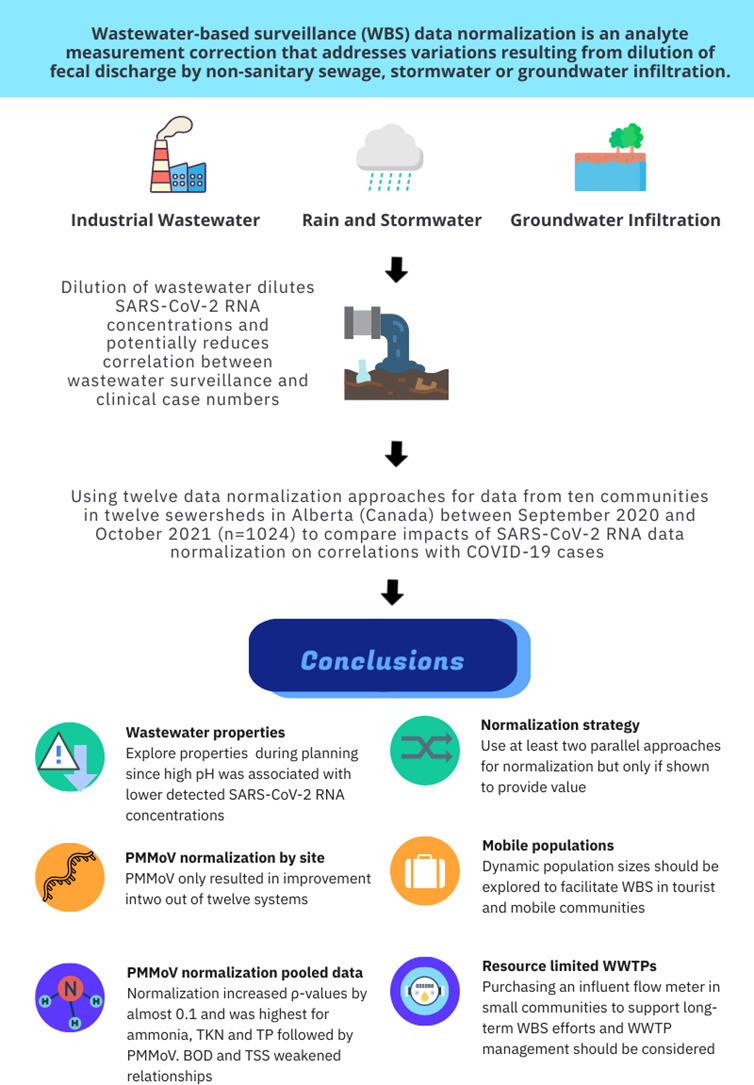
Abstract
Wastewater-based surveillance (WBS) data normalization is an analyte measurement correction that addresses variations resulting from dilution of fecal discharge by non-sanitary sewage, stormwater or groundwater infiltration. No consensus exists on what WBS normalization parameters result in the strongest correlations and lead time between SARS-CoV-2 WBS data and COVID-19 cases. This study compared flow, population size and biomarker normalization impacts on the correlations and lead times for ten communities in twelve sewersheds in Alberta (Canada) between September 2020 and October 2021 (n = 1024) to determine if normalization by Pepper Mild Mottle Virus (PMMoV) provides any advantages compared to other normalization parameters (e.g., flow, reported and dynamic population sizes, BOD, TSS, NH3, TP). PMMoV concentrations (GC/mL) corresponded with plant influent flows and were highest in the urban centres. SARS-CoV-2 target genes E, N1 and N2 were all negatively associated with wastewater influent pH, while PMMoV was positively associated with temperature. Pooled data analysis showed that normalization increased ρ-values by almost 0.1 and was highest for ammonia, TKN and TP followed by PMMoV. Normalization by other parameters weakened associations. None of the differences were statistically significant. Site-specific correlations showed that normalization of SARS-CoV-2 data by PMMoV only improved correlations significantly in two of the twelve systems; neither were large sewersheds or combined sewer systems. In five systems, normalization by traditional wastewater strength parameters and dynamic population estimates improved correlations. Lead time ranged between 1 and 4 days in both pooled and site-specific comparisons. We recommend that WBS researchers and health departments: a) Investigate WWTP influent properties (e.g., pH) in the WBS planning phase and use at least two parallel approaches for normalization only if shown to provide value; b) Explore normalization by wastewater strength parameters and dynamic population size estimates further; and c) Evaluate purchasing an influent flow meter in small communities to support long-term WBS efforts and WWTP management.
Keywords: Wastewater-based epidemiology (WBE), COVID-19, Normalization, Industrial wastewater, Combined sewers, Mobile populations
Graphical abstract
1. Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) wastewater-based surveillance (WBS) has complemented clinical surveillance of Coronavirus disease 2019 (COVID-19) by providing scientific evidence to support public health decision making for pandemic management (Howard et al., 2020). Unlike clinical diagnostic tests, which are invasive, costly, time-consuming and affected by testing policies and behaviours, wastewater monitoring of SARS-CoV-2 RNA provides pooled testing of all COVID-19 cases relying on a centralized wastewater collection and treatment system (Daughton, 2020a; Daughton, 2020b). WBS can also provide an early indication of changes in community-level trends compared to clinical data (Ahmed et al., 2020a; Gerrity et al., 2021; Gonzalez et al., 2020; Peccia et al., 2020; Randazzo et al., 2020; Weidhaas et al., 2021). Lead time reports vary substantially with early COVID-19 trend detection ranging between 0–2 days and 2–3 weeks (Bibby et al., 2021). Various studies have compared analytical methods used in COVID-19 WBS (Ahmed et al., 2020c; Kevill et al., 2022; LaTurner et al., 2021; Philo et al., 2021; Qiu et al., 2022; Sapula et al., 2021; Vadde et al., 2022; Zhan et al., 2022). Fewer studies have explored the role of SARS-CoV-2 data normalization on correlations between WBS and community prevalence of COVID-19 (Isaksson et al., 2022; Sakarovitch et al., 2022).
