient efficacy [3]. This emphasizes the need for discovering alternative therapeutic opportunities. For this purpose, at least two aspects should be addressed: (i) identification of novel molecular targets for anti-angiogenic therapy development; and (ii), ideally, specific effects of the anti-angiogenic therapy for a particular endothelial cell (EC) subtype or condition.
There are around 20,000 protein-coding genes in the human genome and millions of cells in any given tissue, making it a complex, multi-dimensional problem [4]. Single-cell sequencing approaches attempt to solve this problem by characterizing cell subtypes and identifying cell type-specific marker genes at different biological scales (transcriptome, epigenome, proteome, metabolome, interactome) [5]. However, true biological function results from the complex interplay between these different scales. Integrative approaches like network prediction methodologies and machine learning (ML) can help mitigate these challenges. Therefore, the angiogenesis field can benefit from a shift of focus towards integrating complex, multi-omic, biological big datasets for target / mechanism discovery.
Mathematical, statistical and ML models can be used to integrate such high dimensional datasets. However, most of the available studies either use mathematical modeling to simulate (in vitro) biomechanical changes in angiogenesis via proangiogenic stimuli, their effects on angiogenic morphological phenotypes (migration, vessel sprouting, shear stress, etc.) or implement statistical ML models to predict dysfunctional vasculature from imaging studies [6], [7], [8]. Very few studies focus on the prediction and discovery of angiogenic gene signatures using high throughput ‘omics’ datasets [9]. This review will provide a brief overview of overall developments in single-cell characterization of angiogenic cellular heterogeneity, computational tools that predict mechanisms using single-cell abundance information, tools that integrate multiple omics sources at a single-cell level, and techniques that can help with prioritizing important genes. This review does not aim to provide depth and technical detail into specific tools or methodologies for specialized analyses of high throughput data, but rather overviews, in a user-friendly manner for the vascular biologist who is not an expert in computational biology, a breadth of techniques that can be used to identify targets for anti-angiogenic therapy development. This overview will serve as a springboard for integrative research and target discovery in the angiogenesis field and should be regarded as an open invitation for this field to consider and exploit the enormous potential of these computational approaches.
2. Single-cell omics in characterizing vascular heterogeneity
Endothelial cells (ECs), the main cellular players of angiogenesis, form new blood vessels under the stimulus of pro-angiogenic factors secreted by tissues requiring vascularization. EC phenotypes are heterogeneous and vary across different organs, within the vascular loop segments of an organ, and even between neighboring ECs and physiological functions [10]. High-throughput transcriptomics has been successfully used to identify novel clusters of ECs, map the evolution of EC states and identify changes in EC subtype-specific mechanisms based on the similarities or differences between their transcriptomes. Such techniques allow fine resolution in characterizing EC populations by providing transcriptomic snapshots of tissue-level changes in samples isolated from different biological conditions [11]. This has led to the initiation of multiple single-cell atlases that have characterized EC populations in various conditions. We will discuss them briefly in the following sections.
2.1. Adult healthy organs/tissues
Early bulk RNA sequencing studies discovered that ECs from different organs are transcriptionally heterogeneous, suggesting tissue-specific functions [12]. However, bulk RNA transcriptomics averages global expression and does not give more information about the cells that represent a tissue/organ. To get more insight and to better dissect the heterogeneity between and within organs, a tissue-wide EC atlas based on high-throughput single-cell transcriptomics analysis identified 78 unique groups of EC subpopulations across 11 distinct tissue (organ) types in mice [13]. This study disclosed profound differences at the single-cell level in overall gene expression and transcription factor expression levels, where multiple arterial, venous, and lymphatic EC markers were shared across organs (demonstrating cross-organ, tissue phenotype homogeneity, and a cross-linked network). In contrast, capillary ECs exhibited primarily an organ-restricted, heterogeneous phenotype dependent on the organ-specific metabolic and physiological needs. Similar atlases focused on adult, healthy (human/murine) organs like the liver [14], heart [15], brain [16], lung [17] and kidney [18] were able to identify distinct EC subpopulations.
2.2. Developing tissues
Single-cell RNA sequencing was also used to characterize EC populations in developing tissues, namely developing mouse embryos [19], zebrafish skeletal muscle [20] and embryonic stem cell differentiation [21]. EC heterogeneity and lineage relationships during early vascular development were resolved by applying single-cell RNA sequencing and lineage tracing methodologies to a time window where key vascular and angiogenic events occur in human and mouse embryos [22]. Analysis of primordial ECs in mice showed that ECs have distinctive characteristics that were described as branching out from mesodermal cells during vascular development and having allantois- and non-allantois-derived cell subtypes [23].
2.3. Disease
Apart from healthy organs, EC populations also differ between diseases through multiple levels of heterogeneity [11]. Tumor ECs are one group of key components in the tumor microenvironment that play an essential role in tumor progression and metastasis, showing both angiogenic and anti-angiogenic properties. Tumor EC heterogeneity has been reported in multiple cancer types including lymphoma [24], glioblastoma [25], breast [26], liver [27], [28], lung [29], [30], cervical [31], colorectal cancer [32], [33], [34], pancreatic [35], gastric [36] and renal [37], [38], [39] cancer. For instance, in human non-small cell lung cancer (NSCLC), a direct comparison of tumor versus non-malignant ECs revealed that Myc targets were the most enriched signatures in tumor ECs [40]. This finding is consistent with previous evidence of c-Myc's role in tumor angiogenesis. A human spatial transcriptomic atlas could reveal a loss of endothelial arteriovenous zonation in malformed brain vasculature compared to normal brain vasculature with an emergence of a transcriptomic state characterized by increased angiogenic potential and immune cell cross-talk [41]. Furthermore, a shift in the ratio of certain EC subtypes was found to be another type of EC heterogeneity in diseases. This is particularly evident in idiopathic pulmonary fibrosis (IPF), where out of the 5 ECs subtypes identified, a specific subtype (known as peribronchial) was highly prevalent in IPF samples compared to another pulmonary disease (i.e., obstructive pulmonary disease) [42].
2.4. Studies focusing on identifying angiogenic targets
Very few studies have used single-cell biomolecular abundance to identify angiogenic targets for anti-angiogenic therapy development. Focusing on NSCLC, freshly isolated tumoral and peri-tumoral ECs were profiled for their transcriptomes to identify novel tip tumor EC subtypes (“tip” ECs lead the vessel sprout [43], [44]) and further integrated with multi-omics data to identify conserved phenotypes and markers across patients, tumor / tissue types, species and animal models [29]. This integrative analyses led to the prioritization of potential candidates for anti-angiogenic therapy, validated for their roles through in vitro vessel sprouting experiments. In the context of age-related macular degeneration (AMD), which is characterized by the formation of leaky blood vessels, integrative computational analyses (meta-analysis with the above lung tumor EC atlas, available bulk RNA sequencing datasets and genome-scale metabolic modeling of EC proliferation) on single-cell (normal vs neovascularized) choroidal EC populations isolated from pre-clinical mouse models were successful in identifying potential metabolic anti-angiogenic candidates [45]. Silencing the selected metabolic enzyme targets in vitro and in vivo demonstrated an evident reduction in vessel sprouting and blood vessel area, thereby validating the predictions. The above studies showcase the strength of integrative computational analyses in identifying experimentally verifiable anti-angiogenic targets.
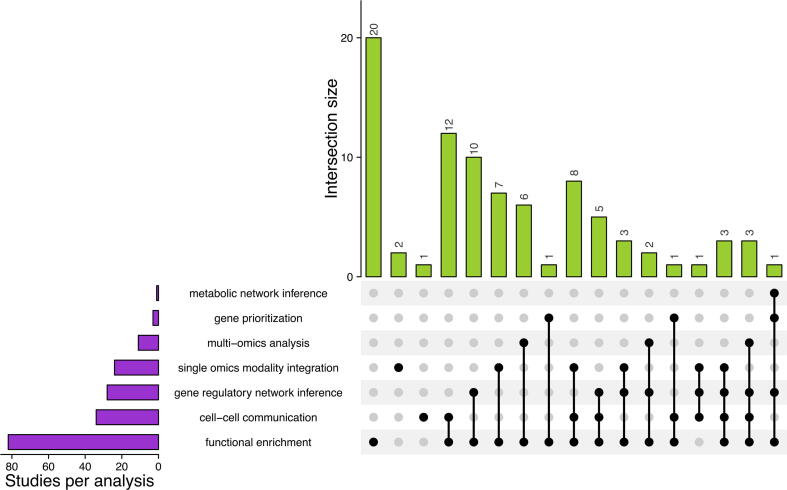
With the help of EndoDB (an EC-specific transcriptomics database [46] and keyword-based searches), we curated around 87 datasets that explicitly characterized EC heterogeneity and compared these studies based on the computational analyses performed to extract biological knowledge (Fig. 1, Supplementary Table 1). Despite the availability of detailed single-cell atlases, most of the above studies typically focus on resolving populations of ECs and perform functional enrichment to identify / predict biological processes based on pre-defined gene sets. Specialized analysis for the systematic prediction of biological networks, integration of multi-omics datasets or prioritization of essential genes is rarely performed. In the subsequent sections, we introduce the readers to the specialized computational arsenal that might provide depth to the biological interpretations and AAT target identification, in addition to the routine analyses. Table 1 provides a summary of the different classes of techniques that perform specialized downstream analyses and the publicly available tools that provide formal implementation of the analyses. Table 2 enlists web-based applications belonging to the different classes of techniques that can be used by non-expert users to obtain a simple and quick hands-on experience of the different techniques. A glossary of different terms (techniques) is also provided in Box 1 to introduce the non-expert readers to various concepts.
Fig. 1.
UpSet Plot showing the classification of studies characterizing single-cell EC heterogeneity with respect to the applied computational techniques. A total of 87 studies detailed in Supplementary Table 1, characterize single-cell EC heterogeneity with the distribution of studies that use different task-specific computational techniques. Performing differential expression of biomolecular abundances between conditions and subsequent coupling with functional enrichment techniques are commonly used to discover novel biological knowledge in single-cell ECs (82 studies). This is followed by the use of biological network inference techniques to identify novel biomolecular interactions from changes in gene expression (18 studies). Within biological network inference approaches, most studies intend to predict cell–cell communication through ligand-receptor interactions followed by inference of gene-regulatory networks. Only one study focused on predicting varying pathway activity using genome-scale metabolic networks. Also, biological network inference studies are only used complementary to functional enrichment techniques (overlap between biological network-based studies and functional enrichment). Among integration-based approaches, most studies fuse single-cell transcriptomes from multiple datasets laterally as compared to vertical fusion of multiple omics data types. Automated gene-prioritization for the identification of AAT targets is the least explored (only 3 studies have attempted prioritization of genes). The bar plot in the bottom left shows comparison of the number of studies which use a particular technique. The bar plots on the top indicate the number of studies that have used a combination of different tools for analysis. The filled dots and lines in the matrix visually represent studies that use different combinations of the tools enlisted in the rows.
Table 1.
Computational tools for knowledge discovery and target prioritization.
| Class | Methodology | Tools |
|---|---|---|
| ||
| Over-Representation Analysis | identifies enriched gene-sets based on the strength of overlap between user-defined gene list and reference gene sets | g:Profiler; Panther; Enrichr |
| Gene Set Enrichment Analysis | enriches gene sets based on the degree / significance of relative gene expression changes | clusterProfiler; GenePattern; GSEA tool, BIOMEX |
| Gene Set Variation Analysis | estimates varying gene-sets across samples by generating gene-sets vs samples scoring matrix | GSVA package, BIOMEX |
| ||
| Differential Combination Methods | use differentially expressed ligands and receptors to identify interactions between clusters of cells. | CellTalker; iTALK; PyMINEr |
| Expression Permutation Tools | statistical scoring of each ligand-receptor pair based on permutation test-based filtering, non-parametric tests with a null model or defined empirical rules | CellChat; CellPhoneDB; Giotto; ICELLNET; SingleCellSignalR |
| Network-Based Methods | uses networks of interactions between ligands, receptors, and downstream targets to prioritize ligand-receptor interactions | CCCExplorer; NicheNet; SoptSC; SpaOTsc |
| Tensor-Based Methods | help to generate a hypergraph (network representing many-to-many relationships) of ligands and receptors from co-expression data. | scTensor |
| ||
| GRN Inference Methods | prediction of activation / inhibition relationships based on co-expression of transcription factors and their targets (or transcription-factor target promotor binding) across conditions or time dependent changes. | GENIE3; SCENIC; AR1MA1; SCODE |
| ||
|
Genome-Scale Metabolic Reconstruction |
mathematical model of whole cell metabolism that can be tailored to predict condition-specific metabolic fluxes using uptake and ‘omics’ abundance constraints | COBRA toolbox, COBRApy, RAVEN toolbox |
| Flux Balance Analysis (FBA) | a method to estimate pseudo steady-state metabolic fluxes in a genome-scale metabolic reconstruction that is required to optimize the synthesis of specific metabolites | |
| Single-cell data-based tailoring | modification of optimization solver to account for cell–cell metabolic variation | scFEA |
| ||
| Joint Dimensionality Reduction | captures cell–cell correspondence by identifying shared feature associations between paired or unpaired modalities | Seurat V3; BindSC; MOFA+; MATCHER |
| Network-Based Fusion Approaches | captures cell–cell correspondence by identifying conserved cluster structures between paired or unpaired modalities | Seurat V4; CiteFuse |
| Statistical Modeling | uses the Bayesian framework of modeling to scale and map different modalities | BREM-SC; Clonealign |
| Deep learning representations | uses auto-encoders to identify non-linear relationships between features and modalities to make interpretations | TotalVI; GLUE |
| ||
| Raw Fusion | an early integration technique, where the fusion of several data sources takes place at the raw data level | |
|
Transitional Fusion |
an intermediate integration technique, where different data sources are fused while learning | |
| Decision Fusion | a late integration technique, where each data source is modeled separately and integrates the data at the decision level through decision aggregation | ScanCluster |
|
Partial Least-Squares Discriminant Analysis |
reduces data dimensionality while remaining fully aware of the class labels and can be used for classification purposes | MixOmics; MINT; DIABLO |
| ||
| One-class classification (OCC) | OCC aims at identifying data elements of a given class among all objects by learning mostly from a training set that only contains objects of that class. | |
| PU Learning | similar to one-class classification, PU-Learning focuses on one-class. However, in PU learning, two sets of examples are supposed to be accessible for training: a positive set P and an unlabeled set, which is expected to contain both positive and negative examples. In PU learning, a binary classifier is trained in a semi-supervised manner from solely positive and unlabeled sample points. | GuiltyTargets; n2a-SVM; Node2vec; DeepPVP |
| ML-Based Gene Prioritization | detecting disease-associated genes through ML technologies. | exTasy; Endeavour; Genehound |
Table 2.
