Abstract
mRNA and lipid nanoparticles have emerged as powerful systems for the preparation of vaccines against severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) infection. The emergence of novel variants or the necessity of cold chain logistics for approved mRNA vaccines undermines the investigation of next-generation systems that could preserve both potency and stability. However, the correlation between lipid nanoparticle composition and activity is not fully explored. Here, we screened a panel of ionizable lipids in vivo and identified lead lipid nanoparticles with a branched-tail lipid structure. Buffer optimization allowed the determination of lyophilization conditions, where lipid nanoparticle-encapsulated mRNA encoding SARS-CoV-2 spike protein could induce robust immunogenicity in mice after 1 month of storage at 5°C and 25°C. Intramuscularly injected lipid nanoparticles distributed in conventional dendritic cells in mouse lymph nodes induced balanced T helper (Th) 1/Th2 responses against SARS-CoV-2 spike protein. In nonhuman primates, two doses of 10 or 100 μg of mRNA induced higher spike-specific binding geometric mean titers than those from a panel of SARS-CoV-2-convalescent human sera. Immunized sera broadly inhibited the viral entry receptor angiotensin-converting enzyme 2 (ACE2) from binding to the spike protein in all six strains tested, including variants of concern. These results could provide useful information for designing next-generation mRNA vaccines.
Keywords: MT: Oligonucleotides: Therapies and Applications, lipid nanoparticles, SARS-CoV-2 spike protein, mRNA, viral entry receptor, vaccines, COVID-19, mRNA vaccines, ionizable lipid, lyophilization, freeze-drying
Graphical abstract
We screened ionizable lipids and identified lead lipid nanoparticles with a branched-tail lipid structure. Intramuscularly injected lipid nanoparticles distributed in dendritic cells in mouse lymph nodes induced balanced Th1/Th2 responses against SARS-CoV-2 spike protein. In nonhuman primates, two doses of mRNA induced higher spike-specific binding titers.
Introduction
Out of the several COVID-19 vaccines developed, a new class of vaccines, mRNA-based vaccines, mRNA-12731 and BNT162b2,2 have shown high efficacy against SARS-CoV-2. These two vaccines are composed of two components: synthetic mRNA molecules encoding the antigen that triggers immune responses and lipid nanoparticles (LNPs) that can encapsulate and deliver mRNA into cells.3 An early clinical trial revealed that a rabies mRNA vaccine administered in solution form produced no immunogenicity unless dispensed with a high-pressure intradermal injection device, demonstrating the crucial role of LNPs for formulating mRNA vaccines.4 Among the components of LNPs (ionizable lipid, cholesterol, phospholipid, and PEG-lipid), the key component, ionizable lipid, mediates cytosolic delivery of nucleic acids by enabling endosomal escape.5 Tremendous efforts have been dedicated to optimizing ionizable lipid for systemic administration of small interfering RNA (siRNA)6, 7, 8, 9, 10 and mRNA.11, 12, 13 However, there is limited understanding regarding which ionizable lipid structures are crucial for locally administered mRNA.14,15 Hassett et al. reported that protein expression and immunogenicity induced by LNP-delivered mRNA are not necessarily correlated, indicating that a design of ionizable lipid structure unique to mRNA vaccines is needed.15 Understanding these structures that can induce potent immunogenicity is important to discover next-generation mRNA vaccines. In addition to the potency, the requirement of a cold or ultra-cold supply chain (chain logistics) to transport approved mRNA vaccines also poses challenges for distribution worldwide.16 Therefore, it is highly desirable to develop mRNA vaccines that can be stored at 2°C–8°C or higher.
We previously reported novel ionizable lipid structures17,18 that are chemically distinct from ionizable lipids (SM-102, ALC-0315) used in two approved mRNA vaccines.19,20 Our designed LNPs showed safe and potent siRNA delivery via intravenous administration in mice and nonhuman primates.21 In this study, we describe the design and lyophilization of lipid nanoparticles for the mRNA vaccine. We created a panel of ionizable lipids and elucidated the importance of a branched-tail lipid structure for potent immunogenicity. Buffer optimization technique allowed us to search for an ideal lyophilization condition, where we found that LNPs could induce robust immunogenicity in mice after 1 month of storage at 5°C and 25°C. In nonhuman primates, the LNP-encapsulated mRNA encoding SARS-CoV-2 spike protein induced robust, durable, and broad antibody responses against the six strains, including variants of concern (Alpha, Beta, Gamma, and Delta variants).
Results
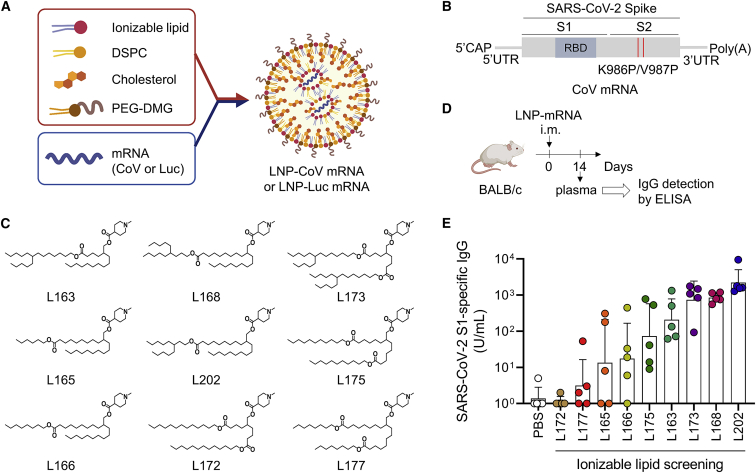
Ionizable lipid screening for mRNA vaccine
We initially screened molecules for preparing LNP formulations to induce potent antigen-specific immunoglobulin (Ig) G antibodies in vivo (Figure 1A). We designed nucleoside-modified mRNA that encodes prefusion-stabilized SARS-CoV-2 spike (S) proteins with two proline substitutions (2P), coronavirus (CoV) mRNA22 (Figure 1B). Based on our previous experience with siRNA delivery,17 we designed nine new ionizable lipids with a cyclic head consisting of N-methyl-piperidine but different lipid tail structures (Figure 1C). First, we confirmed the expression of spike protein in mRNA-transfected cells (Figure S1). Next, CoV mRNA was encapsulated into nine LNPs containing each ionizable lipid (Table S1). All lipids showed pKa 6–7, which was reported to be important for efficient endosomal escape.7,12 While L175 showed low mRNA encapsulation of about 50%, the remaining eight lipids showed over 85% of mRNA encapsulation (Table S1). All formulations were dosed at 2.5 μg mRNA per mouse intramuscularly and analyzed for the presence of SARS-CoV-2 S1-specific IgG antibodies in the plasma after 2 weeks (Figures 1D and 1E). Interestingly, the chemical structure of the top four lipids (L202, L168, L173, L163) possessed branched-tail lipid at the left side of ester linkage, while the other five lipids had linear-tail lipid (Figure 1C). The results of immunogenicity and LNP size concluded that superior formulations had a size in the range 95–115 nm (Figure S2). To assess how protein expression affects immunogenicity, we formulated mRNA that encodes the luciferase enzyme into nine LNPs containing each ionizable lipid and tested luciferase expression in vitro (Figures S3A and S3B). The top three LNPs were composed of L202, L168, and L173, which were in the same order of mice immunogenicity (Figure S3B). There was a good positive correlation between in vitro luciferase expression and mouse immunogenicity among nine lipids (correlation coefficient, R = 0.790) (Figure S3C). Based on the results of the highest immunogenicity in mice, we selected L202 lipid for further assessment.
Figure 1.
Ionizable lipid screening for mRNA vaccine
(A) Schematic illustration of LNP formulation for mRNA vaccine. mRNA is encapsulated in LNP that consists of ionizable lipid, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC), cholesterol, and polyethylene glycol dimyristoyl glycerol (PEG2K-DMG). (B) The construct of CoV mRNA expressing pre-fusion stabilized SARS-CoV-2 spike antigen with two proline substitutions (S-2P). CoV mRNA was chemically modified with N1-methylpseudouridine (N1mΨ). (C) A panel of newly created nine ionizable lipids. All lipids have an identical head but a different lipid tail. (D and E) In vivo screening of the potent mRNA vaccine. CoV mRNA was incorporated into LNPs containing each type of ionizable lipid. Plasma IgG (in arbitrary units) binds to the recombinant SARS-CoV-2 S1 14 days after the intramuscular injection of 2.5 μg of mRNA of each formulation into BALB/c mice (n = 5/group). Data are presented as a geometric mean titer (GMT) ± geometric SD. The number indicates the length of the carbon (C) chain.
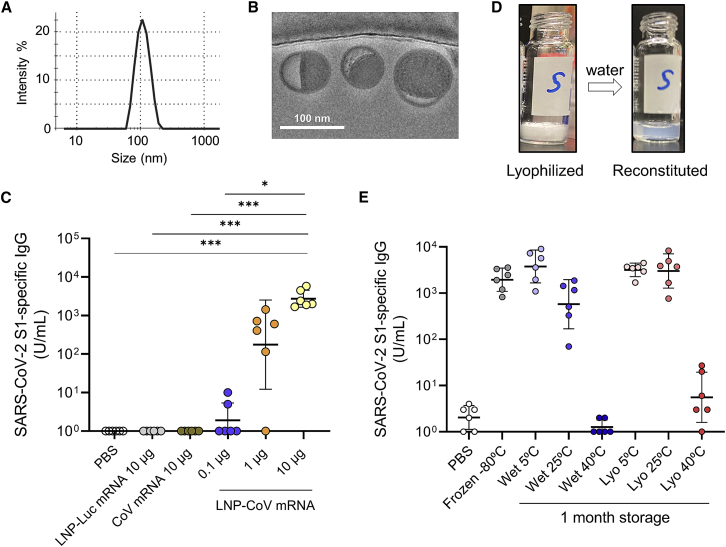
Lyophilization of LNP-CoV mRNA containing lead L202
We first characterized lead L202 lipid. LNP-CoV mRNA containing L202 was homogeneous (PDI 0.08), 103 nm in diameter, with 97% mRNA encapsulation efficiency (Figure 2A). Cryoelectron microscopy (cryo-EM) indicated that LNP-CoV mRNA has an electron-dense core with bilayer “blebs”23 (Figure 2B). In mice, LNP-CoV mRNA induced dose-dependent SARS-CoV-2 S1-specific binding antibodies at a single mRNA dose of 0.1, 1, or 10 μg, while LNP-encapsulated irrelevant mRNA (LNP-Luc mRNA) or CoV mRNA without LNP formulation showed no immunogenicity (Figure 2C).
