Abstract
Traditional Chinese medicine, as a complementary and alternative medicine, has been practiced for thousands of years in China and possesses remarkable clinical efficacy. Thus, systematic analysis and examination of the mechanistic links between Chinese herbal medicine (CHM) and the complex human body can benefit contemporary understandings by carrying out qualitative and quantitative analysis. With increasing attention, the approach of network pharmacology has begun to unveil the mystery of CHM by constructing the heterogeneous network relationship of “herb-compound-target-pathway,” which corresponds to the holistic mechanisms of CHM. By integrating computational techniques into network pharmacology, the efficiency and accuracy of active compound screening and target fishing have been improved at an unprecedented pace. This review dissects the core innovations to the network pharmacology approach that were developed in the years since 2015 and highlights how this tool has been applied to understanding the coronavirus disease 2019 and refining the clinical use of CHM to combat it.
Keywords: Chinese traditional medicine, Herbal medicine, Network pharmacology, Compound identification, COVID-19
1. Introduction
Traditional Chinese medicine (TCM), as a treasure of the Chinese nation and a critical component of China’s medical and health care system, plays an essential role in the field of healthcare for the Chinese people [1]. Characterized by holistic, personalized, and rich experience-based therapy, TCM, including modalities such as Chinese herbal medicine (CHM) and acupuncture, has broad applications for the systematic control of complex diseases [2]. Due to the complexity of CHM which comprises a crucial part of TCM, traditional reductionism method remains difficult to simplify the interplay between the multiple compounds present in an herbal formula and the multiple targets on which they act; this has become a major obstacle to TCM’s modernization and its incorporation into modern healthcare [3].
Systems biology is a new frontier in biological research which provides a framework for assembling models of biological systems from systematic measurements. Further, bioinformatics is conceptualizing biology from a molecular perspective and applying “informatics techniques,” including applied mathematics, computer science and statistics, to extract knowledge from biological data for large-scale analysis, prediction, imaging and visualization [4]. Cheminformatics is an emerging frontier in the field of information technology, focusing on the collection, storage, analysis and operation of chemical data [5]. Hence, with the development and integration of fields such as systems biology, bioinformatics, cheminformatics, artificial intelligence, and “big data,” research on the mechanisms of CHM has shifted from investigating single, isolated compounds to a new multi-faceted and systematic research approach [6]. One of these breakthroughs is network pharmacology, which is used to explore the molecular mechanisms of CHM from the perspective of a complex biomolecular network. Utilizing this network approach generates an unprecedented opportunity for systematic research into CHM. In the last five years, network pharmacology studies of CHM have increased rapidly [7], and it is evolving as a systematic paradigm and the leading edge in research and development of CHM [8]. At the same time, computational methodologies and high-quality databases play an essential role in satisfying the data-driven aspects of network pharmacology. Therefore, a concise overview of the use of network pharmacology in CHM research is urgent.
This review is structured into two main sections. In the first section, the cutting-edge CHM network pharmacology studies published between 2015 and 2021 that established strategies for active compound screening, target prediction, and network analysis are reviewed and summarized alongside the specialized databases on which these techniques depend. In the second section, the application of network pharmacology in mechanistic investigation and repositioning of CHM against coronavirus disease 2019 (COVID-19) is highlighted.
2. Network pharmacology for CHM research
2.1. Strategies for compound screening
Lack of certainty in the bioactive compounds responsible for the actions of CHM is one of the key issues that makes CHM research difficult. It is extremely time-consuming and labor-intensive to obtain the chemical profiles of CHM formulas following traditional chemical methods (i.e., isolation, identification and evaluation). Many natural product databases are open-source, although it can be difficult to extract and screen the active compounds present in CHM from these databases, as they contain vast amounts of data. In the modern drug discovery procedure, there is a high failure rate for converting candidate active compounds into effective drugs; this is primarily caused by undesirable absorption, distribution, metabolism, elimination and toxicity (ADMET) profiles of the target compound. Thus, it is practical to include ADMET data in parallel with information on CHM that can be gathered from natural product databases. Insufficiency of data is, however, often a problem in this approach. To fill in some of these information gaps, some researchers prefer to use their own experiments to profile compounds that are present in CHM, combined with ADMET filtering. For instance, ultra-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF-MS) was used for the chemical profiling of Isatis indigotica roots and leaves, and the active constituents present in I. indigotica were then screened using the prediction of gastrointestinal absorption and drug-likeness (DL) analysis in SwissADME (https://www.swissadme.ch) [9]. The active components in ginsenoside H dripping pills were identified by UPLC-Q-TOF-MS and then filtered based on oral bioavailability (OB) and DL [10]. To detect the volatile compounds from Scutellaria baicalensis, gas chromatography-MS (GC-MS) was performed. The active compounds were further screened by OB, druggability and blood brain barrier (BBB) screening [11]. Likewise, the bioactive constituents in Morus alba L. leaves were detected using GC–MS and then screened for DL and their topological polar surface area [12]. Further, GC–MS was used to detect chemical constituents from Hibiscus cannabinus L. leaves and Ganoderma lucidum, and then they were filtered according to Lipinski’s rule through the SwissADME to identify their DL [13], [14]. Liu et al. [15] used OB, DL, human colonic adenocarcinoma cells, and BBB criteria to screen the potential active compounds of Guanxinshutong Capsule that had been identified through LC-MS and GC-MS profiling.
Experimental errors in the datasets, poor-quality models and the idea of applicability domain are major concerns related to the reliability of ADMET predictions. Compared to the compounds isolated from herbs, considering the CHM constituents that are absorbed in vivo could reduce the rate of false-positive results from ADMET prediction [16]. For example, based on the serum/plasma pharmacochemical evaluation, the compounds from formulas (Shentong Zhuyu Decoction, Xiaokewan, and Shenzhi Jiannao Formula) or single herbs (Viticis fructus, Poria cocos [Schw.] Wolf, and Cyclocarya paliurus [Batal.] Iljinskaja leaves) that could enter the serum were considered to be potential active compounds for network pharmacology analysis [17], [18], [19], [20], [21], [22]. The compounds present in other biological samples, such as urine and tissues, could be also used for network pharmacology analysis [23], [24]. If metabolites have potential biological activities, they should be included with prototype compounds in the network pharmacology analysis. Zhang et al. [25] used phellodendrine and its main in vivo metabolites to explore the potential pharmacological network to address diabetes mellitus. Arctiin and prim-O-glucosylcimifugin and their in vivo metabolites were used for network pharmacology analysis [26], [27]. The in vivo metabolites of Achyranthes bidentata Blume saponins and their targets associated with rheumatic arthritis were used to construct a multi-layer network [28]. The prototype constituents plus metabolites of Schisandra chinensis (Turcz.) Baill. fruits and Paeonia lactiflora Pall. roots that were retrieved from rat plasma were considered to be bioactive ingredients used for network pharmacology analysis [29], [30].
It is also useful to identify suitable pharmacokinetic marker(s) of CHM as bioactive compounds. For instance, network pharmacology was conducted by selecting the pharmacokinetic markers from Phlomis brevidentata H.W.Li extract [31]. Further, an everted gut sac model, coupled with UPLC-Q-TOF-MS, was used to screen and identify the active compounds of Xijiao Dihuang Decoction combined with Yinqiao Powder [32]. Considering the pivotal role of gut microbial transformation, three phenylethanoid glycosides from Cistanche deserticola Y.C.Ma stems and their in vitro metabolites transformed by intestinal bacteria were forwarded to network pharmacology analysis [33].
Overall, integrating computational and analytical methods serves as a credible method for identifying preliminary bioactive compounds present in CHM.
2.2. Strategies for target prediction
Compound-target interaction (CTI) is the core part of network pharmacology to understand comprehensive mechanisms of CHM [34]. The traditional way to identify CTIs is to quantitatively determine the inhibitory or activation values between compounds and targets by in vitro or in vivo assays [35]. However, it is not feasible to determine all possible CTIs present in the thousands of CHMs [36]. The development of various computational methods, such as molecular docking-based [37], pharmacophore-based [38], chemical similarity-based [39], machine learning-based [40], and network-based [41] methods, has provided valuable strategies for the systematic prediction of potential CTIs. Several data sources for screening and prediction of CTIs are introduced in Table 1 [42], [43], [44], [45], [46], [47], [48], [49], [50], [51].
Table 1.