Data normalization is an analyte measurement correction that addresses variations in fecal strength and dilution in the sewershed. Estimating the concentration of any biological or chemical analyte in wastewater is complicated by flow changes and dilution events (e.g., stormwater, industrial discharges, hauled wastes, groundwater infiltration and inflow), analyte shedding rates and patterns, the size of the population served and the stability and transport mechanics of the biomarker being monitored in the sewershed (Arabzadeh et al., 2021; Mazumder et al., 2022; Wade et al., 2022a). While there is currently no consensus on what WBS normalization parameters result in the strongest correlations or longest lead time between SARS-CoV-2 WBS data and COVID-19 case numbers, the majority of approaches incorporate corrections by flow, population size and endogenous fecal biomarkers. To compare viral concentrations in wastewater over time, monitoring loading estimates (i.e., gene copies (GC) per day) by multiplying viral RNA concentrations by daily wastewater flows to account for changes in sanitary sewer contributions has been recommended (Centre for Disease Control and Prevention, 2022; Greenwald et al., 2021b; Gudra et al., 2022; Rusiñol et al., 2021; Sakarovitch et al., 2022). Traditional wastewater physico-chemical properties and dynamic population size estimates have also been used to account for fecal strength changes but studies have often been challenged by lack of critical daily influent data or limited sample sizes (Cluzel et al., 2022; Greenwald et al., 2021b; Sweetapple et al., 2021; Wade et al., 2022b). Dynamic population estimates are of interest when participating wastewater treatment plants (WWTPs) receive influent from adjacent towns or have mobile populations with high tourism, commuters or part-time residents (Gudra et al., 2022; Isaksson et al., 2022) making census population size data up to 50 % inaccurate (Castiglioni et al., 2013). Dynamic population estimates calculate the size of the population contributing to the sewershed by using influent wastewater contaminant loading (e.g., ammonia, phosphorus) and the concentration of the specific contaminant shed per capita providing data in the units GC per capita to facilitate comparison across sites. Normalizing SARS-CoV-2 RNA concentrations by endogenous human fecal markers by assaying organisms or compounds specific to human feces in wastewater to estimate its human fecal content or fecal strength is also common (American Public Health Laboratories, 2022; Been et al., 2014; Centre for Disease Control and Prevention, 2022; Choi et al., 2019). Explored biomarkers include Pepper Mild Mottle Virus (PMMoV), Bacteroides HF183, F-specific RNA bacteriophages, human 18S rRNA, crAssphage, fecal coliforms and β-2-microglobulin (Ai et al., 2021; Duvallet et al., 2022; Feng et al., 2021; LaTurner et al., 2021; Sakarovitch et al., 2022; Vadde et al., 2022; Wilder et al., 2021; Zhan et al., 2022). Hypothetically, normalization of quantitative virus data with these endogenous fecal controls can improve correlations between RNA concentrations and case numbers by accounting for variability in human fecal content provided that the endogenous wastewater marker shedding, fate and transport mimics that of SARS-CoV-2 (Crank et al., 2022; Gutierrez et al., 2021). Several studies have reported that normalization by biomarkers does not consistently improve correlations with cases at the community level (Duvallet et al., 2022; Feng et al., 2021; Greenwald et al., 2021a; Sakarovitch et al., 2022; Zhan et al., 2022). PMMoV is not an endogenous fecal marker but is frequently used for SARS-CoV-2 RNA data normalization. Several studies have advocated normalizing SARS-CoV-2 concentrations by the PMMoV (D'Aoust et al., 2021b; Wu et al., 2022), especially for normalization of SARS-CoV-2 concentrations in solids. Despite being recovered in high concentrations from feces and untreated wastewater, PMMoV concentrations in wastewater are highly reliant on availability and consumption of infected capsicums and some other solanaceous plants and their products (Holm et al., 2022; Symonds et al., 2018; Wu et al., 2022; Zhang et al., 2006), access to which is influenced by geographical and socio-cultural community characteristics.
The objective of this study is to compare the impacts of flow, population size and biomarker normalization on correlations and lead time between SARS-CoV-2 WBS data and COVID-19 case rates using a large, long-term dataset (n = 1024) and a published and validated analytical method to provide recommendations to WBS practitioners. The analysis is divided into four sections: a) Comparing the loading of three SARS-CoV-2 gene targets (N1, N2 and E) and PMMoV in all participating communities; b) Investigating wastewater properties that impact SARS-CoV-2, PMMoV detection and HCoV-229E recovery rates to better understand WBS data limitations; c) Exploring if data normalization provides better correlations with active population infection trends when data is pooled; and d) Comparing impacts of normalizing averaged SARS-CoV-2 gene targets E, N1 and N2 by PMMoV concentrations, wastewater influent flows, reported and dynamic population sizes, wastewater strength chemical parameters (biochemical oxygen demand (BOD), total suspended solids (TSS), ammonia (NH3), total Kjeldahl nitrogen (TKN), total phosphorus (TP) and total organic nitrogen (TON)) on correlations and lead times with active cases at each site. We provide a statistically powerful assessment of an extensive and unique number of normalization parameters in twelve wastewater systems with high rates of COVID-19 clinical testing among sewershed subpopulation. Our hypothesis was that normalization by PMMoV would not always improve correlations or lead times with COVID-19 clinical cases and that using other classical wastewater strength parameters for normalization would provide comparable value. Given the constant changes in COVID-19 testing algorithms, we recognize that correlations with clinical data is not a gold standard for determining validity of WBS (Qiu et al., 2022). However, correlations with reported cases can be useful to determine if WBS data reflects broad trends of infection in the population (Duvallet et al., 2022).
2. Materials and methods
2.1. Wastewater treatment plants samples and populations served
Twelve WWTPs in Alberta (Canada) participated in the study. All participating plants with the exceptions of High River (aerated lagoon) were traditional secondary treatment facilities with biological nutrient removal. All collection systems were sanitary except for Edmonton which includes some combined stormwater sewers. Edmonton's Gold Bar WWTP and Calgary Plant A were the largest two facilities receiving >200 megalitres per day (MLD) and serving populations larger than one million residents. All other plants received <100 MLD. Each utility identified the communities within the boundaries of its WWTP sewershed which was mapped to corresponding Local Geographic Areas (LGA), created by Alberta Health and Alberta Health Services based on census boundaries. This information was used to determine the number of new and active COVID-19 cases and case rates per 100,000 in the population served by each respective WWTP from the COVID-19 surveillance database of the Government of Alberta. Active cases were defined as number of cases within 14 days from onset.
New cases were defined as daily new cases reported by Alberta Health Services. COVID-19 epidemic waves in Alberta were defined as the first (May–Sept 2020), second (Oct 2020–Feb 2021), third (March–June 2021) and fourth (July–October 2021). Given that sample storage methods were changed in Wave 2 (Qiu et al., 2022), the correlations only include data collected in Waves 2–4 between September 30, 2020 to October 3, 2021.
Post-grit screening raw influent samples were collected two to three times per week using hourly or flow-based composite samplers from all twelve participating WWTPs. Most 24-hour composite samplers ran from midnight to midnight and the same composite sample was subsampled for both physico-chemical analyses and virological assays. Physico-chemical analyses and data sources are provided in Section 2.2. For virological assays, a 500 mL subsample from the post-grit 24-hour composite samples was collected 2–3 days per week from May 15, 2020 to October 3, 2021. Wastewater subsamples were stored at 4 °C after collection, shipped weekly to the analytical virology laboratory and processed within 72 h in accordance with other publications (Ahmed et al., 2020b; Isaksson et al., 2022). Table 1 provides descriptions of participating wastewater treatment plants and their served communities in twelve sewersheds in Alberta between May 2020 and October 2021.
Table 1.