Figure 2.
Immunogenicity and lyophilization of LNP-CoV mRNA containing lead L202
(A) The representative particle size of LNP-CoV mRNA containing L202, as determined by dynamic light scattering. (B) Cryo-EM image of LNP-CoV mRNA containing L202. (C) The dose-dependent activity of LNP-CoV. BALB/c mice (n = 6/group) received a single intramuscular injection of PBS, LNP-encapsulated luciferase mRNA in LNP (LNP-Luc mRNA), CoV mRNA alone, or LNP-encapsulated CoV mRNA ranging between 0.1 and 10 μg. (D) The representative appearance of lyophilized LNP-CoV mRNA and its reconstitution with water. (E) Immunization activity of wet or lyophilized formulations of LNP-CoV mRNA that were stored at 5°C, 25°C, or 40°C for 1 month. Mice (n = 6/group) received a single intramuscular injection of each LNP-CoV mRNA at 3 μg of mRNA dose. Plasma samples were collected 14 days after administering a single dose and were assessed for SARS-CoV-2 S1-specific IgG by ELISA (C and E). Data are presented as a GMT ± geometric SD. Groups were compared by a Kruskal-Wallis test with Dunn’s multiple comparison test (C). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001; ns, not significant.
We next assessed whether LNP-CoV mRNA could be lyophilized. Previous reports suggested that, among potential mRNA degradation pathways, hydrolysis plays a major role in mRNA stability.20,24 We, therefore, hypothesized that the removal of water by lyophilization improves storage stability. We first screened for buffer solutions that could protect LNPs from potential damage during the freeze-drying process, where sucrose was selected as a cryoprotectant. The concentration of sucrose in the solution varied (0%, 4%, 8%, 12%, and 16% w/v) with 20 mM Tris at pH 7.5. LNP formulations were lyophilized using a lyophilizer. The lyophilization cycle consisted of a freezing step at −40°C for 4 h under vacuum, a primary drying step at −40°C for 45 h under vacuum (∼3 Pa), and a secondary drying step at +10°C for 66 h under vacuum (∼3 Pa). The pressure within the shelf was returned to atmospheric pressure using dry nitrogen to obtain lyophilized samples. A solid cake was formed after freeze-drying LNP-CoV mRNA with at least 4% sucrose, and no aggregation was observed after reconstitution with water (Figures 2D, S4A, and S4B). We selected a 16% sucrose formulation for further analysis because it showed the lowest increase in LNP size (from 109 to 136 nm) without aggregation and mRNA leakage (Figure S4C).
Next, to test the storage stability, we stored LNP-CoV mRNA (16% sucrose buffer) with or without lyophilization (i.e., lyophilized formulation and wet formulation) at 5°C, 25°C, and 40°C, and performed physicochemical characterization at 2 weeks and 1 month. For 1-month storage at 5°C, both wet and lyophilized formulations showed no significant change in all parameters tested (Figure S5). However, for accelerated storage conditions at 25°C and 40°C, the mRNA integrity of wet formulations decreased more rapidly compared with that of the lyophilized formulations (Figure S5A). Here, only mRNA integrity decreased without any significant change in mRNA concentration and encapsulation efficiency, which were quantified by RiboGreen assay using fluorescent dyes (Figures S5B and S5C), suggesting that mRNA was degrading inside the nanoparticles.
Finally, samples stored for 1 month were immunized intramuscularly once in mice, and their plasmas were evaluated for the presence of SARS-CoV-2 S1-specific binding IgG on day 14 (Figure 2E). In a comparable control group, we employed a frozen sample that was not processed for lyophilization. The wet 5°C, lyophilized 5°C, and lyophilized 25°C formulations after 1 month of storage induced comparable immunogenicity with that of the control group (Figure 2E). The immunogenicity correlated well with mRNA integrity (Figures 2E and S5A). Altogether, the immunogenicity and physicochemical data demonstrated that lyophilized formulation has an improved thermostability over wet formulation.
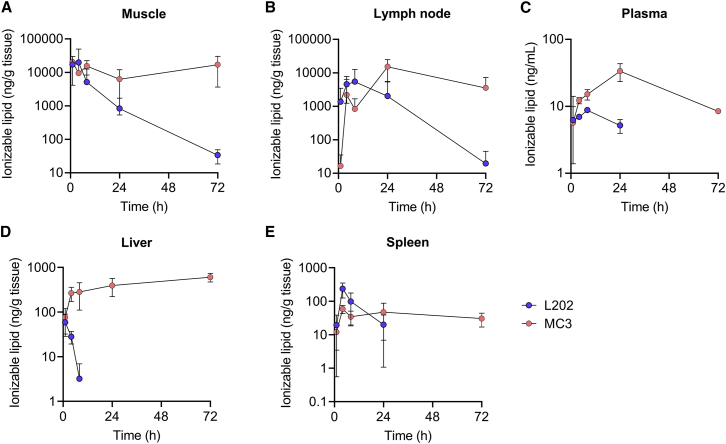
Biodegradability of L202 allows its rapid clearance in mice
L202 was designed to possess biodegradable properties by introducing ester linkage in the lipid tail.25 To evaluate the biodegradability of L202, we measured the lipid levels after intramuscular administration. We selected the most clinically advanced LNP containing the ionizable lipid MC3 as the benchmark.26 CoV mRNA was encapsulated in LNP containing L202 or MC3 (Table S2), which were intramuscularly injected into mice at a dose of 2 μg of mRNA. Mass spectrometry analysis revealed that MC3 remained at the muscle and lymph node injection site after 72 h post injection and accumulated in the liver and spleen through blood circulation (Figure 3). In contrast, L202 showed a significant rapid clearance through the muscle or lymph node, and no accumulation was observed in the liver 24 h post injection (Figure 3).
Figure 3.
Pharmacokinetics of LNP-CoV mRNA containing L202 and MC3 after intramuscular administration in mice
Lipid concentration after intramuscular administration of LNP-CoV mRNA containing L202 (blue) or MC3 (orange) in (A) the muscle at injection site, (B) draining inguinal lymph node, (C) plasma, (D) liver, and (E) spleen at 1, 4, 8, 24, and 72 h post injection (n = 3–4 per group per time point). Data are presented as mean ± SD.
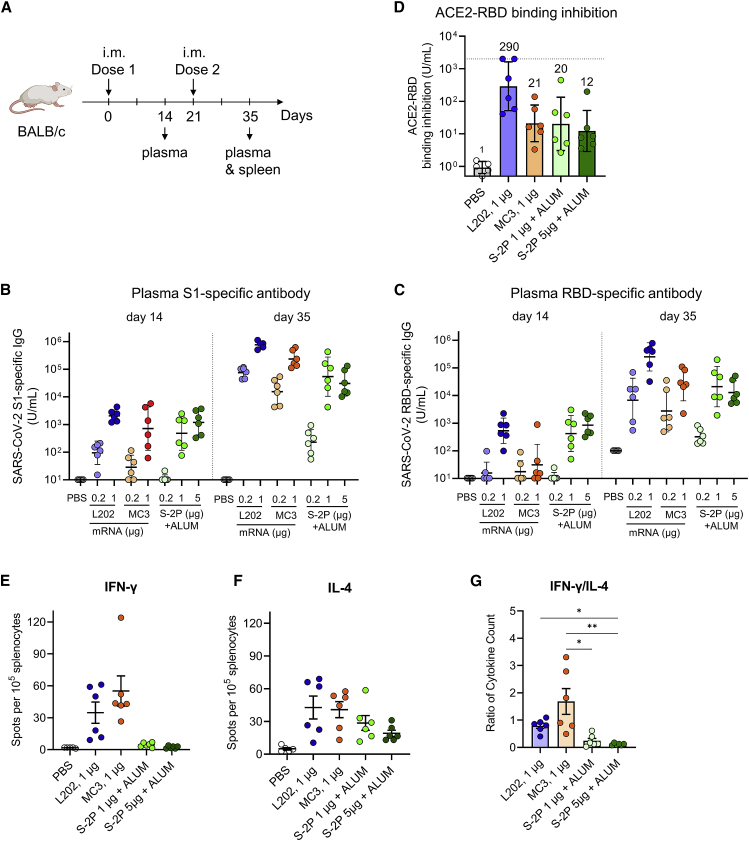
Robust antibody response of LNP-CoV mRNA containing L202 in mice
To assess the immunogenicity of LNP-CoV mRNA containing L202, we selected two benchmarks: LNP-CoV mRNA containing MC3 as mRNA-based vaccine and SARS-CoV-2 S-2P recombinant protein adjuvanted in alum as the protein-based vaccine. BALB/c mice were immunized intramuscularly twice with three samples at a dose of 0.2–1 μg of mRNA or 0.2–5 μg of protein dose on day 0 and day 21 (Figure 4A). Plasma assessment on day 14 and day 35 revealed that LNP-CoV mRNA containing L202 induced higher S1-specific (Figure 4B) and receptor-binding domain (RBD)-specific (Figure 4C) IgG compared with both benchmarks at each equivalent dose. Based on induced IgG levels, we performed a further analysis with 1 μg of mRNA-treated and 1–5 μg of the protein-treated group. A critical mechanism for SARS-CoV-2 infection in vivo is the interaction of the RBD with host ACE2, which is the entry receptor for the virus.22 We then determined whether plasma from immunized mice could inhibit the interaction between RBD and angiotensin-converting enzyme 2 (ACE2) using a commercially validated enzyme-linked immunosorbent assay (ELISA) kit. Indeed, plasma treated with LNP-CoV mRNA containing L202 had an inhibitory activity 13.8 times as high as that in plasma treated with MC3 and 14.3 times as high as that in plasma treated with protein, based on geometric mean titer at 1 μg of mRNA or protein dose (Figure 4D). Finally, single-dose tolerability of LNP-CoV mRNA containing L202 was investigated in BALB/c mice (n = 5/group) after intramuscular injection on day 0 (20 μg of mRNA dose; saline was used as a control) (Figure S6A). The body weights in the LNP-CoV mRNA-treated group decreased transiently on day 1 and recovered to the same levels as that of the saline-treated group (Figure S6B). The plasma cytokines and chemokines of the LNP-CoV mRNA-treated group sharply increased on day 1 and returned to the same levels as those of the saline-treated group on day 4 (Figure S6C). The level of aspartate transaminase (AST) and alanine aminotransferase (ALT) were similar in both groups on day 1 and day 4 (Figure S6D). These data demonstrated that a single dose of LNP-CoV mRNA containing L202 is tolerated at 20 μg of mRNA dose.