Several representative databases and web servers for screening and prediction of CTI data.
| Database and web server | Website | Contents and main features | Quantitative activity values | Reference |
|---|---|---|---|---|
| Binding MOAD | https://www.bindingmoad.org | Including 23,269 complexes and 8156 binding affinities. | Yes | [42] |
| DrugCentral | https://drugcentral.org/ | Integrating structure, bioactivity, regulatory and pharmacologic actions, and indications for active pharmaceutical compounds. | Yes | [43] |
| IUPHAR/BPS Guide to PHARMACOLOGY | https://www.guidetopharmacology.org/ | Including approximately 9000 ligands, 15,000 binding constants, 6000 papers and 1700 human proteins. | Yes | [44] |
| PubChem BioAssay | https://www.ncbi.nlm.nih.gov/pcassay/ | Covering 5000 protein targets and 30,000 gene targets, and providing over 130 million bioactivity outcomes. | Yes | [45] |
| Therapeutic Target Database | https://bidd.nus.edu.sg/group/ttd/ttd.asp | Providing the known and explored therapeutic protein and nucleic acid targets, the targeted diseases, pathway information and corresponding drugs directed at each of these targets. | No | [46] |
| SIDER | https://sideeffects.embl.de/ | Containing marketed medicines and their recorded side effects, as well as drug-target associations. | No | [47] |
| SwissTargetPrediction | https://www.swisstargetprediction.ch/ | Inferring the targets of small molecules based on the combination of 2D and 3D similarity values with known ligands. | No | [48] |
| DGIdb 3.0 | https://dgidb.org/ | Containing > 40,000 genes and > 10,000 drugs involved in > 100,000 drug-gene interactions. | No | [49] |
| TargetNet | https://targetnet.scbdd.com/ | Netting or predicting the binding of multiple targets for any given molecule. | No | [50] |
| HIT 2.0 | https://hit2.badd-cao.net/ | A comprehensive searching and curation platform for CTI information based on literature evidence. | No | [51] |
2D: two dimensions; 3D: three dimensions; CTI: compound-target interaction.
Multi-omics technologies (e.g., transcriptomics and metabolomics) could pave the discovery of potential CTIs. For example, Dai et al. [52] combined the target information collected from publicly available databases and their own transcriptomics data of celastrol treatment for osteoarthritis. Liu et al. [53] screened the potential targets of Danggui Buxue Decoction against anemia by integrating the data-mined upstream proteins of the differential metabolites from metabonomics and the anemia-associated targets obtained from GeneCards database (https://www.genecards.org/). Similarly, metabolic proteins related to potential biomarkers from metabolomics and predicted proteins of cantharidin were all introduced into String database (https://string-db.org/) to conduct a protein–protein interaction analysis. The hub targets were then filtered by mean degree value and selected for network pharmacology analysis [54]. Thus, it is feasible to unveil the potential CTIs of CHM via computational methods integrated with multi-omics strategy.
2.3. Strategies for network analysis
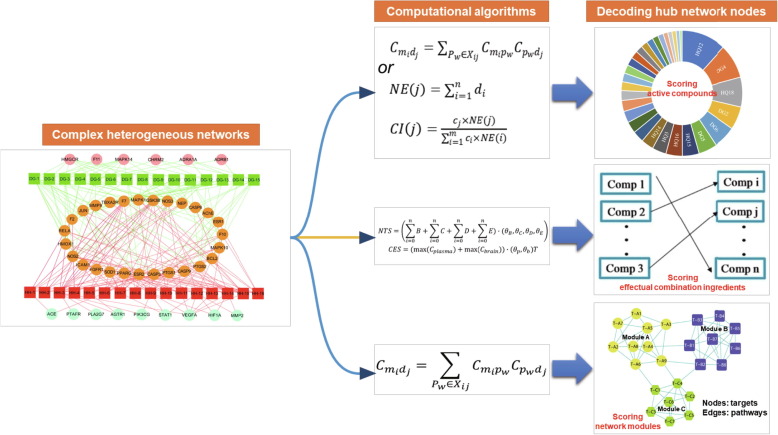
Network thinking has contributed a number of important unanticipated insights on the complex mechanisms underlying CHM, so how to extract key information from the heterogeneous networks is the main goal. Many network-based computational approaches have been conducted to excavate effective components and hub targets (Fig. 1 ).
Fig. 1.
Network-based computational algorithms to excavate effective components and underlying mechanisms of Chinese herbal medicine.
2.3.1. Scoring active compounds
As a demonstrative example, we proposed a contribution index (CI) to estimate each active compound’s contribution to the efficacy of CHM based on network topology property (NE) and efficacy weight. The CI was proposed and calculated by equations (1) and (2):
| (1) |
| (2) |
where n is the target number of compound j in the compound-target network; di is the target i’s degree of compound j in the target-pathway network; ci is the number of disease-related literature of compound i; m is the number of compounds; NE(j) is the NE of compound j; CI(j) is the CI of compound j [55], [56].
Likewise, Wang et al. [57] proposed a contribution score (CS) to evaluate the effectiveness of each compound of two herbs in a Chinese patent medicine. CAi and CBi represent the degree of each compound only in compound-target network of herb A and herb B, respectively; Cedge and Tedge represent the edge of compounds and targets in compound-target network, respectively; P represents the degree of each protein; Aij is the index of affinity determined from the ωei value as shown in equations (3), (4), (5).
| (3) |
| (4) |
| (5) |
Gao et al. [58] let m denote compounds, n denote the specific disease genes, Xij (i = 1,…, m; j = 1,…, n) represent a compound-target interacting score (Xij ranged from 0 to 1), and Cj represent the correlation coefficient (specifically, Cj = 0.1 [degree < 5], Cj = 0.2 [5 ≤ degree < 10], Cj = 0.3 [10 ≤ degree < 20], and Cj = 0.4 [degree ≥ 20]). The anti-aging score of compound i (AAi) was calculated by the following formula, i.e., equation (6):
| (6) |
Wang et al. [59] performed a computational algorithm with Fisher’s exact test method (equation [7]) to investigate and rank the active compounds of a prescription in treating specific disease: specifically, if compounds have no known targets, n = 20 and k was the number of disease-related genes in n; if compounds have s known targets, n = 20 + s, and k was s plus the number of disease-related genes in n. N was the number of protein-coding genes in the constructed network and K was the number of all disease-related genes in the network. P value was calculated and adjusted by Benjamini-Hochberg method, for ranking all compounds.
| (7) |
Zhang et al. [60] developed an index of effective rate, indicating the possibility of a compound affecting a specified function (see equation [8]). The outdegree and sub-outdegree mean the number of putative targets for each compound and the number of putative targets for a specific function, respectively. This algorithm has also been successfully applied in our previous network pharmacology study of Danggui Buxue Decoction [61].
| (8) |
Suo et al. [62] assumed that if one unit of information comes to a node of degree kj, it flows downstream through kj − 1 branch, each of which transfers 1/kj − 1 unit of the original information. Then, the scoring scheme for the active ingredients can be evaluated as equations (9)−(11),
| (9) |
| (10) |
| (11) |
where V(i) is the protein nodes between n and m in path i; Ii(m → n) is the effectiveness of ingredient m on target n; I(m → n) gives the specificity of ingredient m to target n; and I(m) shows the overall effectiveness of ingredient m on the disorder under investigation.
Considering that CHM compounds vary dramatically in content, we proposed another CI based on both the intrinsic properties (active components’ content and OB) of CHM and the rank-sum ratio (RSR) of integrated network topology parameters (including degree, closeness, betweenness, eccentricity, neighborhood connectivity and average shortest path length) of active compounds in the heterogeneous network [63], as equation (12):
| (12) |
where CIi,j is the CI of component j in herb i, mi is the weight of herb i in a formula, n is the total count of herbs in a formula, Ci,j is the content of component j in herb i, and Mj is the molecular weight of component j; OBj represents the OB value of compound j retrieved from the TCMSP database (https://old.tcmsp-e.com/tcmsp.php); and RSRj is the RSR of component j in the compound-target-pathway network.
2.3.2. Scoring effectual-combination ingredients
With the attempt to find the effectual-combination ingredients (ECIs) from CHM, a strategy was proposed by Liu et al. [64]; it was defined as metabolic exposure-oriented network regulation for identification of ECIs, including network topology score (NTS) and component exposure score (CES) (see equations [13] and [14]). NTS represents the topological importance of a compound’s targets in CHM-regulated network, by calculating “Betweenness” (B), “Closeness” (C), “Degree” (D), and “Eigenvector centrality” (E) of all regulated genes through principal component analysis (PCA). CES represents the metabolic exposure of each compound by PCA integrating the Cmax of plasma and brain of each compound. Finally, the combinatory compounds of which the NTS or CES was above 2, 1 and 0 were selected as candidate ECIs.
| (13) |
| (14) |
Similarly, Luo et al. [65] screened ECIs of a formula by combination of NTS and variable importance in projection (VIP) value. The VIP values of differential absorbed components in plasma were revealed by metabolomics-driven strategy coupled with the orthogonal-partial-least-squares-discrimination analysis. The combinatory compounds of which the VIP was above 1.5, 1.25 and 1.0 or NTS was above 2, 1 and 0 were selected as candidate ECIs.
2.3.3. Scoring network modules
To estimate the intensity of associating a specific network module with a specific disease, Zuo et al. [66] used an algorithm in the “targets-(pathways)-targets” network, as equation (15):
| (15) |
where Xij is a subset of P and refers to the pathways that are relevant to mi and dj simultaneously; Cmidj refers to the CS of mi to dj, which is the sum of the contribution of mi to dj through all its relevant pw in Xij. The value of Cmidj varies from 0 to 1: the higher the value, the greater the contribution mi might make to dj; and all the modules contribute 1 to a particular disease.