Description of participating wastewater treatments and their served communities in twelve sewersheds in Alberta with corresponding average values (Ave) and standard deviations (±SD) of plant post-grit influent temperature (°C), total suspended solids (TSS, mg/L), biochemical oxygen demand (BOD, mg/L) and flow rates (10 m3/day) between May 2020 and October 2021.
| Wastewater treatment plant | # of samples (n) | Flow rate (1000 m3/day or MLD) |
Total suspended solids (TSS, mg/L) |
Biochemical oxygen demand (BOD, mg/L) |
Influent temperature (°C) |
City/town/county served | Population in the sewershed |
|---|---|---|---|---|---|---|---|
| Ave ± SD | Ave ± SD | Ave ± SD | Ave ± SD | ||||
| EPCOR Gold Bar | 118 | 306 ± 124 | 342.6 ± 365.3 | 274.9 ± 171 | 15.6 ± 5.6 | Edmonton, Leduc, Beaumont | 1,115,021 |
| The City of Calgary A | 106 | 346 ± 44 | 274 ± 38.4 | 251.9 ± 37.1 | 14.4 ± 2.2 | Calgary North, Cochrane, Airdrie | 1,104,208 |
| The City of Calgary B | 97 | 34 ± 9 | 295.4 ± 86.4 | 288.9 ± 57.1 | 15.2 ± 2.1 | Calgary South | 90,922 |
| The City of Calgary C | 106 | 89 ± 13 | 239.1 ± 57.8 | 207.3 ± 32.8 | 15.3 ± 2.5 | Calgary South | 307,622 |
| Alberta Capital Region Wastewater Commission (ACRWC) | 108 | 76 ± 11 | 353.8 ± 82.3 | 256.2 ± 44 | 13 ± 2 | Fort Saskatchewan, St. Alberta, Spruce Grove, Strathcona County, Sturgeon County, Stoney Plain, Morinville, Bon Accord; Gibbons | 326,497 |
| The City of Red Deer | 96 | 56 ± 8 | 250.1 ± 50.5 | 259.3 ± 57.5 | 15.3 ± 1.5 | Red Deer, Sylvan Lake, Olds, Lacombe, Innisfail | 187,857 |
| The City of Lethbridge | 72 | 40 ± 3 | 8 ± 4.7 | 4.3 ± 1.9 | 18.9 ± 3.4 | Lethbridge | 100,655 |
| Aquatera | 82 | 19 ± 3 | 285.9 ± 86.7 | 324.1 ± 83.9 | 13.2 ± 2.1 | Grande Prairie | 74,245 |
| The City of Medicine Hat | 67 | 24 ± 1 | 208.4 ± 23 | 206.4 ± 25 | 16.2 ± 8.8 | Medicine Hat | 68,115 |
| High River Treatment Facility | 53 | 4 ± 0.3 | 154.7 ± 77.2 | 315.3 ± 147.8 | 12.4 ± 3.1 | Town of High River | 16,922 |
| EPCOR Canmore | 60 | 9 ± 4 | 227.5 ± 341.1 | 229.2 ± 96.6 | 10.8 ± 2.0 | Town of Canmore | 16,547 |
| The Town of Banff | 82 | 6 ± 2 | 155.3 ± 43.9 | 160.6 ± 35.5 | 10.9 ± 1.7 | Town of Banff | 13,451 |
2.2. Physico-chemical water quality parameters
All participating WWTPs provided influent flows in 1000 m3 per day or megaliters per day (MLD), temperature (°C), biochemical oxygen demand (BOD, mg/L) and total suspended solids (TSS, mg/L) for the dates on which samples for virological analyses were taken. Influent flow and temperature data was measured in real-time using appropriate controllers and recorded in the WWTP Supervisory Control and Data Acquisition (SCADA) systems. Reported physico-chemical laboratory analyses were conducted by WWTP wastewater laboratories located on-site or shipped out to accredited commercial laboratories. Results were recorded in the Laboratory Information Management Systems (LIMS) or in Excel. Additional physico-chemical influent data was collected from the City of Calgary Plant A and Edmonton's Gold Bar WWTP. Both facilities have laboratories accredited by the Canadian Association for Laboratory Accreditation (CALA) and use Standard Methods for the Examination of Water and Wastewater (American Public Health Association and American Water Works Association, 2017). Additional parameters included: pH, Total Ammonia Nitrogen (NH3), Biochemical Oxygen Demand (BOD), Total Phosphorus (TP), Total Suspended Solids (TSS), Total Kjeldahl Nitrogen (TKN). To estimate total organic nitrogen concentrations, ammonia values were subtracted from TKN values for the same composite sample. All samples were run with blanks with every batch analyzed on the same date. Instruments were calibrated with prepared or purchased standards solutions for each instrument run. 10 % of samples were run in duplicate.
2.3. Processing and testing wastewater samples for SARS-CoV-2 RNA
The optimized laboratory protocol for processing of wastewater samples was published in Qiu et al. (2022). All reported RNA concentrations and QA/QC data were produced by the same laboratory at the University of Alberta. Briefly, 100 mL of sample was centrifugated at 4500 ×g for 10 min. The supernatant was collected, transferred to the Centricon filter cup (30-kDa MWCO, Millipore) and centrifuged. The concentrated sample was made up to a final volume of 1 mL with phosphate buffered saline. The total nucleic acid (NA) was extracted from 400 μL of the concentrated sample using MagMAX-96 viral RNA isolation kit on the automated KingFisher™ Flex instrument and eluted at a final volume of 100 μL. One-step real time RT-qPCR assay was performed in duplicates to detect E, N1 and N2 genes of SARS-CoV-2 on an ABI 7500 PCR instrument. For quantification, an external standard curve was prepared by 10-fold serial dilutions. The limit of detection was 80 copies per 100 mL for all three targets and was determined as described in Pabbaraju et al. (Pabbaraju et al., 2021).
For every run, a matrix spike was prepared by spiking a known concentration of cultured human coronavirus 229E (hCoV-229E, 100 μL, VR-740, ATCC) into each unprocessed sample as previously described (Qiu et al., 2022). The recovery rate of the virus in wastewater (%) = amount of hCoV 229E detected in the spiked sample / amount of virus detected in the baseline sample × 100. Recoveries ranged between 1 and 10 % for the majority of samples tested and were used for the assessment of wastewater variables that showed relationships with measured SARS-CoV-2 RNA concentrations. Salmon DNA (5 μL) was spiked into concentrated sample before the NA extraction and quantified by qPCR to assess the degree of PCR inhibition. Inhibition was defined as a delay of Ct by 3 cycles as compared to a distilled water control spiked with the same amount of salmon DNA and was identified in 7 % of samples. PMMoV was quantified using RT-qPCR alongside the 3 gene targets of SARS-CoV-2. Negative and positive controls were included during sample concentration, nucleic acid extraction and RT-qPCR. All quality control criteria were met before reporting. Positive result (SARS-CoV-2 RNA) was reported when there were two or more positive PCR tests out of the six duplicate PCR runs for each of the three SARS-CoV-2 gene targets.