Figure 4.
LNP-CoV mRNA containing L202 elicits robust humoral and cellular immune responses
(A) BALB/c mice (n = 5–6/group) were immunized intramuscularly at day 0 and day 21 with either PBS, LNP-CoV mRNA containing L202, LNP-CoV mRNA containing MC3, ALUM-adjuvanted SARS-CoV-2 spike protein (S-2P) at 0.2–1 μg of mRNA dose, or 0.2–5 μg of protein dose. Plasma samples were assessed using ELISA for SARS-CoV-2 S1-specific binding IgG at day 14 and day 35 (B), SARS-CoV-2 receptor-binding domain (RBD)-specific binding IgG at day 14 and day 35 (C), and inhibition of angiotensin-converting enzyme 2 (ACE2) binding to the RBD at day 35 (D). Spleen samples collected on day 35 were analyzed for cellular T cell responses using ELISpot assay. Splenocytes were isolated and re-stimulated with pools of overlapping peptides from SARS-CoV-2 S protein. After 6 h, intracellular cytokine staining was performed to quantify IFN-γ- (E) and IL-4-positive (F) T cell responses. The ratio of IFN-γ/IL-4 cytokine count was calculated (G). Data are presented as GMT ± geometric SD (B–D) or mean ± SEM (E–G). The dotted line indicates the limit of detection (D). GMT values are written above the bars (D). Groups were compared by Kruskal-Wallis test with Dunn’s multiple comparison test (G). ∗p < 0.05, ∗∗p < 0.01.
Th1/Th2-balanced immune response of LNP-CoV mRNA containing L202
Next, we evaluated the balance of T helper (Th) 1 and Th2 cells because studies have raised safety concerns regarding vaccine-associated immune enhancement of respiratory disease (VAERD) associated with Th2-biased responses in the measles virus, RSV, and SARS-CoV vaccine development.27, 28, 29 First, we assessed SARS-CoV-2-specific cellular responses using ELISpot assay. After restimulation with peptide pools of spike protein, splenocytes (Figure 4A) in mRNA vaccine-immunized mice produced a higher number of IFN-γ+ cells on day 35 compared with those from PBS-treated or protein vaccine-immunized mice (Figure 4E). Consequently, the ratio of IFN-γ+/IL4+ cells induced by the mRNA vaccines was higher than that from protein vaccines (Figures 4E–4G). We also measured levels of S1-specific IgG2a and IgG1 antibodies, which are surrogates of Th1 and Th2 responses. Both mRNA-based vaccines induced balanced IgG1 and IgG2a subclass S-binding antibodies, indicating a balanced Th1/Th2 response (Figures S7A and S7B). In contrast, protein-based vaccines mainly induced IgG1 without sufficient IgG2a expression, indicating a Th2-biased response (Figures S7A and S7B). These results indicate that LNP-CoV mRNA containing L202 induces a balanced Th1/Th2 immune response as against the Th2-biased response induced by S protein adjuvanted with alum, suggesting that LNP-CoV mRNA lowers the safety concern of VAERD.
Intramuscularly injected LNP-CoV mRNA is localized in antigen-presenting cells in the draining lymph nodes and elicits robust germinal center B cell responses
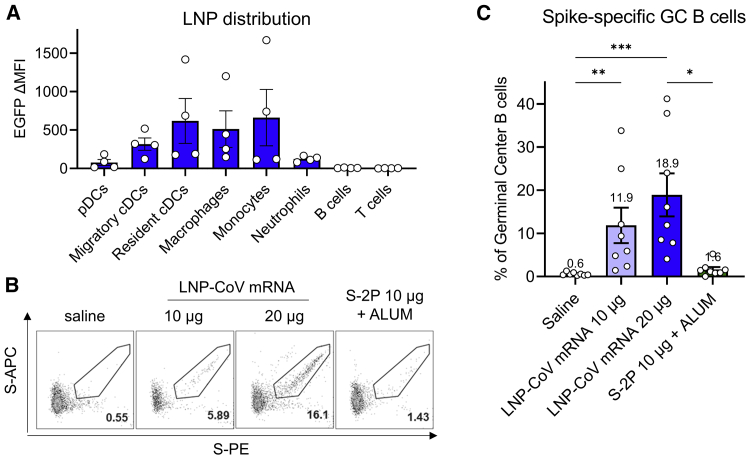
To elucidate robust immunogenicity by LNP-CoV mRNA containing L202, we evaluated its distribution in immune cell populations using mRNA encoding fluorescence protein EGFP. After an intramuscular injection of LNP-EGFP mRNA into the mouse thigh, draining inguinal lymph node was collected 24 h after dosing. Flow cytometry analysis revealed that the fluorescence signal of EGFP was most abundant in monocytes, macrophages, and conventional dendritic cells (DCs) and to a lesser degree in neutrophils (Figure 5A). No signal was detected in B cells, plasmacytoid DCs, CD4+ T cells, and CD8+ T cells (Figure 5A).
Figure 5.
LNP-CoV mRNAs localized in the antigen-presenting cells in the lymph nodes and induce strong antigen-specific germinal center B cell responses
(A) Flow cytometry analysis of LNP-EGFP mRNA distribution following the intramuscular injection of LNPs in mice (n = 4/group). Immune cells in draining inguinal lymph nodes were analyzed 24 h after injection of LNP-encapsulated EGFP mRNA at a 10-μg mRNA dose. (B) Mice (n = 8/group) were immunized intramuscularly with either saline, LNP-CoV mRNA at a 10-20-μg mRNA dose, or ALUM-adjuvanted SARS-CoV-2 spike protein (S-2P) at a 10-μg protein dose. GC B cells in inguinal lymph nodes were measured 7 days post immunization. Representative flow cytometry plots showing S-specific GC B cells, defined as live CD45+B220+CD19+Fas+GL7+S-PE+S-APC+ cells. (C) Frequency of spike-specific GC B cells in total GC B cell populations at 7 days post immunization (n = 8/group). LNPs in this study are formulated with L202. Data are presented as mean ± SEM (A and C). Groups were compared by Kruskal-Wallis test with Dunn’s multiple comparison test (C). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.
Germinal center (GC) B cells have been recognized for giving rise to high-quality and robust antibody responses.30 To confirm antigen-specific GC B cell formation, BALB/c mice were immunized intramuscularly once with 10–20 μg of LNP-CoV mRNA containing L202 or with 10 μg of recombinant SARS-CoV-2 S-2P adjuvanted in alum, respectively. We evaluated spike protein-specific GC B cells in draining inguinal lymph nodes 7 days after administering the dose using flow cytometry. The protein-based vaccine induced negligible spike-specific GC B cells (Figures 5B and 5C), with 1.6% mean expression in total GC B cell populations at a 10-μg protein dose. In contrast, LNP-CoV mRNA induced robust spike-specific GC B cells in a dose-dependent manner, with 11.9% mean expression at the 10-μg mRNA dose and 18.9% mean expression at the 20-μg mRNA dose (Figures 5B and 5C).
LNPs serve as potent adjuvants
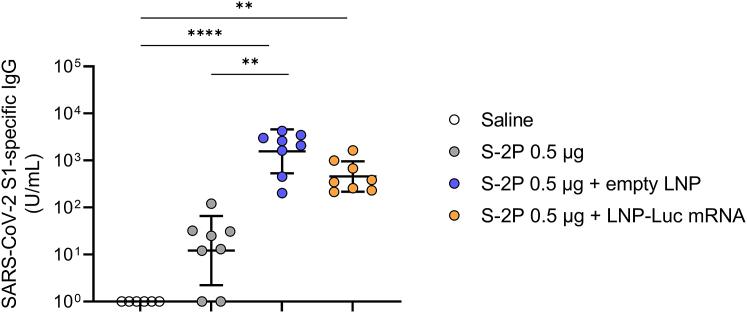
How LNP-CoV mRNA containing L202 induces robust immune responses without conventional adjuvant remains unclear.31 As previous reports have addressed these questions by checking whether their LNPs work as adjuvants,32,33 we have also hypothesized that either our developed LNP or LNP-mRNA serves as an adjuvant. To test this hypothesis, LNPs containing L202 were formulated with or without mRNA (empty LNP). To avoid spike protein-specific IgGs being produced by LNP-CoV mRNA, we used an irrelevant, luciferase mRNA and prepared LNP-Luc mRNA. BALB/c mice were immunized once intramuscularly with 0.5 μg of recombinant SARS-CoV-2 spike protein (S-2P) alone or mixed with 10 μg of empty LNP or LNP-Luc mRNA, and immune responses were evaluated at 14 days after dosing. S-2P alone induced negligible antibody responses (Figure 6). Notably, S-2P mixed with either empty LNP or LNP-Luc mRNA induced robust antigen-specific IgG (Figure 6). This result indicates that LNP with or without mRNA functions as an adjuvant in the protein-based vaccine.
Figure 6.
