Abstract
Mitochondrial cytopathies, among which the Leigh syndrome (LS), are caused by variants either in the mitochondrial or the nuclear genome, affecting the oxidative phosphorylation process. The aim of the present study consisted in defining the molecular diagnosis of a group of Tunisian patients with LS.
Six children, belonging to five Tunisian families, with clinical and imaging presentations suggestive of LS were recruited. Whole mitochondrial DNA and targeted next-generation sequencing of a panel of 281 nuclear genes involved in mitochondrial physiology were performed. Bioinformatic analyses were achieved in order to identify deleterious variations.
A single m.10197G>A (p.Ala47Thr) variant was found in the mitochondrial MT-ND3 gene in one patient, while the others were related to autosomal homozygous variants: two c.1412delA (p.Gln471ArgfsTer42) and c.1264A>G (p.Thr422Ala) in SLC19A3, one c.454C>G (p.Pro152Ala) in SLC25A19 and one c.122G>A (p.Gly41Asp) in ETHE1.
Our findings demonstrate the usefulness of genomic investigations to improve LS diagnosis in consanguineous populations and further allow for treating the patients harboring variants in SLC19A3 and SLC25A19 that contribute to thiamine transport, by thiamine and biotin supplementation. Considering the Tunisian genetic background, the newly identified variants could be screened in patients with similar clinical presentation in related populations.
Keywords: Leigh syndrome, mitochondrial cytopathies, NGS, North Africa, Tunisia
Introduction
Mitochondrial diseases (MD) are multisystemic, encompassing a wide phenotypic spectrum, and can be manifested at any age [1,2]. MD affect approximately 1 in 5000 and are characterized by the alteration of the mitochondrial oxidative phosphorylation system (OXPHOS), composed of five complexes embedded in the mitochondrial inner membrane [3]. They are caused either by variants in the mitochondrial DNA (mtDNA) and nuclear DNA (nDNA), with >300 nuclear genes involved in the composition of the OXPHOS complexes, in the biogenesis and structure of the mitochondria, the mtDNA maintenance and in metabolic pathways [4,5].
Leigh syndrome (LS), also known as subacute necrotizing encephalo-myelopathy, was first described by Archibald Leigh in 1951, as a progressive psychomotor retardation or regression, induced by a symmetrical necrosis in the brain stem, thalamus and basal ganglia. Other LS-associated symptoms are hypotonia, muscle weakness, cerebellar ataxia, spasticity, nystagmus, optic atrophy, dysarthria, failure to thrive due to dysphagia, and elevated lactate levels in blood and in the cerebrospinal fluids. The disease usually appears in the first year of life and death may occur after respiratory muscle failure [6,7].
LS inheritance is complex since it involves maternal, autosomal recessive and dominant, and X-link modes of transmission. MtDNA variations are found in approximately 20% of cases, affecting mainly the respiratory complexes I, IV and V, and among them, the m.8993T>G in MT-ATP6 is the most frequent, altering the complex V efficiency. The other 80% of LS cases involve variations in nuclear genes, altering essentially the function or assembly of the OXPHOS complexes, mostly resulting in complex I deficiency [8,9].
In Tunisia, several genetic studies on LS patients were performed leading to the identification of variants in both mtDNA and nDNA. For example, the m.8993T>G variant in MT-ATP6 was detected with variable heteroplasmic loads in all affected members of a single family [10]. Two novel mitochondrial variants, m.5523T>G and m.5559A>G located in MT-TW affecting conserved regions of the tRNAtrp, were also identified [11] as well as the m.9478T>C missense variant (V91A) in COXIII [12]. Alternatively, variants in SURF1 were identified, either as a homozygous splice site c.516-517delAG or as compound heterozygous c.752-18A>C plus c.751+16G>A, most probably altering SURF1 mRNA splicing and consequently the abundance of SURF1 protein [13].
The aim of this study was to further characterize genetic alterations in the mitochondrial and nuclear genomes in a cohort of Tunisian LS patients.
Patients and methods
Patient’s descriptions
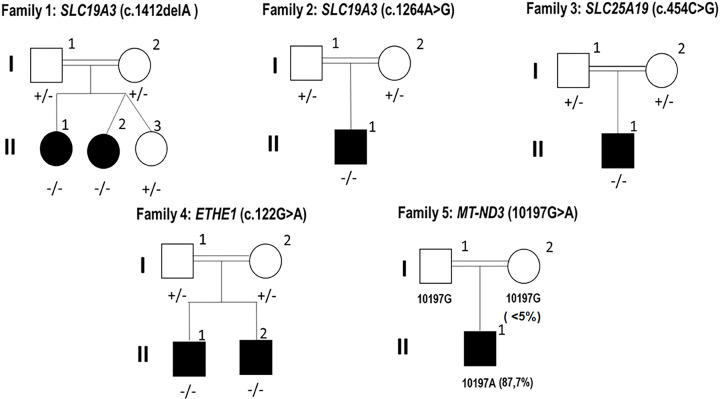
The present study was conducted according to the declaration of Helsinki and following the IRB recommendations and was approved by the Ethical Committee of The Institut Pasteur de Tunis Tunisia (Registration number IRB00005445, FWA00010074, Reference 2017/28/ILR16IPT). Written informed consent was obtained from parents for all the patients under the age of 18 years. Blood samples were obtained from five Tunisian families with six children having clinical, radiological and biochemical data suggestive of LS (Figure 1). They were recruited from the Department of Pediatric Neurology (National Institute Mongi Ben Hmida of Neurology, Tunis).
Figure 1. Pedigrees of the investigated families and segregation of their pathogenic variants.
Double lines indicates consanguineous parents; +/- indicates heterozygous for the variant; +/+ homozygous for the normal allele and -/- homozygous for the deleterious variant. The position of each variation is in-between brackets.