2.4. Statistical analyses
2.4.1. Statistical analyses of wastewater SARS-CoV-2 concentrations, trends and relationships
All data analyses and statistics were performed in JMP® (Version 16.1. SAS Institute Inc., Cary, NC, 1989–2021). For all statistical analyses, seven-day moving averages were used for all active and new COVID-19 cases as recommended in the literature (Ai et al., 2021; Johns Hopkins Coronavirus Resource Center, 2022). In addition, seven-day moving averages were used for SARS-CoV-2 RNA concentrations to smooth daily variability of the dataset as recommended by Li et al. (2022) (Li et al., 2022). For smoothing case numbers and RNA concentration data, we calculated seven-day moving averages using an exponential function and averaged values for seven days starting with the value recorded on the date of sample collection (i.e., right-aligned window). Exponential smoothing models use a weighted average of the observed value yt − 1 and the forecasted value ŷt − 1 as the forecast for time period t, which allows us to give more recent values more weight. The forecasting equation is
where w is called the weight or smoothing constant for the exponential smoothing model and is traditionally assumed have a range of 0 ≤ w ≤ 1. In our calculations, t = 7 and the smoothing constant was automatically set by JMP to 1.0.
Data related to influent wastewater samples characteristics were averaged and presented as means and standard deviations. Quartile range outlier analysis (tail quantile = 0.1; Q = 3) was used to detect outliers for all parameters that fell outside the mean by more than three standard deviations. The outlier analysis identified seven SARS-CoV-2 outliers and an additional 25 flow and chemical parameter outliers, which were excluded from the remainder of the analyses following visual confirmation. Comparing gene target concentrations was performed using a Mann–Whitney U test, where p < 0.05 was considered significant. Relationships between wastewater properties and SARS-CoV-2 gene target loading were examined using response screening, which relies on bivariate regressions but adjusts for False Discovery Rates (FDR), outliers and missing values. SARS-CoV-2 gene targets, PMMoV concentrations and hCoV-229E recoveries were used as the Y variables, while plant influent flow, BOD, TSS, temperature, NH3, TP and TKN were used as the predictors. SARS-CoV-2 gene target concentrations were hCoV-229E recovery adjusted by multiplying the gene target concentrations by 100 and dividing by the corresponding matrix spike recovery result. SARS-CoV-2 flow adjustment (or loading) required multiplying the SARS-CoV-2 gene target concentration by plant influent flow for the date of composite sampler collection.
2.4.2. Normalization comparison based on influent properties and other parameters
To improve our understanding of data normalization impacts, we compared correlations between seven-day moving averages of active and new COVID case rates and averaged normalized and unnormalized seven-day moving averages of SARS-CoV-2 gene target concentrations for both the pooled dataset and by individual WWTPs. Recognizing that the SARS-CoV-2 data did not follow a normal distribution and was highly autocorrelated with case rates and wastewater properties, we used Spearman correlations, a non-parametric statistical correlation approach (Greenwald et al., 2021b). Since all p-values were very low for these analyses (p < 0.001), we reported Spearman's ρ throughout the results to characterize the strength of the associations between both variables. Spearman's ρ values were interpreted as provided by Von Sperling et al. (2020): a strong correlation had a ρ-value higher than 0.7; a moderate correlation had a ρ-value between 0.4 and 0.7; and a ρ-value between 0 and 0.4 was considered a weak association (Von Sperling et al., 2020). Differences between normalized and unnormalized ρ-values were presented. To compare normalized and unnormalized correlations statistically, a multiple comparison rank sum test was used where the control was set as the correlation between cases and unnormalized SARS-CoV-2 concentrations (Steel, 1959). We also used a six-day lag function for new and active case rates to explore the effect of normalized SARS-CoV2 data on lead time determination using the same Spearman correlations. A lag function in JMP is used to access data from previous rows for a defined number of rows for statistical calculations. The lag function can be edited to become a lead function and access subsequent rows. The lag function therefore allows calculating the Spearman correlations between RNA concentrations and case rates on subsequent dates to identify number of days of lead time where correlations are strongest. Averaged SARS-CoV-2 RNA concentrations were normalized by PMMoV, plant flow in MLD (Gene targets averaged*Flow), plant loading per capita (Gene targets averaged*Flow/Population), BOD (Gene targets averaged/BOD), TSS (Gene targets averaged/TSS), plant loading per capita as determined by a dynamic population calculation by BOD (Gene targets averaged*Flow/BOD Dyn Pop) and plant loading per capita as determined by a dynamic population calculation by TSS (Gene targets averaged*Flow/TSS Dyn Pop) for all plants. To calculate dynamic population estimates as previously described (Sweetapple et al., 2021; Wade et al., 2022b), site-specific concentrations of wastewater physico-chemical parameters, literature reported per capita excretion estimates for the specific biomarker per capita and flow rates were used. The dynamic population size was calculated by multiplying the daily measured parameter concentrations (Xd) and daily wastewater flow rates (Qd) and dividing by literature reported per capita excretion estimates for the specific biomarker (x mg per capita per day). When dividing SARS-CoV-2 RNA loading by dynamic population size, plant flow and mg/L cancel out and the final unit is GC per capita.
Literature derived daily discharge per capita (x mg per capita per day) estimates were as follows: 1.8 g of TP/capita.day of (Alexander and Stevens, 1976); 50–118 g of BOD/capita.day (Metcalf et al., 2014) with an average of 84 g BOD/capita.day; 8.1 g of NH3/capita.day (Been et al., 2014), 59–150 g of TSS/capita.day (Metcalf et al., 2014) with an average of 105 g TSS/capita.day; and 9.1–22.7 g TKN/capita.day (Metcalf et al., 2014) with an average of 15.9 g TKN/capita.day. When ranges were provided by the source, the average value was used.
Daily influent concentrations of NH3, TP, TKN and total organic nitrogen (TON) were only available for Calgary Plant A and the Edmonton plant. Thus, for Calgary Plant A and Edmonton, averaged SARS-CoV-2 RNA concentrations were normalized by all the previously mentioned parameters, as well as NH3 (Gene targets averaged/NH3), TKN (Gene targets averaged/TKN), TON (Gene targets averaged/TON), TP (Gene targets averaged/TP), plant loading per capita as determined by a dynamic population calculation by NH3 (Gene targets averaged*Flow/NH3 Dyn Pop), plant loading per capita as determined by a dynamic population calculation by TKN (Gene targets averaged*Flow/TKN Dyn Pop) and plant loading per capita as determined by a dynamic population calculation by TP (Gene targets averaged*Flow/TP Dyn Pop). The previously described approach in Sweetapple et al. (2021) was used.