Web-based applications for knowledge discovery and target prioritization.
Box 1. Glossary.
Artificial Neural Networks (ANNs): A machine learning network of neurons (typically referred as nodes or units) that learns and finds patterns in data. Like neurons in the nervous system, each node receives an input, performs some computation and passes the signal onto the next node. Separate sets of nodes are typically classified into input, hidden and output layers. For example, if our aim is to classify genes into different biological processes based on gene expression variation across single cells, an ANN will be designed, such that: (i) the input layer nodes use gene expression across different single cells as attributes, (ii) the hidden layer nodes will provide weights (confidence) to gene expression values from each single cell, and (iii) at the output layer, the weights of the gene from different nodes will be summed. These cumulative weight values will be used for classifying genes into its known biological process. This procedure is iteratively repeated multiple times so that the network can learn the training data accurately (by adjusting the weights) and predict their associated biological processes. ANNs form the basis of deep learning methodologies, where deep learning ANNs consists of multiple hidden layers that improve learning (Fig. 4D).
Autoencoders: Deep ANNs that learn an encoded representation for a set of data (by transforming real, high dimensional data to low dimensional representations) and uses a decoder that maps the coded representation to reconstruct the output. They are well-suited for unsupervised learning (Fig. 3F).
Bayesian inference or probabilistic modeling: Probabilistic modeling is a statistical technique used to consider the impact of random events or actions in predicting the potential occurrence of future outcomes, given that randomness or uncertainty plays a role in predicting outcomes. Probabilistic models are a powerful idiom to describe the world, using random variables as building blocks held together by probabilistic relationships. Bayesian inference methods typically generate probabilistic models that update the probability of a hypothesis when more evidence or information becomes available. These methods estimate prior and posterior probabilities to improve confidence over a hypothesis. Bayesian statistics use the data and consider parameters (e. g. mean, standard deviation of gene expression) to be random variables with a distribution that can be inferred from data. Bayesian methods enable the estimation of uncertainty in predictions, extracting crucial information from small data sets and handling missing data. A prior probability is the probability that an observation may belong to a group before performing a classification task (for instance, the prior assumption that a cell belongs to a single-cell cluster before considering the underlying patterns within the data). Usually, prior probability distributions are the known probability distributions that can be used for transforming the input data (for example, uniform distribution, beta distribution, Dirichlet distribution, etc.). A posterior probability is the probability of assigning observations to groups given the patterns in the data (for example, posterior classification of cells to correct single-cell given the mapping of prior probabilities to raw single-cell gene expression). For instance, when integrating two modalities (transcriptomics and proteomics) to identify cell clusters, both transcriptomics and proteomics would have different data distributions as they measure different biological features. With the known prior probability distributions that randomly assign cells to clusters, the transcriptomic and proteomic abundances are tuned such that the shared cell-specific random effects (relationships) between the omics data types are estimated. This can be used to identify the posterior probabilities that the cells actually belong to specific clusters (Fig. 3E).
Ensemble learning: ML strategy in which numerous learning models are trained to tackle a classification or regression task, and their outputs are integrated to maximize the accuracy of predictions as compared to the individual learning models.
Graph: Graphs are mathematical structures that embody the pairwise relationships between objects (e. g. biological features like genes, proteins (Fig. 2E)). A graph is made up of nodes (which represents genes, proteins, cells) and the edges or vertices that connect the nodes represent a relationship. Graphs can be directed where the edges unidirectionally start from one node and end in the other node; or undirected where the edges do not represent any direction. Graph-based methods automatically generate graphs from data to gain new information about mechanistic (e. g. the use of directed graphs for representing biological networks (Fig. 2E)) or associative relationships (e. g. co-expression-based graphs).
Graph Neural Network (GNN): While ANNs typically learn information of individual data points per sample, GNNs learn the structure of multiple data points from an n-dimensional attribute space. For instance, when using unsupervised clustering of single-cells, based on their transcriptional profiles, single cells are the biological instances and genes are the attributes of the biological measurement. Graphs (networks) can be created based on the similarity of transcriptional profiles between cells (Fig. 3D). These graphs can be transformed into a low dimensional space by a technique called graph embedding. In a supervised setting, GNNs can learn these graph embedding representations to classify such cell similarity graphs. GNNs can also be used for unsupervised learning using auto-encoders, where the output clusters can be decoded from the encoded graph embedding.
Heuristic Approaches: Practical and scalable methods that produce solutions based on a trial-and-error, rule of thumb or an educated guess. Such solutions may not be optimal, perfect or rational, but are sufficient for getting short-term solutions or approximations.
Manifolds: Manifolds represent a wide variety of geometric surfaces in mathematics (Fig. 3C). In ML, data can come from a variety of spaces (e.g., the single-cell transcriptome represents the single-cell gene expression space, the single-cell proteome represents the single-cell protein abundance space, etc.). Each of these spaces are multi-dimensional in nature (e.g., multiple genes represent the multiple dimensions in a single-cell transcriptomic dataset). High dimensional representations cannot always be visualized. However, data can come from a subset of points (e.g., subset of single cells) in space that can represent a manifold. In other words, features having similar patterns across omics modalities can represent a common manifold. In case of single-cell multi-omics data integration, manifolds are generated from pairwise omics modalities (e.g., transcriptome and epigenome) and are aligned together to identify conserved clusters of single-cells.
Modality: An omics modality indicates the type of omics data under consideration. Each omics modality represents a different characteristic of the underlying biology. Genomics, transcriptomics, proteomics, epigenomics, metabolomics, lipidomics, kinomics; each represents different modalities.
Multiple Kernel Learning (MKL): MKL uses a pre-defined set of kernel functions for learning data distributions as part of a classification or a regression task. Kernels or kernel functions are mathematical functions that transform the non-linearly arranged real-world attributes of data points (characteristics of genes like gene expression) to higher dimensions for (linear) separation of data points into groups within this newly generated high dimensional space. Thus, kernel functions generate transformed kernel matrices that represent linear or non-linear covariance/correlation matrix that contains sample (e.g., single cell) similarities in their corresponding input space. Kernel functions like the linear kernel, polynomial kernel, radial basis kernel, etc. help ML algorithms like support vector machines to linearly classify non-linear data albeit in a high dimension space (see below).
Non-Negative Matrix Factorization (NMF): NMF is a method that can reveal the component parts of a non-negative signal. A non-negative signal can be any data distribution (for example, distribution of cells in an m-dimensional gene expression space and n-dimensional protein abundance space where m, n = biological attributes from a two different omics data types) and the components of this non-negative distribution are mapped onto a low dimensional space (called latent space). When, for example, using single-cell multi-omics data integration (Fig. 3B), the assumption is that two different omics data types (e.g., attributes from epigenome and transcriptome) are components of the same underlying biological signal. Hence, some patterns emerging from each omics data should be conserved in a common “latent” space. In other words, NMF maps biological features from the two omics components onto a low-dimensional common latent factor space. Each latent factor is a linear combination of correlated epigenomic and transcriptomic attributes.
Permutation tests: Random re-assignment of sample labels (e.g., cell labels, assigning genes to processes) frequently used to compute null (background) models in biological systems. Permutation tests are used for gene set enrichment analysis, cell–cell communication inference to prioritize enriched processes or ligand-receptor pairs. For example, in ligand-receptor communication inference, labels representing single-cells are permuted and the probability of a ligand-receptor to undergo an interaction across permuted cell types is calculated to generate a random background distribution. Comparison of this background score to the actual ligand-receptor communication score leads to the identification of significant ligand-receptor pairs between a pair of cells.
Randomforests: An ensemble-learning algorithm that operates by constructing a forest of decision trees on different samples for classification or regression. Each decision tree is a hierarchical network of nodes and connections where each node represents a decision rule for each attribute, using which every biological feature (e.g., the gene phosphofructokinase) can be split into two groups at a time. The decision rules start with a root node (first decision rule - for example, log-normalized counts, an attribute of transcript abundance can be used to split genes into two groups based on cut-offs) and moves further downwards with a second node (the second decision rule for splitting genes – for example, number of genes correlated with a given gene). This iteratively continues for all attributes until each group of genes cannot be split further and each set represents a known set (e.g., phosphofructokinase belongs to glycolysis). The threshold cut-offs for splitting are directly determined from the training data distribution. Random forests are a randomly generated bunch of decision trees bundled together, where every tree in the decision forest helps in classifying a subset of training examples (genes) into its classes (biological processes) that were randomly sampled using a bagging approach (where a sampling with replacement bootstrap approach picks random training examples from the entire training dataset to generate a decision tree). In the next step, each data point (gene) is assigned to a class (biological process) based on a majority vote across decision trees. Along with bagging, random forests can also find true biological attributes that are required to find the best split possible, thereby performing an automated attribute selection. Instead of the majority voting procedure for the classification task which involves voting based on predicted class across decision trees, the regression task involves averaging the value of each attribute across decision trees.
scATAC-sequencing: Like the traditional ATAC (assay for transposase-accessible chromatin with sequencing) sequencing, single cell ATAC sequencing (scATAC) uses transposase-mediated insertion of sequencing primers into open chromatin regions for capturing profiles of accessible chromatin regions at a single-cell resolution. These chromatin-accessible regions are indicative of active regulatory regions within the genome.
Support Vector Machines (SVMs): a subset of supervised ML methods commonly used for classification, regression, and outlier detection. When aiming to classify biological instances (genes) into classes (e.g., pro-angiogenic and anti-angiogenic) based on different attributes (e.g., gene expression across different single cells), SVMs attempt to generate an imaginary hyperplane that can divide data points (e.g., genes) into two (or multiple) groups/classes based on their attributes (e.g., gene expression in every single cell). When there are two attributes calculated for every data point, we have a two-dimensional (X-Y) plane, where each data point (or gene) is represented by the values of attributes X and Y (e.g., gene expression across the two single cells). In this 2-D space, a line can classify the data points into two groups. In a given n-dimensional space, the SVM procedure generates an n-1 dimensional hyperplane for classifying the data points. The distance between the hyperplane and the nearest data points from each class to the hyperplane (support vectors (SVs)) is called a margin. SVM iteratively generates multiple hyperplanes that can classify data points into two groups. Then, the classification aims at finding the hyperplane with maximum possible margin. Moreover, it is difficult to classify data points in many real-world scenarios using a linear hyperplane. Therefore, SVM typically exploits non-linear kernel functions (e.g., polynomial and radial basis kernels) to transform data inputs into a space with higher dimensions so that the data inputs become separable.
Tensor-based methods: Tensor methods (in the context of cell-cell communications) help to decompose a ligand-receptor co-expression matrix into multiple components to generate a hypergraph. A hypergraph is a special form of graph that can capture many-to-many ligand-receptor relationships instead of a standard graph which can only capture pairwise relationships. Tensor-based methods capture the many-to-many ligand-receptor relationships across single-cells or clusters of single-cell.
Trajectory Inference: determine the pattern of a dynamic process experienced by cells and then arrange cells based on their progression.
3. Mechanism discovery in single-cell datasets
High-throughput (single-cell) omics studies provide a snapshot of the changes in abundance of biomolecules (genes/proteins/metabolites) between biological samples, which directly result from synchronous changes occurring in various cellular processes. Specialized downstream analyses use this abundance of information to discover correlative or cause-effect relationships between different biomolecules to find underlying biological mechanisms.
3.1. Functional enrichment-based methods
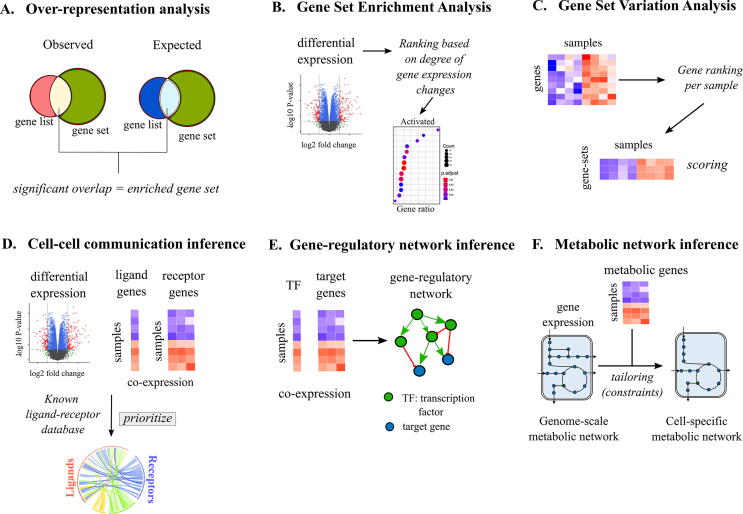
Most single-cell studies that characterized EC heterogeneity preferred a functional enrichment-based analysis to predict biological functions (Fig. 1). Enrichment-based methods typically assume that genes with similar expression changes across conditions should belong to similar functions. Over-representation analysis (ORA), gene set enrichment analysis (GSEA) and gene set variation analysis (GSVA) are the most commonly used methods to identify enriched processes in endothelial single-cell datasets (Table 1, Table 2, Fig. 2A). ORA identifies whether the overlap between the test gene list and a reference gene set is unlikely due to random chance (Fig. 2A) [47]. Online tools, such as g:Profiler [48], Panther [49] and Enrichr [50] perform ORA on a given list of genes. To overcome the assumption of ORA that all genes are equal regardless of their magnitude of differential expression, functional class scoring methods, like GSEA, rank genes based on the expression differences between control and case samples (or clusters) calculated by any differential metric (e. g. log-fold change, P-value, product of log-fold change sign and -log10(P-value), etc.) [47]. Subsequently, the association between members of a given gene set and the control-case phenotypes is measured by calculating an ‘enrichment score’ that uses the rank information of overlapping genes with a given gene set to score a biological process (Fig. 2B). Many tools like the ‘clusterProfiler’ R package [51], GenePattern [52] and the GSEA tool developed by the Broad Institute, implement gene set enrichment analysis. GSVA, on the other hand, performs an unsupervised estimation of pathway activity variation across samples by converting the log-normalized gene expression matrix (genes vs samples) into a GSVA score matrix (gene sets vs samples), where the GSVA score represents the overall activity of the gene set within a sample (Fig. 2C). GSVA is implemented in the GSVA package in R [53]. BIOMEX, a bioinformatics software suite developed for non-expert users, contains state-of-the-art implementations of these popular enrichment-based methods for multi-omics data interpretation [54].
Fig. 2.