LNPs with or without mRNA serve as a potent adjuvant
Mice (n = 6–8/group) were immunized with a single intramuscular injection of 0.5 μg of SARS-CoV-2 spike protein (S-2P) alone or in combination with empty LNP or LNP-encapsulated luciferase mRNA at a 10-μg mRNA dose. Plasma samples were collected 14 days after administering the dose and were assessed for SARS-CoV-2 S1-specific IgG using ELISA. LNPs in this study are formulated with L202. Data are presented as a GMT ± geometric SD. Groups were compared by Kruskal-Wallis test with Dunn’s multiple comparison test. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
LNP-CoV mRNA elicits immunogenicity in nonhuman primates
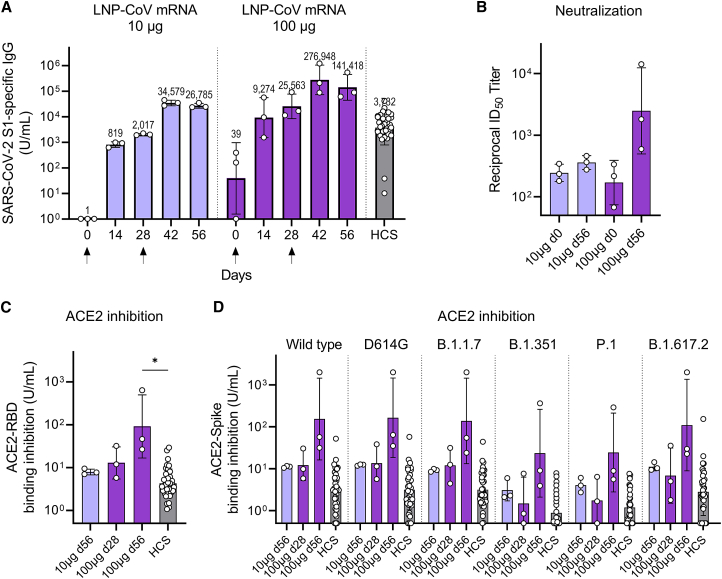
To assess the immunogenicity in nonhuman primates, we intramuscularly injected two groups of cynomolgus monkeys with 10 or 100 μg of LNP-CoV mRNA containing L202 on day 0 (dose 1) and day 28 (dose 2). S1-binding IgG was observed on day 14 after dose 1, and levels further increased 14 days after dose 2 (day 42) (Figure 7A). On day 42, the geometric mean titer (GMT) of S1-binding IgG was 34,579 units/mL at the 10-μg mRNA dose and 276,948 units/mL at the 100-μg mRNA dose. For comparison, the GMT of a panel of 36 SARS-CoV-2 human convalescent serum (HCS) was 3,732 units/mL, which is lower than the GMT of the immunized monkeys after two doses (Figure 7A). Similarly, robust RBD-binding IgG was also induced in a dose-dependent manner (Figure S8). Pseudovirus neutralization assay showed that monkey sera immunized twice with 100 μg of mRNA induced high neutralizing activity (Figure 7B). Furthermore, monkey sera at three critical, selected time points (day 56 for the 10-μg-treated group, day 28 and day 56 for the 100-μg-treated group) more strongly inhibited ACE2 binding to RBD than HCS (Figure 7C). Given concerns about reduced activity by mutations in SARS-CoV-2, we assessed antibody responses against six virus strains, including variants of concern (VOCs): wild-type, D614G, B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta). Notably, monkey sera at three selected time points inhibited ACE2 binding to the spike protein of all six strains compared with HCS, demonstrating the broad cross-reactivity of LNP-CoV mRNA (Figure 7D).
Figure 7.
Antibody responses after LNP-CoV mRNA immunization in nonhuman primates
Cynomolgus monkeys (n = 3/group) were injected with LNP-CoV mRNA containing L202 at a dose of 10 μg (blue) or 100 μg (purple) of mRNA intramuscularly on day 0 and day 28 (arrows below the x axes indicate the day of injection). Results were compared with the antibody responses in a panel of HCS specimens (n = 36). Plasma samples were assessed using ELISA for SARS-CoV-2 S1-specific binding to IgG (A), pseudovirus neutralization (B), inhibition of ACE2 binding to RBD (C), and inhibition of ACE2 binding to the spike protein of six strains including wild-type, D614G, B.1.1.7 (Alpha), B.1.351 (Beta), P.1 (Gamma), and B.1.617.2 (Delta) (D). On the day of immunization, plasma samples were collected before dose administration. Data are presented as a GMT ± geometric SD. Groups were compared by a Kruskal-Wallis test with Dunn’s multiple comparison test (B). ∗p < 0.05.
Antibody responses persist over 6 months and are further enhanced by the third dose
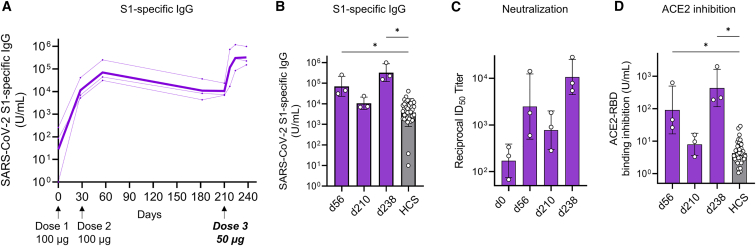
Finally, we assessed the durability of antibody responses after two doses and enhancement of antibody responses after a third dose. We used cynomolgus monkeys (n = 3/group) that received two 100-μg doses of LNP-CoV mRNA containing L202. Considering immunological memory, a lower dose of 50 μg was selected for the third dose. After 6 months from dose 2, monkeys received 50 μg of LNP-CoV mRNA intramuscularly as dose 3 on day 210 (Figure 8A). The GMT of S1-binding IgG at 6 months after dose 2 (day 210, day of dose 3) was 6.6-fold lower than the GMT of IgG at 1 month after dose 2 (day 56); however, both IgG levels were higher than the GMT measured for an HCS panel of 36 SARS-CoV-2-affected individuals (Figure 8B). Furthermore, the GMT of S1-binding IgG at 1 month after dose 3 (day 238) was further increased and was 4.6-fold higher than the GMT of IgG at 1 month after dose 2 (day 56) (Figure 8B). The corresponding neutralization activity was confirmed for those monkey sera by pseudovirus neutralization assay (Figure 8C). In addition, those monkey sera strongly inhibited the binding of ACE2 to RBD compared with that observed for HCS panels (Figure 8D). Similarly, robust antibody responses across all six strains were confirmed using an ACE2-spike inhibition assay (Figure S9).
Figure 8.
LNP-CoV mRNA containing L202 elicits durable antibody responses over 6 months after two doses and further induces potent responses following a third dose
Cynomolgus monkeys (n = 3) immunized with two doses (each dose of 100 μg) were used in a third dose study. Six months after dose 2, a 50-μg third dose (dose 3) was administered intramuscularly (in Figure 8A, arrows below the x axes indicate the day of injection. Faint lines represent individual animals, and bold lines represent the GMT for three animals). Plasma samples were assessed using ELISA for SARS-CoV-2 S1-specific binding IgG (A and B), pseudovirus neutralization (C), and inhibition of ACE2 binding to RBD (D). Results were compared with the antibody responses in a panel of HCS specimens (n = 36). On the day of immunization, plasma samples were collected before administration. Data are presented as a GMT ± geometric SD. Groups were compared by Kruskal-Wallis test with Dunn’s multiple comparison test (B and D). ∗p < 0.05.
Discussion
The components of LNP (i.e., ionizable lipid, phospholipid, cholesterol, PEG-lipids) exert an influence on efficacy and tolerability.5 This study screened a panel of ionizable lipids and identified lead LNPs with branched-tail lipids. Despite its importance, knowledge on ionizable lipid structures for locally administered mRNA vaccine has remained limited.15 We chemically designed nine ionizable lipids with a cyclic head of N-methylpiperidine based on our previous siRNA delivery material.17 The top two lipids, L202 and L168, have a chemical structure similar to that of L165 and L166, except for the branched-tail lipid at the left side of the ester linkage shown in Figure 1C. Accordingly, four lipids showed similar pKa (6.04–6.29), mRNA encapsulation (>90%), and nanoparticle size (99–136 nm). However, L202 and L168 with branched-tail lipids induced approximately 100-fold higher antibody response compared with L165 and L166 with linear-tail lipids (Figure 1E). The result demonstrates that slight differences in lipid tail structure can significantly affect immunogenicity.34,35 To improve clearance, we incorporated an ester linkage in the lipid tail of L202. We and other groups previously reported that intravenously21,25 or intramuscularly15 injected LNPs containing biodegradable lipid resulted in rapid clearance, leading to improved tolerability. We showed that L202 is rapidly cleared from all tissues tested while maintaining robust immunogenicity compared with MC3. Based on previous findings that ionizable lipid rather than other lipid components drive inflammation,33,36 lower local and systemic exposure of L202 may help improve tolerability. Another component, PEG-lipids, may affect efficacy due to anti-PEG responses that have been described for intravenously injected LNP-siRNA37 and LNP-mRNA.38 To date, there have been no reports on the anti-PEG response by intramuscularly injected LNP. Given that a robust increase in titers occurs after booster shots, we do not consider that anti-PEG antibody negatively affected in vivo activity.
Lyophilization of lipid-based nanoparticles remains challenging because physical stresses imposed by the freeze-drying process destabilize the fragile particle, leading to aggregation or drug leakage.39,40 This study revealed that the LNP-based mRNA vaccine can be lyophilized through buffer optimization. There are few reports on the lyophilization of lipid-based nanoparticles for mRNA delivery.41,42 Zhao et al. reported that lyophilization of mRNA nanoparticles using lipid-like material maintained in vitro efficacy but abolished in vivo efficacy, possibly due to nanostructure change during the lyophilization process.41 Muramatsu et al. reported that lyophilization of the LNP-mRNA vaccine showed preserved physicochemical properties and mice immunogenicity for 12 weeks of storage at room temperature and 24 weeks of storage at 4°C.42 To protect LNPs from physical stress, we employed sucrose as a cryoprotectant, the commonly used excipient in lyophilized liposomes.43 Sucrose acts as a cushion by replacing water molecules between particles and protects the particles from physical stress to maintain integrity.39 Lyophilization and not freeze-thawing increased particle size across all formulations tested, indicating that the major stress stems from drying and reconstitution (Figure S4). Interestingly, only particle size was increased without aggregation and mRNA leakage during lyophilization and storage (Figures S4 and S5D). We speculate that a trace amount of water in the cake triggered particle size increase through Ostwald ripening.44
LNP-CoV mRNA containing L202 elicited a balanced Th1/Th2 response, as against a Th2-biased response by protein with alum adjuvant. LNP-mRNA is primarily distributed in antigen-presenting cells (APCs) in lymph nodes, not in T cells and B cells. It remains unclear why LNPs were preferably localized to APCs. Several groups reported that LNPs containing a class of ionizable lipids were taken up into APCs via the endocytosis pathway45 or ApoE-independent pathway,46 while being taken up into hepatocytes via the ApoE-dependent pathway.47,48 Furthermore, our data showed that EGFP expression was abundant in migratory and resident conventional dendritic cells (cDCs) that play major roles in cross-presentation but was not detectable in plasmacytoid dendritic cells (pDCs) that are important for secreting type I interferons.49 Similarly, Uemura et al. reported that intravenously injected LNP-siRNA reduced target CD45 expression in DCs but not in pDCs.50 Preferable antigen expression in cDC subsets may contribute to a powerful immune response.51 Besides, germinal center B cells allow the production of high-quality persistent antibodies in mice52 and humans.53 LNP-CoV mRNA containing L202 slightly increased the GC B cell population (Figure S10) and significantly induced antigen-specific GC B cells (Figures 5B and 5C). The superior antibody responses to LNP-CoV mRNA compared with protein-based vaccine stems from robust formation of GC B cells.