Recently, in order to integrate the target score information of the TCM prescription as well as the disease, Xiong et al. [67] used two iterations of PageRank algorithm to obtain the PageRank value of targets in prescription-disease system shown in equations (16), (17), (18).
| (16) |
| (17) |
| (18) |
where V0 is a target score vector of a prescription or a disease; V1 and V2 are the target score vectors after the first and second iterations; N denotes the total number of targets; M is a symmetric adjacency stochastic matrix which denotes target interaction network; α ∈ (0, 1) is a constant representing the importance of the network while ranking targets. With target score vector Vp of a TCM prescription and target interaction matrix M, PageRank score vector Vrank1 was achieved; with target score vector Vd of a disease and matrix M, PageRank score vector Vrank2 was achieved; finally the average PageRank score vector Vavg was achieved.
2.3.4. Network cluster/subgroup analysis
Generally, a network can be analyzed from three different levels: individual, subgroup and whole network. Clusters/subgroups refer to highly interconnected regions distilled from different, complex objects with similar underlying properties. It is of great significance to divide the biological regulatory network into subgroups/clusters to analyze and identify key node groups. Currently, various methods have been reported to dissect the cluster/subcluster structure of networks. Some of these methods are graph theory-based (spectral dichotomy and Kernighan-Lin algorithm), such as sociological-based methods (-plexes, -cores, and maximal clique algorithms) and cluster-based methods (optimization correlation algorithms and similarity correlation methods) [68]. For example, Song et al. [69] conducted a cluster analysis on the network of Maxing Shigan Decoction in treating asthma, and found that it involved 5 functional clusters such as gene expression, silencing and replication, DNA/RNA damage repair and transcriptional regulation, and inflammatory immune response. Therefore, the cluster/subgroup analysis of CHM network pharmacology will help to identify active ingredient and key target groups for disease prevention and treatment.
Overall, the development of network analysis methodology is critically important for finding effective components in the discovery pipelines and generating systematic insights into the mechanism of action of CHM in the treatment of diseases.
2.4. Databases and web servers
2.4.1. Web servers for Gene Ontology enrichment and pathway analysis
Since TCM involves using multi-compound, multi-target agents, annotating their targets in the context of networks can help reveal its mechanisms of action. Gene Ontology resource (GO, https://geneontology.org) is the most comprehensive knowledge base concerning the functions of genes/targets. It provides three major categories of controlled terms for describing gene products: molecular function (activity of gene products at the molecular level), cellular component (location of gene product activity relative to biological structures), and biological process (larger biological programs that exploit gene molecular function) [70]. Identifying GO terms within a given gene list can provide a better understanding of the genes involved in these functions and further elucidate the role of CHM in regulating genes involved in improving disease processes. Furthermore, pathway analysis has become the preferred choice for gaining insights into the underlying biology of differentially expressed genes and proteins. The action of drugs is not only related to target proteins, but also affected by the biological pathways of the target proteins, especially for multi-target CHM. Table 2 lists several web sources for GO enrichment and pathway analysis [71], [72], [73], [74], [75], [76]. Among them, the Reactome knowledgebase provides molecular details of signal transduction, transport, DNA replication, metabolism and other cellular processes as an ordered network of molecular transformations in a single consistent data model [71]; and the STRING database aims to integrate all known and predicted associations between proteins, including both physical interactions and functional associations, which relies on the annotated proteomes maintained by Swiss-Prot (https://www.expasy.org/sprot/ and https://www.ebi.ac.uk/swissprot/) [72].
Table 2.
Several web sources for GO enrichment and pathway analysis.
| Web server | Website | Contents and main features | Reference |
|---|---|---|---|
| Reactome | https://reactome.org | Including molecular details of signal transduction, transport, DNA replication, metabolism, and other cellular processes as an ordered network of molecular transformations—an extended version of a classic metabolic map, in a single consistent data model. | [71] |
| STRING | https://string-db.org/ | Aims to integrate all known and predicted associations between proteins, including both physical interactions and functional associations. | [2] |
| Gene Ontology Annotation Database | https://www.ebi.ac.uk/GOA | Including evidence-based GO annotations to proteins in the UniProt knowledgebase; supplies 368 million GO annotations to almost 54 million proteins in more than 480,000 taxonomic groups. | [73] |
| GOATOOLS | https://github.com/tanghaibao/goatools | A Python-based library, making it more efficient to stay current with the latest ontologies and annotations; performs gene ontology enrichment analyses to determine over- and under-represented terms, and organizes results for greater clarity and easier interpretation using a novel GOATOOLS GO grouping method. | [4] |
| PANTHER | https://pantherdb.org | A multifaceted data resource for classification of protein sequences by evolutionary history, and by function. | [75] |
| Metascape | https://metascape.org/ | A web-based portal designed to provide a comprehensive gene list annotation and analysis resource for experimental biologists, including functional enrichment, interactome analysis, gene annotation, and membership search. | [76] |
GO: Gene Ontology.
2.4.2. Databases for CHM network pharmacology
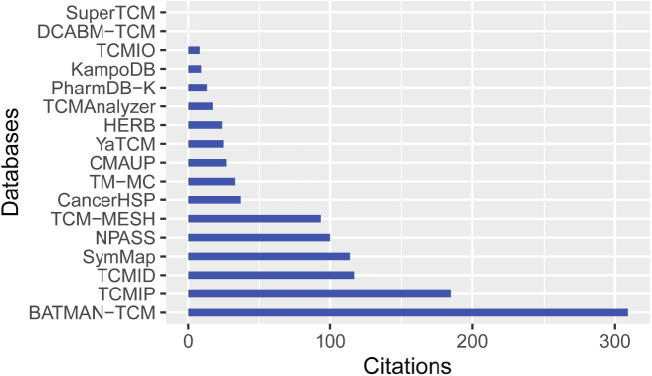
Network pharmacology is a new frontier that is becoming a paradigm for investigating the therapeutic mechanism of CHM from a systemic and molecular perspective. During the past five years, with the aid of cheminformatics and big data science, many high-quality databases have been curated to support CHM network pharmacology research and CHM repositioning. Some of the important databases are introduced in Table 3 [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], [90], [91], [92], [93]. From the citation metrics in Fig. 2 , four specialized databases in network pharmacology were cited over 100 times. More importantly, the majority of them were built via curating TCMSP, a well-known database for CHM network pharmacology with 1346 citations as of the time of this writing. TCMSP includes 29,384 ingredients, 3311 targets and 837 associated diseases, as well as 12 ADME-related properties involving DB, DL, intestinal epithelial permeability and BBB, etc. [94]. The majority of them, however, suffer from common limitations, such as lack of experimental evidence in the cited literature, as well as lack of ingredient composition of CHM from high-performance liquid chromatography or mass spectrographic analysis. In addition, it is worth paying attention to how to reasonably analyze the large number of targets predicted from the databases. On the one hand, a goal of CHM network pharmacology is to predict the potential efficacy or disease spectrum of CHM, so further analysis of the whole genome predicted by CHM is appropriate; on the other hand, it is to explore the active ingredients and mechanism of action of CHM in the treatment of specific diseases, so focusing on drug targets is also needed. More importantly, these databases are slow to update, thereby lagging behind the latest development of CHM research. Therefore, a more comprehensive and complete database with powerful web services is necessary and will more effectively promote the network pharmacology research of CHM.
Table 3.
Several representative databases for CHM network pharmacology research.