Family 1 patient 1
An 8-year-old girl born from first-degree consanguineous parents. She had no perinatal asphyxia and a normal psychomotor development. Walking unaided was acquired at the age of 18 months. She was admitted to the department of pediatric neurology at the age of 5 years for acute gait disorder, dysarthria and abnormal posture. Examination revealed a right hemiparesis and generalized dystonia with dystonic dysarthria. Brain magnetic resonance imaging (MRI) revealed a bilateral necrosis of striatum, T2 weighted hyperintensity of the midbrain, with subcortical white matter and mammillary tubercles. The spectroscopy showed doublets of lactate. She had high levels of lactate in blood (2.82 mmol/l; normal values 0.5–2.2 mmol/l) but normal in the cerebrospinal fluid (1.38 mmol/l; normal values <2 mmol/l). The lactate and pyruvate analysis revealed an elevated lactate-to-pyruvate ratio of 36.78, with a plasma lactate levels 2.06 mmol/l and pyruvate concentration of 0.056 mmol/l. This patient does not have other disorder than this diagnosis and no family history for LS.
Family 1 patient 2
She is a 6-year-old girl born from a twin pregnancy. She is the younger sister of patient 1. Her dizygotic twin sister is healthy. She had no perinatal suffering and normal psychomotor development. The onset of the disease began at the age of 3 years and 8 months with acute abdominal pain followed by a gait instability, bilateral divergent strabismus and drowsiness without fever. Clinical examination revealed a drowsy patient spastic tetraparesis, an unsteady gait, axial hypotonia, pyramidal signs in four limbs, generalized dystonia, bilateral nystagmus, divergent strabismus and ptosis. Computerized Tomography scan (CT-scan) showed a hypodensity of caudate nuclei and thalami. The MRI revealed T2 and FLAIR weighted hyperintensity of thalami, striatum, vermis, peri-aqueducal region and fronto-temporo-parietal cortex without enhancement and without restriction of diffusion. She had no lactate peak on spectroscopy. Her CSF lactate levels were normal (1.4 mmol/l). The lactate and pyruvate ratio appeared to be normal (18.16), with a plasma lactate level of 2.18 mmol/l and pyruvate concentration of 0.12 mmol/l. No other genetic disorder is diagnosed for this patient.
Family 2 patient 1
He is a 7 years old boy born from first-degree consanguineous parents. He had a personal history of perinatal asphyxia. He had a normal psychomotor development. He presented at the age of 14 months abnormal posture of upper limbs and gait disorders that rapidly worsened until a complete loss of the ability to walk. He had concomitant flu syndrome. Clinical examination showed that the patient was apyretic, drowsy and irritable. He had quadri-pyramidal syndrome and generalized dystonia. The first brain MRI performed at 7 days of evolution and showed abnormalities in subcortical white matter and in basal ganglia. The diagnosis of acute disseminated encephalomyelopathy (ADEM) was evoked initially. He was hospitalized in the department of pediatric neurology. A second brain and spinal MRI was performed and revealed T2 and FLAIR weighted hyperintensity in basal ganglia with necrosis of the central area of the striatum suggestive of LS. Spectroscopy showed a lactate peak. CSF lactate was 1.47 mmol/l. Electromyoneurography (EMNG) was in favor of axonal sensory neuropathy of the lower limbs. He was treated as ADEM with corticosteroids and thiamin. The evolution was marked by improvement of consciousness and contact. However, he kept a severe generalized dystonia. The MRI performed 4 years later and showed a striatum atrophy with persistent hyperintensity. The lactate-to-pyruvate ratio was normal (17.2), with lactate plasma levels 1.38 mmol/l and pyruvate concentration 0.08 mmol/l. No other genetic disease is found and no family history for LS for this patient.
Family 3 patient 1
He is a 4-year-old boy born from first-degree consanguineous parents. As for family history he had three aunts that died for an unknown reason between the ages of 4 and 5 years old. He had initially a normal psychomotor development. Walking was acquired at 14 months. He had a language delay. At the age of 14 months, he was hospitalized with 39°C fever, generalized epileptic seizures, sleep disturbance and psychomotor regression with loss of head control, sitting station, standing and walking. Examination showed generalized hypotonia and irritability. The diagnosis of encephalitis was evoked initially and the patient was treated by acyclovir and sodium valproate. One month later, he presented dyspnea and dehydration. Biochemical analysis showed cytolysis. Pyruvate blood levels before and after meal were 0.145 and 0.217 mmol/L, respectively. Brain MRI showed thrombosis of the left lateral sinus, and bilateral T2 and FLAIR weighted hyperintensity of striatum were observed. MRI spectroscopy showed a peak of lactate. He was switched to phenobarbital and received heparinotherapy during 3 months. The evolution was marked by improvement of motor skills. However, he presented recurrent episodes of drowsiness. Brain MRI performed 3 years later showed striatum necrosis with peak of lactate suggestive of LS. The lactate-to-pyruvate ratio was elevated 40.12, with lactate plasma levels 5.34 mmol/l and pyruvate levels 0.133 mmol/l. No other genetic disorder is diagnosed for this patient.
Family 4 patient 1
A 5-year-old boy born from second-degree consanguineous parents. He had a brother with similar clinical signs who died at the age of 7 years. He was admitted to the hospital at the age of 5 months with chronic glairy diarrhea without fever. Examination showed axial hypotonia associated with spastic tetraparesis and generalized dystonia. He started walking at the age of 2 years and 2 months and his language skills were at monosyllable stage at the age of 2 years. MRI revealed lacunar lesion in the putamen and the anterior limb of the internal capsule. CSF lactate level was high (3.13 mmol/l). The lactate-to-pyruvate ratio was elevated 23.06, with lactate plasma levels 3.69 mmol/l and pyruvate levels 0.16 mmol/l. This patient does not have other disorder than this diagnosis.