3. Results
3.1. SARS-CoV-2 gene target and PMMoV loading across participating WWTPs
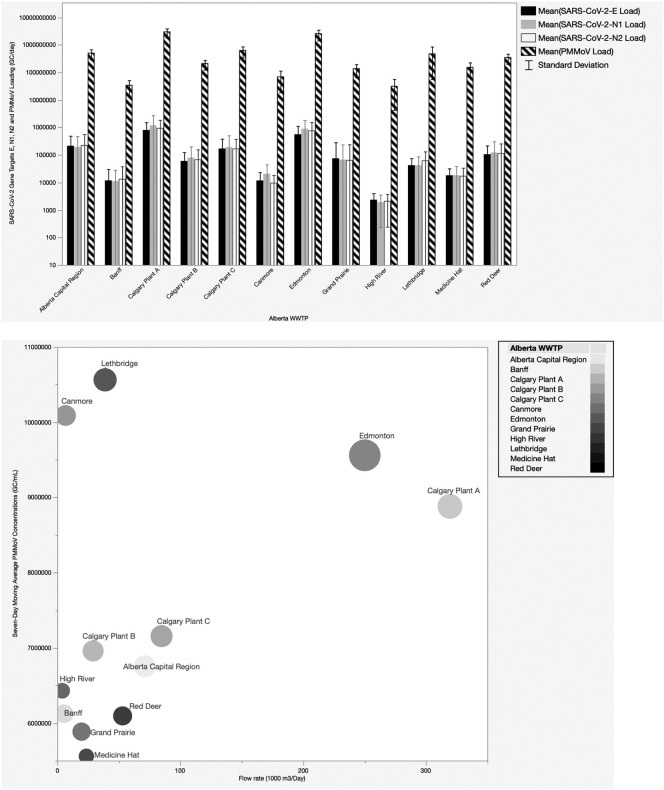
Fig. 1 highlights the differences in SARS-CoV-2 gene target loading and PMMoV loading over the study period for all participating plants in the top panel. Within each site, the three SARS-CoV-2 gene target loadings (GC per day) were not statistically significantly different from each other (Mann Whitney U test results p-values > 0.05 for all sites). PMMoV RNA loadings were statistically significantly higher than SARS-CoV-2 concentrations (p-value < 0.01), which is to be expected. PMMoV concentrations (GC/mL) mostly corresponded with plant influent flows (Fig. 1 bottom panel) and were highest in the urban centres (i.e., Edmonton and Calgary Plant A). Despite serving the same population size and having similar COVID-19 case rates as Canmore, High River had the lowest SARS-CoV-2 RNA loadings possibly reflecting the difficulty of collecting a representative wastewater influent sample at aerated lagoons or the mobile population in Canmore. Detailed RNA loading data is provided in Supplementary Appendix A.
Fig. 1.
Mean SARS-CoV-2 target genes E, N1 and N2 and PMMoV RNA loading (GC/day) in post-grit raw influent samples between May 2020 and October 2021 collected from twelve WWTPs in Alberta. The limit of detection was 80 copies per 100 mL for all three SARS-CoV-2 gene targets (top panel). Bottom panel shows PMMoV concentrations by plant and flow rate (1000 m3/Day).
3.2. Wastewater properties impacting SARS-CoV-2 and PMMoV RNA loading into WWTPs
Relationships between wastewater quality parameters (influent temperature, BOD, TSS, pH, NH3, TP and TKN), SARS-CoV-2 loading (GC/day) and PMMoV loading (GC/day) at all twelve participating WWTPs by response screening are shown in Table 2 . The response screening only revealed four statistically significant relationships. SARS-CoV-2 loading for the three gene targets were negatively impacted by wastewater pH with the effect size ranging between 11 % and 14 %. Highest recovered SARS-CoV-2 loads were seen when pH ranged between 7.2 and 7.6. pH values were only reported by six plants and ranged between 6.1 and 8.2 with a mean value of 7.59 ± 0.17. The dataset in the low and high pH range was very limited but likely reflected industrial inputs into the sewershed suggesting the importance of industrial interference. The effect size was most pronounced on the N1, E and N2 genes, respectively, but followed the same trend. PMMoV loading increased with increasing wastewater temperatures possibly reflecting the availability and price of peppers in the market. PMMoV loads plateau at approximately 25 °C. All other relationships were not statistically significant, including relationships between hCoV-229E recoveries and wastewater parameters (p-values > 0.05). Two scatterplots detailing the relationships between SARS-CoV-2 and pH and between PMMoV and temperature can be found in Appendix 2.
Table 2.
Response screening assessing the relationships between various wastewater quality parameters, matrix spike recovery adjusted SARS-CoV-2 loading (GC/Day) and PMMoV RNA loading (GC/Day). Reported results include the sample size, p-value associated with the regressions, the FDR Log Worth, effect size and R squared.
| Y | X | Count | p-Value | LogWorth | FDR p-Value | FDR LogWorth | Effect size | Rank fraction | RSquare |
|---|---|---|---|---|---|---|---|---|---|
| SARS-CoV-2-E | pH | 236 | 0.007 | 2.164 | 0.083 | 1.078 | 0.124 | 0.063 | 0.031 |
| SARS-CoV-2-N1 | pH | 254 | 0.010 | 1.982 | 0.083 | 1.078 | 0.137 | 0.125 | 0.026 |
| SARS-CoV-2-N2 | pH | 259 | 0.016 | 1.790 | 0.086 | 1.063 | 0.110 | 0.188 | 0.022 |
| PMMoV | Temp | 883 | 0.026 | 1.578 | 0.106 | 0.976 | 0.078 | 0.250 | 0.006 |
3.3. Spearman correlations and lead times between SARS-CoV-2 RNA WBS data and COVID-19 case rates
3.3.1. Impacts of normalization on correlations between WBS and clinical SARS-CoV-2 data pooled for all twelve communities
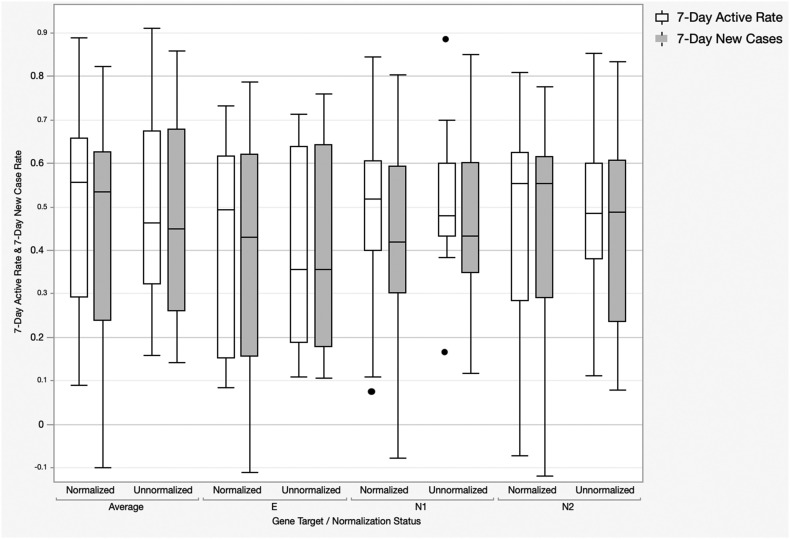
Fig. 2 summarizes the results of the Spearman correlation ρ-values to describe the strength of the associations between all normalized and unnormalized, raw SARS-CoV-2 gene targets concentrations with active and new COVID-19 case rates for all twelve participating wastewater treatment plants. ρ-values were used instead of Spearman p-values because all p-values were <0.001 and statistically significant. A strong correlation had a ρ-value higher than 0.7; a moderate correlation had a ρ-value between 0.4 and 0.7; and a ρ-value between 0 and 0.4 was considered a weak association (Von Sperling et al., 2020). The data shows that SARS-CoV-2 WBS data had higher correlations with active COVID-19 case rates than new case rates but none of the differences were statistically significant (Mann Whitney U test p-value > 0.05). In addition, normalization reduced the differences in correlations between WBS data and active case rates regardless of the gene target. The data highlights that normalization improves correlations by increasing ρ-values up to by approximately 0.1. Given that there was no statistically significant difference between gene target loading at each site, relationships between gene target loading and wastewater properties, or correlations with case numbers, for the remainder of the paper concentrations of E, N1 and N2 results will be averaged.