| Database and web server | Website | Contents and main features | Reference |
|---|---|---|---|
| BATMAN-TCM | https://bionet.ncpsb.org/batman-tcm/ | Including (1) ingredients’ target prediction; (2) functional enrichment analyses of targets; (3) the visualization of ingredient-target-pathway/disease association network and KEGG pathway; (4) comparison analysis of multiple CHMs. | [77] |
| TCMIP (including ETCM) | https://www.tcmip.cn/TCMIP/index.php; https://www.nrc.ac.cn:9090/ETCM/ | Including 403 herbs, 3962 formulae, 7274 herbal ingredients, 2266 validated or predicted drug targets, and 3027 related diseases. | [78] |
| TCMID | https://47.100.169.139:8000/tcmid/ | Containing approximately 47,000 prescriptions, 8159 herbs, 25,210 compounds, 6828 drugs, 17,521 targets and 3791 diseases. | [79] |
| SymMap | https://www.symmap.org/ | Focusing on TCM symptoms and their relationships to herbs and diseases. | [80] |
| NPASS | https://bidd2.nus.edu.sg/NPASS/ | Providing 35,032 natural products, 25,041 species, 5863 targets; containing 222,092 natural product-target pairs and 288,002 natural product-species pairs. | [81] |
| TCM-Mesh | https://mesh.tcm.microbioinformatics.org/ | Containing 6235 herbs, 383,840 compounds, 14,298 genes, 6204 diseases, 144,723 gene-disease associations, and a web-based software to construct a network between herbs and treated diseases. | [82] |
| CancerHSP | https://lsp.nwsuaf.edu.cn/CancerHSP.php | Including 2439 anticancer herbs, 2439 active compounds, and activity data based on 492 cancer cell lines. | [83] |
| TM-MC | https://informatics.kiom.re.kr/compound/ | Including 536 medicinal materials, 14,492 compounds, and 24,154 links between them. | [84] |
| CMAUP | https://bidd2.nus.edu.sg/CMAUP/ | Including 47,645 active ingredients against 646 targets in 234 KEGG pathways associated with 2473 gene ontologies and 656 diseases. | [85] |
| YaTCM | https://cadd.pharmacy.nankai.edu.cn/yatcm/home | Containing 6220 herbs, 47,696 herbal compounds, 18,697 targets, 1907 predicted targets, 390 pathways and 1813 prescriptions. | [86] |
| HERB | https://herb.ac.cn/ | Linking 7263 herbs and 49,258 ingredients to 12,933 targets and 28,212 diseases, and providing six pairwise relationships among them. | [87] |
| TCMAnalyzer | https://www.rcdd.org.cn/tcmanalyzer | Allowing to (1) identify the potential compounds that are responsible for the bioactivities for a CHM through scaffold-activity relation search techniques, (2) investigate the molecular mechanism for a CHM at the systemic level, and (3) explore the potentially targeted bioactive herbs. | [88] |
| PharmDB-K | https://pharmdb-k.org | Containing 262 traditional medicines, 7815 drugs, 32,373 proteins, 3721 diseases, and 1887 side effects. | [89] |
| KampoDB | https://wakanmoview.inm.u-toyama.ac.jp/kampo/ | Containing 42 traditional medicines, 54 drugs, 1230 compounds, 460 known targets, and 1369 potential targets, together with biological pathways and molecular function annotations. | [90] |
| TCMIO | https://tcmio.xielab.net/ | Including the data of TCM on immuno-oncology. | [91] |
| DCABM-TCM | https://bionet.ncpsb.org.cn/dcabm-tcm/#/Home | Including 4206 blood constituents, 194 herbs and 192 prescriptions. | [92] |
| SuperTCM | https://tcm.charite.de/supertcm | Providing the information about 6516 CHMs with 5372 botanical species, 55,772 active ingredients against 543 targets in 254 KEGG pathways associated with 8634 diseases. | [93] |
CHM: Chinese herbal medicine; KEGG: Kyoto encyclopedia of genes and genomes; TCM: traditional Chinese medicine.
Fig. 2.
The citation metrics of databases for Chinese herbal medicine (CHM) network pharmacology research and CHM repositioning. The citations were curated from Google Scholar on November 15, 2021.
3. Network pharmacology for mechanism elucidation of CHM against COVID-19
COVID-19 was initially reported at the end of 2019 and spread rapidly around the world. Hundreds of millions of lives have been affected during this pandemic [95], and the overwhelmed healthcare systems greatly impacted the global economy. As of January 16, 2022, there have been reported over 323 million confirmed cases and over 5.5 million deaths worldwide [96].
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) belongs to the same family of coronaviruses responsible for the severe acute respiratory syndrome in 2003 and Middle East respiratory syndrome in 2012 [97]. The emergence of SARS-CoV-2 variants is continuing to surge, causing a large number of virus replications. Since the outbreak, researchers are developing vaccines and targeted antiviral drugs. At the moment, vaccines are still one of the most important means to alleviate the COVID-19 pandemic. However, for some underdeveloped countries and regions, it remains difficult to achieve vaccination for the whole population. Thus, people are struggling to seek alternative ways to protect themselves from this virus or to reduce its severity.
The World Health Organization reported that > 80% of the world’s population rely on herbal medicines (involving TCM, Persian medicine, and traditional Indian medicine) for a particular aspect of their primary health care needs. TCM has a long history and plays a unique role in the prevention and treatment of major infectious diseases. During the SARS epidemic in 2003, TCM was shown to provide remarkable therapeutic effects [98]. TCM was included in the Chinese guideline on diagnosis and treatment of COVID-19 since the beginning, and abundant clinical usage demonstrated that CHM exerted positive effects against COVID-19 through increasing the cure rate of Western medications, and decreasing severity and concomitant symptoms [99]. The national percentage of treating confirmed cases of COVID-19 patients with CHM-integrated therapy exceeds 90% with the support from over 4900 TCM practitioners all over China [100]. And modern pharmacological studies have shown that CHM was effective in clearing heat and toxicity, as well as in dispelling dampness, thereby inhibiting the virus [101]. In most cases, CHM inhibits the virus-mediated inflammatory response by regulating the immune function of the body, which corresponds to the multi-target role of CHM in the overall regulation of the body’s systems [102]. Despite the widespread use and promising results of CHM in clinical practice, proving its effectiveness via scientific trials and dissecting the molecular mechanisms are still big challenges.
There is increasing evidence indicating the reliability and effectiveness of network pharmacology in expanding the rationale and mechanism for the clinical efficacy of CHM against COVID-19 [103]. In this pandemic, three herbal formulas have been key players used during different COVID-19 stages. Yang et al. [104] performed network pharmacology combined with experimental study on Qingfei Paidu Decoction and Maxing Shigan Decoction in treating COVID-19, revealing that the therapeutic effects against COVID-19 may be attributed to their anti-inflammatory effects via the thrombin and Toll-like receptor signaling pathway. Zhao et al. [105] also conducted a network pharmacological study to illustrate the immune regulation, anti-infection, anti-inflammation, and multi-organ protection mechanisms of Qingfei Paidu Decoction against COVID-19, and results showed that 88 high-confidence targets affected by SARS-CoV-2 infection of 12 active compounds in Qingfei Paidu Decoction were identified and involved in biological processes related with COVID-19 development, such as pattern recognition receptor signaling, interleukin signaling, cell growth and death, hemostasis, and injuries of the nervous, sensory, circulatory, and digestive systems. Zheng et al. [106] employed a network pharmacology approach and found that Lianhua Qingwen formula has the most relationship to the respiratory system, indicating specific effects in lung diseases, and modulates the inflammatory process, exerts antiviral effects and repairs lung injury. Moreover, it also relieves the “cytokine storm” and improves angiotensin-converting enzyme 2 (ACE2)-expression-disorder-caused symptoms. Ai et al. [107] performed network pharmacology on “Fei Yan No. 1,” a specific formula against COVID-19 recommended by the Health Commission of Hubei Province, and revealed that it may exert antiviral and immune response-regulatory effects through multiple pathways, also affecting influenza A, hepatitis B, hepatitis C, Kaposi sarcoma-associated herpesvirus infection, human cytomegalovirus infection, viral carcinogenesis and human immunodeficiency virus 1 infection.
Notably, network pharmacology shows an immense advantage in the feasibility analysis of drug repositioning, especially in the analysis of CHM ingredients. Wang et al. [108] applied network pharmacology (integrated network proximity and network diffusion) to quantify the relationship between CHM ingredients’ targets and COVID-19 disease targets in the protein–protein-interaction network, thereby predicting new anti-COVID-19 ingredients. Quercetin, luteolin, acacetin and kaempferol were screened as anti-COVID-19 candidates. Other CHM ingredients such as berberine [109], emodin [110], astragaloside IV [111], matrine [112], puerarin [113], glycyrrhizic acid [114], hesperidin, isorhapontigenin and gallocatechin-7-gallate [115] were also repurposed as therapeutic candidates against COVID-19 based on network pharmacology analysis (Fig. 3 ), and some typical compounds (berberine, emodin and glycyrrhizic acid, etc.) have been validated by molecular docking and dynamics simulation as potential inhibitors for different proteins of SARS-CoV-2 or a drug in treating COVID-19 cytokine storm [116], [117], [118]. In preclinical explorations on cell or animal models, luteolin, quercetin, emodin, glycyrrhizic acid, hesperidin, isorhapontigenin and gallocatechin-7-gallate have been successfully validated to block the SARS-CoV-2 replication or the spike protein and ACE2 interaction [115], [119].
Fig. 3.
Rich resources of Chinese herbal medicine ingredient repositioning via network pharmacology for coronavirus disease 2019 treatment mainly targeting SARS-CoV-2 replication, ACE2 receptor and/or cytokine storm. ACE2: angiotensin-converting enzyme 2; 3CLpro: 3-chymotrypsin-like protease; CXCL: chemokine (CXC motif) ligand 1; IL-6: interleukin-6; PLpro: P-like protease; RBD: receptor-binding domain; RdRp: RNA-dependent RNA polymerase; SARS-CoV-2: severe acute respiratory syndrome coronavirus 2; TNF-α: tumor necrosis factor-α.
Although network pharmacology research on CHM against COVID-19 is expanding, there are still many limitations that need to be discussed and resolved [99]. For example, the CTIs obtained by database-based strategy should be experimentally validated. TCM as adjunctive therapy to Western medication remains the mainstream treatment of COVID-19 in China, so a systematic network-based model should be built to better understand how integrated Chinese and Western medicine works together. Last but not least, it is necessary to combine network pharmacology-based identification, experimental validation and clinical data.