Techniques for specialized mechanism discovery. The commonly used tools for mechanism predictions are based either on functional enrichment (A to C) or biological network inference (D to F). (A) Over-representation analysis (ORA): ORA compares the fraction of observed list of genes overlapping with known gene sets (observed) versus the fraction of total list of genes within an organism’s genome that overlaps with known gene sets (expected) to identify enriched gene sets. The overlaps are indicated by Venn diagrams. (B) Gene-set enrichment analysis (GSEA): GSEA ranks genes based on differential expression between control and case samples (indicated by red dots in the Volcano plot) and subsequently, uses the ranks of overlapping genes between the observed and expected cases to score the membership of a gene list to each of the known gene sets (shown as dot plot in the figure). The statistical significance of the enrichment score per gene set is calculated using permutation tests (Box 1). (C) Gene-set variation analysis: GSVA converts the log-normalized gene expression matrix (genes vs samples) into a GSVA score matrix (gene sets vs samples) by ranking genes per sample. (D) Cell-cell communication inference (CCI): CCI methods use the information of differentially expressing (indicated by the red dots in the Volcano plot) or co-expressing ligands and receptors (indicated by heatmap) and compare them with a database of known ligand-receptor interactions to prioritize potential ligand-receptor interactions in a given condition (indicated by a Circos plot connecting ligands to receptors). (E) Gene-regulatory network (GRN) inference: GRN inference methods use the information of transcription factor (TF) expression profile and expression profile of their downstream target genes (indicated by heatmap vectors) to find meaningful co-expressing pairs, which are represented as a network of TF-target interactions. (F) Metabolic network inference: Active, condition-specific metabolic networks are derived by using metabolic gene expression data (heatmap) as biochemical constraints for tailoring a generic genome-scale metabolic network of an organism. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Even though functional enrichment analyses provide a quick and easy overview into the biological processes that are associated with a list of genes, most analyses are affected by overlapping genes and variable distributions of differentially regulated genes in gene sets. Differing gene set sizes, sample sizes and an imbalanced number of samples per group may also impact the analyses [47]. Apart from these technical problems, there are also concerns in applying the above enrichment methods to single-cell sequencing data that may lead to false positives, which may occur due to the measured proportion of genes being lower or in situations where an overall gene count is imbalanced across conditions [55]. Therefore, caution is advised against solely using enrichment analyses to draw biological interpretations and conclusions. Biological network enrichment methods that also use the information of underlying biological mechanisms, can be an effective alternative to the above methods [47].
3.2. Biological network prediction-based methods
Prediction of active biological networks using transcript abundance information can complement functional enrichment analyses in identifying (associative or causal) biological interactions and hence, interpretations. Typically, cell-specific ligand-receptor interaction (cell–cell communication), active gene regulatory and metabolic networks can be predicted using single-cell transcript relative abundance estimated from angiogenic single-cell datasets (Table 1, Table 2, Fig. 2).
3.2.1. Cell-cell communication inference
Recent advances in single-cell and spatial omics have drastically increased the resolution at which we can study biological systems. These next-generation tools yield unprecedented opportunities to go beyond a mere description of cell types and states, allowing us to better study the dynamics of biological systems, an important aspect of which is defined by how cells interact with each other to establish tissue functioning. Since 2019, computational biology has witnessed a steep increase in the number of tools available to study several aspects of cell–cell communication (CCC). Early methods such as CCCExplorer [56] and CMN (community-wide molecular network) [57] were developed for bulk gene expression data. However, since the introduction of single-cell transcriptomics, the number of CCC modeling tools has drastically increased. Armingol et al. summarize the recent CCC literature and organize the methods into four categories, depending on their approach [58]. CCC methods use differential expression or co-expression information of different ligands and receptors across conditions to predict and prioritize ligand-receptor interactions (Fig. 2D). Differential combination-based methods such as CellTalker [59], iTALK [60] and PyMINEr [61] use differentially expressed ligands and receptors to identify interactions between clusters of cells. Expression permutation-based tools, such as CellChat [62], CellPhoneDB [63], Giotto [64], ICELLNET [65] and SingleCellSignalR [66] score each ligand-receptor pair, and subsequently perform filtering based on permutation tests (Box 1), non-parametric tests with a null model, or empirical methods. Network-based methods, such as CCCExplorer [56], NicheNet [67], SoptSC [68] and SpaOTsc [69], use networks of interactions between ligands, receptors and downstream targets to prioritize ligand-receptor interactions, some of them even taking into account spatial information, such as SpaOTsc. The fourth category of tensor-based methods (Box 1), exemplified by scTensor [70], generalizes the graph-based methods (Box 1) – which could be equivalently formulated as matrix-based methods – even further to a tensor-based setting.
While many tools have been developed, evaluation and benchmarking of all these tools to reveal their respective strengths and weaknesses is still in its infancy. Recently, Dimitrov et al. [71] performed a comparative study, revealing a large heterogeneity in the output of these methods, even though many of them use similar resources. This poses a formidable challenge to biologists who have to interpret the varying outcomes of these tools, requiring necessary biological follow-up and validation experiments.
3.2.2. Gene regulatory network inference
Inferring the dynamics of gene regulation is a powerful approach to understand how biological systems are controlled. Gene regulatory network (GRN) inference methods aim to infer how transcription factor combinations control downstream target genes. Historically, GRN inference methods were developed concurrently with large-scale gene expression profiling methods [72]. In this context, GRN inference methods typically infer gene regulatory networks, where edges between transcription factors and target genes are predicted from gene expression compendia (Fig. 2E). A landmark algorithm in this field has been the GENIE3 algorithm [73], which elegantly decomposes the network inference problem as a series of feature importance estimation problems. For every gene, a random forest model (Box 1) is built, which is subsequently used to perform feature (i.e. transcription factor) ranking, in this way identifying the most important transcription factors, based on whose expression profile the expression profile of the target gene can be predicted. Both in early benchmarks [72], as well as more recent ones [74], the GENIE3 algorithm has shown consistently good performance. Furthermore, it forms the basis of many subsequent developments, including dynamic versions of GENIE3 to infer dynamic GRNs from time series data [75] and single-cell GRN inference methods such as SCENIC [76].
However, expression data alone is not sufficient to accurately model gene regulation. Current approaches include other types of data such as epigenomics (e.g. scATAC-sequencing (Box 1)) and the presence of binding motifs to enhance GRN inference [77]. The advent of single-cell transcriptomics data has led to an explosion of new methods to infer GRNs, some of which focus more on cell type-specific GRNs, while others are more dedicated to inferring the dynamics of GRNs over time [78]. Several novel types of GRN inference can be distinguished here. Condition-specific methods (sometimes also referred to as differential network inference) refer to a class of methods that infer one network for each condition. Examples of such methods include case-specific random forests [79] and Bayesian Pólya trees (Box 1) [80]. Dynamic network inference methods use additional time series information (e.g. obtained by trajectory inference) to obtain a dynamic network, where edges might be present only in a specific time window. Examples of such approaches include AR1MA1 [81] and SCODE [82]. It can be expected that novel advances in single-cell sequencing technologies, such as high-throughput CRISPR/Cas perturbations, will significantly impact GRN inference methods, leading to better methods that will reconstruct gene regulation at a much higher resolution.
3.2.3. Single-cell metabolic network inference
As metabolic changes are challenging to observe at the single-cell transcriptome level, innovative techniques that post-process transcriptome abundance to predict genome-scale metabolic pathway states are instrumental. Genome-scale metabolic models (GEMs) are mathematical libraries of whole cell metabolism that can be easily tuned using extracellular metabolite uptake conditions and integrated with condition-specific biological ‘omics’ datasets, to predict optimal genome-scale metabolic routes required for fulfilling cellular demand [83]. Rohlenova et al. tailored a generic human genome-scale metabolic reconstruction by integrating bulk and single-cell transcriptomic profiles of proliferating choroidal ECs (CECs) and subsequently conducted a stepwise elimination procedure to systematically remove metabolic genes (reactions) with low or no expression (activity) and predicted a minimal constraint-based GEM for proliferating CECs [45]. This study was the first to integrate endothelial single-cell transcriptomic abundance with GEMs (Fig. 2F). Applying flux balance analysis (a method to estimate pseudo steady-state metabolic fluxes in a genome-scale metabolic network given a cellular objective function; e. g. biomass [83]) to this CEC-tailored GEM, core metabolic enzymes that play an essential role in maximal production of biomass and extracellular matrix collagen synthesis during choroidal neovascularization were predicted and these predictions were also validated experimentally. The integration of omics data with metabolic networks to predict condition-specific metabolism is challenging as different types of data (transcriptomics, proteomics, metabolomics) indirectly measure changes in either substrate or enzyme, representing different biological constraints that need to be tailored differently within GEMs [84].
Apart from the above application, methods that predict active metabolic networks across cell clusters by optimizing the agreement of flux distributions with single-cell expression distributions are slowly being applied to single-cell datasets [85]. An interesting study by Alghamdi et al. implemented scFEA, a novel graph neural network-based optimization solver that identifies cell groups sharing similar metabolic variations (correlated to the changes in single-cell transcriptome abundances) and validated their methodology on datasets with tissue-level targeted metabolomics profiling [86]. Tools like COBRA toolbox [87], COBRApy [88], and RAVEN toolbox [89] facilitate the construction of GEMs and seamless integration of omics data with metabolic models as constraints. Such applications can pave the way for the prediction of single-cell metabolic changes in ECs from transcriptomic abundance and thereby help understand cell-type or subtype-specific metabolic functions. Supplementary to these computational approaches, single-cell metabolomics technologies are slowly expanding to facilitate comprehensive validation of single-cell metabolic states [90], [91].
4. Multi-omics data fusion methods for single-cell datasets
In order to discover meaningful biological mechanisms, it is essential to sample information about different biomolecules (e.g., DNA, RNA, protein, metabolites) from a given tissue of interest. Single-cell omics technologies are rapidly expanding their scope to measure multiple modalities like the genome, transcriptome, epigenome, proteome and metabolome in both temporal and spatial scales for obtaining deeper insights and resolution into biological variations between cell types, phenotypes, markers and processes [92]. Developing technologies that simultaneously assay multiple omics layers has further advanced this inquiry. Although multi-omics single-cell fusion methodologies are already being applied to cancer biology, most of the single-cell studies in the field of angiogenesis (or EC) research either focus on generating / analyzing datasets belonging to a single modality (single omics data type such as transcriptomics data) (Box 1, Fig. 1) or simply comparing modalities by meta-analysis (e.g., proteome with single-cell transcriptome [29]) without systematic integration to identify common cell-clusters or relationships. ML techniques provide suitable frameworks for integrating multiple omics datasets, as they use the multi-dimensional information of genes and cells, which are inherently heterogeneous across biological scales. According to the availability of reference omics datasets with known cell annotations, multi-omics fusion methods can be classified into unsupervised (no prior knowledge of reference cell types), supervised (reference cell annotations from single-cell atlases), and semi-supervised (when cell annotations from samples are limited due to the usage of noisy data, erroneous annotations or the availability of label information only for a part of the data) methods (Table 1, Table 2).
4.1. Unsupervised omics data fusion
Unsupervised data fusion techniques are applied when no prior knowledge of reference cell types is available. This makes unsupervised fusion techniques most suited for data integration and discovery in single-cell omics datasets. Many statistical and mathematical approaches have been developed for unsupervised data fusion, depending on whether biomolecules from different compartments are profiled within the same cell (paired datasets) or from different cells and experiments (unpaired datasets). Unsupervised approaches aim to identify cell–cell or cluster–cluster correspondence across omics layers. Unsupervised fusion methods can be classified into multiple methods based on the underlying mathematical/statistical concept used for omics data integration.
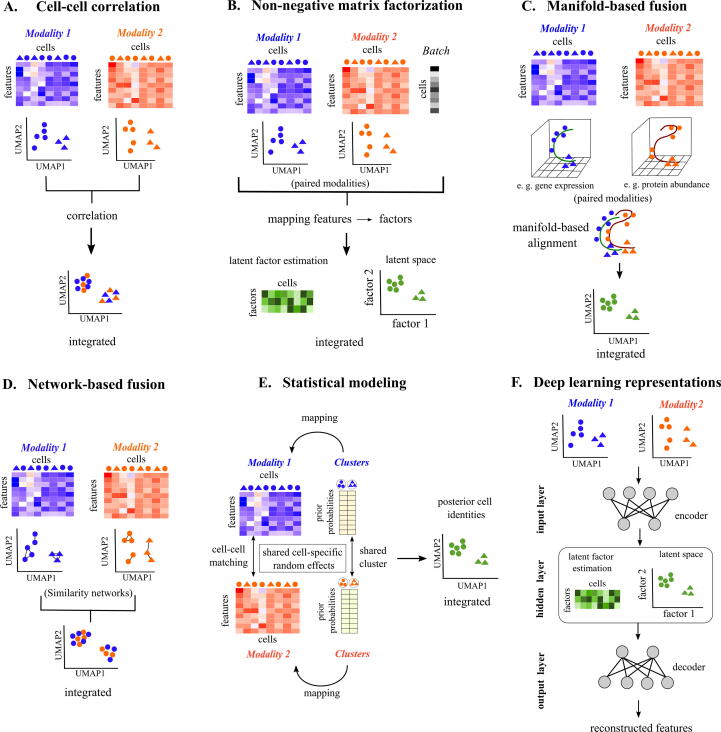
Joint Dimensionality reduction (jDR)-based methods are among the most popular single-cell multi-omics fusion methods. Cell-cell correlation-based methods like Seurat v3 [93] and bindSC [94] examine the linear association between different modalities/datasets to identify linear combinations that capture cell–cell correspondence across unpaired modalities (Table 1, Fig. 3A). Non-negative matrix factorization (NMF; Box 1)-based fusion methods like MOFA+ identify common clusters across modalities by assuming a pre-existing underlying relationship between cells [95]. As NMF methods assume that two different omics modalities (e. g. attributes from epigenome and transcriptome) are components of the same underlying biological signal, they identify a common latent space (Box 1, Fig. 3B) where there are conserved clusters of cells. NMF methods also correct for experimental batch effects as they can explicitly model experimental batches as a separate component of the underlying biological signal (Box 1, Fig. 2B). Manifold (Box 1)-based methods like MATCHER create low-dimensional representations (or manifolds) for paired modalities and align these manifolds in a shared space where the datasets become comparable (Fig. 3C) [96]. An important caveat of jDR approaches is that a specific modality can be given more weight (unless properly normalized) because of higher feature dimensions and scales than another modality (e.g., chromatin accessible regions in scATAC approaches vs transcript abundance from scRNA-seq).
Fig. 3.