We found that LNP without mRNA serves as a potent adjuvant. An early study by Swaminathan et al. demonstrated that LNPs with their ionizable lipids work as an adjuvant of subunit antigens,54 and recent reports revealed that the ionizable lipid in LNPs elicits inflammatory effects.33,36 It would be intriguing to explore the mechanism by which LNPs or ionizable lipid L202 can induce adjuvant activity. Recent studies showed that ionizable lipid or lipid-like material of nanoparticles induced innate immune responses through pattern recognition receptors of TLR455,56 and STING.57 Because cytokines, induced by adjuvants, trigger differentiation of Th cells and Th1/Th2 responses,31,58 a balanced Th1/Th2 immune response may stem from the L202 structure. We are currently exploring immune pathways that play crucial roles in the adjuvant action of ionizable lipids.
In nonhuman primates, LNP-CoV mRNA containing L202 elicited potent and broad antibody responses against all six strains tested. The broad responses could be attributed to the encoding of a full-length spike that should be digested to produce various peptide pieces for antigen presentation. Furthermore, two 100-μg doses elicited durable antibody responses over 6 months, which was further enhanced by the third injection of the lowered 50-μg dose. Notably, the antibody response levels 14 days after a single 100-μg dose were higher than a GMT of HCS, suggesting that efficacy may appear at least 2 weeks post dosing. To predict immunogenicity in humans, we used cynomolgus macaques, which have been proved to be an appropriate model reflecting human SARS-CoV-2 infection.59,60 Meta-analysis of seven COVID-19 vaccines tested in phase III trials revealed a robust correlation between neutralizing antibody titer and efficacy (the Spearman’s rank correlation coefficient ρ = 0.79) and spike protein-binding antibody titer and efficacy (ρ = 0.93).61 Given that both neutralizing and binding antibodies showed a better correlate of protection (CoP) in the mRNA-1273 COVID-19 vaccine trial,62 we believe that LNP-CoV mRNA containing L202 elicits protective immunity against SARS-CoV-2 infection.
In summary, we created a new LNP for mRNA vaccines that can be lyophilized and can elicit robust immune responses. New knowledge on ionizable lipid structures that can induce immunogenicity and lyophilization should open the gates to developing next-generation mRNA-based vaccines.
Materials and methods
Materials and animals
Materials used in the present study are summarized in the Key Resources table in the supplemental information (Table S3). BALB/c mice and cynomolgus monkeys were purchased from Jackson Laboratory (Japan) and HAMRI (Japan), respectively. Animal care and experimental procedures were performed in an animal facility accredited by the Health Science Center for Accreditation of Laboratory Animal Care and Use of the Japan Health Sciences Foundation. All protocols were approved by the Institutional Animal Care and Use Committee of Eisai and performed in accordance with the Animal Experimentation Regulations of Eisai.
Preparation of LNP-mRNA
A nucleoside-modified mRNA encoding prefusion-stabilized SARS-CoV-2 S-2P protein (wild type with amino acid substitutions of K968P and V987P, based on GenBank MN908947) was designed based on a previous report22 and synthesized at TriLink BioTechnologies (United States). In the designed mRNA (CoV mRNA), uridine was completely replaced by N1-methyl-pesuouridine, with the Cap 1 structure (TriLink #N-7113), the 5′ and 3′ untranslated regions (UTRs), and a poly(A) tail. Chemically modified luciferase mRNA (TriLink #L-7202) and EGFP mRNA (TriLink #L-7201) were commercially available. The nine ionizable lipids were synthesized based on the procedures described in our previous patent (WO2017222016A1). Lipid nanoparticles were prepared by mixing an ethanol phase containing lipids with mRNA in an aqueous phase.17 In general, mRNA was dissolved in 50 mM citric acid at pH 3.5, while ionizable lipid, 1,2-distearoyl-sn-glycero-3-phosphocholine (DSPC) (Nippon Fine Chemical), cholesterol (Nippon Fine Chemical), and mPEG2000-DMG (NOF SUNBRIGHT GM-020) (approximately 50:10:38.5:1.5 molar ratio) were dissolved in ethanol. The mRNA/total lipid ratio was approximately 0.05 (wt/wt). The mRNA and lipid solutions were mixed at a flow ratio of 3:1 using NanoAssembler (Precision Nanosystems, Canada). Using 100-kDa dialysis tubes (Spectrum Labs #G235035), solutions were first dialyzed with PBS (pH 7.5) and then with 8% sucrose/20 mM Tris buffer (pH 7.5) overnight in the refrigerator. The resulting solution was filtered using a 0.22-μm membrane filter to produce LNPs. Except for the lyophilization study, LNPs were used immediately or cryopreserved at −70°C before in vivo study.
Characterization of LNPs
The particle size and polydispersity index were determined by dynamic light scattering using Zetasizer Nano ZS (Malvern Panalytical, UK). The mRNA integrity was assayed by electrophoresis using Agilent 4200 TapeStation system (Agilent Technologies, United States). As previously described, free and total mRNA concentrations in LNPs were determined using the Quant-iT Ribogreen RNA assay kit (Invitrogen #R11491) and SpectraMAX M2 fluorescence microplate reader (Molecular Device, United States).17 Encapsulation efficiency (EE, %) was calculated as follows:
All lyophilized samples were reconstituted with water prior to the characterization of LNPs.
Cryo-EM analysis of LNPs
A droplet of 3 μL of LNP-CoV mRNA solution was applied to a glow-discharged holey carbon grid (Cu R1.2/1.3, 300 mesh, Quantifoil Micro Tools, #M2955C-1-300). The grid was blotted for 15 s with a blot force of 0 and flash-frozen in liquid ethane using Vitrobot Mark IV (Thermo Fisher Scientific, United States) at 18°C and 100% humidity. Data were collected on a Talos Arctica electron microscope (Thermo Fisher Scientific, United States) equipped with a Falcon 3 direct electron detector at 200 kV. Images were acquired at a defocusing of −1 μm with a nominal magnification of 92,000 at a pixel size of 1.13 Å.
In vitro expression of mRNA encoding SARS-CoV-2 spike protein
Human embryonic kidney 293 cells (HEK-293) (ATCC #CRL-1573) were transfected with mRNA encoding SARS-CoV-2 spike protein (CoV mRNA) using Lipofectamine MessengerMAX Transfection Reagent (Invitrogen #LMRNA003). After 24 h, the cells were resuspended in fluorescence-activated cell sorting buffer: D-PBS(−) (Wako #045-29795), 2% (v/v) FBS (Gibco #10270-106), 2 mM EDTA (Invitrogen #15575-038). According to the manufacturer’s instructions, the cells were fixed and permeabilized in Fixation/Permeabilization Buffer (BD Biosciences #51-2090KZ) and Perm/Wash Buffer (BD Biosciences #51-2091KZ). Subsequently, the cells were stained with 10 μg/mL CR3022 (CST #37475S) in Perm/Wash Buffer (BD Biosciences #51-2091KZ) for 30 min at room temperature. Afterward, the cells were washed in Perm/Wash Buffer and incubated with anti-human IgG Fc PE-conjugated antibody (Jackson ImmunoResearch #709-116-149) in Perm/Wash Buffer for 30 min at room temperature. Finally, the cells were washed in Perm/Wash Buffer and resuspended in fluorescence-activated cell sorting buffer. Data acquisition was performed on ZE5 Cell Analyzer (Bio-Rad Laboratories, United States) and analyzed using FlowJo software (BD Biosciences).
In vitro expression of mRNA encoding firefly luciferase
In 96-well culture plates, 80 μL/well of 1 × 104 Hep3B cells (ATCC #HB-8064) were seeded and treated with 20 μL/well of each LNP containing firefly luciferase mRNA (10 ng mRNA/well). After 24 h, firefly luciferase expression and cell viability were measured with Steady-Glo (Promega #E2510) and Cell-Titer Glo (Promega #G9241), respectively. The luminescence was quantified using Nivo multi-plate reader (PerkinElmer). The results, expressed in the relative light unit (RLU), were normalized as the ratio of luciferase expression to cell viability. The correlation coefficient (Pearson’s r) of expression and immunogenicity was determined by GraphPad Prism.
Lyophilization process of LNPs
The final LNP solution with 20 mM Tris (Sigma-Aldrich #T1503-1KG) and varying sucrose (Wako #192-00017) concentrations (0%, 4%, 8%, 12%, and 16% w/v) at pH 7.5 was prepared by ultrafiltration (Millipore, #UFC810024) and aliquoted into glass vials. The vials containing 0.2 mL of LNP of 0.10 mg/mL mRNA were placed on the inside shelf of the lyophilizer DRC-1100 (Tokyo Rikakikai, Japan) connected to a vacuum pump. In general, the shelf temperature was preset to −40°C, and the vials were equilibrated for 4 h. The primary drying at −40°C lasted for 45 h under vacuum. The temperature was gradually increased from −40°C to +10°C. The secondary drying was completed at +10°C for 66 h under vacuum. The pressure within the shelf was returned to atmospheric pressure using dry nitrogen. The obtained vials with a rubber cap were subjected to a storage stability study.
Mouse immunogenicity of stored samples
LNP-CoV mRNA was prepared using 16% sucrose/20 mM Tris buffer at pH 7.5. Samples were either frozen at −80°C (i.e., frozen samples), used in liquid form (i.e., wet formulations), or lyophilized (i.e., lyophilized samples). The wet or lyophilized formulation in glass vials with a rubber cap was stored at 5°C, 25°C, or 40°C on a temperature-controlled shelf. After 2 weeks and 1 month, all samples were collected, and lyophilized formulations were reconstituted with water. For samples stored for 1 month, BALB/c mice (n = 6/group) were immunized with a single intramuscular injection of each sample at a dose of 3 μg of mRNA. Plasma was collected 14 days after dosing and assessed for SARS-CoV-2 S1-specific binding IgG antibody.