Family 5 patient 1
A 5-year-old boy born from third-degree consanguineous parents. As for family history he had a 7-year-old maternal cousin who had diabetes and developed psychomotor regression at 5 years old. Pregnancy and delivery were uneventful. Psychomotor development was normal in the first year. His language acquisition was at babbling stage. The disease started at the age of one year with a motor regression followed by a complete loss of ambulation. He was unable to sit or stand independently. He was hospitalized at 15 months of age for investigation. Examination showed axial hypotonia, quadripyramidal syndrome and dystonia of the upper limbs. Brain MRI showed a T2 and FLAIR weighted hyperintensity of basal ganglia of the periaqueductal region and the substantia nigra with a decrease in apparent diffusion coefficient (ADC). Spectroscopy revealed a doublet of lactate. Visual evoked potential, brain stem evoked response, cardiac and abdominal ultrasound were normal. EMNG was myogenic. CSF lactate levels were elevated (3.30 mmol/l). Evolution was marked by improvement of motor skills with appearance of generalized dystonia. He also developed epilepsy with tonic seizures treated by lamotrigine with improvement. This patient does not have other disorder than this diagnosis.
Methods
Targeted next-generation sequencing
Total DNA was extracted from blood using the FlexiGene DNA kit (Qiagen). DNA quality was assessed using nano-drop spectrophotometer (Thermofisher scientific) and Qubit 2.0 (ThermoFisher scientific). As previously described by [14], whole mitochondrial genome was amplified using two 8.5 kilo base (kb) over-lapping fragments. Ion Plus Fragment Library kit (Cat.no.4471269) was used in order to prepare the mtDNA library. mtDNA rearrangement and deletions were establish using eKLIPse software [15].
A customized NGS panel of 281 nuclear genes involved in the maintenance of the mitochondrial genome, the assembly and function of the OXPHOS complexes, and the mitochondrial biogenesis was used to screen pathogenic variants for the six patients and their parents. Library preparation, sequencing and bioinformatics analysis were processed as described elsewhere [14].
Confirmation of variants by Sanger sequencing
Sanger sequencing was performed to confirm all the identified variants and to test the family segregation. Exons with candidate variants were amplified by PCR and sequenced on an ABI PRISM 3100- Avant automated DNA sequence using the BigDye Terminator Cycle sequencing reaction kit v1.1. Sequences were compared with the updated Cambridge sequence of the mtDNA (GenBank accession number: NC_012920). Sequence analyses were performed using SeqScape.
Results
Mitochondrial DNA screening for variants and deletions
The mitochondrial genome from blood was screened for the six index patients. Results revealed only in the index case of family 5, the presence of m.10197G>A (p.Ala47Thr) variant in MT-ND3 with a heteroplasmic rate of 87.7%. This variant is considered as probably damaging and deleterious according to PolyPhen and Combined Annotation-Dependent Depletion (CADD), respectively. This variant has previously been reported with complex I deficiency disorders but newly identified for the Tunisian population in our patient with LS. The data of maternal heteroplasmy are <5%. Indeed, we performed whole mitochondrial DNA sequencing for the mother we did not find the mutation. We checked for the coverage of the gene region and it was sufficiently covered. We performed Sanger sequencing, we found that the mother was wild-type for the mutation. Consequently, the heteroplasmy rate for the mutation is <5% since it cannot be detected in Sanger sequencing performed on DNA extracted from blood.
Analysis of mtDNA sequences of all patients using eKLIPse software did not reveal rearrangement or deletion (data not shown).
Screening nuclear DNA for variations
Next generation sequencing of a panel of 281 genes involved in mitochondrial physiology was performed for the five patients devoid of mtDNA variant. Using a filtering step [14], four pathogenic variations in SLC19A3, SLC25A19 and ETHE1 genes (Table 1) were identified.
Table 1. Variants identified in Tunisian patients with LS.
| Family and Patients | G | Age | Gene | ORF variant | Position | Protein change | PolyPhen | Sift | Mutation Taster | LRT | Mutation Assessor | MetaRNN | rs number |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Family 1 | |||||||||||||
| Patient 1 | F | 6 | SLC19A3 |
NM_025243.4: c.1412delA |
Chr2: 228552192 | p.Gln471ArgfsTer42 | – | – | – | – | – | – | – |
| Patient 2 | F | 5 | SLC19A3 |
NM_025243.4: c.1412delA |
Chr2: 228552192 | p.Gln471ArgfsTer42 | – | – | – | – | – | – | – |
| Family 2 | |||||||||||||
| Patient 1 | M | 5 | SLC19A3 |
NM_025243: c.1264A>G |
Chr2: 228552932 | p.Thr422Ala | Damaging | Damaging | Disease causing | Deleterious | High | Damaging | rs121917884 |
| Family 3 | |||||||||||||
| Patient 1 | M | 4 | SLC25A19 |
NM_001126122.1: c.454C>G |
Chr17: 73279509 | p.Pro152Ala | Possibly damaging | Damaging | Disease causing | Deleterious | Medium | Damaging | – |
| Family 4 | |||||||||||||
| Patient 1 | M | 3 | ETHE1 |
NM_014297: c.122G>A |
Chr19: 44030771 | p.Gly41Asp | Probably damaging | Damaging | Disease causing | Deleterious | Medium | Damaging | – |
| Family 5 | |||||||||||||
| Patient 1 | M | 3 | MT-ND3 | m. 10197G>A | mitochondria | p.Ala47Thr | – | – | – | – | – | – | – |
G: gender; M: male; F: female.
Both sisters from family 1 (patient 1 and 2) harbored the same homozygous frameshift deletion c.1412delA in SLC19A3 exon 6. This novel variant was heterozygous in both parents and in the dizygotic twin sister that leads to a frameshift p.Gln471ArgfsTer42 classified as likely pathogenic = PP1/PM2/PM4 according to the ACMG classification (the American college of medical genetics). A second variation c.1264A>G in SLC19A3 exon 5 was found in family 2 patient 1. Both parents were heterozygous for this variant. This variation is predicted to be pathogenic = PS1/PS3/PP1/PM2/PM3 according to the ACMG classification and was linked to biotin-thiamine responsive basal ganglia disease (BTRBGD) as a founder mutation for the Saudi Arabian population. And previous functional studies support our findings by demonstrating that this variant down-regulate the thiamine transport activity of THTR2.