4. Concluding remarks
Network pharmacology presents an immense scope for exploring traditional knowledge to find solutions for the modernization of TCM. In this review, we mainly introduced many pioneering explorations on active compound identification, CTI prediction and network topology analysis embedded in the workflow of network pharmacology as well as specialized databases, thereby providing reference for deciphering the exact mechanisms of CHM against diseases. The spotlight of network pharmacology in the mechanistic investigation and repositioning of CHMs against COVID-19 was also discussed.
Recently, the first international standard for evaluating network pharmacology—“Network Pharmacology Evaluation Method Guidance” (2021) was released by the World Federation of Chinese Medicine Societies, to promote more standardized implementation of network pharmacology by result verification from the perspective of computer models, experimental models, and clinical data [120]. From this, we conclude that (1) the best future direction for network pharmacology is to integrate the post-network analysis (e.g., molecular docking and simulation) and the experimental and clinical data, such as analytic and multi-omics data; (2) the reliability and repeatability of the network pharmacology results should be improved; (3) more robust computing algorithms/softwares should be developed for the systematic screening, integration, and processing of data on various compounds, genes, and proteins; (4) the specialized database development with high data quality and quantity along with constant updating and regulation is very necessary. Collectively, by integrating reductionist and systems approaches as well as computational and experimental methods, network pharmacology will accelerate the modernization process of TCM in the future.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No. 81903786), the Shaanxi Natural Science Foundation of China (No. 2021JQ-731), and Subject Innovation Team of Shaanxi University of Chinese Medicine (No. 2019-YL10).
Authors’ contributions
SJY and YPT conceived of and proposed the idea. SJY and YXW designed the study. YXW, ZY and WXW reviewed the literature. SJY, YXH, QZ, JJL and YPT contributed to writing, revising and proof-reading the manuscript. All authors read and approved the final manuscript.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Huang Y.X., Xu D.Q., Yue S.J., Chen Y.Y., Tao H.J., Fu R.J., et al. Deciphering the active compounds and mechanisms of Qixuehe Capsule on qi stagnation and blood stasis syndrome: a network pharmacology study. Evid Based Complement Alternat Med. 2020;2020:5053914. doi: 10.1155/2020/5053914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li S., Fan T., Jia W., Lu A., Zhang W. Network pharmacology in traditional Chinese medicine. Evid Based Complement Alternat Med. 2014;2014 doi: 10.1155/2014/138460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao J., Yang J., Tian S., Zhang W. A survey of web resources and tools for the study of TCM network pharmacology. Quant Biol. 2019;7(1):17–29. [Google Scholar]
- 4.Babu M.M., Mori H. Computational and systems biology. Mol Biosyst. 2009;5(12):1391. doi: 10.1039/b921381n. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y., Kirchmair J. Cheminformatics in natural product-based drug discovery. Mol Inform. 2020;39(12):e2000171. doi: 10.1002/minf.202000171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lai X., Wang X., Hu Y., Su S., Li W., Li S. Editorial: network pharmacology and traditional medicine. Front Pharmacol. 2020;11:1194. doi: 10.3389/fphar.2020.01194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X., Wang Z.Y., Zheng J.H., Li S. TCM network pharmacology: a new trend towards combining computational, experimental and clinical approaches. Chin J Nat Med. 2021;19(1):1–11. doi: 10.1016/S1875-5364(21)60001-8. [DOI] [PubMed] [Google Scholar]
- 8.Lee W., Lee C.Y., Kim Y.S., Kim C.E. The methodological trends of traditional herbal medicine employing network pharmacology. Biomolecules. 2019;9(8):362. doi: 10.3390/biom9080362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deng J., Ma Y., He Y., Yang H., Chen Y., Wang L., et al. A network pharmacology-based investigation to the pharmacodynamic material basis and mechanisms of the anti-inflammatory and anti-viral effect of Isatis indigotica. Drug Des Dev Ther. 2021;15:3193–3206. doi: 10.2147/DDDT.S316701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L., Guo R., Cao N., Lin Y., Yang W., Pei S., et al. An integrative pharmacology-based pattern to uncover the pharmacological mechanism of ginsenoside H dripping pills in the treatment of depression. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.590457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z., Cheng L., Shang Z., Li Z., Zhao Y., Jin W., et al. Network pharmacology for analyzing the key targets and potential mechanism of wogonin in gliomas. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.646187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh K., Adnan M., Cho D.H. Network pharmacology study on Morus alba L. leaves: pivotal functions of bioactives on RAS signaling pathway and its associated target proteins against gout. Int J Mol Sci. 2021;22(17):9372. doi: 10.3390/ijms22179372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oh K.K., Adnan M., Ju I., Cho D.H. A network pharmacology study on main chemical compounds from Hibiscus cannabinus L. leaves. RSC Adv. 2021;11(19):11062–11082. doi: 10.1039/d0ra10932k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oh K.K., Adnan M., Cho D.H. A network pharmacology analysis on drug-like compounds from Ganoderma lucidum for alleviation of atherosclerosis. J Food Biochem. 2021;45(9):e13906. doi: 10.1111/jfbc.13906. [DOI] [PubMed] [Google Scholar]
- 15.Liu F., Du X., Liu P., Sun Y., Zhang Y. Screening and analysis of key active constituents in Guanxinshutong Capsule using mass spectrum and integrative network pharmacology. Chin J Nat Med. 2018;16(4):302–312. doi: 10.1016/S1875-5364(18)30060-8. [DOI] [PubMed] [Google Scholar]
- 16.Xi D., Bao T., Chen Q., Chen S., Cheng Y.C., Cullen J., et al. State of the science: cancer complementary and alternative medicine therapeutics research—NCI strategic workshop highlights of discussion report. J Natl Cancers Inst Monogr. 2017;2017(52):lgx003. doi: 10.1093/jncimonographs/lgx003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tian D., Gao Q., Lin J., Chang Z., Wang Y., Shi Y., et al. Uncovering the mechanism of the Shenzhi Jiannao formula against vascular dementia using a combined network pharmacology approach and molecular biology. Phytomedicine. 2021;90 doi: 10.1016/j.phymed.2021.153637. [DOI] [PubMed] [Google Scholar]
- 18.Chen X., Wang X., Ma L., Fang S., Li J., Boadi E.O., et al. The network pharmacology integrated with pharmacokinetics to clarify the pharmacological mechanism of absorbed components from Viticis fructus extract. J Ethnopharmacol. 2021;278 doi: 10.1016/j.jep.2021.114336. [DOI] [PubMed] [Google Scholar]
- 19.Wang L., Pu X., Nie X., Wang D., Jiang H., Chen Y., et al. Integrated serum pharmacochemistry and network pharmacological analysis used to explore possible anti-rheumatoid arthritis mechanisms of the Shentong-Zhuyu decoction. J Ethnopharmacol. 2021;273 doi: 10.1016/j.jep.2021.113988. [DOI] [PubMed] [Google Scholar]
- 20.Feng G., Li S., Liu S., Song F., Pi Z., Liu Z. Targeted screening approach to systematically identify the absorbed effect substances of Poria cocos in vivo using ultrahigh performance liquid chromatography tandem mass spectrometry. J Agric Food Chem. 2018;66(31):8319–8327. doi: 10.1021/acs.jafc.8b02753. [DOI] [PubMed] [Google Scholar]
- 21.Zhai L., Ning Z., Huang T., Wen B., Liao C.H., Lin C.Y., et al. Cyclocarya paliurus leaves tea improves dyslipidemia in diabetic mice: a lipidomics-based network pharmacology study. Front Pharmacol. 2020;9:973. doi: 10.3389/fphar.2018.00973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu C., Cai T., Jin Y., Chen J., Liu G., Xu N., et al. Artificial intelligence and network pharmacology based investigation of pharmacological mechanism and substance basis of Xiaokewan in treating diabetes. Pharmacol Res. 2020;159 doi: 10.1016/j.phrs.2020.104935. [DOI] [PubMed] [Google Scholar]
- 23.Zhang F.X., Li Z.T., Li M., Yuan Y.L.L., Cui S.S., Chen J.X., et al. Dissection of the potential anti-influenza materials and mechanism of Lonicerae Japonicae Flos based on in vivo substances profiling and network pharmacology. J Pharm Biomed Anal. 2021;193 doi: 10.1016/j.jpba.2020.113721. [DOI] [PubMed] [Google Scholar]
- 24.Gao Y., Shi W., Yao H., Ai Y., Li R., Wang Z., et al. An integrative pharmacology based analysis of refined Liuweiwuling against liver injury: a novel component combination and hepaprotective mechanism. Front Pharmacol. 2021;12 doi: 10.3389/fphar.2021.747010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang F.X., Yuan Y.L.L., Cui S.S., Li M., Li R.M. Characterization of metabolic fate of phellodendrine and its potential pharmacological mechanism against diabetes mellitus by ultra-high-performance liquid chromatography-coupled time-of-flight mass spectrometry and network pharmacology. Rapid Commun Mass Spectrom. 2021;35(18):e9157. doi: 10.1002/rcm.9157. [DOI] [PubMed] [Google Scholar]