ELISpot assay
BALB/c mice (n = 5–6/group, 5 weeks old) were intramuscularly immunized at day 0 and day 21 with PBS, LNP-CoV mRNA containing L202, LNP-CoV mRNA containing MC3, or ALUM-adjuvanted (Thermo Scientific #77161) SARS-CoV-2 spike protein (S-2P) (R&D Systems #10549-CV-100) with mRNA and protein doses of 0.2–1 μg and 0.2–5 μg, respectively. Mice were euthanized at day 35 and spleens were harvested in RPMI-1640 medium (Wako #189-02025). Subsequently, the spleens were filtered through a 70-μm-mesh cell strainer (Corning #352350) and treated in BD Pharm Lyse Lysing Buffer (BD Biosciences #555899) to remove red blood cells and isolate mouse splenocytes. Detection of IFN-γ- and IL-4-positive splenocytes was performed according to the manufacturer’s instructions for the FluoroSpot assay (Mabtech AB #FSX-41A-1 and #FSX-46B-1). Briefly, a 96-well plate was coated with both anti-mouse IFN-γ (kit component) and IL-4 (kit component) monoclonal antibodies overnight at 4°C. The plate was washed with D-PBS(−) five times and was blocked using blocking buffer (RPMI-1640 (Wako #189-02025), 10% (v/v) FBS (Gibco #10270-106), 100 units/mL penicillin and 100 μg/mL streptomycin (Wako #168-23191), 50 μM 2-mercaptoethanol (Invitrogen #21985-023)) at room temperature for 30 min. Subsequently, 1 × 105 cells of isolated mouse splenocytes per well were stimulated at 37°C for 16 h using 0.1 μg/mL peptide pool of the SARS-CoV-2 Wuhan wild-type spike protein (Miltenyi Biotec #130-127-951). After washing, detection antibodies with dilution buffer (D-PBS(−) and 0.1% [w/v] BSA; Wako #017-22231) were added and incubated at room temperature for 2 h. After washing, fluorophore-conjugated reagents (kit component) with dilution buffer were added and incubated at room temperature for 1 h. Finally, the plates were washed, and the fluorescence spots of IFN-γ (green) and IL-4 (red) were detected under a BZ-X810 fluorescence microscope (Keyence Corporation, Japan). Two individuals counted the number of fluorescence spots, and the average was calculated.
Antibody measurements
A 96-well streptavidin plate (MSD #L-15SA-1) was blocked using blocking buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl, 5% [w/v] skim milk [Wako #190-12865], 0.2% [v/v] ProClin 150 [Sigma-Aldrich #49376-U], and 0.01% [v/v] Tween 20 [Bio-Rad Laboratories #1706531]) at room temperature for 1 h. The plates were washed using the wash buffer (Sigma-Aldrich #T9039) three times and coated with 25 ng/well of biotinylated SARS-CoV-2 S1 (ACROBiosystems #S1N-C82E8) or RBD (ACROBiosystems #SPD-C82E9) protein diluted with D-PBS(−) at room temperature for 1 h. After washing, standard, immunized serum, or plasma diluted with blocking buffer were added to the plates and incubated at room temperature for 1 h. After washing the incubated mixture, approximately 1 μg/mL of ruthenium-conjugated (MSD #R91AO-1) anti-IgG antibodies (Jackson ImmunoResearch #115-005-164) diluted with blocking buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5% [w/v] skim milk [Wako #190-12865], 0.2% [v/v] ProClin 150 [Sigma-Aldrich #49376-U], and 0.01% [v/v] Tween 20 [Bio-Rad Laboratories #1706531]) were added, and the plates were incubated at room temperature for 1 h. Finally, the plates were washed, and the binding signals were detected with reading buffer (MSD #R92TC-3) using MSD sector imager 6000 (Meso Scale Diagnostics, United States).
ACE2 binding inhibition was measured using a commercially available kit (V-PLEX SARS-CoV-2 Panel 17 [ACE2] Kit, MSD #K15527U-2) by following the manufacturer’s instructions. Plates were precoated with the SARS-CoV-2 RBD or the spike protein of each strain. Immunized serum or plasma was used on 1:50 diluted samples. Binding was detected with SLUFO-TAG-labeled ACE2 (kit component).
Pseudovirus neutralization assay
Pseudoviruses were produced by co-transfection of plasmids encoding a luciferase reporter in retrovirus vector (pMXs-ires-Puro, Cell Biolabs #RTV-014) and S protein (D614G, d19) into Plat-GP (Cell Biolabs #RV-103). Serum samples were mixed with pseudoviruses, incubated, and then added to ACE2 and TMPRSS2-expressing HEK293T cells (GeneCopoeia #SL222). After 48 h, cells were lysed, and luciferase activity (RLU) was measured. The infection rate was normalized, considering uninfected cells as 0% and cells infected with only pseudovirus as 100%. ID50 titers were determined using an inhibitor versus normalized response (variable slope) nonlinear function.
Lipid pharmacokinetic analysis in mice
BALB/c mice (n = 3–4/time point, 5 weeks old) were immunized with a single intramuscular injection of either LNP-CoV mRNA containing MC3 or LNP-CoV mRNA containing L202 at a dose of 2 μg of mRNA. At 0.5, 1, 4, 8, 24, and 72 h post injection, mice were euthanized and the plasma, site of injection in muscle, draining lymph node, liver, and spleen were harvested. All samples were stored at −80°C prior to analysis using liquid chromatography-tandem mass spectroscopy (LC-MS/MS). All samples except for plasma were weighed and homogenized in homogenization buffer (60 mM Na2HPO4, 900 mM NaCl, and 0.24% [v/v] Tween 20 at pH 7.4). The plasma samples (10 μL) and tissue homogenates (10 μL) were extracted with 200 μL of acetonitrile containing 1 ng/mL of an internal standard lipid. The samples were mixed with a vortexer and centrifuged at 2,095 × g for 10 min at 4°C. The supernatant was transferred to 96-well plastic plates and analyzed using LC-MS/MS. For the ionizable lipids, a mobile phase with a gradient composition of 0.1% formic acid in water (A) and 0.1% formic acid in acetonitrile (B), were used at a flow rate of 0.4 mL/min. The mobile phase flow was programmed as follows: 1% of B for 0.5 min, followed by a linear gradient of B to 99% over 0.5 min, held at 99% of B for 4 min, and back to 1% of B in 0.01 min, followed by equilibration to reach the initial conditions. The total run time was 6.0 min. Separation was carried out using an ACQUITY UPLC (2.1 × 50 mm) (Waters #186002350), and eluted fractions were directly passed through Xevo TQ-XS system (Waters, United States) equipped with an electrospray ionization source operating in the positive ion mode. The ionization mode, parent ion, and product ion were as follows: L202 m/z = 650.3 [M + H]+ to 143.8, MC3 m/z = 642.9 [M + H]+ to 131.8, and the internal standard lipid m/z = 519.5 [M + H]+ to 83.9.
Single dose tolerability in mice
Female BALB/c mice (n = 5/group, four groups in total) were immunized intramuscularly on day 0 with saline (two groups) or LNP-CoV mRNA containing L202 (2 groups) at a dose of 20 μg of mRNA. Mice were weighed during the study and sacrificed on day 1 and day 4 to collect plasma. Cytokines and chemokines in plasma were measured using a commercially available kit (U-PLEX Mouse customized multiplex assay kit, MSD #K15069L-1) by following the manufacturer’s instructions. Clinical chemistry parameters were measured on the Hitachi 7180 clinical analyzer (Hitachi).
Cell-type-specific distribution analysis
Distribution was measured by cellular GFP expression with EGFP-encoding mRNA. BALB/c mice (n = 4/group, 5 weeks old) were immunized intramuscularly with saline or LNP-encapsulated EGFP mRNA containing L202 at a dose of 10 μg of mRNA. After 24 h, mouse inguinal lymph nodes were collected and digested with RPMI-1640 medium (Wako #189-02025) containing 0.3 mg/mL Liberase DL (Sigma-Aldrich #05401160001) and 0.2 mg/mL DNase I (Roche #10104159001) at 37°C for 30 min and then filtered through a 70-μm-mesh cell strainer (Corning #352350). Cells were pre-incubated with Mouse BD Fc Block (BD Biosciences #553142) for 10 min and stained with fluorophore-conjugated antibodies as follows: anti-CD45 (clone: 3F-11, BD Biosciences #565967), B220 (clone: RA3-6B2, BioLegend #103210), CD3ε (clone: 17A2, BD Biosciences #612803), CD11b (clone: M1/70, BD Biosciences #624294), CD11c (clone: N418, BioLegend #117339), IA/IE (clone: M5/114.15.2, BD Biosciences #5660866), CD317 (clone: 129C1, BD Biosciences #127025), Ly6C (clone: HK1.4, BD Biosciences #128044), Ly6G (clone: 1A8, BD Biosciences #560601), and CD169 (clone: 3D6.112, BioLegend #142421). Data acquisition was performed on a BD FACSymphony Flow Cytometer (BD Biosciences) and data were analyzed by FlowJo software (BD Biosciences).
Germinal center B cell analysis
BALB/c mice (n = 8/group, 9 weeks old) were immunized intramuscularly with either saline, LNP-CoV mRNA at a dose of 10–20 μg of mRNA, or SARS-CoV-2 spike protein (S-2P) (R&D Systems #10549-CV-100) adjuvanted with alum (Thermo Scientific #77161) at a dose of 10 μg of protein. After 7 days, mouse inguinal lymph nodes were isolated and dissolved in fluorescence-activated cell sorting buffer (D-PBS(−) [Wako #045-29795], 2% [v/v] FBS [Gibco #10270-106], 2 mM EDTA [Invitrogen #15575-038]). The cells were pre-incubated with Mouse BD Fc Block (BD Biosciences #553142) for 10 min and stained with 4′,6-diamidino-2-phenylindole (Dojinkagaku #341–07881) and fluorophore-conjugated antibodies as follows: anti-CD45 (clone: 30-F11, BD Biosciences # 565,967), CD19 (clone: 1D3/CD19, BioLegend #152403), B220 (clone: RA3-6B2, BioLegend #103253), GL7 (clone: GL7, BioLegend #144609), Fas (clone: Jo2, BD Biosciences #557653), spike protein (R&D Systems #10549-CV-100), PE conjugated (abcam #ab102918), and spike protein (R&D Systems #10549-CV-100) APC conjugated (abcam #ab201807). Finally, the cells were washed in fluorescence-activated cell sorting buffer. Data acquisition was performed on FACSymphony (BD Biosciences) and analyzed using FlowJo software (BD Biosciences).
HCS
Thirty-six HCS (ReproCELL #S-100) panels were purchased from ReproCELL (Japan) and used as a benchmark for nonhuman primate serology. The participants were of diverse races aged 18–59 years and had symptoms such as fever or flu-like symptoms but had not stayed in the intensive care unit. The sera were collected at least 10 days after being diagnosed as COVID-19 positive based on polymerase chain reaction (PCR) tests and after being diagnosed as COVID-19 negative. All participants provided written informed consent. All samples were de-identified after collection.