Fig. 2.
Spearman ρ-values describing the strength of the associations between all normalized and unnormalized, raw SARS-CoV-2 gene targets concentrations (E, N1, N2 and the aaverage of all three) with active and new COVID-19 case rates for all twelve participating wastewater treatment plants (n = 1024). Spearman p-values were all <0.001 and statistically significant.
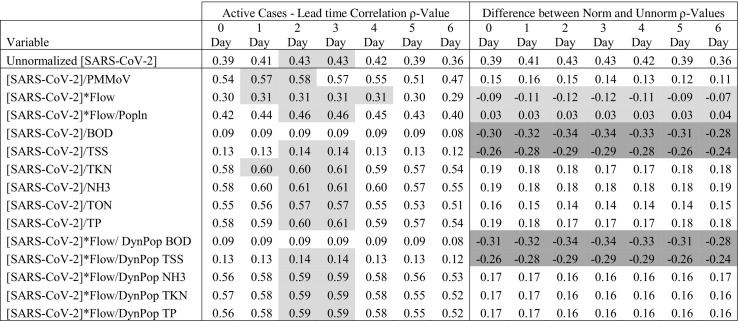
Table 3 breaks down the normalization parameters to provide additional insight into which parameters improved ρ-values for SARS-CoV-2 concentrations and case rates or lead times by depicting the difference in ρ-values. The table reports ρ-values and the change in ρ-values; the latter was calculated by subtracting unnormalized from normalized ρ-values. Negative values indicate a weakened association due to normalization, while positive values reflect improved associations. The results show that normalization with ammonia, TKN and TP provide the highest difference in ρ-values, followed by PMMoV and dynamic population estimates by the same parameters. Meanwhile normalization by BOD, TSS, BOD and TSS dynamic population estimates, flow, and by flow and population weakened associations. Regardless of ρ-values, the difference between normalization and unnormalized associations was not statistically significant as shown by the multiple comparison rank sum test (p-values > 0.2). Number of days of lead time was selected as the day on which the ρ-values between WBS data and clinical data was strongest. Lead time ranged between 1 and 4 days for most normalization parameters. Mean BOD and TSS dynamic population calculations and their standard deviations can be found in Appendix C.
Table 3.
Spearman ρ-values comparing strength of association between normalized and unnormalized average SARS-CoV-2 RNA concentrations and active COVID-19 case rates pooled for all twelve participating plants (n = −1024) over a six-day lead time. The table includes difference between normalized and unnormalized ρ-values. Spearman p-values were all <0.001 and statistically significant. Light grey in ρ-value columns designate lead times. Light and dark grey in the difference column designate rows where normalization weakened associations or had no impact.
3.3.2. Impacts of normalizations on correlations and early detection of active cases at each site
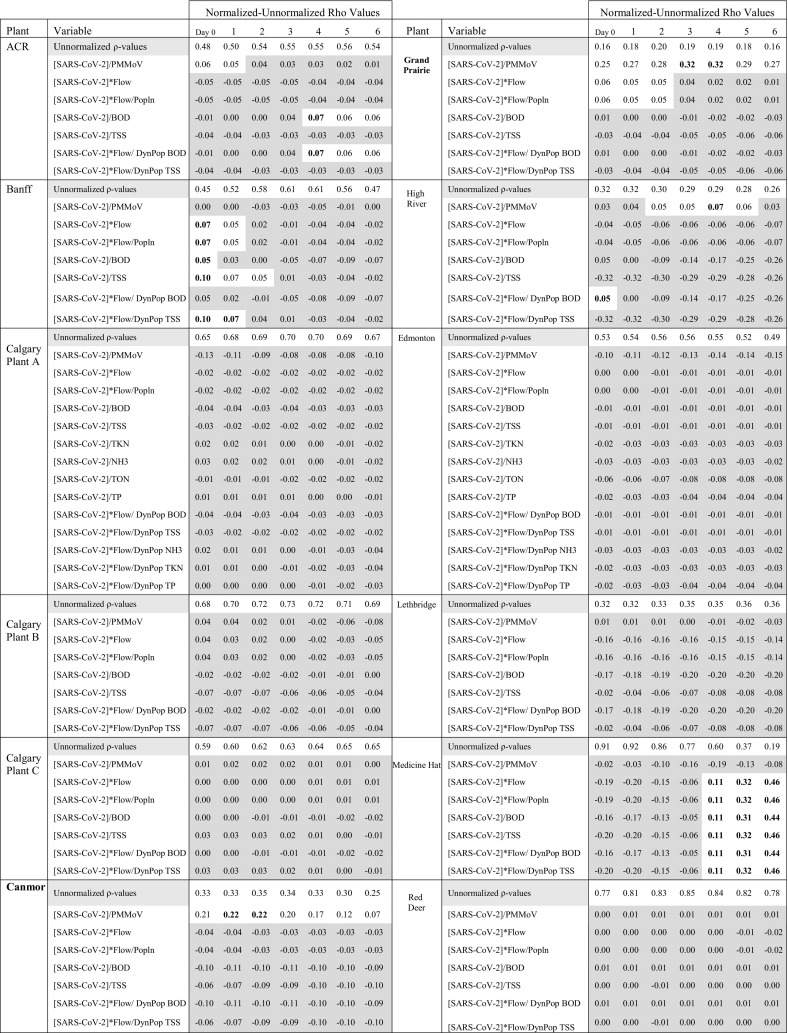
To gain more insight into whether the pooling of results yielded generalizable WBS recommendations, we examined site-specific correlations between SARS-CoV-2 WBS and active COVID-19 case rates. Since all Spearman correlation p-values were statistically significant (<0.001), Spearman ρ-values were used to assess association strength. Similar to Table 3, Table 4 shows the difference between unnormalized and normalized ρ-values for correlations between 7-day moving averages of SARS-CoV-2 concentrations and 7-day moving averages of active case rates by wastewater treatment plant with a six-day lag to identify lead times between WBS data and clinical cases.
Table 4.
Spearman ρ-values comparing strength of association between normalized and unnormalized average SARS-CoV-2 RNA concentrations (shown as [Average] in the table)and active COVID-19 case rates for each participating plants (n = 1024) over a six-day lead time. The table shows differences between normalized and unnormalized ρ-values. Spearman p-values were all <0.001 and statistically significant. Light grey designates parameters where difference between normalized and unnormalized parameter ρ-values were ≤ 0.04. Bolded numbers are the highest differences.