- 26.Zhang F.X., Li Z.T., Li M., Yuan Y.L.L., Cui S.S., Wang G.H., et al. An integrated strategy for revealing the pharmacological changes based on metabolites profiling and network pharmacology: arctiin as an example. J Chromatogr B. 2020;1157 doi: 10.1016/j.jchromb.2020.122270. [DOI] [PubMed] [Google Scholar]
- 27.Zhang F.X., Yuan Y.L.L., Cui S.S., Wang G.H., Li R.M. Revealing the potential pharmacological mechanism of traditional Chinese medicine by integrating metabolite profiling of a Q-marker and network pharmacology, prim-O-glucosylcimifugin as an example. New J Chem. 2021;45:15571. [Google Scholar]
- 28.Fu J., Wu H., Wu H., Ran D., Sun M. Deciphering the metabolic profile and pharmacological mechanisms of Achyranthes bidentata Blume saponins using ultra-performance liquid chromatography quadrupole time-of-flight mass spectrometry coupled with network pharmacology-based investigation. J Ethnopharmacol. 2021;274 doi: 10.1016/j.jep.2021.114067. [DOI] [PubMed] [Google Scholar]
- 29.Li X., Yang H., Xiao J., Zhang J., Zhang J., Liu M., et al. Network pharmacology based investigation into the bioactive compounds and molecular mechanisms of Schisandrae Chinensis Fructus against drug-induced liver injury. Bioorg Chem. 2020;96 doi: 10.1016/j.bioorg.2019.103553. [DOI] [PubMed] [Google Scholar]
- 30.Luo Z., Liu Y., Han X., Yang W., Wang G., Wang J., et al. Mechanism of Paeoniae Radix Alba in the treatment of non-alcoholic fatty liver disease based on sequential metabolites identification approach, network pharmacology, and binding affinity measurement. Front Nutr. 2021;8 doi: 10.3389/fnut.2021.677659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang C., Liu C., Qu Y., Cao Y., Liu R., Sun Y., et al. LC-MS-based qualitative analysis and pharmacokinetic integration network pharmacology strategy reveals the mechanism of Phlomis brevidentata H.W.Li treatment of pneumonia. ACS. Omega. 2021;6(6):4495–4505. doi: 10.1021/acsomega.0c06201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo R., Zhao M., Liu H., Su R., Mao Q., Gong L., et al. Uncovering the pharmacological mechanisms of Xijiao Dihuang Decoction combined with Yinqiao Powder in treating influenza viral pneumonia by an integrative pharmacology strategy. Biomed Pharmacother. 2021;141 doi: 10.1016/j.biopha.2021.111676. [DOI] [PubMed] [Google Scholar]
- 33.Guo Y., Cui Q., Ren S., Hao D., Morikawa T., Wang D., et al. The hepatoprotective efficacy and biological mechanisms of three phenylethanoid glycosides from Cistanches Herba and their metabolites based on intestinal bacteria and network pharmacology. J Nat Med. 2021;75:784–797. doi: 10.1007/s11418-021-01508-y. [DOI] [PubMed] [Google Scholar]
- 34.Luo T.T., Lu Y., Yan S.K., Xiao X., Rong X.L., Guo J. Network pharmacology in research of Chinese medicine formula: methodology, application and prospective. Chin J Integr Med. 2020;26(1):72–80. doi: 10.1007/s11655-019-3064-0. [DOI] [PubMed] [Google Scholar]
- 35.Wu Z., Li W., Liu G., Tang Y. Network-based methods for prediction of drug-target interactions. Front Pharmacol. 2018;9:1134. doi: 10.3389/fphar.2018.01134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang J., Wu Z., Cai C., Wang Q., Tang Y., Cheng F. Quantitative and systems pharmacology. 1. In silico prediction of drug-target interactions of natural products enables new targeted cancer therapy. J Chemical Inf Model. 2017;57(11):2657–2671. doi: 10.1021/acs.jcim.7b00216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hsin K.Y., Matsuoka Y., Asai Y., Kamiyoshi K., Watanabe T., Kawaoka Y., et al. SystemsDock: a web server for network pharmacology-based prediction and analysis. Nucleic Acids Res. 2016;44(W1):W507–W513. doi: 10.1093/nar/gkw335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X., Pan C., Gong J., Liu X., Li H. Enhancing the enrichment of pharmacophore-based target prediction for the polypharmacological profiles of drugs. J Chemical Inf Model. 2016;56(6):1175–1183. doi: 10.1021/acs.jcim.5b00690. [DOI] [PubMed] [Google Scholar]
- 39.Wang C., Kurgan L. Survey of similarity-based prediction of drug-protein interactions. Curr Med Chem. 2020;27(35):5856–5886. doi: 10.2174/0929867326666190808154841. [DOI] [PubMed] [Google Scholar]
- 40.Cheng R., Liu X., Jin S., Lin J., Liu J. Machine learning for drug-target interaction prediction. Molecules. 2018;23(9):2208. doi: 10.3390/molecules23092208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang L., You Z.H., Chen X., Xia S.X., Liu F., Yan X., et al. A computational-based method for predicting drug-target interactions by using stacked autoencoder deep neural network. J Computational Biol. 2018;25(3):361–373. doi: 10.1089/cmb.2017.0135. [DOI] [PubMed] [Google Scholar]
- 42.Ahmed A., Smith R.D., Clark J.J., Dunbar J.B., Jr, Carlson H.A. Recent improvements to Binding MOAD: a resource for protein-ligand binding affinities and structures. Nucleic Acids Res. 2015;43(Database issue):D465–D469. doi: 10.1093/nar/gku1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ursu O., Holmes J., Bologa C.G., Yang J.J., Mathias S.L., Stathias V., et al. DrugCentral 2018: an update. Nucleic Acids Res. 2019;47(D1):D963–D970. doi: 10.1093/nar/gky963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harding S.D., Sharman J.L., Faccenda E., Southan C., Pawson A.J., Ireland S., et al. The IUPHAR/BPS Guide to PHARMACOLOGY in 2018: updates and expansion to encompass the new Guide to IMMUNOPHARMACOLOGY. Nucleic Acids Res. 2018;46(D1):D1091–D1106. doi: 10.1093/nar/gkx1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y., Bryant S.H., Cheng T., Wang J., Gindulyte A., Shoemaker B.A., et al. PubChem BioAssay: 2017 update. Nucleic Acids Res. 2017;45(D1):D955–D963. doi: 10.1093/nar/gkw1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Y.H., Yu C.Y., Li X.X., Zhang P., Tang J., Yang Q., et al. Therapeutic target database update 2018: enriched resource for facilitating bench-to-clinic research of targeted therapeutics. Nucleic Acids Res. 2018;46(D1):D1121–D1127. doi: 10.1093/nar/gkx1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuhn M., Letunic I., Jensen L., Bork P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016;44(D1):D1075–D1079. doi: 10.1093/nar/gkv1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Daina A., Michielin O., Zoete V. SwissTargetPrediction: updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 2019;47(W1):W357–W364. doi: 10.1093/nar/gkz382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cotto K.C., Wagner A.H., Feng Y.Y., Kiwala S., Coffman A.C., Spies G., et al. DGIdb 3.0: a redesign and expansion of the drug-gene interaction database. Nucleic Acids Res. 2018;46(D1):D1068–D1073. doi: 10.1093/nar/gkx1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao Z.J., Dong J., Che Y.J., Zhu M.F., Wen M., Wang N.N., et al. TargetNet: a web service for predicting potential drug-target interaction profiling via multitarget SAR models. J Comput Aided Mol Des. 2016;30(5):413–424. doi: 10.1007/s10822-016-9915-2. [DOI] [PubMed] [Google Scholar]
- 51.Yan D., Zheng G., Wang C., Chen Z., Mao T., Gao J., et al. HIT 2.0: an enhanced platform for Herbal Ingredients’ Targets. Nucleic Acids Res. 2022;50(D1):D1238–D1243. doi: 10.1093/nar/gkab1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dai S., Wang H., Wang M., Zhang Y., Zhang Z., Lin Z. Comparative transcriptomics and network pharmacology analysis to identify the potential mechanism of celastrol against osteoarthritis. Clin Rheumatol. 2021;40(10):4259–4268. doi: 10.1007/s10067-021-05726-3. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y., Ju Y., Qin X. Studies on the compatibility mechanism and material basis of Danggui Buxue Decoction against anemia mice using metabonomics and network pharmacology. J Pharm and Pharmacol. 2021;73(6):767–777. doi: 10.1093/jpp/rgab016. [DOI] [PubMed] [Google Scholar]
- 54.He T., Liu J., Wang X., Duan C., Li X., Zhang J. Analysis of cantharidin-induced nephrotoxicity in HK-2 cells using untargeted metabolomics and an integrative network pharmacology analysis. Food Chem Toxicol. 2020;146 doi: 10.1016/j.fct.2020.111845. [DOI] [PubMed] [Google Scholar]
- 55.Yue S.J., Liu J., Feng W.W., Zhang F.L., Chen J.X., Xin L.T., et al. System pharmacology-based dissection of the synergistic mechanism of Huangqi and Huanglian for diabetes mellitus. Front Pharmacol. 2017;8:694. doi: 10.3389/fphar.2017.00694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yue S.J., Xin L.T., Fan Y.C., Li S.J., Tang Y.P., Duan J.A., et al. Herb pair Danggui-Honghua: mechanisms underlying blood stasis syndrome by system pharmacology approach. Sci Rep. 2017;7:40318. doi: 10.1038/srep40318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang C., Ren Q., Chen X.T., Song Z.Q., Ning Z.C., Gan J.H., et al. System pharmacology-based strategy to decode the synergistic mechanism of Zhi-zhu Wan for functional dyspepsia. Front Pharmacol. 2018;9:841. doi: 10.3389/fphar.2018.00841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao L., Duan D.D., Zhang J.Q., Zhou Y.Z., Qin X.M., Du G.H. A bioinformatic approach for the discovery of antiaging effects of baicalein from Scutellaria baicalensis Georgi. Rejuvenation Res. 2016;19(5):414–422. doi: 10.1089/rej.2015.1760. [DOI] [PubMed] [Google Scholar]