Family 3 patient 1 carried a novel homozygous missense c.454C>G variation in SLC25A19 exon 5, responsible for proline to alanine change in position 152. This variant was predicted to be deleterious according to Sift, PolyPhen and Mutation Taster. This variant was also classified according to the ACMG classification criteria as likely pathogenic (PM2, PM5 and PP3). No functional studies for this variant have been reported to support the likely pathogenic effect which still present some ambiguity about this impact for this variant, which is recurrently the case with novel variants in rare disease genes, so further functional studies need to be performed to showcase the impact of this variant. Both parents were heterozygous for this variant.
Finally, a novel homozygous missense variation c.122G>A variant in ETHE1 was identified in patient 1 from family 4. This caused an amino acid substitution of glycine to aspartate at the protein position 41 (p.Gly41Asp). Sanger sequencing revealed that both parents were heterozygous for this variation, which is predicted to be damaging and disease causing by PolyPhen, Sift and Mutation Taster. And according to the ACMG classification criteria it was predicted as uncertain significance (PM2 and PP2). Biochemical investigation of the respiratory complexes’ activities and cellular respiration of a patient biopsy showed normal values (data not shown).
Discussion
Impact of NGS/TGS on the diagnosis of LS patients
In the present study, we report five consanguineous Tunisian families with six children affected by LS. Mitochondrial genome sequencing revealed a novel m.10197G>A variation in family 5, with high heteroplasmic rate (87.7%), which has never been reported before in Tunisia, but well in patients affected by encephalo-myopathy [16], Leigh like syndrome [17], mitochondrial diseases with complex I deficiency [18], progressive generalized dystonia of childhood, Leber hereditary optic neuropathy with dystonia [19] and in another patient with stroke like episodes [20]. In addition, three patients with LS or MELAS/LS overlapping syndromes harbored this m.10197G>A variant. High heteroplasmic rates of this variant can alter complex 1 activity and decrease cell respiration by 30% [21,22], and may explain the clinical features of our patient.
NGS sequencing of 281 genes implicated in mitochondrial diseases identified four variations, among them three novels in three different genes. These novel variants were checked in 100 unrelated Tunisian controls and were not found. The first variation c.1412delA (p.Gln471ArgfsTer42) in SLC19A3 in the two sisters of family 1 was homozygous and led to a highly similar clinical phenotype. This variant created a new nonsense variant further downstream destroying the initial stop codon. This variant will lead to an abnormal protein with longer peptide chain altering the 3D structure and subsequently its function. Added to that since the alteration occurred on the 3’ untranslated region it will alter the regulatory function of this region [23]. Although further functional studies need to be performed to assess the pathogenicity of this variant and its impact on the protein expression and the patho-mechanism associated with LS. The variant c.1264A>G in SLC19A3 found in family 2 results in a Threonine to Alanine change at position 422, altering a transmembrane domain. It was already reported in three families originated from Saudi Arabia [24] leading to decreased thiamine transporter 2 (hTHTR2) activity and down-regulation of biotin accumulation [25]. Furthermore, this c.1264A>G variation was identified as a founder variant in the Saudi Arabian population for BTRBGD [26].
The SLC19A3 gene encodes for hTHTR2, an ubiquitously expressed thiamine transporter, composed of 12 transmembrane domains [25]. Additional SLC19A3 variations were described in Leigh and Leigh-like syndromes [27,28], in BTRBGD disease [29,30] and in Wernicke’s encephalopathy [31]. To date there is a total of 62 disease-causing variants identified in the SLC19A3 gene with our findings, which are associated with a wide spectrum of disorders from mild-to-severe clinical manifestations. Among these variants 37 are missense, 18 truncating mutations (nonsense or frameshift), four gross deletions of either exons, promoter region or the whole gene and 2 splicing mutations [32]. A novel homozygous missense variant c.958G>C (p.Glu320Gln) was found in four Japanese patients with a clinical presentation and radiological traits that were classified as LS. The p.Glu320Gln variant decreased of 63% the thiamine uptake [33]. Considering a case with adult-onset Wernicke’s-like encephalopathy a compound heterozygous variants Glu320 Gln combined with Lys44Glu were identified leading to a mild clinical manifestation [31]. Other than that, the hot spot variant identified Thr422Ala in the Saudi Arabian result in a somewhat mild phenotypes with a good prognosis [32,34]. As for the truncated variants, a Turkish patient harbored the SLC19A3 homozygous frameshift variant c.982del (p.Ala328Leufs*10), resulting in a truncated protein. This loss-of-function mutation reported in a case of early onset Leigh like syndrome and rapidly fatal prognosis [27]. The SLC19A3 homozygous c.20C>A variant causing a premature stop codon was found in two Moroccan siblings with LS, and then in 17 other LS patients, nine of these patients soon died after diagnosis [35]. Pyruvate dehydrogenase activity from a muscle biopsy of one LS patient was virtually null, but the addition of thiamine pyrophosphate (TPP) did restore this activity [35]. Still genotype–phenotype remains unclear but usually homozygous loss-of-function variants are associated with more severe phenotypes with infantile disease onset and a poor vital prognosis compared with missense variants.
The second thiamine transporter SLC25A19 gene was found mutated in family 3, disclosing for the first time the c.454C>G (p.Pro152Ala) variant in a Tunisian patient. Alterations of SLC25A19 gene have been linked to the bilateral striatal necrosis and neuropathy characterized by a truncal ataxia and hypotonia, episodes of encephalopathy, swallowing problems and recurrent episodes of flaccid paralysis and progressive polyneuropathy [36] and with the Amish lethal microencephalopathy. This latter disease is characterized by an apparent congenital microcephaly, high levels of α-ketoglutarate in urine, brain malformations and psychomotor retardation with encephalopathy episodes [37]. Li et al. identified the same variant c.454C>A p.(Pro152Thr) but not the same amino acid change, in a Chinese LS patient, as compound heterozygous together with another variation c.194C>T (p.Ala65Val) in the same gene [38]. Variants of SLC25A19 play a major role on the etiology of LS, since neurological LS symptoms are convergent toward a patho-mechanism of diseases related to thiamine transport. The members of thiamine transporters are expressed ubiquitously and thiamine plays a major role in the proper maintenance of the nervous, cardiovascular and motor systems [39,40]. But still, further functional studies need to be conducted in order to show beyond doubt the impact of this variant on LS pathophysiology since it is a bit ambiguous to demonstrate the effect for novel variant on their classification as disease causing.