Nonhuman primate immunization
LNP-CoV mRNA containing L202 at 10 or 100 μg mRNA was intramuscularly injected into the thigh muscles of male cynomolgus monkeys (4–6 years old). The low-dose (10 μg) and high-dose (100 μg) groups (n = 3/group) received two doses on day 0 and day 28. Serum samples were collected on days 0 (dose 1), 14, 28 dose 2), 42, and 56. On the day of immunization, serum was collected before dose administration. For a third dose study, monkeys (n = 3) that had already received two 100-μg doses were used. Six months after the second immunization, a 50-μg dose of LNP-CoV mRNA (dose 3) was injected intramuscularly. Serum samples were obtained on days 182, 210 (dose 3), 217, 224, and 238.
Statistical analysis
GraphPad Prism (GraphPad Software, United States) was used to perform Kruskal-Wallis test with Dunn’s multiple comparisons for non-parametric data (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001).
Acknowledgments
We thank the members of the cryo-EM facility at the High Energy Accelerator Research Organization (KEK) (Japan) for cryo-EM data collection. The cryo-EM research was supported by the Platform Project for Supporting Drug Discovery and Life Science Research (Basis for Supporting Innovative Drug Discovery and Life Science Research (BINDS)) from AMED under grant number JP21am0101071. We thank Muneo Aoyama of Eisai for technical advice regarding ELISA, Yoshinori Takahashi of Eisai for providing lipids, and Yosuke Watanabe of Eisai for technical advice regarding the lyophilization process. The figures were created with BioRender.com.
Author contributions
Y.S., conceptualization, methodology, investigation, writing – original draft, and writing – review & editing; T.M., methodology and investigation; H.M., methodology and investigation; K.M., methodology and investigation; Y.M., visualization and writing – review & editing; R.W., methodology and investigation; S.S., methodology and investigation; K.K., methodology and investigation; S.T., methodology and investigation; K.Y., methodology and investigation; M.I., project administration, resources, supervision, and writing – review & editing; K.T., project administration, resources, supervision, and writing – review & editing.
Declaration of interests
All authors except for S.T. and K.Y. were employees of the Eisai Co., Ltd. during the execution of this research project.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.omtn.2022.09.017.
Supplemental information
Data availability
The data that support the findings of this study are available from the corresponding author, upon reasonable request.
References
- 1.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., Diemert D., Spector S.A., Rouphael N., Creech C.B., et al. Efficacy and safety of the mRNA-1273 SARS-CoV-2 vaccine. N. Engl. J. Med. Overseas. Ed. 2021;384:403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Pérez Marc G., Moreira E.D., Zerbini C., et al. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pardi N., Hogan M.J., Porter F.W., Weissman D. mRNA vaccines - a new era in vaccinology. Nat. Rev. Drug Discov. 2018;17:261–279. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Alberer M., Gnad-Vogt U., Hong H.S., Mehr K.T., Backert L., Finak G., Gottardo R., Bica M.A., Garofano A., Koch S.D., et al. Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet. 2017;390:1511–1520. doi: 10.1016/S0140-6736(17)31665-3. [DOI] [PubMed] [Google Scholar]
- 5.Kulkarni J.A., Cullis P.R., van der Meel R. Lipid nanoparticles enabling gene therapies: from concepts to clinical utility. Nucleic Acid Ther. 2018;28:146–157. doi: 10.1089/nat.2018.0721. [DOI] [PubMed] [Google Scholar]
- 6.Semple S.C., Akinc A., Chen J., Sandhu A.P., Mui B.L., Cho C.K., Sah D.W.Y., Stebbing D., Crosley E.J., Yaworski E., et al. Rational design of cationic lipids for siRNA delivery. Nat. Biotechnol. 2010;28:172–176. doi: 10.1038/nbt.1602. [DOI] [PubMed] [Google Scholar]
- 7.Jayaraman M., Ansell S.M., Mui B.L., Tam Y.K., Chen J., Du X., Butler D., Eltepu L., Matsuda S., Narayanannair J.K., et al. Maximizing the potency of siRNA lipid nanoparticles for hepatic gene silencing in vivo. Angew. Chem. Int. Ed. Engl. 2012;51:8529–8533. doi: 10.1002/anie.201203263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whitehead K.A., Dorkin J.R., Vegas A.J., Chang P.H., Veiseh O., Matthews J., Fenton O.S., Zhang Y., Olejnik K.T., Yesilyurt V., et al. Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun. 2014;5:4277. doi: 10.1038/ncomms5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato Y., Hashiba K., Sasaki K., Maeki M., Tokeshi M., Harashima H. Understanding structure-activity relationships of pH-sensitive cationic lipids facilitates the rational identification of promising lipid nanoparticles for delivering siRNAs in vivo. J. Control. Release. 2019;295:140–152. doi: 10.1016/j.jconrel.2019.01.001. [DOI] [PubMed] [Google Scholar]
- 10.Rajappan K., Tanis S.P., Mukthavaram R., Roberts S., Nguyen M., Tachikawa K., Sagi A., Sablad M., Limphong P., Leu A., et al. Property-driven design and development of lipids for efficient delivery of siRNA. J. Med. Chem. 2020;63:12992–13012. doi: 10.1021/acs.jmedchem.0c01407. [DOI] [PubMed] [Google Scholar]
- 11.Kauffman K.J., Dorkin J.R., Yang J.H., Heartlein M.W., DeRosa F., Mir F.F., Fenton O.S., Anderson D.G. Optimization of lipid nanoparticle formulations for mRNA delivery in vivo with fractional factorial and definitive screening designs. Nano Lett. 2015;15:7300–7306. doi: 10.1021/acs.nanolett.5b02497. [DOI] [PubMed] [Google Scholar]
- 12.Sabnis S., Kumarasinghe E.S., Salerno T., Mihai C., Ketova T., Senn J.J., Lynn A., Bulychev A., McFadyen I., Chan J., et al. A novel amino lipid series for mRNA delivery: improved endosomal escape and sustained pharmacology and safety in non-human primates. Mol. Ther. 2018;26:1509–1519. doi: 10.1016/j.ymthe.2018.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao L., Lin J., Huang Y., Li L., Delcassian D., Ge Y., Shi Y., Anderson D.G. Synergistic lipid compositions for albumin receptor mediated delivery of mRNA to the liver. Nat. Commun. 2020;11:2424. doi: 10.1038/s41467-020-16248-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oberli M.A., Reichmuth A.M., Dorkin J.R., Mitchell M.J., Fenton O.S., Jaklenec A., Anderson D.G., Langer R., Blankschtein D. Lipid nanoparticle assisted mRNA delivery for potent cancer immunotherapy. Nano Lett. 2017;17:1326–1335. doi: 10.1021/acs.nanolett.6b03329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T., et al. Optimization of lipid nanoparticles for intramuscular administration of mRNA vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crommelin D.J.A., Anchordoquy T.J., Volkin D.B., Jiskoot W., Mastrobattista E. Addressing the cold reality of mRNA vaccine stability. J. Pharm. Sci. 2021;110:997–1001. doi: 10.1016/j.xphs.2020.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suzuki Y., Hyodo K., Tanaka Y., Ishihara H. siRNA-lipid nanoparticles with long-term storage stability facilitate potent gene-silencing in vivo. J. Control. Release. 2015;220:44–50. doi: 10.1016/j.jconrel.2015.10.024. [DOI] [PubMed] [Google Scholar]
- 18.Ishigooka H., Katsumata H., Saiga K., Tokita D., Motoi S., Matsui C., Suzuki Y., Tomimatsu A., Nakatani T., Kuboi Y., et al. Novel complement C5 small-interfering RNA lipid nanoparticle prolongs graft survival in a hypersensitized rat kidney transplant model. Transplantation. 2022 doi: 10.1097/TP.0000000000004207. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki Y., Ishihara H. Difference in the lipid nanoparticle technology employed in three approved siRNA (Patisiran) and mRNA (COVID-19 vaccine) drugs. Drug Metab. Pharmacokinet. 2021;41:100424. doi: 10.1016/j.dmpk.2021.100424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schoenmaker L., Witzigmann D., Kulkarni J.A., Verbeke R., Kersten G., Jiskoot W., Crommelin D.J.A. mRNA-lipid nanoparticle COVID-19 vaccines: structure and stability. Int. J. Pharm. 2021;601:120586. doi: 10.1016/j.ijpharm.2021.120586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki Y., Hyodo K., Suzuki T., Tanaka Y., Kikuchi H., Ishihara H. Biodegradable lipid nanoparticles induce a prolonged RNA interference-mediated protein knockdown and show rapid hepatic clearance in mice and nonhuman primates. Int. J. Pharm. 2017;519:34–43. doi: 10.1016/j.ijpharm.2017.01.016. [DOI] [PubMed] [Google Scholar]
- 22.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brader M.L., Williams S.J., Banks J.M., Hui W.H., Zhou Z.H., Jin L. Encapsulation state of messenger RNA inside lipid nanoparticles. Biophys. J. 2021;120:2766–2770. doi: 10.1016/j.bpj.2021.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fabre A.L., Colotte M., Luis A., Tuffet S., Bonnet J. An efficient method for long-term room temperature storage of RNA. Eur. J. Hum. Genet. 2014;22:379–385. doi: 10.1038/ejhg.2013.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J., et al. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Akinc A., Maier M.A., Manoharan M., Fitzgerald K., Jayaraman M., Barros S., Ansell S., Du X., Hope M.J., Madden T.D., et al. The Onpattro story and the clinical translation of nanomedicines containing nucleic acid-based drugs. Nat. Nanotechnol. 2019;14:1084–1087. doi: 10.1038/s41565-019-0591-y. [DOI] [PubMed] [Google Scholar]
- 27.Fulginiti V.A., Eller J.J., Downie A.W., Kempe C.H. Altered reactivity to measles virus. Atypical measles in children previously immunized with inactivated measles virus vaccines. JAMA. 1967;202:1075–1080. doi: 10.1001/jama.202.12.1075. [DOI] [PubMed] [Google Scholar]