Techniques for unsupervised fusion of single-cell multi-omics modalities. In all the figure panels, Modality 1 (red in color), Modality 2 (blue in color) represent two omics modalities. Heatmaps represent variation in feature across cells. Paired modality integrations are illustrated in green color, whereas unpaired modality integration are represented by mixture of blue and orange colors. Colored dots and triangles represent different types of cells. (A) Cell-cell correlation: Cells from modalities 1 and 2 are integrated by measuring correlation between the features from the two omics modalities. (B) Non-negative matrix factorization (NMF): NMF methods map features from two paired modalities and cell-level batch effects to latent factors (Box 1). The number of latent factors being less compared to original number of genes in the figure signifies dimensionality reduction. Cluster identities are assigned to common cells in this latent space (Box 1). (C) Manifold-based fusion: Manifold fusion methods map the input feature dimensions from modalities 1 and 2 to a low-dimensional manifold space (In the figure, 9 row-wise features are mapped to 3 dimensions). The manifolds (Box 1) generated for each paired modality are aligned with each other to identify common cells between modalities. (D) Network-based fusion: Similarity networks are generated for the unpaired modalities 1 and 2. Cells with similar feature profiles are connected to each other within this network. The conserved connections between the two networks are used for integration. (E) Statistical modeling: Statistical modeling methods identify shared clusters and common cells between paired modalities 1 and 2 by generating a probabilistic model (Box 1). As the same prior probability distribution is used for clusters in both modalities to tune the model, shared cell-specific random effects are captured, which are useful for finding posterior cell identities. (F) Deep learning representations: Deep learning for unsupervised omics integration is performed using autoencoders (Box 1), which contains an encoder-decoder scheme. In theory, any of the methods (A to E) can be combined in the hidden layer of the autoencoder scheme to predict cell clusters. Here, the NMF method is shown as an example. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Network-based fusion approaches like Seurat v4 [97] and CiteFuse [98] use similarity-based network models inferred from each modality to identify a common representation space (Fig. 3D). This similarity allows for identifying affinities between cells across unpaired or paired modalities. Network fusion approaches integrate datasets with the assumption that each modality discovers the same cell types, which might not be the case in all biological conditions. Statistical approaches like BREM-SC [99] and Clonealign [100] systematically integrate multi-omics data using a Bayesian framework for probabilistic modeling (Fig. 3E). Such methods model relationships between features across modalities. Although relatively simple to implement, these approaches only focus on statistical integration without considering biological variance in different contexts.
The aforementioned mathematical/statistical concepts can also be integrated with deep learning representations like autoencoders to identify non-linear relationships between features and modalities by transforming them into interpretable, common, low-dimensional subspaces. Typically, autoencoders have input, hidden, and output layers (Fig. 3F). The input layer is an encoder that transforms data from high-dimension to low-dimension cell states. The hidden middle layer stores the information about the low-dimensional space shared by different modalities, thus, performing integration and clustering. The output layer decodes the low-dimensional information at the hidden layer to reconstruct the input. Tools like totalVI [101] and GLUE [102] combine NMF and graph-based embedding with autoencoders to fuse multiple paired modalities. To acquire a comprehensive list of tools and techniques regarding unsupervised data fusion techniques, we suggest the readers refer to additional review articles [92], [103], [104], [105].
4.2. Supervised/semi-supervised omics data fusion
Unsupervised learning assumes that all observations are produced by a set of common, latent variables. In contrast, supervised learning assumes that one set of data, termed inputs, is the source of another set of observations, called outputs. Supervised learning finds a mapping function that translates the input data to the label information given the input data and output labels. Then the mapping function is applied to a set of input data without label information. Identifying the label of unseen data is called prediction. Depending on the output types of the problem of interest, this prediction can be seen as classification when the output information is discrete labels, regression when the output information is continuous labels, and prioritization when the output label is a ranking list of input data. Neural networks (NNs), Support Vector Machines (SVMs), and random forests are among the most popular and successful ML approaches in supervised learning. Similar to unsupervised settings, it has been shown that, in the context of supervised learning, integrating multiple complementary inputs (biological data) leads to more robust models and more accurate predictions for a biological problem of interest. Supervised approaches have been mainly applied to integrate several genomics data sets and sometimes incorporate multiple bulk transcriptomic datasets to predict a phenotype or function of interest. However, they are less prevalent in single-cell data fusion because of the limited availability of accurate annotations for genes and cells together. As a result, unsupervised-based methods dominate contemporary omics data integration in single-cell cancer research. While it is believed that unsupervised-based integration approaches also deliver an unbiased representation of fused omics, they sometimes fail to provide a stable and realistic picture of the underlying data. Recently, with the availability of more annotation and phenotypic information for genes, supervised and semi-supervised omics data fusion has slowly gained growing attention in cancer research. For example, Dietrich and colleagues integrate genomics, transcriptome, and DNA methylome data to understand the mechanisms of drug response to Chronic Lymphocytic Leukaemia [106]. Here, we focus on different strategies for integrating several omics data sets using various ML algorithms.
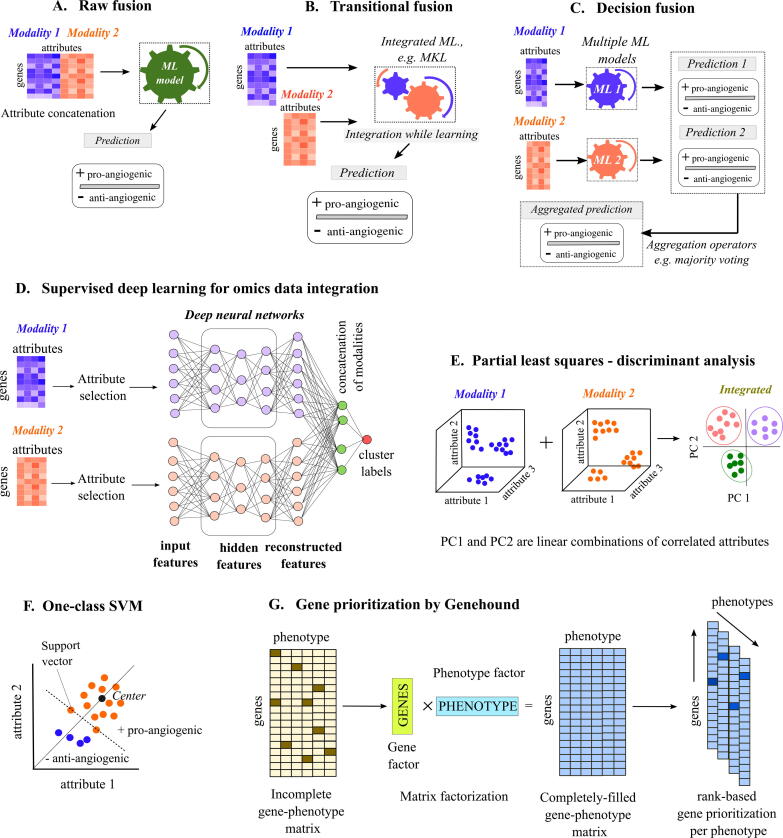
In a supervised manner, data fusion can be divided into three categories: raw fusion, transitional fusion, and decision fusion (Table 1, Fig. 4A–C). One of the most prevalent strategies for integrating biological data sources is raw fusion, also called early integration (Fig. 4A). The fusion of several data sources takes place at the raw data level (attribute concatenation). After that, the learning algorithm is applied to the concatenated data set, which yields a single result. Nonetheless, the heterogeneity of omics data sources makes this data fusion technique difficult. In transitional fusion, also called intermediate integration, different data sources are fused throughout the learning process (Fig. 4B). Transitional-based fusion approaches apply the same learning structure to each data source separately to address constraints and difficulties in coping with heterogeneous data. In several intermediate integration methods, such as those dedicated to Multiple Kernel Learning (MKL) (Box 1) [107], [108], [109], the parameter learning step is dependent on the learning structure level. In contrast, this step is independent of how the structure was constructed in some methods, such as Geometric Kernel Fusion (GFK) [110]. Individual structures are eventually integrated into one structure in both scenarios, resulting in a single outcome based on all data sources.
Fig. 4.
Techniques for ML-based supervised fusion of attributes from various data sources. To commonly explain multiple ML techniques, we use a representative example where the aim is to classify genes as pro-angiogenic (+ class) and anti-angiogenic (− class) based on different attributes measured from multiple data sources. (A) Raw fusion: A supervised fusion method that first concatenates attributes from data modalities 1 and 2 (blue and orange colors) and subsequently uses the concatenated dataset for machine learning and classification. (B) Transitional fusion: Here, a structure or pattern is generated for each modalities 1 and 2 separately but they are integrated while learning. The integrated structure is used for classification. (C) Decision fusion: Unlike transitional fusion, the data structures are generated independently for independent learning and only prediction outcomes of + and − class are fused based on majority voting. (D) Supervised deep learning for omics data integration: Deep neural networks (Box 1) are generated for each modality separately. Attributes for each modality are reconstructed an compared with input to evaluate learning performance. The reconstructed features from each omics modality are concatenated finally providing information of cluster labels. (E) Partial least squares-discriminant analysis (PLS-DA): PLS-DA integrates the different attributes from two modalities (blue and orange colors) into PC1 and PC2 and learns the cluster information during integration, and, hence, is an example of intermediate integration. Each PLS-DA component (PC1, PC2) represents a linear combination of correlated attributes from each data source. (F) One-class support vector machine (one-class SVM): Unlike binary SVM (Box 1), in a one-class SVM, different sets of data points are classified into high (large number of points with orange color) or low density regions (low number of points with blue color). The support vectors are then chosen from the high density region depending upon the distance from the center of the high density region to form a hyperplane that is farther from the origin. Based on the labelled information from + pro-angiogenic class, it can predict genes that belong to the - anti-angiogenic class. (G) Gene prioritization by Genehound: Genehound employs a gene prioritization strategy that transforms a gene by phenotype matrix into a completely-filled gene by phenotype matrix using matrix factorization to decompose the gene (green box) and phenotype information (cyan box) as latent factors (Box 1). This completely-filled matrix is used to prioritize genes based on ranking for each phenotype. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A separate model is learned for each data source in decision fusion, also called late integration (Fig. 4C). Each data source might be subjected to different ML algorithms in the decision fusion scheme, and data integration occurs at the decision level. Then, various computational methods are used to combine and aggregate the results. This data fusion method successfully merges the results acquired from several learning algorithms, especially when each data source has a different underlying data structure, requiring distinct learning methods for each data source. In a supervised learning manner, this type of integration is often regarded as a natural way to deal with heterogeneous biological data. This type of fusion can also use a single source of data or a limited number of data sources to boost the learning algorithm's performance. For example, ensemble-based approaches (Box 1) employ several learning algorithms to achieve higher predictive performance than any individual learning algorithm could.
Kernel-based methods are one of the most adaptable and successful ML algorithms for developing appropriate data fusion integration frameworks at all levels of data realization. They are particularly well suited to intermediate integration [111]. In particular, by representing the data as a kernel matrix, kernel approaches detach the original data from the ML algorithms, making them available and more manageable for various data integration strategies. Also, deep learning through various deep algorithms and different architectures successfully exploits the different structures in multiple omics data types and offers a practical and scalable framework for data fusion at all levels of data realization [112] (Fig. 4D). Data fusion methods also provide a flexible framework for combining supervised and unsupervised learning to deliver more accurate single-cell RNA-seq clustering and annotation. For example, scAnCluster [113] offers an end-to-end cell deep-supervised clustering and annotation model that exploits cell type labels accessible from reference data to assist cell clustering and annotation on unlabeled target data.
While principal component analysis (PCA) achieves dimensionality reduction in an unsupervised manner, Partial Least Squares Discriminant Analysis (PLS-DA) reduces dimensionality while remaining fully aware of the class labels and can be used for classification purposes. PLS-DA has recently gained increasing attention for multi-omics integration because of its efficiency in dealing with data with high dimensional attributes and missing or noisy data [114]. In particular, MixOmics [115] formulates and implements several algorithms for integrating multi-omics using PLS-DA. It can be considered an intermediate data fusion approach through which the most informative attributes from different omics are chosen with the constraint of correlation between their first PLS-DA components (Fig. 4E). In particular, MINT [116] presents integration across samples, akin to batch effect correction, while DIABLO [117] performs data integration across omics attributes, which are two of the most popular MixOmics approaches.
As an extension, it is also possible to adapt such supervised approaches of (multi-)omics data integration for gene prioritization tasks. The following section will focus on supervised gene prioritization and discuss the possible advantages of combining multiple heterogeneous omics in the gene prioritization task using various data fusion strategies.
5. Gene prioritization methods for target identification
Identifying disease-associated genes is critical to understand the disease phenotype. The current surge in high-throughput omics methodologies has provided access to a vast array of information that can help explore candidate genes for a biological process of interest in pathophysiological angiogenesis. Thousands of candidate genes can potentially underlie a complex biological process, like vessel sprouting. Experimentally confirming the roles of all these potential genes is impractical, since it is a time-consuming procedure with costly wet-lab tests to evaluate which of those candidates is truly promising. Hence, it is essential to perform a prioritization step before testing the genes for their roles experimentally. The gene prioritization task entails identification of biologically relevant genes from a wide list of potential genes for subsequent examination and study. While candidate gene prioritization seems to be an intelligent strategy, it is challenging due to the noisy nature of omics data, our limited knowledge of the phenotypic roles of genes, their manifestations in different pathological conditions, and their relationships with other genes.
Prioritizing candidate genes using ML techniques allow formal integration of heterogeneous attributes and samples (instances) for classification or regression. This provides a much more efficient solution by evaluating only the most promising genes, rather than all candidate genes. Although ML methods are routinely used in prioritizing genes in various fields [118], [119], [120], [121], to the best of our knowledge, they have never been applied to prioritizing genes in the context of angiogenesis. ML methods rely on a suitable training dataset (set of seed genes and biological samples) as most of these techniques exploit the “guilt-by-association” principle for setting up a prioritization model. Typically, prior knowledge of positive and negative training classes is required to train most supervised and semi-supervised ML methods, such as Support Vector Machines, Deep Neural Networks, random forests, etc., and then test the models using cross-validation strategies. For example, if the aim is to prioritize genes essential for growth, it is imperative to design a prior set of essential and non-essential genes with measured attributes, while model training, and test it on a new set of genes for which the role in survival is unknown.