Family 4 patient 1 presented a novel ETHE1 homozygous variant c.122G>A,p.(Gly41Asp). ETHE1 variants have been previously described as responsible for the development of ethylmalonic encephalopathy (EE), which is characterized by chronic diarrhea, recurrent petechiae, neurological degeneration, psychomotor delay, hypotonia, spastic tetraplegia, orthostatic acrocyanosis and Leigh-like syndrome [41,42]. EE was mistakenly diagnosed as a LS, since both presentations have common clinical and radiological features, such as psychomotor regression, brain lesions, pyramidal and quadri-pyramidal syndromes, hypotonia and ataxia. According to the literature one patient was diagnosed as LS at the beginning, then after the apparition of petechia, orthostatic acrocyanosis, mild hepatomegaly and high ethylmalonate in blood and urine levels, the diagnosis was reconsidered to EE. Importantly, enzymatic activities of the mitochondrial respiration complexes from EE patient fibroblasts were normal [43], as we found in the case of family 4.
Since all of our patients are born from consanguineous parents and the Tunisian population is known for a higher rate of consanguinity suggesting that autosomal recessive inheritance is a predominant mode of inheritance (62.9% of disease are autosomal recessive in the Tunisian population) [44], it would be more appropriate to screen the nuclear genome first and then if it is required the mitochondrial genome.
Founder mutations in LS
With respect to founder effects in LS, previous studies have identified the SLC19A3 c.20C>A variation in the Northern region of Morocco, in three unrelated families originated from the Al Houceima province [35]. Other founder variants were reported for LS patients in the literature. Variants with founder effect in NDUFS4 gene was described in the Moroccan, Algerian and Ashkenazi Jewish population [45]. Homozygous variant in PET100 was reported in Lebanese patients [46]. Variants in LRPPRC gene were found in patients originated from Northern Québec with LS French-Canadian type [47,48]. Variant in NDUFS4 gene present in five patients with LS is suggestive of a founder effect for the Hutterite population [49]. A new founder variant in USGM5 was identified for Ashkenazi Jewish patients affected with LS [50]. These LS founder variants were not found in our Tunisian LS patients. Nevertheless, the SLC19A3 variation c.1264A>G rs121917884 observed in our patient (family 2 patient 1), was identified as a founder variant for the Saudi Arabian population for BTRBGD. These latter variants could be screened as founder variants for LS Tunisian patients.
Strategies for treatments
In the context of LS, for which no efficient treatment is available in most cases, patients harboring variants in the thiamine transporter genes, like SLC19A3 and SLC25A19 can be treated by thiamine and biotin supplementation, in order to increase thiamine intra cellular and mitochondrial concentrations [25]. Thiamine enters the cytosol through the hTHTR2, then is converted to TPP, an active coenzyme, by the pyrophosphokinase 1 (TPK-1). Then, TPP is transported across the mitochondrial membrane by the thiamine pyrophosphate carrier encoded by SLC25A19. TPP stimulates the production of acetyl-COA through the conversion of pyruvate to fuel the Krebs cycle [39,51]. Thus, high doses of thiamine associated with biotin can be indicated as a treatment for LS patients with alteration of thiamine transporters in particular for those with variant in SLC19A3 or SLC25A19 [52]. Taking into consideration these recommendations, our Tunisian patients with SLC19A3 and SLC25A19 variants affecting thiamine transport were supplemented with high doses of thiamine: 250 mg/day and biotin 5–10 mg/kg/day. This led to a notable improvement and stabilization of the course of the disease, which was reached for patient 1 harboring the SLC19A3 c.1412delA variant after only 11 days of treatment. As regards to patient 2 family 1, we observed a total recovery after treatment for 12 days. After 3 months of treatment, the patient 1 from family 2 has partially recovered and did not present any novel episodes of encephalopathy. Lastly, patient 1 from family 3 started the treatment late, we observed a gradual improvement by the acquisition of walking and talking despite another encephalitic episode.
Regarding patients with ETHE1 variants, it was reported that they can be treated with metronidazole and neomycine in alternance, in addition to N-acetylcysteine in order to decrease H2S levels. Alternatively, liver transplant can also expand the life span of children with EE [53,54].
Conclusion
The present study reports for the first time in Tunisia a NGS analysis of five families with children affected with LS. These patients were mutated in LS genes (MT-ND3 and SLC19A3) and also in syndrome Leigh-like (SLC25A19 and ETHE1). Taking into account the broad spectrum of identified variants in our cohort, patients with similar phenotype for Leigh or Leigh-like syndrome should be screed for the common variant previously identified for the sake of cost-effectiveness and to improve prognosis for treatable cases. And for patients without genetic diagnosis a wider genetic analysis should be performed such as whole mitochondrial sequencing, targeted gene sequencing and in some cases whole exome sequencing.
Consent for Participation
All subjects gave written informed consent.
Acknowledgements
We would like to thank the patients and their families. We would to express our acknowledgement also to Ismail Gouiza for his help in some experiements.
Abbreviations
- ADC
apparent diffusion coefficient
- BTRBGD
biotin-thiamine responsive basal ganglia disease
- CADD
combined annotation-dependent depletion
- EE
ethylmalonic encephalopathy
- EMNG
electromyoneurography
- LS
Leigh syndrome
- MD
mitochondrial diseases
- OXPHOS
oxidative phosphorylation system
- TPP
thiamine pyrophosphate
Data Availability
Materials and raw data can be requested from the authors upon request.
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
Funding
This work was funded by the Ministry of Higher Education and Scientific Research [grant number LR16IPT05]; the University of Carthage in Tunisia and by the Institut National de la Santé et de la Recherche Médicale (INSERM); Centre National de la Recherche Scientifique (CNRS); the Université d'Angers, the University Hospital (CHU) of Angers; and the Région Pays de Loire and Angers Loire Métropole in France.