- 28.Kim H.W., Canchola J.G., Brandt C.D., Pyles G., Chanock R.M., Jensen K., Parrott R.H. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am. J. Epidemiol. 1969;89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 29.Bolles M., Deming D., Long K., Agnihothram S., Whitmore A., Ferris M., Funkhouser W., Gralinski L., Totura A., Heise M., Baric R.S. A double-inactivated severe acute respiratory syndrome coronavirus vaccine provides incomplete protection in mice and induces increased eosinophilic proinflammatory pulmonary response upon challenge. J. Virol. 2011;85:12201–12215. doi: 10.1128/JVI.06048-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lederer K., Castaño D., Gómez Atria D., Oguin T.H., 3rd, Wang S., Manzoni T.B., Muramatsu H., Hogan M.J., Amanat F., Cherubin P., et al. SARS-CoV-2 mRNA vaccines foster potent antigen-specific germinal center responses associated with neutralizing antibody generation. Immunity. 2020;53:1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Iwasaki A., Omer S.B. Why and how vaccines work. Cell. 2020;183:290–295. doi: 10.1016/j.cell.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pardi N., Hogan M.J., Naradikian M.S., Parkhouse K., Cain D.W., Jones L., Moody M.A., Verkerke H.P., Myles A., Willis E., et al. Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J. Exp. Med. 2018;215:1571–1588. doi: 10.1084/jem.20171450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alameh M.-G., Tombácz I., Bettini E., Lederer K., Sittplangkoon C., Wilmore J.R., Gaudette B.T., Soliman O.Y., Pine M., Hicks P., et al. Lipid nanoparticles enhance the efficacy of mRNA and protein subunit vaccines by inducing robust T follicular helper cell and humoral responses. Immunity. 2021;54:2877–2892.e7. doi: 10.1016/j.immuni.2021.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hajj K.A., Ball R.L., Deluty S.B., Singh S.R., Strelkova D., Knapp C.M., Whitehead K.A. Branched-tail lipid nanoparticles potently deliver mRNA in vivo due to enhanced ionization at endosomal pH. Small. 2019;15:e1805097. doi: 10.1002/smll.201805097. [DOI] [PubMed] [Google Scholar]
- 35.Han X., Zhang H., Butowska K., Swingle K.L., Alameh M.-G., Weissman D., Mitchell M.J. An ionizable lipid toolbox for RNA delivery. Nat. Commun. 2021;12:7233. doi: 10.1038/s41467-021-27493-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ndeupen S., Qin Z., Jacobsen S., Bouteau A., Estanbouli H., Igyártó B.Z. The mRNA-LNP platform's lipid nanoparticle component used in preclinical vaccine studies is highly inflammatory. iScience. 2021;24:103479. doi: 10.1016/j.isci.2021.103479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki T., Suzuki Y., Hihara T., Kubara K., Kondo K., Hyodo K., Yamazaki K., Ishida T., Ishihara H. PEG shedding-rate-dependent blood clearance of PEGylated lipid nanoparticles in mice: faster PEG shedding attenuates anti-PEG IgM production. Int. J. Pharm. 2020;588:119792. doi: 10.1016/j.ijpharm.2020.119792. [DOI] [PubMed] [Google Scholar]
- 38.Besin G., Milton J., Sabnis S., Howell R., Mihai C., Burke K., Benenato K.E., Stanton M., Smith P., Senn J., Hoge S. Accelerated blood clearance of lipid nanoparticles entails a biphasic humoral response of B-1 followed by B-2 lymphocytes to distinct antigenic moieties. Immunohorizons. 2019;3:282–293. doi: 10.4049/immunohorizons.1900029. [DOI] [PubMed] [Google Scholar]
- 39.Franzé S., Selmin F., Samaritani E., Minghetti P., Cilurzo F. Lyophilization of liposomal formulations: still necessary, still challenging. Pharmaceutics. 2018;10:E139. doi: 10.3390/pharmaceutics10030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohammady M., Mohammadi Y., Yousefi G. Freeze-drying of pharmaceutical and nutraceutical nanoparticles: the effects of formulation and technique parameters on nanoparticles characteristics. J. Pharm. Sci. 2020;109:3235–3247. doi: 10.1016/j.xphs.2020.07.015. [DOI] [PubMed] [Google Scholar]
- 41.Zhao P., Hou X., Yan J., Du S., Xue Y., Li W., Xiang G., Dong Y. Long-term storage of lipid-like nanoparticles for mRNA delivery. Bioact. Mater. 2020;5:358–363. doi: 10.1016/j.bioactmat.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muramatsu H., Lam K., Bajusz C., Laczkó D., Karikó K., Schreiner P., Martin A., Lutwyche P., Heyes J., Pardi N. Lyophilization provides long-term stability for a lipid nanoparticle-formulated, nucleoside-modified mRNA vaccine. Mol. Ther. 2022;30:1941–1951. doi: 10.1016/j.ymthe.2022.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.FDA Package Insert, VYXEOS™ (Daunorubicin and Cytarabine) Liposome. https://wwwaccessdatafdagov/drugsatfda_docs/label/2017/209401s000lblpdf
- 44.Gindy M.E., Feuston B., Glass A., Arrington L., Haas R.M., Schariter J., Stirdivant S.M. Stabilization of Ostwald ripening in low molecular weight amino lipid nanoparticles for systemic delivery of siRNA therapeutics. Mol. Pharm. 2014;11:4143–4153. doi: 10.1021/mp500367k. [DOI] [PubMed] [Google Scholar]
- 45.Basha G., Novobrantseva T.I., Rosin N., Tam Y.Y.C., Hafez I.M., Wong M.K., Sugo T., Ruda V.M., Qin J., Klebanov B., et al. Influence of cationic lipid composition on gene silencing properties of lipid nanoparticle formulations of siRNA in antigen-presenting cells. Mol. Ther. 2011;19:2186–2200. doi: 10.1038/mt.2011.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kawase W., Kurotaki D., Suzuki Y., Ishihara H., Ban T., Sato G.R., Ichikawa J., Yanai H., Taniguchi T., Tsukahara K., Tamura T. Irf5 siRNA-loaded biodegradable lipid nanoparticles ameliorate concanavalin A-induced liver injury. Mol. Ther. Nucleic Acids. 2021;25:708–715. doi: 10.1016/j.omtn.2021.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R., et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suzuki Y., Ishihara H. Structure, activity and uptake mechanism of siRNA-lipid nanoparticles with an asymmetric ionizable lipid. Int. J. Pharm. 2016;510:350–358. doi: 10.1016/j.ijpharm.2016.06.124. [DOI] [PubMed] [Google Scholar]
- 49.Ali S., Mann-Nüttel R., Schulze A., Richter L., Alferink J., Scheu S. Sources of type I interferons in infectious immunity: plasmacytoid dendritic cells not always in the driver's seat. Front. Immunol. 2019;10:778. doi: 10.3389/fimmu.2019.00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Uemura Y., Naoi T., Kanai Y., Kobayashi K. The efficiency of lipid nanoparticles with an original cationic lipid as a siRNA delivery system for macrophages and dendritic cells. Pharm. Dev. Technol. 2019;24:263–268. doi: 10.1080/10837450.2018.1469149. [DOI] [PubMed] [Google Scholar]
- 51.Mildner A., Jung S. Development and function of dendritic cell subsets. Immunity. 2014;40:642–656. doi: 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 52.Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd, et al. A single immunization with nucleoside-modified mRNA vaccines elicits strong cellular and humoral immune responses against SARS-CoV-2 in mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Turner J.S., O'Halloran J.A., Kalaidina E., Kim W., Schmitz A.J., Zhou J.Q., Lei T., Thapa M., Chen R.E., Case J.B., et al. SARS-CoV-2 mRNA vaccines induce persistent human germinal centre responses. Nature. 2021;596:109–113. doi: 10.1038/s41586-021-03738-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Swaminathan G., Thoryk E.A., Cox K.S., Meschino S., Dubey S.A., Vora K.A., Celano R., Gindy M., Casimiro D.R., Bett A.J. A novel lipid nanoparticle adjuvant significantly enhances B cell and T cell responses to sub-unit vaccine antigens. Vaccine. 2016;34:110–119. doi: 10.1016/j.vaccine.2015.10.132. [DOI] [PubMed] [Google Scholar]
- 55.Zhang H., You X., Wang X., Cui L., Wang Z., Xu F., Li M., Yang Z., Liu J., Huang P., et al. Delivery of mRNA vaccine with a lipid-like material potentiates antitumor efficacy through Toll-like receptor 4 signaling. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2005191118. e2005191118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.de Groot A.M., Thanki K., Gangloff M., Falkenberg E., Zeng X., van Bijnen D.C.J., van Eden W., Franzyk H., Nielsen H.M., Broere F., et al. Immunogenicity testing of lipidoids in vitro and in silico: modulating lipidoid-mediated TLR4 activation by nanoparticle design. Mol. Ther. Nucleic Acids. 2018;11:159–169. doi: 10.1016/j.omtn.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miao L., Li L., Huang Y., Delcassian D., Chahal J., Han J., Shi Y., Sadtler K., Gao W., Lin J., et al. Delivery of mRNA vaccines with heterocyclic lipids increases anti-tumor efficacy by STING-mediated immune cell activation. Nat. Biotechnol. 2019;37:1174–1185. doi: 10.1038/s41587-019-0247-3. [DOI] [PubMed] [Google Scholar]
- 58.O'Hagan D.T., Fox C.B. New generation adjuvants--from empiricism to rational design. Vaccine. 2015;33(Suppl 2):B14–B20. doi: 10.1016/j.vaccine.2015.01.088. [DOI] [PubMed] [Google Scholar]
- 59.Salguero F.J., White A.D., Slack G.S., Fotheringham S.A., Bewley K.R., Gooch K.E., Longet S., Humphries H.E., Watson R.J., Hunter L., et al. Comparison of rhesus and cynomolgus macaques as an infection model for COVID-19. Nat. Commun. 2021;12:1260. doi: 10.1038/s41467-021-21389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urano E., Okamura T., Ono C., Ueno S., Nagata S., Kamada H., Higuchi M., Furukawa M., Kamitani W., Matsuura Y., et al. COVID-19 cynomolgus macaque model reflecting human COVID-19 pathological conditions. Proc. Natl. Acad. Sci. USA. 2021;118 doi: 10.1073/pnas.2104847118. e2104847118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Earle K.A., Ambrosino D.M., Fiore-Gartland A., Goldblatt D., Gilbert P.B., Siber G.R., Dull P., Plotkin S.A. Evidence for antibody as a protective correlate for COVID-19 vaccines. Vaccine. 2021;39:4423–4428. doi: 10.1016/j.vaccine.2021.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gilbert P.B., Montefiori D.C., McDermott A.B., Fong Y., Benkeser D., Deng W., Zhou H., Houchens C.R., Martins K., Jayashankar L., et al. Immune correlates analysis of the mRNA-1273 COVID-19 vaccine efficacy clinical trial. Science. 2022;375:43–50. doi: 10.1126/science.abm3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author, upon reasonable request.