5.1. Single-class ML methods
In gene prioritization, we can produce a list of, for example, cell-specific or function-specific genes as positive training genes using biological annotation-based or literature-based data sources. However, choosing negative training genes for a cell type of interest is more complicated and requires focused experimental scrutiny. In fact, our current biological knowledge does not allow us to produce a consensus theoretical ground for determining the actual set of cell types or functions in which a gene is involved. This observation led researchers to focus on ML algorithms designed to learn from only positive data, such as one-class SVM [122]. The one-class SVM strategy transforms the typical binary classification problem into a one-class learning problem by modeling regions using a function that classifies regions with higher density of points (typically genes with known biological functions) as the positive class and the lower density of points as the negative class (Fig. 4F). This approach works under the assumption that genes with similar biological functions will have similar attributes. Then, the decision values of the one-class SVM models are employed to rank genes, i.e., genes are prioritized based on their importance in defining cell types or functions. A study by Yu et al. [123] uses the one-class supervised SVM approach for prioritizing disease-candidate genes based on text mining from various biomedical databases.
5.2. Semi-supervised ML-based approaches
Alternatively, the gene prioritization task is tracked by learning from both positive (P) and unlabeled (U) data, also called the PU learning approach. Mordelet and Vert [120] use the bagging approach (Box 1) to randomly sample genes from the unknown class and treat them as negative. Another approach, proposed by Fusilier and colleagues [124], first treats all unknown data (genes) as negatives and trains a classifier for positive (seed genes) vs unknown (genes). Then, the model iteratively reduces the negative data set from within the unknown data (genes) by focusing on the most dissimilar genes to the seed genes. Wenric and Shemirani [125] extended the PU learning framework using a random forest classifier to rank genes in a case-control RNA-Seq experiment. Similarly, GuiltyTargets [126] uses PU learning for training a logistic regression model on a protein–protein interaction network annotated with disease-specific differential gene expression. N2A-SVM [127] employs SVM and PU learning to prioritize Parkinson-associated genes, profiled from an autoencoder-based low dimension representation of protein–protein interaction networks—obtained via node2vec [128]. DeepPVP [129] uses deep neural networks and automated inference to detect potential causal variations in whole exome or whole-genome sequence data. Although simplistic, this method has its limitations. A typical simplification adopted in PU learning is dealing with the unlabeled set as negative and assessing the model as if it were fully supervised. In particular, when the available positives (training seeds) are not a representative subset of all positives, including known and unknown positives, they are not an unbiased or random sample. Moreover, considering unlabeled data as negative could introduce false negatives into the model’s training process. These issues are exacerbated when the amount of positive training data is limited, and hence a method that penalizes false negatives needs to be developed.
5.3. ML-based gene prioritization using multi-omics data
Traditionally, a heuristic-based integrated analysis (Box 1) is a straightforward and commonly used approach to prioritize genes. For example, a study used a heuristic integrated analysis based on combining single-cell RNA sequencing with orthogonal datasets from other studies for prioritizing metabolic targets that affect vessel sprouting in choroidal ECs [45]. Even though the strategy was able to identify important targets that could be experimentally validated, such heuristic analyses are not flexible enough to be generalized for a new set of seed genes, as there is no systematic integration to capture correlations amongst multiple omics layers. Although less available in the context of angiogenesis, systematic integration of multi-omics datasets for gene prioritization tasks has been growing steadily in other contexts (Table 1, Table 2). Endeavor [118] combines similarity-based ML models for prioritizing disease-candidate genes in each omics data separately and provides a global ranking by combining the ranking of the genes in each modality using order statistics. Similarly, eXtasy [130] uses a random forest classifier to rank non-synonymous single nucleotide variants given a specific biological disease phenotype. Likewise, a graph-based approach was used to construct an integrated network, combining gene regulatory, protein–protein interaction, text mining, and co-expression data to prioritize growth regulators in Arabidopsis thaliana. Subsequently, supervised ML methods were used to show that the local topological properties of the integrated network improve gene prioritization [131].
Kernel-based strategies are among the most robust techniques to integrate multi-omics data at different levels of data realization. In particular, kernel fusion-based SVM can exploit different prioritization strategies, such as one-class classification [123], [132], [133] and PU learning [120]. For example, De Bie and colleagues [132] introduced the first kernel-based multiple-omics data fusion approach for gene prioritization. After transforming all omics into kernels using a Radial Basis Function (RBF), they proposed an MKL (i.e., learning the weights of each omics-associated kernel in the fused kernel) formulation for one-class SVM to prioritize disease-associated genes. Subsequently, to handle noise from different omics data sources, another study introduced a kernel fusion-based gene prioritization approach using geometrically-inspired kernel integration that captures the complementary nature between multiple omics modalities [133]. Furthermore, a gene prioritization strategy for the prediction of human phenotype ontology (HPO) terms using late-integration operators (e.g., ordered weighted averaging), to combine several annotation-based omics data sources, was also proposed [134]. While most of the prioritization tools, as mentioned earlier, model each trait separately, Genehound [121] uses a multi-task approach to prioritize genes. Genehound formulates the gene prioritization task as the factorization of an incompletely filled gene-phenotype matrix to impute the unknown values to identify common patterns across various phenotypes (Fig. 4G). Then, to deliver a more accurate prediction, it incorporates phenotypic side data and multiple genomic side data simultaneously into the process of factorization.
6. Exemplary computational pipeline for the prediction of promising anti-angiogenic targets
Although the above techniques from sections 3 – 5 can be used individually for specific applications, we propose the unification of these different techniques into a conceptual workflow that can optimize the discovery of novel anti-angiogenic targets (Fig. 5). First, single-cell omics datasets (publicly available or in-house newly generated) belonging to different modalities can be merged into a unified dataset. Performing quality control (e.g. elimination of low quality cells, features, doublets, etc.) and subsequent normalization, a feature selection for highly variable features and dimensionality reduction, needs to be performed. These transformed datasets can be fused using single (datasets belonging to the same omics datatype) or cross-modality (datasets belonging to different omics datatypes) fusion techniques, depending on the research question. Cross-modality fusion can be performed depending upon the kind of modalities (whether they are paired or unpaired). For paired modalities, techniques like NMF, manifold or statistical fusion can be used. For unpaired modalities, techniques like network-based fusion can be used. The fused datasets (either from single or cross-modality fusion) can be used for unsupervised clustering and cell-type annotations. If the clustering does not represent biologically relevant clusters, steps from feature selection and dimensionality reduction need to be repeated. In order to take this decision, clusters can be visualized using the t-SNE/UMAP cluster plots. Once the fusion is successful to capture biologically relevant clusters, features between different clusters or conditions can be compared using differential feature analysis techniques. The differential features between clusters or conditions can be visualized using heatmaps and volcano plots. These differential features can be used for functional enrichment and/or network discovery techniques. Enriched processes can be visualized using dot plots and tree plots. Predicted biological networks can be visualized using various network layouts. The normalized data, functional enrichment scores (e.g. GSVA scores) and connectivity metrics of different genes/proteins within the discovered networks can be used as processed features and fused into a (unsupervised/supervised) machine learning fusion strategy for gene prioritization. The prioritized targets thus identified, can be used for experimental validation.
Fig. 5.
A potential pipeline for discovering novel anti-angiogenic targets from single-cell multi-omics datasets. This pipeline showcases a potential workflow that can seamlessly integrate the discussed techniques for anti-angiogenic target discovery. The olive green boxes represent the data fusion and knowledge discovery techniques. The light blue boxes highlight the use of unsupervised and supervised data fusion techniques for integrating heterogeneous data sources. The green box highlights the target predicted from this workflow and its subsequent follow-up with experimental validation. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
7. Future directions
As enlisted above, a plethora of computational tools is available for the integration of multi-omics datasets, prioritization of important genes and mechanism discovery. Multi-omics data integration is already being applied in cancer biology for prognosis, biomarker identification, anti-cancer drug response, identifying mechanisms and survival predictions [135]. Multi-omics integration methods successfully identified biological mechanisms specific to patients affected by renal cell carcinoma, glioblastoma and lung adenocarcinoma [136], [137], [138]. Moreover, recent applications of novel deep-learning technologies are helping to stratify patients suffering from lung adenocarcinoma, neuroblastoma, breast cancer, and bladder cancer into different cancer subtypes [139], [140], [141], [142], [143], [144], [145]. Thus, computational multi-omics approaches have tremendous potential to provide insights into precision treatments, drug resistance and relapse treatment. However, these techniques are (to date) seldom applied in the context of angiogenesis research.
Angiogenesis is a complex biological process, involving multiple signals at different levels, including secreted angiogenic signals, inter- and intracellular signals, environmental cues, cell-intrinsic signals, and others, which can all interact with each other. Mapping and uncovering novel multilevel attributes of pathophysiological angiogenesis from multi-omics data can greatly advance our ability to probe into and interpret these complex signals by elucidating functional cellular networks. As more mechanistic details are incorporated into complex systems biology models, computational methods in large-scale models should be incorporated into existing single-cell datasets to assist in angiogenic target discovery [45]. To be able to apply such computational tools in routine angiogenesis research, user-friendly frameworks, benchmarking studies that compare these tools in different biological scenarios and biologically intuitive visualizations of high dimensional data are necessary. User-friendly intuitive analytical and visualization tools (like BIOMEX and EndoDB [46], [54]) and integration frameworks (Endeavour [146], PriorityIndex [147], TargetMine [148]) are already being applied to high-throughput bulk datasets in general disease biology. Similar software workflows that can include automated prioritization of targets, mechanism discovery and multi-omics integration which will formidably benefit this cause. Introducing the computational tools enlisted in Table 1 within formal workflows (similar to our proposed strategy (Fig. 5)), can help to interpret, analyze and implement omics angiogenesis data.
ML methodologies are contributing to many promising biomedical discoveries [149]. Although applied for morphological blood-vessel image analysis in certain cases, there is a severe lack of ML applications on high-throughput molecular datasets of angiogenesis. This is surprising given the surge of endothelial omics datasets/atlases at the bulk and single-cell scales of cells, organs and tissues. Therefore, sufficient emphasis must be given to the development of novel ML approaches that can flexibly integrate high-throughput data systematically generated from different experimental platforms for the prediction of novel genes, biological processes and their association with EC types. In addition, multiple challenges like sparsity in single-cell data, missing data during identification of variation, batch correction, reference annotation of cell types and reference annotation of biological processes might affect target predictions and should be considered / corrected for. Hence, sufficient benchmarking studies that compare the performance of tools concerning the above scenarios on both synthetic and real-world (angiogenesis) datasets need to be developed. Furthermore, the integration of existing single-cell datasets with prior knowledge of biological networks (gene-regulatory, metabolic and protein–protein interactions), drug-protein interactions, protein structural information, disease-specific mutations, disease/gene ontologies and vessel morphology based on image data will immensely assist anti-angiogenic target discovery.
In order to obtain a higher success rate in the prediction of suitable targets, the quality of the chosen single-cell datasets is paramount. The availability of gold-standard “ground-truth” datasets with non-subjective cell-type/gene-level/process-level annotations for testing and comparing tools will help in this regard [150], [151]. Also, distinct label information that characterizes the identity of genes to different classes of annotations is vital for supervised ML approaches associated with various purposes. Therefore, such gold-standard datasets need to provide metadata with curated, cell-level and gene/process-level annotations. For generating gold-standard single-cell annotations, it is essential to integrate various available single-cell atlases and to generate a database of integrated omics datasets containing curated cell-level annotations with the option of user-friendly rectification of cell-type annotations.
Most importantly, predictions from these diverse computational methodologies need to be backed up with experimental validations of the roles of prioritized genes/biological processes. Experimental assays that capture the changes in abundance of the biomolecules monitor detrimental effects of target inhibition in different biological conditions both in vitro and in vivo. Validation of biological roles by quantitative measurements of morphological, physiological, molecular changes and therapeutic effects of drugs on normalization of dysfunctional vessels are all required to meet this challenge.
Single-cell RNA-sequencing studies of ECs revealed the presence of novel EC subtypes, such as immunomodulatory ECs (IMECs) [29], [152], which might play a more important role in anti-cancer immunity than previously realized. In fact, several tumor ECs have an immunosuppressive gene signature [152], [153], yet up to nearly a third of the human coding genes lack any solid functional annotation and are only minimally described in the literature [154]. It remains to be explored whether “smart” computational techniques can be developed to demystify the mystery genome expressed in IMECs and gene prioritization methods can be designed to rank genes important for IMECs’ role in anti-cancer immunity.
We envision that generating appropriate ground-truth datasets, multiple levels of information, systematic integration of this information into flexible computational (ML) workflows, sufficient benchmarking and experimental validations will help develop hybrid computational-experimental pipelines that will ultimately provide targeted solutions to diseases/disorders involving severe vascular dysfunction. We anticipate that the use of integrative ML frameworks for identifying novel targets and the therapeutic effects on specific EC subtypes will help to decipher novel biological roles of endothelial cells (like immune function) other than their conventional role in vessel formation.
CRediT authorship contribution statement
Abhishek Subramanian: Methodology, Formal analysis, Investigation, Data curation, Writing – original draft, Visualization, Project administration. Pooya Zakeri: Writing – original draft, Visualization. Mira Mousa: Data curation, Writing – original draft, Visualization. Halima Alnaqbi: Data curation, Visualization. Fatima Yousif Alshamsi: Data curation, Writing – original draft, Visualization. Leo Bettoni: Data curation. Ernesto Damiani: Supervision. Habiba Alsafar: Supervision. Yvan Saeys: Writing – original draft. Peter Carmeliet: Conceptualization, Writing – original draft, Supervision, Project administration, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The figures in the graphical abstract and Fig. 2 were created using BioRender.com. P.C. is supported by Grants from Methusalem funding (Flemish government), the Fund for Scientific Research-Flanders (FWO-Vlaanderen), the European Research Council (ERC) Advanced Research Grant EU-ERC74307 and the NNF Laureate Research Grant from Novo Nordisk Foundation (Denmark). This work was funded in part by Khalifa University as part of the Khalifa University and VIB-KU Leuven Biomedical Science Discovery Program.
Footnotes
Supplementary Table 1 contains a list of single cell sequencing studies that characterized endothelial cell heterogeneity at a cellular subtype level, along with information of specific computational techniques used in each study. Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2022.09.019.
Contributor Information
Yvan Saeys, Email: yvan.saeys@ugent.be.