CRediT Author Contribution
Meriem Hechmi: Conceptualization, Software, Formal analysis, Validation, Investigation, Methodology, Writing—original draft. Majida Charif: Conceptualization, Formal analysis, Validation, Writing—review & editing. Ichraf Kraoua: Validation, Investigation, Writing—review & editing. Meriem Fassatoui: Validation, Investigation. Hamza Dallali: Validation. Valerie Desquiret-Dumas: Formal analysis. Céline Bris: Formal analysis. David Goudenège: Software, Formal analysis. Cyrine Drissi: Formal analysis. Saïd Galaï: Data curation, Formal analysis. Slah Ouerhani: Conceptualization, Writing—review & editing. Vincent Procaccio: Validation, Visualization, Writing—review & editing. Patrizia Amati-Bonneau: Validation, Visualization, Writing—review & editing. Sonia Abdelhak: Writing—review & editing. Ilhem Ben Youssef-Turki: Validation, Writing—review & editing. Guy Lenaers: Conceptualization, Resources, Funding acquisition, Validation, Project administration, Writing—review & editing. Rym Kefi: Conceptualization, Resources, Funding acquisition, Writing—review & editing.
Ethics Approval
This study was conducted according to the declaration of Helsinki and the approach was approved by the Ethical Committee of The Institut Pasteur de Tunis Tunisia (Registration number IRB00005445, FWA00010074, Reference 2017/28/ILR16IPT).
References
- 1.Schaefer A.M., McFarland R., Blakely E.L., He L., Whittaker R.G., Taylor R.W.et al. (2008) Prevalence of mitochondrial DNA disease in adults. Ann. Neurol. 63, 35–39 10.1002/ana.21217 [DOI] [PubMed] [Google Scholar]
- 2.DiMauro S., Schon E.A., Carelli V. and Hirano M. (2013) The clinical maze of mitochondrial neurology. Nat. Rev. Neurol. 9, 429–444 10.1038/nrneurol.2013.126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thorburn D.R. (2004) Mitochondrial disorders: prevalence, myths and advances. J. Inherit. Metab. Dis. 27, 349–362 10.1023/B:BOLI.0000031098.41409.55 [DOI] [PubMed] [Google Scholar]
- 4.Dard L., Blanchard W., Hubert C., Lacombe D. and Rossignol R. (2020) Mitochondrial functions and rare diseases. Mol. Aspects Med. 71, 1–28 10.1016/j.mam.2019.100842 [DOI] [PubMed] [Google Scholar]
- 5.Russell O.M., Gorman G.S., Lightowlers R.N. and Turnbull D.M. (2020) Mitochondrial diseases: hope for the future. Cell 181, 168–188 10.1016/j.cell.2020.02.051 [DOI] [PubMed] [Google Scholar]
- 6.Ruhoy I.S. and Saneto R.P. (2014) The genetics of leigh syndrome and its implications for clinical practice and risk management. Appl. Clin. Genet. 7, 221–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feigin I. and Kim H.S. (1977) Subacute necrotizing encephalomyelopathy in a neonatal infant. J. Neuropathol. Exp. Neurol. 36, 364–372 10.1097/00005072-197703000-00010 [DOI] [PubMed] [Google Scholar]
- 8.Chen L., Cui Y., Jiang D., Ma C.Y., Tse H.F., Hwu W.L.et al. (2018) Management of Leigh syndrome: current status and new insights. Clin. Genet. 93, 1131–1140 10.1111/cge.13139 [DOI] [PubMed] [Google Scholar]
- 9.Schubert M.B. and Vilarinho L. (2020) Molecular basis of Leigh syndrome: a current look. Orphanet J. Rare Dis. 15, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mkaouar-Rebai E., Chaari W., Younes S., Bousoffara R., Sfar M.T. and Fakhfakh F. (2009) Maternally inherited Leigh syndrome: T8993G mutation in a Tunisian family. Pediatr. Neurol. 40, 437–442 10.1016/j.pediatrneurol.2009.01.004 [DOI] [PubMed] [Google Scholar]
- 11.Mkaouar-Rebai E., Chamkha I., Kammoun F., Kammoun T., Aloulou H., Hachicha M.et al. (2009) Two new mutations in the MT-TW gene leading to the disruption of the secondary structure of the tRNATrp in patients with Leigh syndrome. Mol. Genet. Metab. 97, 179–184 10.1016/j.ymgme.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 12.Mkaouar-Rebai E., Ellouze E., Chamkha I., Kammoun F., Triki C. and Fakhfakh F. (2011) Molecular-clinical correlation in a family with a novel heteroplasmic leigh syndrome missense mutation in the mitochondrial cytochrome c oxidase III gene. J. Child Neurol. 26, 12–20 10.1177/0883073810371227 [DOI] [PubMed] [Google Scholar]
- 13.Maalej M., Kammoun T., Alila-Fersi O., Kharrat M., Ammar M., Felhi R.et al. (2018) Cytochrome C oxydase deficiency: SURF1 gene investigation in patients with Leigh syndrome. Biochem. Biophys. Res. Commun. 497, 1043–1048 10.1016/j.bbrc.2018.02.169 [DOI] [PubMed] [Google Scholar]
- 14.Felhi R., Sfaihi L., Charif M., Desquiret-Dumas V., Bris C., Goudenège D.et al. (2019) Next generation sequencing in family with MNGIE syndrome associated to optic atrophy: Novel homozygous POLG mutation in the C-terminal sub-domain leading to mtDNA depletion. Clin. Chim. Acta 488, 104–110 10.1016/j.cca.2018.11.003 [DOI] [PubMed] [Google Scholar]
- 15.Goudenège D., Bris C., Hoffmann V., Desquiret-Dumas V., Jardel C., Rucheton B.et al. (2019) eKLIPse: a sensitive tool for the detection and quantification of mitochondrial DNA deletions from next-generation sequencing data. Genet. Med. 21, 1407–1416 10.1038/s41436-018-0350-8 [DOI] [PubMed] [Google Scholar]