- 59.Wang T., Wu Z., Sun L., Li W., Liu G., Tang Y. A computational systems pharmacology approach to investigate molecular mechanisms of herbal formula Tian-Ma-Gou-Teng-Yin for treatment of Alzheimer’s disease. Front Pharmacol. 2018;9:668. doi: 10.3389/fphar.2018.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang M., Wu H., Guo F., Yu Y., Wei J., Geng Y., et al. Identification of active components in Yixinshu Capsule with protective effects against myocardial dysfunction on human induced pluripotent stem cell-derived cardiomyocytes by an integrative approach. Mol Biosyst. 2017;13(8):1469–1480. doi: 10.1039/c6mb00813e. [DOI] [PubMed] [Google Scholar]
- 61.Shi X.Q., Yue S.J., Tang Y.P., Chen Y.Y., Zhou G.S., Zhang J., et al. A network pharmacology approach to investigate the blood enriching mechanism of Danggui Buxue Decoction. J Ethnopharmacol. 2019;235:227–242. doi: 10.1016/j.jep.2019.01.027. [DOI] [PubMed] [Google Scholar]
- 62.Suo T., Liu J., Chen X., Yu H., Wang T., Li C., et al. Combining chemical profiling and network analysis to investigate the pharmacology of complex prescriptions in traditional Chinese medicine. Sci Rep. 2017;7:40529. doi: 10.1038/srep40529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang X.Y., Wang W.X., Huang Y.X., Yue S.J., Zhang B.Y., Gao H., et al. Network pharmacology-based dissection of the active ingredients and protective mechanism of the Salvia miltiorrhiza and Panax notoginseng herb pair against insulin resistance. ACS Omega. 2021;6(27):17276–17288. doi: 10.1021/acsomega.1c01209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu X.G., Cheng C.Y., Wang J.X., Luo H., Tu L.F., Lin L., et al. A metabolic exposure-oriented network regulation strategy for the identification of effective combination in the extract of Ginkgo biloba L. J Pharm Biomed Anal. 2018;149:151–159. doi: 10.1016/j.jpba.2017.11.004. [DOI] [PubMed] [Google Scholar]
- 65.Luo K., Xing Y., Wang M. Identifying the effectual-combination ingredients of Zhi-zi-Hou-po decoction based on metabolic difference-oriented network regulation strategy. J Chromatogr B Analyt Technol Biomed Life Sci. 2021;1184 doi: 10.1016/j.jchromb.2021.122980. [DOI] [PubMed] [Google Scholar]
- 66.Zuo H., Zhang Q., Su S., Chen Q., Yang F., Hu Y. A network pharmacology-based approach to analyse potential targets of traditional herbal formulas: an example of Yu Ping Feng decoction. Sci Rep. 2018;8(1):11418. doi: 10.1038/s41598-018-29764-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xiong Z., Chan W.K., Kuan C., Hu Y., Chan W. A method of mechanism analysis about a traditional Chinese medicine prescription on a disease based on PageRank algorithm and network pharmacology. Pharmacol Res Mod Chin Med. 2021;3 [Google Scholar]
- 68.Yang M., Chen J.L., Xu L.W., Ji G. Navigating traditional Chinese medicine network pharmacology and computational tools. Evid Based Complement Alternat Med. 2013;2013 doi: 10.1155/2013/731969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Song W., Ni S., Fu Y., Wang Y. Uncovering the mechanism of Maxing Ganshi Decoction on asthma from a systematic perspective: a network pharmacology study. Sci Rep. 2018;8(1):17362. doi: 10.1038/s41598-018-35791-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.The Gene Ontology Consortium The Gene Ontology Resource: 20 years and still GOing strong. Nucleic Acids Res. 2019;47(D1):D330–D338. doi: 10.1093/nar/gky1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., et al. The Reactome pathway knowledgebase. Nucleic Acids Res. 2020;48(D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Szklarczyk D., Gable A.L., Nastou K.C., Lyon D., Kirsch R., Pyysalo S., et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49(D1):D605–D612. doi: 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huntley R.P., Sawford T., Mutowo-Meullenet P., Shypitsyna A., Bonilla C., Martin M.J., et al. The GOA database: Gene Ontology annotation updates for 2015. Nucleic Acids Res. 2015;43(Database issue):D1057–D1063. doi: 10.1093/nar/gku1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Klopfenstein D.V., Zhang L., Pedersen B.S., Ramírez F., Warwick Vesztrocy A., Naldi A., et al. GOATOOLS: a python library for Gene Ontology analyses. Sci Rep. 2018;8(1):10872. doi: 10.1038/s41598-018-28948-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mi H., Ebert D., Muruganujan A., Mills C., Albou L.P., Mushayamaha T., et al. PANTHER version 16: a revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021;49(D1):D394–D403. doi: 10.1093/nar/gkaa1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu Z., Guo F., Wang Y., Li C., Zhang X., Li H., et al. BATMAN-TCM: a bioinformatics analysis tool for molecular mechANism of traditional Chinese medicine. Sci Rep. 2016;6:21146. doi: 10.1038/srep21146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu H.Y., Zhang Y.Q., Liu Z.M., Chen T., Lv C.Y., Tang S.H., et al. ETCM: an encyclopaedia of traditional Chinese medicine. Nucleic Acids Res. 2019;47(D1):D976–D982. doi: 10.1093/nar/gky987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Huang L., Xie D., Yu Y., Liu H., Shi Y., Shi T., et al. TCMID 2.0: a comprehensive resource for TCM. Nucleic Acids Res. 2018;46(D1):D1117–D1120. doi: 10.1093/nar/gkx1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wu Y., Zhang F., Yang K., Fang S., Bu D., Li H., et al. SymMap: an integrative database of traditional Chinese medicine enhanced by symptom mapping. Nucleic Acids Res. 2019;47(D1):D1110–D1117. doi: 10.1093/nar/gky1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zeng X., Zhang P., He W., Qin C., Chen S., Tao L., et al. NPASS: natural product activity and species source database for natural product research, discovery and tool development. Nucleic Acids Res. 2018;46(D1):D1217–D1222. doi: 10.1093/nar/gkx1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang R., Yu S., Bai H., Ning K. TCM-Mesh: the database and analytical system for network pharmacology analysis for TCM preparations. Sci Rep. 2017;7(1):2821. doi: 10.1038/s41598-017-03039-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tao W., Li B., Gao S., Bai Y., Shar P.A., Zhang W., et al. CancerHSP: anticancer herbs database of systems pharmacology. Sci Rep. 2015;5:11481. doi: 10.1038/srep11481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Kim S., Nam S., Jang H., Kim A., Lee J. TM-MC: a database of medicinal materials and chemical compounds in Northeast Asian traditional medicine. BMC Complement Altern Med. 2015;15:218. doi: 10.1186/s12906-015-0758-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zeng X., Zhang P., Wang Y., Qin C., Chen S., He W., et al. CMAUP: a database of collective molecular activities of useful plants. Nucleic Acids Res. 2019;47(D1):D1118–D1127. doi: 10.1093/nar/gky965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Li B., Ma C., Zhao X., Hu Z., Du T., Xu X., et al. YaTCM: yet another traditional Chinese medicine database for drug discovery. Comput Struct Biotechnol J. 2018;16:600–610. doi: 10.1016/j.csbj.2018.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fang F., Dong L., Liu L., Guo J., Zhao L., Zhang J., et al. HERB: a high-throughput experiment- and reference-guided database of traditional Chinese medicine. Nucleic Acids Res. 2021;49(D1):D1197–D1206. doi: 10.1093/nar/gkaa1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu Z., Du J., Yan X., Zhong J., Cui L., Lin J., et al. TCMAnalyzer: a chemo- and bioinformatics web service for analyzing traditional Chinese medicine. J Chem Inf Model. 2018;58(3):550–555. doi: 10.1021/acs.jcim.7b00549. [DOI] [PubMed] [Google Scholar]
- 89.Lee J.H., Park K.M., Han D.J., Bang N.Y., Kim D., Na H., et al. PharmDB-K: Integrated bio-pharmacological network database for traditional Korean medicine. PLoS One. 2015;10(11):e0142624. doi: 10.1371/journal.pone.0142624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sawada R., Iwata M., Umezaki M., Usui Y., Kobayashi T., Kubono T., et al. database of predicted targets and functional annotations of natural medicines. Sci Rep. 2018;8(1):11216. doi: 10.1038/s41598-018-29516-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Liu Z., Cai C., Du J., Lin B., Cui L., Fan X., et al. TCMIO: a comprehensive database of traditional Chinese medicine on immuno-oncology. Front Pharmacol. 2020;11:439. doi: 10.3389/fphar.2020.00439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu X, Liu J, Xu F, Li R, Xing L, Yuan L, et al. DCABM-TCM, a database of constituents absorbed into blood and metabolites of traditional Chinese medicine. PREPRINT (Version 1). (2021-04-14) [2022-01-01]. https://doi.org/10.21203/rs.3.rs-409645/v1.