Medicine Hat, Red Deer, and Calgary Plants B and C had the strongest relationships between SARS-CoV-2 RNA in wastewater and active case rates (ρ > 0.6). Normalization by PMMoV reduced or insignificantly impacted ρ-values for all plants except for Canmore and Grand Prairie where correlation ρ-values were increased by 0.2 and 0.3, respectively. In both cases, correlations between SARS-CoV-2 WBS data and COVID-19 case rates remained low (ρ < 0.6). There were five cases where normalization by other parameters (flow, population, chemical parameters or dynamic population) improved correlations by 0.1 (ACR, Grand Prairie, High River, Banff, Medicine Hat) but overall changes in ρ-values were insignificant. For most plants, active case lead times ranged between 2 and 3 days, with Calgary Plants A and C having lead times up to 4–5 days. A full list of Spearman correlations is provided in Appendix D.
4. Discussion
Wastewater surveillance is increasingly being incorporated into public health surveillance to respond to the COVID-19 pandemic. To obtain the maximum benefit from WBS, detected analyte concentrations need to correlate with relevant epidemiological parameters, represent the spatial and longitudinal changes in pathogen prevalence in wastewater and be reported in a standardized manner (McClary-Gutierrez et al., 2021a; McClary-Gutierrez et al., 2021b). There is also a need to understand correlations with case numbers in smaller towns and communities where populations are mobile and fluctuate significantly over time and where clinical case numbers reported may not represent the true residents in the sewershed. Similar to illicit drug use and environmental exposure monitoring, normalizing SARS-CoV-2 RNA concentrations by WWTP flow, population served per capita or by the estimated amount of human fecal material in the samples (i.e., fecal strength) is needed to compare results across different temporal and spatial scales (Feng et al., 2021; Greenwald et al., 2021b; Thai et al., 2019). The present study compared the correlation between COVID-19 cases and normalized and unnormalized SARS-CoV-2 concentrations in the WWTP influent of twelve wastewater systems in Alberta (Canada) between September 2020 and October 2021 using multiple approaches cited in the literature. While this approach is limited by accuracy of COVID-19 clinical data, testing accessibility and test seeking behaviour, exploring correlations between SARS-CoV-2 WBS and clinical data remains a common approach to evaluate normalization efficacy (Duvallet et al., 2022; Isaksson et al., 2022).
In the present study, SARS-CoV-2 RNA loading varied significantly between participating plants regardless of whether they had high (>200 MLD) or low (<100 MLD) flows. There were no statistically significant differences between the three target gene concentrations and all three had the same limit of detection. This finding differs from the literature that reports that N1 and N2 usually have the highest SARS-CoV-2 positivity rate and outperform the E gene target in WBS (Acosta et al., 2021; Ai et al., 2021; Gerrity et al., 2021; La Rosa et al., 2021; Shah et al., 2022). It suggests that initial WBS exploratory studies to confirm target gene suitability in new participating communities may not be necessary. Meanwhile, PMMoV results were highest at the two large, urban WWTPs and stable throughout the study similar to reports by other (Hamza and Bibby, 2019; Kitajima et al., 2018). Interestingly, unlike PMMoV, SARS-CoV-2 RNA loads (GC/day) did correspond with sewershed size and population size served. The lowest concentrations of SARS-CoV-2 were recovered from High River as can be seen in Fig. 1, which is the only system that is an aerated lagoon. This highlights the importance of collecting a representative sample in more challenging and rural systems. This finding corresponds with D'Aoust et al. who recommended that sampling from lagoons should be avoided when possible in rural communities (D'Aoust et al., 2021c). The difference in PMMoV and SARS-CoV-2 persistence, decay and recoveries could be explained by differences in vulnerability of the protein capsid and envelope of SARS-CoV-2 making it easier to rupture compared to the sole protein capsid of PMMoV or by differences in virus forms after conveyance in the sewers system upon sample collection (e.g., free RNA versus intact virus) (Greaves et al., 2020; LaTurner et al., 2021).
In addition to having different trends, SARS-CoV-2 and PMMoV were associated with different wastewater variables. Hamouda et al. (2021) reported that persistence and detection of RNA of SARS-CoV-2 in wastewater is mediated by presence of microorganisms and physico-chemical properties of the sample (e.g., pH, solids, disinfectants), which impact the integrity of the genetic material of the virus making it more difficult to detect (Hamouda et al., 2021). Our results highlighted that wastewater with higher pH significantly decreased the detected concentrations of all three SARS-CoV-2 targets. This corresponds with Amoah et al. (2022) who reported that the highest SARS-CoV-2 RNA concentrations are detected when wastewater samples have a pH between 7.1 and 7.4 (Amoah et al., 2022). Sapula et al. (2021) also showed that SARS-CoV-2 could not be detected from a wastewater plant with high pH (8.80 to 9.35). The results also have implications for WWTPs considering pursuit of WBS that receive waste with high pH (pH > 7.75) from lagoons, septic tanks and industrial operations, which would have a significant impact on virus adsorption to particles and recovered concentrations. The present study did not show an association between SARS-CoV-2 loads and TSS or influent temperature. This differs from studies that report that SARS-CoV-2 decay increases with increasing temperatures (Ahmed et al., 2020b; Bivins et al., 2020) but the difference may be due to the limited range in wastewater influent temperatures (only 10.8–18.9 °C) at the 12 participating WWTPs even during winter. PMMoV RNA concentrations, however, had a positive association with temperature, potentially reflecting increased availability and consumption of peppers in the summer season and the pitfalls of using a normalization parameter that is closely linked to diet and confounded by urbanization.
Population mobility increases the difficulty of interpreting WBS data (Gudra et al., 2022; Isaksson et al., 2022). In this study, Banff and Canmore are tourist towns with highly mobile populations. Calgary Plant A receives flows from Northern Calgary, in addition to flows from Cochrane, Cochrane Lake, Airdrie, and Elbow Valley, and the Northern part of the Tsuu T'ina First Nation community. Edmonton receives additional flows from Alberta Capital Region and includes some combined sewers. Thus, it is possible that the reported COVID-19 cases in these communities are not reflective of the concentrations of SARS-CoV-2 RNA measured in the influent of the plants and dynamic population estimates were explored. Dynamic population size estimated by BOD and BOD normalized data often outperformed TSS. The population estimates for Lethbridge were the least accurate (reported to be 100,000 estimated by BOD and TSS to only be 1880 and 2761, respectively). This could be due to the highly diluted plant influent during the study period with BOD and TSS being unusually low. Similarly, the calculated population sizes for Calgary Plant C and Edmonton also differed from the reported population size served. The only plant where the calculated population size exceeded reported population size was Canmore. Canmore has a highly mobile population with full-time and part-time residents, as well as tourists. Banff, the other tourist town, did not show a similar trend but the use of dynamic population estimates for mobile populations could be of interest and should be explored further (Gudra et al., 2022; Isaksson et al., 2022).
Pooled data from all twelve WWTPs highlighted the value of normalization by traditional wastewater properties (NH3, TKN and TP) and dynamic population sizes similar to what was reported by some (Isaksson et al., 2022; Sakarovitch et al., 2022). Despite the observed improvement in Spearman correlations, none of the normalization approaches result in statistically significant changes in correlations between WBS and clinical data. Even site-specific normalizations only revealed two cases where normalization by PMMoV improved correlations significantly compared to other parameters. In addition, site-specific normalization by dynamic population size did not significantly outperform normalization by more traditional wastewater strength parameter and even unnormalized SARS-CoV-2 concentrations. Overall, our results seem to suggest that normalization of data has limited impact on improving correlations between WBS and clinical data.