- 16.Swalwell H., Kirby D.M., Blakely E.L., Mitchell A., Salemi R., Sugiana C.et al. (2011) Respiratory chain complex i deficiency caused by mitochondrial DNA mutations. Eur. J. Hum. Genet. 19, 769–775 10.1038/ejhg.2011.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kirby D.M., Salemi R., Sugiana C., Ohtake A., Parry L., Bell K.M.et al. (2004) NDUFS6 mutations are a novel cause of lethal neonatal mitochondrial complex I deficiency. J. Clin. Invest. 114, 837–845 10.1172/JCI20683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lebre A.S., Rio M., Faivre D'Arcier L., Vernerey D., Landrieu P., Slama A.et al. (2011) A common pattern of brain MRI imaging in mitochondrial diseases with complex I deficiency. J. Med. Genet. 48, 16–23 10.1136/jmg.2010.079624 [DOI] [PubMed] [Google Scholar]
- 19.Wang K., Takahashi Y., Gao Z.L., Wang G.X., Chen X.W., Goto J.et al. (2009) Mitochondrial ND3 as the novel causative gene for Leber hereditary optic neuropathy and dystonia. Neurogenetics 10, 337–345 10.1007/s10048-009-0194-0 [DOI] [PubMed] [Google Scholar]
- 20.Chae J.H., Lee J.S., Kim K.J., Hwang Y.S., Bonilla E., Tanji K.et al. (2007) A novel ND3 mitochondrial DNA mutation in three Korean children with basal ganglia lesions and complex I deficiency. Pediatr. Res. 61, 622–624 10.1203/pdr.0b013e3180459f2d [DOI] [PubMed] [Google Scholar]
- 21.Tolomeo D., Rubegni A., Severino M., Pochiero F., Bruno C., Cassandrini D.et al. (2019) Clinical and neuroimaging features of the m.10197G>A mtDNA mutation: New case reports and expansion of the phenotype variability. J. Neurol. Sci. 399, 69–75 10.1016/j.jns.2019.02.010 [DOI] [PubMed] [Google Scholar]
- 22.Galkin A., Meyer B., Wittig I., Karas M., Schägger H., Vinogradov A.et al. (2008) Identification of the mitochondrial ND3 subunit as a structural component involved in the active/deactive enzyme transition of respiratory complex I. J. Biol. Chem. 283, 20907–20913 10.1074/jbc.M803190200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nussbaum R., McInnes R. and Willard H. (2015) Chapter 4: human genetic diversity: mutation and polymorphism. Thompson & Thompson Genetics in Medicine 8th edition, 43–56 [Google Scholar]
- 24.Zeng W.Q., Al-Yamani E., Acierno J.S., Slaugenhaupt S., Gillis T., MacDonald M.E.et al. (2005) Biotin-responsive basal ganglia disease maps to 2q36.3 and is due to mutations in SLC19A3. Am. J. Hum. Genet. 77, 16–26 10.1086/431216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Subramanian V.S., Marchant J.S. and Said H.M. (2006) Biotin-responsive basal ganglia disease-linked mutations inhibit thiamine transport via hTHTR2: Biotin is not a substrate for hTHTR2. Am J Physiol. - Cell Physiol. 291, 851–859 10.1152/ajpcell.00105.2006 [DOI] [PubMed] [Google Scholar]
- 26.Alfadhel M., Almuntashri M., Raafat H.J., Bashiri A.F., Al Rifai M.T., Al Shalaan H.et al. (2013) Biotin-responsive basal ganglia disease should be renamed biotin-thiamine-responsive basal ganglia disease: A retrospective review of the clinical, radiological and molecular findings of 18 new cases. Orphanet J. Rare Dis. 8, 1–8 10.1186/1750-1172-8-83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Van Der Knaap M.S. and Kevelam S.H. (2014) Reply: Infantile Leigh-like syndrome caused by SLC19A3 mutations is a treatable disease. Brain 137, 18–20 10.1093/brain/awu130 [DOI] [PubMed] [Google Scholar]
- 28.Alfadhel M. (2017) Early infantile Leigh-like SLC19A3 gene defects have a poor prognosis: report and review. J. Cent. Nerv. Syst. Dis. 9, 117957351773752 10.1177/1179573517737521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alfadhel M. and Tabarki B. (2018) SLC19A3 gene defects sorting the phenotype and acronyms: review. Neuropediatrics 49, 83–92 10.1055/s-0037-1607191 [DOI] [PubMed] [Google Scholar]
- 30.Savasta S., Bassanese F., Buschini C., Foiadelli T., Trabatti C., Efthymiou S.et al. (2019) Biotin-thiamine responsive encephalopathy: report of an Egyptian family with a novel SLC19A3 mutation and review of the literature. J. Pediatr. Genet. 08, 100–108 10.1055/s-0038-1676603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kono S., Miyajima H., Yoshida K., Togawa A., Shirakawa K. and Suzuki H. (2009) Mutations in a thiamine-transporter gene and Wernicke’s-like encephalopathy. N. Engl. J. Med. 360, 1792–1794 10.1056/NEJMc0809100 [DOI] [PubMed] [Google Scholar]
- 32.Wang J., Wang J., Han X., Liu Z., Ma Y., Chen G.et al. (2021) Report of the largest Chinese cohort with SLC19A3 gene defect and literature review. Front. Genet. 12, 1–9 10.3389/fgene.2021.683255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamada K., Miura K., Hara K., Suzuki M., Nakanishi K., Kumagai T.et al. (2010) A wide spectrum of clinical and brain MRI findings in patients with SLC19A3 mutations. BMC Med. Genet. 11, 171 10.1186/1471-2350-11-171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alfadhel M., Umair M., Almuzzaini B., Alsaif S., AlMohaimeed S.A., Almashary M.A.et al. (2019) Targeted SLC19A3 gene sequencing of 3000 Saudi newborn: a pilot study toward newborn screening. Ann. Clin. Transl. Neurol. 6, 2097–2103 10.1002/acn3.50898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gerards M., Kamps R., Van Oevelen J., Boesten I., Jongen E., De Koning B.et al. (2013) Exome sequencing reveals a novel Moroccan founder mutation in SLC19A3 as a new cause of early-childhood fatal Leigh syndrome. Brain 136, 882–890 10.1093/brain/awt013 [DOI] [PubMed] [Google Scholar]