Peter Carmeliet, Email: peter.carmeliet@kuleuven.be.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Carmeliet P. Angiogenesis in health and disease. Nat Med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- 2.Lopes-Coelho F., Martins F., Pereira S.A., Serpa J. Anti-angiogenic therapy: current challenges and future perspectives. Int J Mol Sci. 2021;22 doi: 10.3390/ijms22073765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lupo G., Caporarello N., Olivieri M., Cristaldi M., Motta C., Bramanti V., et al. Anti-angiogenic therapy in cancer: downsides and new pivots for precision medicine. Front Pharmacol. 2016;7:519. doi: 10.3389/fphar.2016.00519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze A v, Mikheenko A, et al. The complete sequence of a human genome. Science 2022;376:44–53. 10.1126/science.abj6987. [DOI] [PMC free article] [PubMed]
- 5.Kashima Y., Sakamoto Y., Kaneko K., Seki M., Suzuki Y., Suzuki A. Single-cell sequencing techniques from individual to multiomics analyses. Exp Mol Med. 2020;52:1419–1427. doi: 10.1038/s12276-020-00499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peirce S.M. Computational and mathematical modeling of angiogenesis. Microcirculation. 2008;15:739–751. doi: 10.1080/10739680802220331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Y., Wang H., Oliveira R.H.M., Zhao C., Popel A.S. Systems biology of angiogenesis signaling: Computational models and omics. WIREs Mech Dis. 2021 doi: 10.1002/wsbm.1550. e1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vilanova G., Colominas I., Gomez H. A mathematical model of tumour angiogenesis: growth, regression and regrowth. J R Soc Interface. 2017;14 doi: 10.1098/rsif.2016.0918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarischi-Sousa R., Monteiro J.S., Alecrim L.C., Michaloski J.S., Cardeal L.B., Ferreira E.N., et al. A transcriptome-based signature of pathological angiogenesis predicts breast cancer patient survival. PLoS Genet. 2019;15:e1008482. doi: 10.1371/journal.pgen.1008482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aird W.C. Phenotypic heterogeneity of the endothelium: I. Structure, function, and mechanisms. Circ Res. 2007;100:158–173. doi: 10.1161/01.RES.0000255691.76142.4a. [DOI] [PubMed] [Google Scholar]
- 11.Becker L.M., Chen S.-H., Rodor J., de Rooij L.P.M.H., Baker A.H., Carmeliet P. Deciphering endothelial heterogeneity in health and disease at single cell resolution: progress and perspectives. Cardiovasc Res. 2022 doi: 10.1093/cvr/cvac018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nolan D.J., Ginsberg M., Israely E., Palikuqi B., Poulos M.G., James D., et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. 2013;26:204–219. doi: 10.1016/j.devcel.2013.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalucka J., de Rooij L.P.M.H., Goveia J., Rohlenova K., Dumas S.J., Meta E., et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. 2020;180:764–779.e20. doi: 10.1016/j.cell.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 14.Aizarani N., Saviano A., Sagar M.L., Durand S., Herman J.S., et al. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tucker N.R., Chaffin M., Fleming S.J., Hall A.W., Parsons V.A., Bedi K.C.J., et al. Transcriptional and cellular diversity of the human heart. Circulation. 2020;142:466–482. doi: 10.1161/CIRCULATIONAHA.119.045401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vanlandewijck M., He L., Mäe M.A., Andrae J., Ando K., del Gaudio F., et al. A molecular atlas of cell types and zonation in the brain vasculature. Nature. 2018;554:475–480. doi: 10.1038/nature25739. [DOI] [PubMed] [Google Scholar]
- 17.Schupp J.C., Adams T.S., Cosme C.J., Raredon M.S.B., Yuan Y., Omote N., et al. Integrated single-cell atlas of endothelial cells of the human lung. Circulation. 2021;144:286–302. doi: 10.1161/CIRCULATIONAHA.120.052318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dumas S.J., Meta E., Borri M., Goveia J., Rohlenova K., Conchinha N., et al. Single-cell RNA sequencing reveals renal endothelium heterogeneity and metabolic adaptation to water deprivation. J Am Soc Nephrol. 2020;31:118–138. doi: 10.1681/ASN.2019080832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo F.-H., Guan Y.-N., Guo J.-J., Zhang L.-J., Qiu J.-J., Ji Y., et al. Single-cell transcriptome analysis reveals embryonic endothelial heterogeneity at spatiotemporal level and multifunctions of microRNA-126 in mice. Arterioscler Thromb Vasc Biol. 2022;42:326–342. doi: 10.1161/ATVBAHA.121.317093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chestnut B., Casie Chetty S., Koenig A.L., Sumanas S. Single-cell transcriptomic analysis identifies the conversion of zebrafish Etv2-deficient vascular progenitors into skeletal muscle. Nat Commun. 2020;11:2796. doi: 10.1038/s41467-020-16515-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McCracken I.R., Taylor R.S., Kok F.O., de la Cuesta F., Dobie R., Henderson B.E.P., et al. Transcriptional dynamics of pluripotent stem cell-derived endothelial cell differentiation revealed by single-cell RNA sequencing. Eur Heart J. 2020;41:1024–1036. doi: 10.1093/eurheartj/ehz351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hou S., Li Z., Dong J., Gao Y., Chang Z., Ding X., et al. Heterogeneity in endothelial cells and widespread venous arterialization during early vascular development in mammals. Cell Res. 2022;32:333–348. doi: 10.1038/s41422-022-00615-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibarra-Soria X., Jawaid W., Pijuan-Sala B., Ladopoulos V., Scialdone A., Jörg D.J., et al. Defining murine organogenesis at single-cell resolution reveals a role for the leukotriene pathway in regulating blood progenitor formation. Nat Cell Biol. 2018;20:127–134. doi: 10.1038/s41556-017-0013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abe Y., Sakata-Yanagimoto M., Fujisawa M., Miyoshi H., Suehara Y., Hattori K., et al. A single-cell atlas of non-haematopoietic cells in human lymph nodes and lymphoma reveals a landscape of stromal remodelling. Nat Cell Biol. 2022;24:565–578. doi: 10.1038/s41556-022-00866-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie Y., He L., Lugano R., Zhang Y., Cao H., He Q., et al. Key molecular alterations in endothelial cells in human glioblastoma uncovered through single-cell RNA sequencing. JCI. Insight. 2021;6 doi: 10.1172/jci.insight.150861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sun Z, Wang C-Y, Lawson DA, Kwek S, Velozo HG, Owyong M, et al. Single-cell RNA sequencing reveals gene expression signatures of breast cancer-associated endothelial cells. Oncotarget 2018;9:10945–61. 10.18632/oncotarget.23760. [DOI] [PMC free article] [PubMed]
- 27.Massalha H, Bahar Halpern K, Abu-Gazala S, Jana T, Massasa EE, Moor AE, et al. A single cell atlas of the human liver tumor microenvironment. Mol Syst Biol 2020;16:e9682. 10.15252/msb.20209682. [DOI] [PMC free article] [PubMed]
- 28.Thomann S., Weiler S.M.E., Marquard S., Rose F., Ball C.R., Tóth M., et al. YAP orchestrates heterotypic endothelial cell communication via HGF/c-MET signaling in liver tumorigenesis. Cancer Res. 2020;80:5502–5514. doi: 10.1158/0008-5472.CAN-20-0242. [DOI] [PubMed] [Google Scholar]
- 29.Goveia J., Rohlenova K., Taverna F., Treps L., Conradi L.-C., Pircher A., et al. An integrated gene expression landscape profiling approach to identify lung tumor endothelial cell heterogeneity and angiogenic candidates. Cancer Cell. 2020;37:21–36.e13. doi: 10.1016/j.ccell.2019.12.001. [DOI] [PubMed] [Google Scholar]
- 30.Wu F., Fan J., He Y., Xiong A., Yu J., Li Y., et al. Single-cell profiling of tumor heterogeneity and the microenvironment in advanced non-small cell lung cancer. Nat Commun. 2021;12:2540. doi: 10.1038/s41467-021-22801-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li C., Guo L., Li S., Hua K. Single-cell transcriptomics reveals the landscape of intra-tumoral heterogeneity and transcriptional activities of ECs in CC. Mol Ther Nucleic Acids. 2021;24:682–694. doi: 10.1016/j.omtn.2021.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei Z., Feng M., Wu Z., Shen S., Zhu D. Bcl9 depletion modulates endothelial cell in tumor immune microenvironment in colorectal cancer tumor. Front Oncol. 2020;10 doi: 10.3389/fonc.2020.603702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Li Z., Skrzypczynska K.M., Fang Q., Zhang W., O’Brien S.A., et al. Single-cell analyses inform mechanisms of myeloid-targeted therapies in colon cancer. Cell. 2020;181:442–459.e29. doi: 10.1016/j.cell.2020.03.048. [DOI] [PubMed] [Google Scholar]
- 34.Qian J., Olbrecht S., Boeckx B., Vos H., Laoui D., Etlioglu E., et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30:745–762. doi: 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlesinger Y., Yosefov-Levi O., Kolodkin-Gal D., Granit R.Z., Peters L., Kalifa R., et al. Single-cell transcriptomes of pancreatic preinvasive lesions and cancer reveal acinar metaplastic cells’ heterogeneity. Nat Commun. 2020;11:4516. doi: 10.1038/s41467-020-18207-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yin H., Guo R., Zhang H., Liu S., Gong Y., Yuan Y. A dynamic transcriptome map of different tissue microenvironment cells identified during gastric cancer development using single-cell RNA sequencing. Front Immunol. 2021;12 doi: 10.3389/fimmu.2021.728169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Narayanan SP, Mannan R, Raskind G, Wang X, Vats P, et al. Single-cell analyses of renal cell cancers reveal insights into tumor microenvironment, cell of origin, and therapy response. Proc Natl Acad Sci U S A 2021;118. 10.1073/pnas.2103240118. [DOI] [PMC free article] [PubMed]
- 38.Su C., Lv Y., Lu W., Yu Z., Ye Y., Guo B., et al. Single-cell RNA sequencing in multiple pathologic types of renal cell carcinoma revealed novel potential tumor-specific markers. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.719564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y., Zhang Q., Bi H., Lu M., Tan Y., Zou D., et al. Decoding the multicellular ecosystem of vena caval tumor thrombus in clear cell renal cell carcinoma by single-cell RNA sequencing. Genome Biol. 2022;23:87. doi: 10.1186/s13059-022-02651-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lambrechts D., Wauters E., Boeckx B., Aibar S., Nittner D., Burton O., et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24:1277–1289. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 41.Winkler E.A., Kim C.N., Ross J.M., Garcia J.H., Gil E., Oh I., et al. A single-cell atlas of the normal and malformed human brain vasculature. Science. 2022;375 doi: 10.1126/science.abi7377. eabi7377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adams T.S., Schupp J.C., Poli S., Ayaub E.A., Neumark N., Ahangari F., et al. Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv. 2020;6 doi: 10.1126/sciadv.aba1983. eaba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Bock K., Georgiadou M., Carmeliet P. Role of endothelial cell metabolism in vessel sprouting. Cell Metab. 2013;18:634–647. doi: 10.1016/j.cmet.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 44.Carmeliet P., Jain R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rohlenova K., Goveia J., García-Caballero M., Subramanian A., Kalucka J., Treps L., et al. Single-cell RNA sequencing maps endothelial metabolic plasticity in pathological angiogenesis. Cell Metab. 2020;31:862–877.e14. doi: 10.1016/j.cmet.2020.03.009. [DOI] [PubMed] [Google Scholar]
- 46.Khan S., Taverna F., Rohlenova K., Treps L., Geldhof V., de Rooij L., et al. EndoDB: a database of endothelial cell transcriptomics data. Nucleic Acids Res. 2019;47:D736–D744. doi: 10.1093/nar/gky997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maleki F., Ovens K., Hogan D.J., Kusalik A.J. Gene set analysis: challenges, opportunities, and future research. Front Genet. 2020;11:654. doi: 10.3389/fgene.2020.00654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mi H., Thomas P. PANTHER pathway: an ontology-based pathway database coupled with data analysis tools. Methods Mol Biol. 2009;563:123–140. doi: 10.1007/978-1-60761-175-2_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuleshov M v, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res 2016;44:W90-7. 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed]
- 51.Wu T, Hu E, Xu S, Chen M, Guo P, Dai Z, et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Cambridge (Mass)) 2021;2:100141. 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed]
- 52.Kuehn H, Liberzon A, Reich M, Mesirov JP. Using GenePattern for gene expression analysis. Curr Protoc Bioinformatics 2008;Chapter 7:Unit 7.12. 10.1002/0471250953.bi0712s22. [DOI] [PMC free article] [PubMed]
- 53.Hänzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taverna F., Goveia J., Karakach T.K., Khan S., Rohlenova K., Treps L., et al. BIOMEX: an interactive workflow for (single cell) omics data interpretation and visualization. Nucleic Acids Res. 2020;48:W385–W394. doi: 10.1093/nar/gkaa332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Noureen N., Ye Z., Chen Y., Wang X., Zheng S. Signature-scoring methods developed for bulk samples are not adequate for cancer single-cell RNA sequencing data. Elife. 2022;11 doi: 10.7554/eLife.71994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi H., Sheng J., Gao D., Li F., Durrans A., Ryu S., et al. Transcriptome analysis of individual stromal cell populations identifies stroma-tumor crosstalk in mouse lung cancer model. Cell Rep. 2015;10:1187–1201. doi: 10.1016/j.celrep.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 57.Komurov K. Modeling community-wide molecular networks of multicellular systems. Bioinformatics. 2012;28:694–700. doi: 10.1093/bioinformatics/btr718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Armingol E., Officer A., Harismendy O., Lewis N.E. Deciphering cell-cell interactions and communication from gene expression. Nat Rev Genet. 2021;22:71–88. doi: 10.1038/s41576-020-00292-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cillo A.R., Kürten C.H.L., Tabib T., Qi Z., Onkar S., Wang T., et al. Immune landscape of viral- and carcinogen-driven head and neck cancer. Immunity. 2020;52:183–199.e9. doi: 10.1016/j.immuni.2019.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang Y., Wang R., Zhang S., Song S., Jiang C., Han G., et al. iTALK: an R package to characterize and illustrate intercellular communication. BioRxiv. 2019 doi: 10.1101/507871. [DOI] [Google Scholar]
- 61.Tyler S.R., Rotti P.G., Sun X., Yi Y., Xie W., Winter M.C., et al. PyMINEr finds gene and autocrine-paracrine networks from human islet scRNA-Seq. Cell Rep. 2019;26:1951–1964.e8. doi: 10.1016/j.celrep.2019.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jin S., Guerrero-Juarez C.F., Zhang L., Chang I., Ramos R., Kuan C.-H., et al. Inference and analysis of cell-cell communication using Cell Chat. Nat Commun. 2021;12:1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. Cell PhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- 64.Dries R., Zhu Q., Dong R., Eng C.-H.-L., Li H., Liu K., et al. Giotto: a toolbox for integrative analysis and visualization of spatial expression data. Genome Biol. 2021;22:78. doi: 10.1186/s13059-021-02286-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Noël F., Massenet-Regad L., Carmi-Levy I., Cappuccio A., Grandclaudon M., Trichot C., et al. Dissection of intercellular communication using the transcriptome-based framework ICELLNET. Nat Commun. 2021;12:1089. doi: 10.1038/s41467-021-21244-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cabello-Aguilar S., Alame M., Kon-Sun-Tack F., Fau C., Lacroix M., Colinge J. SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 2020;48:e55. doi: 10.1093/nar/gkaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat Methods. 2020;17:159–162. doi: 10.1038/s41592-019-0667-5. [DOI] [PubMed] [Google Scholar]