- 93.Chen Q., Springer L., Gohlke B., Goede A., Dunkel M., Abel R., et al. SuperTCM: a biocultural database combining biological pathways and historical linguistic data of Chinese Materia Medica for drug development. Biomed Pharmacother. 2021;144 doi: 10.1016/j.biopha.2021.112315. [DOI] [PubMed] [Google Scholar]
- 94.Ru J., Li P., Wang J., Zhou W., Li B., Huang C., et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. doi: 10.1186/1758-2946-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang Y., Wang Y., Chen Y., Qin Q. Unique epidemiological and clinical features of the emerging 2019 novel coronavirus pneumonia (COVID-19) implicate special control measures. J Med Virol. 2020;92(6):568–576. doi: 10.1002/jmv.25748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.World Health Organization. Weekly epidemiological update on COVID-19—18 January 2022. (2022-01-18) [2022-01-22]. https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---18-january-2022.
- 97.Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ren J., Zhang A., Wang X. Traditional Chinese medicine for COVID-19 treatment. Pharmacol Res. 2020;155 doi: 10.1016/j.phrs.2020.104743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Huang Y., Wang W., Zhang S., Tang Y., Yue S. The database-based strategy may overstate the potential effects of traditional Chinese medicine against COVID-19. Pharmacol Res. 2020;159 doi: 10.1016/j.phrs.2020.105046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hu YJ, Li QH, Tang XL. Research progress of COVID-19 treated by Chinese medicine. Zhong Yi Wen Xian Za Zhi 2020;38(5):75–9,84. [Chinese with abstract in English].
- 101.Zhao Z., Li Y., Zhou L., Zhou X., Xie B., Zhang W., et al. Prevention and treatment of COVID-19 using traditional Chinese medicine: a review. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ren X., Shao X.X., Li X.X., Jia X.H., Song T., Zhou W.Y., et al. Identifying potential treatments of COVID-19 from traditional Chinese medicine (TCM) by using a data-driven approach. J Ethnopharmacol. 2020;258 doi: 10.1016/j.jep.2020.112932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Pan H., Yao X., Wang W., Lau H., Liu L. Network pharmacological approach for elucidating the mechanisms of traditional Chinese medicine in treating COVID-19 patients. Pharmacol Res. 2020;159 doi: 10.1016/j.phrs.2020.105043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang R., Liu H., Bai C., Wang Y., Zhang X., Guo R., et al. Chemical composition and pharmacological mechanism of Qingfei Paidu Decoction and Ma Xing Shi Gan Decoction against coronavirus disease 2019 (COVID-19): in silico and experimental study. Pharmacol Res. 2020;157 doi: 10.1016/j.phrs.2020.104820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao J., Tian S., Lu D., Yang J., Zeng H., Zhang F., et al. Systems pharmacological study illustrates the immune regulation, anti-infection, anti-inflammation, and multi-organ protection mechanism of Qing-Fei-Pai-Du decoction in the treatment of COVID-19. Phytomedicine. 2021;85 doi: 10.1016/j.phymed.2020.153315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zheng S., Baak J., Li S., Xiao W., Ren H., Yang H., et al. Network pharmacology analysis of the therapeutic mechanisms of the traditional Chinese herbal formula Lian Hua Qing Wen in corona virus disease 2019 (COVID-19), gives fundamental support to the clinical use of LHQW. Phytomedicine. 2020;79 doi: 10.1016/j.phymed.2020.153336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ai Z., Zhou S., Li W., Wang M., Wang L., Hu G., et al. “Fei Yan No. 1” as a combined treatment for COVID-19: an efficacy and potential mechanistic study. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.581277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang H., Zhang J., Lu Z., Dai W., Ma C., Xiang Y., et al. Identification of potential therapeutic targets and mechanisms of COVID-19 through network analysis and screening of chemicals and herbal ingredients. Brief Bioinform. 2022;23(1):bbab373. doi: 10.1093/bib/bbab373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wang Z.Z., Li K., Maskey A.R., Huang W., Toutov A.A., Yang N., et al. A small molecule compound berberine as an orally active therapeutic candidate against COVID-19 and SARS: a computational and mechanistic study. FASEB J. 2021;35(4):e21360. doi: 10.1096/fj.202001792R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Du H.X., Zhu J.Q., Chen J., Zhou H.F., Yang J.H., Wan H.T. Revealing the therapeutic targets and molecular mechanisms of emodin-treated coronavirus disease 2019 via a systematic study of network pharmacology. Aging (Albany NY) 2021;13(11):14571–14589. doi: 10.18632/aging.203098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ge C., He Y. In silico prediction of molecular targets of astragaloside IV for alleviation of COVID-19 hyperinflammation by systems network pharmacology and bioinformatic gene expression analysis. Front Pharmacol. 2020;11 doi: 10.3389/fphar.2020.556984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Peng W., Xu Y., Han D., Feng F., Wang Z., Gu C., et al. Potential mechanism underlying the effect of matrine on COVID-19 patients revealed through network pharmacological approaches and molecular docking analysis. Arch Physiol Biochem. 2020:1–8. doi: 10.1080/13813455.2020.1817944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qin X., Huang C., Wu K., Li Y., Liang X., Su M., et al. Anti-coronavirus disease 2019 (COVID-19) targets and mechanisms of puerarin. J Cell Mol Med. 2021;25(2):677–685. doi: 10.1111/jcmm.16117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li R., Wu K., Li Y., Liang X., Lai K., Chen J. Integrative pharmacological mechanism of vitamin C combined with glycyrrhizic acid against COVID-19: findings of bioinformatics analyses. Brief Bioinform. 2021;22(2):1161–1174. doi: 10.1093/bib/bbaa141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fang J., Wu Q., Ye F., Cai C., Xu L., Gu Y., et al. Network-based identification and experimental validation of drug candidates toward SARS-CoV-2 via targeting virus-host interactome. Front Genet. 2021;12 doi: 10.3389/fgene.2021.728960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Peele K.A., Potla Durthi C., Srihansa T., Krupanidhi S., Ayyagari V.S., Babu D.J., et al. Molecular docking and dynamic simulations for antiviral compounds against SARS-CoV-2: a computational study. Inform Med Unlocked. 2020;19 doi: 10.1016/j.imu.2020.100345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rolta R., Yadav R., Salaria D., Trivedi S., Imran M., Sourirajan A., et al. In silico screening of hundred phytocompounds of ten medicinal plants as potential inhibitors of nucleocapsid phosphoprotein of COVID-19: an approach to prevent virus assembly. J Biomol Struct Dyn. 2021;39(18):7017–7034. doi: 10.1080/07391102.2020.1804457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Elebeedy D., Elkhatib W.F., Kandeil A., Ghanem A., Kutkat O., Alnajjar R., et al. Anti-SARS-CoV-2 activities of tanshinone IIA, carnosic acid, rosmarinic acid, salvianolic acid, baicalein, and glycyrrhetinic acid between computational and in vitro insights. RSC Adv. 2021;11(47):29267–29286. doi: 10.1039/d1ra05268c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Lyu M., Fan G., Xiao G., Wang T., Xu D., Gao J., et al. Traditional Chinese medicine in COVID-19. Acta Pharm Sin B. 2021;11(11):3337–3363. doi: 10.1016/j.apsb.2021.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li S. Network pharmacology evaluation method guidance—draft. World J Tradit Chin Med. 2021;7(1):146–154. [Google Scholar]