- 36.Spiegel R., Shaag A., Edvardson S., Mandel H., Stepensky P., Shalev S.A.et al. (2009) SLC25A19 mutation as a cause of neuropathy and bilateral striatal necrosis. Ann. Neurol. 66, 419–424 10.1002/ana.21752 [DOI] [PubMed] [Google Scholar]
- 37.Kelley R.I., Robinson D., Puffenberger E.G., Strauss K.A. and Holmes Morton D. (2002) Amish lethal microcephaly: a new metabolic disorder with severe congenital microcephaly and 2-ketoglutaric aciduria. Am. J. Med. Genet. 112, 318–326 10.1002/ajmg.10529 [DOI] [PubMed] [Google Scholar]
- 38.Li D., Song J., Li X., Liu Y., Dong H., Kang L.et al. (2020) Eleven novel mutations and clinical characteristics in seven Chinese patients with thiamine metabolism dysfunction syndrome. Eur. J. Med. Genet. 63, 104003 10.1016/j.ejmg.2020.104003 [DOI] [PubMed] [Google Scholar]
- 39.Ganapathy V., Smith S.B. and Prasad P.D. (2004) SLC19: The folate/thiamine transporter family. Pflugers Arch. 447, 641–646 10.1007/s00424-003-1068-1 [DOI] [PubMed] [Google Scholar]
- 40.Tylicki A., Lotowski Z., Siemieniuk M. and Ratkiewicz A. (2018) Thiamine and selected thiamine antivitamins — biological activity and methods of synthesis. Biosci. Rep. 38, 1–23 10.1042/BSR20171148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tiranti V., D'Adamo P., Briem E., Ferrari G., Mineri R., Lamantea E.et al. (2004) Ethylmalonic encephalopathy is caused by mutations in ETHE1, a gene encoding a mitochondrial matrix protein. Am. J. Hum. Genet. 74, 239–252 10.1086/381653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tavasoli A.R., Rostami P., Ashrafi M.R. and Karimzadeh P. (2017) Neurological and vascular manifestations of ethylmalonic encephalopathy. Iran J. Child Neurol. 11, 57–60 [PMC free article] [PubMed] [Google Scholar]
- 43.García-Silva M.T., Ribes A., Campos Y., Garavaglia B. and Arenas J. (1997) Syndrome of encephalopathy, petechiae, and ethylmalonic aciduria. Pediatr. Neurol. 17, 165–170 10.1016/S0887-8994(97)00048-9 [DOI] [PubMed] [Google Scholar]
- 44.Romdhane L. and Abdelhak S. (2011) Genetic diseases in the Tunisian population. Am. J. Med. Genet. Part A. 155, 238–267 10.1002/ajmg.a.33771 [DOI] [PubMed] [Google Scholar]
- 45.Assouline Z., Jambou M., Rio M., Bole-Feysot C., de Lonlay P., Barnerias C.et al. (2012) A constant and similar assembly defect of mitochondrial respiratory chain complex I allows rapid identification of NDUFS4 mutations in patients with Leigh syndrome. Biochim. Biophys. Acta. - Mol. Basis Dis. 1822, 1062–1069 10.1016/j.bbadis.2012.01.013 [DOI] [PubMed] [Google Scholar]
- 46.Lim S.C., Smith K.R., Stroud D.A., Compton A.G., Tucker E.J., Dasvarma A.et al. (2014) A founder mutation in PET100 causes isolated complex IV deficiency in lebanese individuals with Leigh syndrome. Am. J. Hum. Genet. 94, 209–222 10.1016/j.ajhg.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Debray F.G., Morin C., Janvier A., Villeneuve J., Maranda B., Laframboise R.et al. (2011) LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J. Med. Genet. 48, 183–189 10.1136/jmg.2010.081976 [DOI] [PubMed] [Google Scholar]
- 48.Morin C., Mitchell G., Larochelle J., Lambert M., Ogier H., Robinson B.H.et al. (1993) Clinical, metabolic, and genetic aspects of cytochrome C oxidase deficiency in Saguenay-Lac-Saint-Jean. Am. J. Hum. Genet. 53, 488–496 [PMC free article] [PubMed] [Google Scholar]
- 49.Lamont R.E., Beaulieu C.L., Bernier F.P., Sparkes R., Innes A.M., Jackel-Cram C.et al. (2017) A novel NDUFS4 frameshift mutation causes Leigh disease in the Hutterite population. Am. J. Med. Genet. Part A. 173, 596–600 10.1002/ajmg.a.37983 [DOI] [PubMed] [Google Scholar]
- 50.Barca E., Ganetzky R.D., Potluri P., Juanola-Falgarona M., Gai X., Li D.et al. (2018) USMG5 Ashkenazi Jewish founder mutation impairs mitochondrial complex V dimerization and ATP synthesis. Hum. Mol. Genet. 27, 3305–3312 10.1093/hmg/ddy231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lindhurst M.J., Fiermonte G., Song S., Struys E., De Leonardis F., Schwartzberg P.L.et al. (2006) Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc. Natl. Acad. Sci. U.S.A. 103, 15927–15932 10.1073/pnas.0607661103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitford W., Hawkins I., Glamuzina E., Wilson F., Marshall A., Ashton F.et al. (2017) Compound heterozygous SLC19A3 mutations further refine the critical promoter region for biotin-thiamine-responsive basal ganglia disease. Cold Spring Harb. Mol. Case Stud. 3, 1–15 10.1101/mcs.a001909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyer M., Sowa M., Di Meo I., Eftekharian S., Steenari M.R., Tiranti V.et al. (2018) Response to medical and a novel dietary treatment in newborn screen identified patients with ethylmalonic encephalopathy. Mol. Genet. Metab. 124, 57–63 10.1016/j.ymgme.2018.02.008 [DOI] [PubMed] [Google Scholar]
- 54.Mc Kiernan P.J. (2017) Recent advances in liver transplantation for metabolic disease. J. Inherit. Metab. Dis. 40, 491–495 10.1007/s10545-017-0020-z [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Materials and raw data can be requested from the authors upon request.