- 68.Wang S., Karikomi M., MacLean A.L., Nie Q. Cell lineage and communication network inference via optimization for single-cell transcriptomics. Nucleic Acids Res. 2019;47:e66. doi: 10.1093/nar/gkz204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cang Z., Nie Q. Inferring spatial and signaling relationships between cells from single cell transcriptomic data. Nat Commun. 2020;11:2084. doi: 10.1038/s41467-020-15968-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tsuyuzaki K., Ishii M., Nikaido I. Uncovering hypergraphs of cell-cell interaction from single cell RNA-sequencing data. BioRxiv. 2019 doi: 10.1101/566182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Dimitrov D., Türei D., Boys C., Nagai J.S., Ramirez Flores R.O., Kim H., et al. Comparison of resources and methods to infer cell-cell communication from single-cell RNA data. BioRxiv. 2021 doi: 10.1101/2021.05.21.445160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marbach D., Costello J.C., Küffner R., Vega N.M., Prill R.J., Camacho D.M., et al. Wisdom of crowds for robust gene network inference. Nat Methods. 2012;9:796–804. doi: 10.1038/nmeth.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huynh-Thu V.A., Irrthum A., Wehenkel L., Geurts P. Inferring regulatory networks from expression data using tree-based methods. PLoS ONE. 2010;5 doi: 10.1371/journal.pone.0012776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pratapa A., Jalihal A.P., Law J.N., Bharadwaj A., Murali T.M. Benchmarking algorithms for gene regulatory network inference from single-cell transcriptomic data. Nat Methods. 2020;17:147–154. doi: 10.1038/s41592-019-0690-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huynh-Thu V.A., Geurts P. dynGENIE3: dynamical GENIE3 for the inference of gene networks from time series expression data. Sci Rep. 2018;8:3384. doi: 10.1038/s41598-018-21715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.van de Sande B., Flerin C., Davie K., de Waegeneer M., Hulselmans G., Aibar S., et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- 77.Janssens J., Aibar S., Taskiran I.I., Ismail J.N., Gomez A.E., Aughey G., et al. Decoding gene regulation in the fly brain. Nature. 2022;601:630–636. doi: 10.1038/s41586-021-04262-z. [DOI] [PubMed] [Google Scholar]
- 78.Todorov H., Cannoodt R., Saelens W., Saeys Y. Network inference from single-cell transcriptomic data. Methods Mol Biol. 2019;1883:235–249. doi: 10.1007/978-1-4939-8882-2_10. [DOI] [PubMed] [Google Scholar]
- 79.Xu R., Nettleton D., Nordman D.J. Case-specific random forests. J Comput Graph Statistics. 2016;25:49–65. doi: 10.1080/10618600.2014.983641. [DOI] [Google Scholar]
- 80.Filippi S., Holmes C.C. A bayesian nonparametric approach to testing for dependence between random variables. Bayesian Anal. 2017;12:919–938. doi: 10.1214/16-BA1027. [DOI] [Google Scholar]
- 81.Sanchez-Castillo M., Blanco D., Tienda-Luna I.M., Carrion M.C., Huang Y. A Bayesian framework for the inference of gene regulatory networks from time and pseudo-time series data. Bioinformatics. 2018;34:964–970. doi: 10.1093/bioinformatics/btx605. [DOI] [PubMed] [Google Scholar]
- 82.Matsumoto H., Kiryu H., Furusawa C., Ko M.S.H., Ko S.B.H., Gouda N., et al. SCODE: an efficient regulatory network inference algorithm from single-cell RNA-Seq during differentiation. Bioinformatics. 2017;33:2314–2321. doi: 10.1093/bioinformatics/btx194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.O’Brien E.J., Monk J.M., Palsson B.O. Using genome-scale models to predict biological capabilities. Cell. 2015;161:971–987. doi: 10.1016/j.cell.2015.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ramon C., Gollub M.G., Stelling J. Integrating -omics data into genome-scale metabolic network models: principles and challenges. Essays Biochem. 2018;62:563–574. doi: 10.1042/EBC20180011. [DOI] [PubMed] [Google Scholar]
- 85.Hrovatin K., Fischer D.S., Theis F.J. Toward modeling metabolic state from single-cell transcriptomics. Mol Metab. 2022;57 doi: 10.1016/j.molmet.2021.101396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alghamdi N., Chang W., Dang P., Lu X., Wan C., Gampala S., et al. A graph neural network model to estimate cell-wise metabolic flux using single-cell RNA-seq data. Genome Res. 2021;31:1867–1884. doi: 10.1101/gr.271205.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Heirendt L., Arreckx S., Pfau T., Mendoza S.N., Richelle A., Heinken A., et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox vol 3.0. Nat Protoc. 2019;14:639–702. doi: 10.1038/s41596-018-0098-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ebrahim A., Lerman J.A., Palsson B.O., Hyduke D.R. COBRApy: COnstraints-Based Reconstruction and Analysis for Python. BMC Syst Biol. 2013;7:74. doi: 10.1186/1752-0509-7-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang H, Marcišauskas S, Sánchez BJ, Domenzain I, Hermansson D, Agren R, et al. RAVEN 2.0: A versatile toolbox for metabolic network reconstruction and a case study on Streptomyces coelicolor. PLoS Comput Biol 2018;14:e1006541. 10.1371/journal.pcbi.1006541. [DOI] [PMC free article] [PubMed]
- 90.Subramanian A., Becker L.M., Carmeliet P. Endothelial metabolism going single. Nat Metab. 2021;3:593–594. doi: 10.1038/s42255-021-00399-3. [DOI] [PubMed] [Google Scholar]
- 91.Wang G., Heijs B., Kostidis S., Mahfouz A., Rietjens R.G.J., Bijkerk R., et al. Analyzing cell type-specific dynamics of metabolism in kidney repair. Nat Metab. 2022 doi: 10.1038/s42255-022-00615-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Adossa N., Khan S., Rytkönen K.T., Elo L.L. Computational strategies for single-cell multi-omics integration. Comput Struct Biotechnol J. 2021;19:2588–2596. doi: 10.1016/j.csbj.2021.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dou J., Liang S., Mohanty V., Cheng X., Kim S., Choi J., et al. Unbiased integration of single cell multi-omics data. BioRxiv. 2020 doi: 10.1101/2020.12.11.422014. [DOI] [Google Scholar]
- 95.Argelaguet R., Arnol D., Bredikhin D., Deloro Y., Velten B., Marioni J.C., et al. MOFA+: a statistical framework for comprehensive integration of multi-modal single-cell data. Genome Biol. 2020;21:111. doi: 10.1186/s13059-020-02015-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Welch J.D., Hartemink A.J., Prins J.F. MATCHER: manifold alignment reveals correspondence between single cell transcriptome and epigenome dynamics. Genome Biol. 2017;18:138. doi: 10.1186/s13059-017-1269-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hao Y., Hao S., Andersen-Nissen E., Mauck W.M., 3rd, Zheng S., Butler A., et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–3587.e29. doi: 10.1016/j.cell.2021.04.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kim H.J., Lin Y., Geddes T.A., Yang J.Y.H., Yang P. CiteFuse enables multi-modal analysis of CITE-seq data. Bioinformatics. 2020;36:4137–4143. doi: 10.1093/bioinformatics/btaa282. [DOI] [PubMed] [Google Scholar]
- 99.Wang X., Sun Z., Zhang Y., Xu Z., Xin H., Huang H., et al. BREM-SC: a bayesian random effects mixture model for joint clustering single cell multi-omics data. Nucleic Acids Res. 2020;48:5814–5824. doi: 10.1093/nar/gkaa314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Campbell K.R., Steif A., Laks E., Zahn H., Lai D., McPherson A., et al. clonealign: statistical integration of independent single-cell RNA and DNA sequencing data from human cancers. Genome Biol. 2019;20:54. doi: 10.1186/s13059-019-1645-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gayoso A., Steier Z., Lopez R., Regier J., Nazor K.L., Streets A., et al. Joint probabilistic modeling of single-cell multi-omic data with totalVI. Nat Methods. 2021;18:272–282. doi: 10.1038/s41592-020-01050-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cao Z.-J., Gao G. Multi-omics single-cell data integration and regulatory inference with graph-linked embedding. Nat Biotechnol. 2022 doi: 10.1038/s41587-022-01284-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rautenstrauch P., Vlot A.H.C., Saran S., Ohler U. Intricacies of single-cell multi-omics data integration. Trends Genet. 2022;38:128–139. doi: 10.1016/j.tig.2021.08.012. [DOI] [PubMed] [Google Scholar]
- 104.Miao Z., Humphreys B.D., McMahon A.P., Kim J. Multi-omics integration in the age of million single-cell data. Nat Rev Nephrol. 2021;17:710–724. doi: 10.1038/s41581-021-00463-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cantini L., Zakeri P., Hernandez C., Naldi A., Thieffry D., Remy E., et al. Benchmarking joint multi-omics dimensionality reduction approaches for the study of cancer. Nat Commun. 2021;12:124. doi: 10.1038/s41467-020-20430-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Dietrich S., Oleś M., Lu J., Sellner L., Anders S., Velten B., et al. Drug-perturbation-based stratification of blood cancer. J Clin Invest. 2018;128:427–445. doi: 10.1172/JCI93801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lanckriet G.R.G., de Bie T., Cristianini N., Jordan M.I., Noble W.S. A statistical framework for genomic data fusion. Bioinformatics. 2004;20:2626–2635. doi: 10.1093/bioinformatics/bth294. [DOI] [PubMed] [Google Scholar]
- 108.Rakotomamonjy A., Bach F.R., Canu S., Grandvalet Y. SimpleMKL. J Machine Learn Res. 2008;9:2491–2521. [Google Scholar]
- 109.Sonnenburg S., Rätsch G., Schäfer C., Schölkopf B. Large scale multiple kernel learning. J Machine Learn Res. 2006;7:1531–1565. [Google Scholar]
- 110.Zakeri P., Jeuris B., Vandebril R., Moreau Y. Protein fold recognition using geometric kernel data fusion. Bioinformatics. 2014;30:1850–1857. doi: 10.1093/bioinformatics/btu118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Yu S, Tranchevent L-C, de Moor B, Moreau Y. Kernel-based data fusion for machine learning. Studies in Computational Intelligence: Springer Berlin Heidelberg 2011.
- 112.Stahlschmidt S.R., Ulfenborg B., Synnergren J. Multimodal deep learning for biomedical data fusion: a review. Brief Bioinform. 2022;23 doi: 10.1093/bib/bbab569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Chen L, Zhai Y, He Q, Wang W, Deng M. Integrating deep supervised, self-supervised and unsupervised learning for single-cell RNA-seq clustering and annotation. Genes (Basel) 2020;11. 10.3390/genes11070792. [DOI] [PMC free article] [PubMed]
- 114.Lê Cao K.-A., González I., Déjean S. integrOmics: an R package to unravel relationships between two omics datasets. Bioinformatics. 2009;25:2855–2856. doi: 10.1093/bioinformatics/btp515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Rohart F., Gautier B., Singh A., Lê Cao K.-A. mixOmics: An R package for ’omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Rohart F., Eslami A., Matigian N., Bougeard S., Lê Cao K.-A. MINT: a multivariate integrative method to identify reproducible molecular signatures across independent experiments and platforms. BMC Bioinf. 2017;18:128. doi: 10.1186/s12859-017-1553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Singh A., Shannon C.P., Gautier B., Rohart F., Vacher M., Tebbutt S.J., et al. DIABLO: an integrative approach for identifying key molecular drivers from multi-omics assays. Bioinformatics. 2019;35:3055–3062. doi: 10.1093/bioinformatics/bty1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Aerts S., Lambrechts D., Maity S., van Loo P., Coessens B., de Smet F., et al. Gene prioritization through genomic data fusion. Nat Biotechnol. 2006;24:537–544. doi: 10.1038/nbt1203. [DOI] [PubMed] [Google Scholar]
- 119.Eriksson P., Marzouka N.-A.-D., Sjödahl G., Bernardo C., Liedberg F., Höglund M. A comparison of rule-based and centroid single-sample multiclass predictors for transcriptomic classification. Bioinformatics. 2021;38:1022–1029. doi: 10.1093/bioinformatics/btab763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mordelet F., Vert J.-P. ProDiGe: Prioritization Of Disease Genes with multitask machine learning from positive and unlabeled examples. BMC Bioinf. 2011;12:389. doi: 10.1186/1471-2105-12-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zakeri P., Simm J., Arany A., ElShal S., Moreau Y. Gene prioritization using Bayesian matrix factorization with genomic and phenotypic side information. Bioinformatics. 2018;34:i447–i456. doi: 10.1093/bioinformatics/bty289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schölkopf B., Platt J.C., Shawe-Taylor J., Smola A.J., Williamson R.C. Estimating the support of a high-dimensional distribution. Neural Comput. 2001;13:1443–1471. doi: 10.1162/089976601750264965. [DOI] [PubMed] [Google Scholar]
- 123.Yu S., Tranchevent L.-C., de Moor B., Moreau Y. Gene prioritization and clustering by multi-view text mining. BMC Bioinf. 2010;11:28. doi: 10.1186/1471-2105-11-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hernández Fusilier D., Montes-y-Gómez M., Rosso P., Guzmán C.R. Detecting positive and negative deceptive opinions using PU-learning. Inf Process Manag. 2015;51:433–443. doi: 10.1016/j.ipm.2014.11.001. [DOI] [Google Scholar]
- 125.Wenric S., Shemirani R. Using supervised learning methods for gene selection in RNA-Seq case-control studies. Front Genet. 2018;9:297. doi: 10.3389/fgene.2018.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Muslu O., Hoyt C.T., Lacerda M., Hofmann-Apitius M., Frohlich H. GuiltyTargets: prioritization of novel therapeutic targets with network representation learning. IEEE/ACM Trans Comput Biol Bioinform. 2022;19:491–500. doi: 10.1109/TCBB.2020.3003830. [DOI] [PubMed] [Google Scholar]
- 127.Peng J., Guan J., Shang X. Predicting Parkinson’s disease genes based on Node2vec and autoencoder. Front Genet. 2019;10:226. doi: 10.3389/fgene.2019.00226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Grover A, Leskovec J. Node2vec: Scalable Feature Learning for Networks. Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, New York, NY, USA: Association for Computing Machinery; 2016, p. 855–864. 10.1145/2939672.2939754. [DOI] [PMC free article] [PubMed]
- 129.Boudellioua I, Kulmanov M, Schofield PN, Gkoutos G v, Hoehndorf R. DeepPVP: phenotype-based prioritization of causative variants using deep learning. BMC Bioinformatics 2019;20:65. 10.1186/s12859-019-2633-8. [DOI] [PMC free article] [PubMed]
- 130.Sifrim A., Popovic D., Tranchevent L.-C., Ardeshirdavani A., Sakai R., Konings P., et al. eXtasy: variant prioritization by genomic data fusion. Nat Methods. 2013;10:1083–1084. doi: 10.1038/nmeth.2656. [DOI] [PubMed] [Google Scholar]
- 131.Sabaghian E., Drebert Z., Inzé D., Saeys Y. An integrated network of Arabidopsis growth regulators and its use for gene prioritization. Sci Rep. 2015;5:17617. doi: 10.1038/srep17617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.de Bie T., Tranchevent L.-C., van Oeffelen L.M.M., Moreau Y. Kernel-based data fusion for gene prioritization. Bioinformatics. 2007;23:i125–i132. doi: 10.1093/bioinformatics/btm187. [DOI] [PubMed] [Google Scholar]
- 133.Zakeri P, Elshal S, Moreau Y. Gene prioritization through geometric-inspired kernel data fusion. 2015 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), 2015, p. 1559–65. 10.1109/BIBM.2015.7359908.
- 134.Jiang Y., Oron T.R., Clark W.T., Bankapur A.R., D’Andrea D., Lepore R., et al. An expanded evaluation of protein function prediction methods shows an improvement in accuracy. Genome Biol. 2016;17:184. doi: 10.1186/s13059-016-1037-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cai Z., Poulos R.C., Liu J., Zhong Q. Machine learning for multi-omics data integration in cancer. IScience. 2022;25 doi: 10.1016/j.isci.2022.103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Clark D.J., Dhanasekaran S.M., Petralia F., Pan J., Song X., Hu Y., et al. Integrated proteogenomic characterization of clear cell renal cell carcinoma. Cell. 2019;179:964–983.e31. doi: 10.1016/j.cell.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wang L.-B., Karpova A., Gritsenko M.A., Kyle J.E., Cao S., Li Y., et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. 2021;39:509–528.e20. doi: 10.1016/j.ccell.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Gillette MA, Satpathy S, Cao S, Dhanasekaran SM, Vasaikar SV, Krug K, et al. Proteogenomic characterization reveals therapeutic vulnerabilities in lung adenocarcinoma. Cell 2020;182:200-225.e35. 10.1016/j.cell.2020.06.013. [DOI] [PMC free article] [PubMed]
- 139.Chaudhary K., Poirion O.B., Lu L., Garmire L.X. Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res. 2018;24:1248–1259. doi: 10.1158/1078-0432.CCR-17-0853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang L., Lv C., Jin Y., Cheng G., Fu Y., Yuan D., et al. Deep learning-based multi-omics data integration reveals two prognostic subtypes in high-risk neuroblastoma. Front Genet. 2018;9:477. doi: 10.3389/fgene.2018.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Takahashi S., Asada K., Takasawa K., Shimoyama R., Sakai A., Bolatkan A., et al. Predicting deep learning based multi-omics parallel integration survival subtypes in lung cancer using reverse phase protein array data. Biomolecules. 2020;10 doi: 10.3390/biom10101460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zhang X., Wang J., Lu J., Su L., Wang C., Huang Y., et al. Robust prognostic subtyping of muscle-invasive bladder cancer revealed by deep learning-based multi-omics data integration. Front Oncol. 2021;11 doi: 10.3389/fonc.2021.689626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tong L., Mitchel J., Chatlin K., Wang M.D. Deep learning based feature-level integration of multi-omics data for breast cancer patients survival analysis. BMC Med Inform Decis Mak. 2020;20:225. doi: 10.1186/s12911-020-01225-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Malik V., Kalakoti Y., Sundar D. Deep learning assisted multi-omics integration for survival and drug-response prediction in breast cancer. BMC Genomics. 2021;22:214. doi: 10.1186/s12864-021-07524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lee T.-Y., Huang K.-Y., Chuang C.-H., Lee C.-Y., Chang T.-H. Incorporating deep learning and multi-omics autoencoding for analysis of lung adenocarcinoma prognostication. Comput Biol Chem. 2020;87 doi: 10.1016/j.compbiolchem.2020.107277. [DOI] [PubMed] [Google Scholar]
- 146.Tranchevent L.-C., Ardeshirdavani A., ElShal S., Alcaide D., Aerts J., Auboeuf D., et al. Candidate gene prioritization with Endeavour. Nucleic Acids Res. 2016;44:W117–W121. doi: 10.1093/nar/gkw365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Fang H., Knight J.C. Priority index: database of genetic targets in immune-mediated disease. Nucleic Acids Res. 2022;50:D1358–D1367. doi: 10.1093/nar/gkab994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen Y.-A., Tripathi L.P., Mizuguchi K. TargetMine, an integrated data warehouse for candidate gene prioritisation and target discovery. PLoS ONE. 2011;6:e17844. doi: 10.1371/journal.pone.0017844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Goecks J., Jalili V., Heiser L.M., Gray J.W. How Machine Learning Will Transform Biomedicine. Cell. 2020;181:92–101. doi: 10.1016/j.cell.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ziegenhain C., Hendriks G.-J., Hagemann-Jensen M., Sandberg R. Molecular spikes: a gold standard for single-cell RNA counting. Nat Methods. 2022;19:560–566. doi: 10.1038/s41592-022-01446-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Lähnemann D., Köster J., Szczurek E., McCarthy D.J., Hicks S.C., Robinson M.D., et al. Eleven grand challenges in single-cell data science. Genome Biol. 2020;21:31. doi: 10.1186/s13059-020-1926-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Amersfoort J., Eelen G., Carmeliet P. Immunomodulation by endothelial cells - partnering up with the immune system? Nat Rev Immunol. 2022:1–13. doi: 10.1038/s41577-022-00694-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Nagl L., Horvath L., Pircher A., Wolf D. Tumor Endothelial Cells (TECs) as Potential Immune Directors of the Tumor Microenvironment - New Findings and Future Perspectives. Front Cell Dev Biol. 2020;8:766. doi: 10.3389/fcell.2020.00766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wood V., Lock A., Harris M.A., Rutherford K., Bähler J., Oliver S.G. Hidden in plain sight: what remains to be discovered in the eukaryotic proteome? Open Biol. 2019;9 doi: 10.1098/rsob.180241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liao Y., Wang J., Jaehnig E.J., Shi Z., Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47:W199–W205. doi: 10.1093/nar/gkz401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Glaab E., Baudot A., Krasnogor N., Schneider R., Valencia A. EnrichNet: network-based gene set enrichment analysis. Bioinformatics. 2012;28:i451–i457. doi: 10.1093/bioinformatics/bts389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ge S.X., Jung D., Yao R. ShinyGO: a graphical gene-set enrichment tool for animals and plants. Bioinformatics. 2020;36:2628–2629. doi: 10.1093/bioinformatics/btz931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Backes C., Keller A., Kuentzer J., Kneissl B., Comtesse N., Elnakady Y.A., et al. GeneTrail–advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Jain A., Tuteja G. TissueEnrich: Tissue-specific gene enrichment analysis. Bioinformatics. 2019;35:1966–1967. doi: 10.1093/bioinformatics/bty890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Glez-Peña D., Gómez-López G., Pisano D.G., Fdez-Riverola F. WhichGenes: a web-based tool for gathering, building, storing and exporting gene sets with application in gene set enrichment analysis. Nucleic Acids Res. 2009;37:W329–W334. doi: 10.1093/nar/gkp263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Fernandez N.F., Gundersen G.W., Rahman A., Grimes M.L., Rikova K., Hornbeck P., et al. Clustergrammer, a web-based heatmap visualization and analysis tool for high-dimensional biological data. Sci Data. 2017;4 doi: 10.1038/sdata.2017.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Yue Z., Slominski R., Bharti S., Chen J.Y. PAGER Web APP: An Interactive, Online Gene Set and Network Interpretation Tool for Functional Genomics. Front Genet. 2022;13 doi: 10.3389/fgene.2022.820361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Wang Y. talklr uncovers ligand-receptor mediated intercellular crosstalk. BioRxiv. 2020 doi: 10.1101/2020.02.01.930602. [DOI] [Google Scholar]
- 165.Interlandi M., Kerl K., Dugas M. InterCellar enables interactive analysis and exploration of cell-cell communication in single-cell transcriptomic data. Commun Biol. 2022;5:21. doi: 10.1038/s42003-021-02986-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Jakobsson J.E.T., Spjuth O., Lagerström M.C. scConnect: a method for exploratory analysis of cell-cell communication based on single cell RNA sequencing data. Bioinformatics. 2021;37:3501–3508. doi: 10.1093/bioinformatics/btab245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Zhang Y., Liu T., Wang J., Zou B., Li L., Yao L., et al. Cellinker: a platform of ligand-receptor interactions for intercellular communication analysis. Bioinformatics. 2021 doi: 10.1093/bioinformatics/btab036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Liu Y., Li J.S.S., Rodiger J., Comjean A., Attrill H., Antonazzo G., et al. FlyPhoneDB: an integrated web-based resource for cell-cell communication prediction in Drosophila. Genetics. 2022;220 doi: 10.1093/genetics/iyab235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cassan O., Lèbre S., Martin A. Inferring and analyzing gene regulatory networks from multi-factorial expression data: a complete and interactive suite. BMC Genomics. 2021;22:387. doi: 10.1186/s12864-021-07659-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Obayashi T., Kagaya Y., Aoki Y., Tadaka S., Kinoshita K. COXPRESdb v7: a gene coexpression database for 11 animal species supported by 23 coexpression platforms for technical evaluation and evolutionary inference. Nucleic Acids Res. 2019;47:D55–D62. doi: 10.1093/nar/gky1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.van Dam S., Craig T., de Magalhães J.P. GeneFriends: a human RNA-seq-based gene and transcript co-expression database. Nucleic Acids Res. 2015;43:D1124–D1132. doi: 10.1093/nar/gku1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Yang S., Kim C.Y., Hwang S., Kim E., Kim H., Shim H., et al. COEXPEDIA: exploring biomedical hypotheses via co-expressions associated with medical subject headings (MeSH) Nucleic Acids Res. 2017;45:D389–D396. doi: 10.1093/nar/gkw868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Zhu Q, Wong AK, Krishnan A, Aure MR, Tadych A, Zhang R, et al. Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nat Methods 2015;12:211–4, 3 p following 214. 10.1038/nmeth.3249. [DOI] [PMC free article] [PubMed]
- 174.Zhang M., Li Q., Yu D., Yao B., Guo W., Xie Y., et al. GeNeCK: a web server for gene network construction and visualization. BMC Bioinf. 2019;20:12. doi: 10.1186/s12859-018-2560-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Noronha A., Modamio J., Jarosz Y., Guerard E., Sompairac N., Preciat G., et al. The Virtual Metabolic Human database: integrating human and gut microbiome metabolism with nutrition and disease. Nucleic Acids Res. 2019;47:D614–D624. doi: 10.1093/nar/gky992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Robinson J.L., Kocabaş P., Wang H., Cholley P.-E., Cook D., Nilsson A., et al. An atlas of human metabolism. Sci Signal. 2020;13 doi: 10.1126/scisignal.aaz1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Norsigian C.J., Pusarla N., McConn J.L., Yurkovich J.T., Dräger A., Palsson B.O., et al. BiGG Models 2020: multi-strain genome-scale models and expansion across the phylogenetic tree. Nucleic Acids Res. 2020;48:D402–D406. doi: 10.1093/nar/gkz1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hari A., Lobo D. Fluxer: a web application to compute, analyze and visualize genome-scale metabolic flux networks. Nucleic Acids Res. 2020;48:W427–W435. doi: 10.1093/nar/gkaa409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Rowe E., Palsson B.O., King Z.A. Escher-FBA: a web application for interactive flux balance analysis. BMC Syst Biol. 2018;12:84. doi: 10.1186/s12918-018-0607-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Zoppi J., Guillaume J.-F., Neunlist M., Chaffron S. MiBiOmics: an interactive web application for multi-omics data exploration and integration. BMC Bioinf. 2021;22:6. doi: 10.1186/s12859-020-03921-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhou G., Pang Z., Lu Y., Ewald J., Xia J. OmicsNet 2.0: a web-based platform for multi-omics integration and network visual analytics. Nucleic Acids Res. 2022;50:W527–W533. doi: 10.1093/nar/gkac376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Chen J., Bardes E.E., Aronow B.J., Jegga A.G. ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res. 2009;37:W305–W311. doi: 10.1093/nar/gkp427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Radivojac P., Peng K., Clark W.T., Peters B.J., Mohan A., Boyle S.M., et al. An integrated approach to inferring gene-disease associations in humans. Proteins. 2008;72:1030–1037. doi: 10.1002/prot.21989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Kumar A.A., van Laer L., Alaerts M., Ardeshirdavani A., Moreau Y., Laukens K., et al. pBRIT: gene prioritization by correlating functional and phenotypic annotations through integrative data fusion. Bioinformatics. 2018;34:2254–2262. doi: 10.1093/bioinformatics/bty079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Chen Z., Zheng Y., Yang Y., Huang Y., Zhao S., Zhao H., et al. PhenoApt leverages clinical expertise to prioritize candidate genes via machine learning. Am J Hum Genet. 2022;109:270–281. doi: 10.1016/j.ajhg.2021.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Liu Y., Liang Y., Wishart D. PolySearch2: a significantly improved text-mining system for discovering associations between human diseases, genes, drugs, metabolites, toxins and more. Nucleic Acids Res. 2015;43:W535–W542. doi: 10.1093/nar/gkv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Nitsch D., Tranchevent L.-C., Gonçalves J.P., Vogt J.K., Madeira S.C., Moreau Y. PINTA: a web server for network-based gene prioritization from expression data. Nucleic Acids Res. 2011;39:W334–W338. doi: 10.1093/nar/gkr289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Warde-Farley D., Donaldson S.L., Comes O., Zuberi K., Badrawi R., Chao P., et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Biran H., Almozlino T., Kupiec M., Sharan R. WebPropagate: A Web Server for Network Propagation. J Mol Biol. 2018;430:2231–2236. doi: 10.1016/j.jmb.2018.02.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.