In the present study, normalizing SARS-CoV-2 concentrations by PMMoV only improved lead times with active cases significantly for two communities, one of which had other normalization parameters that showed comparable performance. Overall, lead times for active case rates mainly ranged between 2 and 4 days, corresponding with the meta-analysis by Bibby et al. (Bibby et al., 2021). Even though recovered PMMoV concentrations were highest at the two largest WWTPs, our findings contradict findings by D'Aoust et al. (2021a) and Nagarkar et al. (2022) that PMMoV would be particularly useful in surveys of larger populations (D'Aoust et al., 2021a) and findings by Zhan et al. (2022) that suggest PMMoV is suitable for normalizing data from small sewersheds (Zhan et al., 2022). While this may be due to matrix choice (liquid versus solids) and differing analytical methods, it does not change the fact that PMMoV is not an endogenous fecal marker and is highly impacted by dietary, socio-cultural and economic factors.
The present study is part of an increasing number of reports questioning the assumption that normalization by PMMoV improves WBS data standardization and reporting (Ai et al., 2021; Feng et al., 2021; Greenwald et al., 2021a; Nagarkar et al., 2022). We show that in most circumstances using unnormalized concentrations or concentrations normalized by easier, faster and less-costly wastewater strength parameters (e.g., TKN, TP, NH3) could provide equivalent value to the use of PMMoV data, similar to findings by Sakarovitch et al. (2022). Some researchers and health departments suggest that PMMoV normalization is valuable for small and remote WWTPs that have no access to flow data. Given that the cost of running PMMoV is US$ 9–18/run and that collection of samples is recommended 2–3 times per week, the annual cost of running PMMoV is approximately US$ 936–2808. Based on quotes in 2022, the average price of a flow meter is approximately US$ 2000–4000. Thus, it would be helpful for health departments to invest in the purchase of flow meters for small WWTPs that currently lack them to facilitate future WBS efforts and empower utilities to understand and manage their own plant loading better.
5. Conclusions
The present study compared the correlation between COVID-19 cases and normalized and unnormalized SARS-CoV-2 concentrations in the WWTP influent of twelve wastewater systems in Alberta (Canada) between September 2020 and October 2021. To our knowledge, it is the most extensive evaluation of normalization approaches to date. Similar to other studies, we found that normalization of SARS-CoV-2 data by PMMoV only significantly improved correlations between WBS and clinical COVID-19 data in two communities. In most cases, normalization by PMMoV did not improve correlations with case numbers or lead time indication. In the pooled dataset, none of the normalized SARS-CoV-2 concentrations provided statistically significantly stronger associations with active case numbers compared to unnormalized concentrations. Thus, we recommend that researchers explore WWTP influent properties prior to deciding on a sampling and analytical strategy and use at least two parallel approaches to associate WBS and epidemiological data without assuming that normalization will improve correlations. We also propose researchers explore the use of unnormalized SARS-CoV-2 concentrations, as well as flow and traditional wastewater strength parameters for data normalization, when normalization of data is being explored. The use of dynamic population size estimates may be of interest in communities with high tourism or part-time residents. In addition, the cost of running PMMoV annually for two years is almost equivalent to the cost of a flow meter, which provides WWTP with superior data for WBS and other WWTP management practices. The option of helping small, resource-limited WWTPs acquire influent flow meters should be explored if long-term WBS partnerships are desired.
CRediT authorship contribution statement
Rasha Maal-Bared: Conceptualization, Methodology, Formal analysis, Investigation, Visualization, Writing – original draft. Yuanyuan Qiu: Investigation, Writing – review & editing. Qiaozhi Li: Investigation, Writing – review & editing. Tiejun Gao: Project administration, Writing – review & editing. Steve E. Hrudey: Methodology, Writing – review & editing. Sudha Bhavanam: Investigation, Writing – review & editing. Norma J. Ruecker: Methodology, Writing – review & editing. Erik Ellehoj: Writing – review & editing. Bonita E. Lee: Conceptualization, Methodology, Resources, Writing – review & editing. Xiaoli Pang: Conceptualization, Methodology, Resources, Project administration, Funding acquisition, Supervision, Investigation, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We would like to thank the participating wastewater treatment plants in Alberta for their collection and shipping of wastewater samples tirelessly for a long period of time and for providing the required information for analysis for this study. The Alberta Precision Laboratory – Public Health Laboratory and Alberta Health – Analytics Performance & Reporting Branch are acknowledged for their assistance with samples transportation and for established systems sharing with us for daily updates of COVID-19 new and active cases in the province (specially to Allen O'Brien). We are grateful to Emma Zwaigenbaum, Melissa Wilson, and Shiqi Diao for their technical support during COVID-19 pandemic. This study was supported by research grants from the Canadian Institutes of Health Research, Alberta Innovates and Alberta Health [RES0051047].
Editor: Warish Ahmed
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.scitotenv.2022.158964.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Acosta N., Bautista M.A., Hollman J., McCalder J., Beaudet A.B., Man L., et al. A multicenter study investigating SARS-CoV-2 in tertiary-care hospital wastewater. Viral burden correlates with increasing hospitalized cases as well as hospital-associated transmissions and outbreaks. Water Res. 2021;201:1–10. doi: 10.1016/j.watres.2021.117369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O'Brien J.W.…Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728:138764. doi: 10.1016/j.scitotenv.2020.138764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bibby K., Haramoto E., Hewitt J., Huygens F., et al. Decay of SARS-CoV-2 and surrogate murine hepatitis virus RNA in untreated wastewater to inform application in wastewater-based epidemiology. Environ. Res. 2020;191 doi: 10.1016/j.envres.2020.110092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed W., Bertsch P.M., Bivins A., Bibby K., Farkas K., Gathercole A., et al. Comparison of virus concentration methods for the RT-qPCR-based recovery of murine hepatitis virus, a surrogate for SARS-CoV-2 from untreated wastewater. Sci. Total Environ. 2020;739 doi: 10.1016/j.scitotenv.2020.139960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai Y., Davis A., Jones D., Lemeshow S., Tu H., He F., et al. Wastewater SARS-CoV-2 monitoring as a community-level COVID-19 trend tracker and variants in Ohio, United States. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander G., Stevens R. Per capita phosphorus loading from domestic sewage. Water Res. 1976;10:757–764. [Google Scholar]
- American Public Health Association, American Water Works Association . 23rd Edition. American Public Health Association; Washington, DC: 2017. Standard methods for the examination of water and wastewater. [Google Scholar]
- American Public Health Laboratories . APHL; Silver Spring, MD: 2022. SARS-CoV-2 Wastewater Surveillance Testing Guide for Public Health Laboratories. [Google Scholar]
- Amoah I.D., Abunama T., Awolusi O.O., Pillay L., Pillay K., Kumari S., et al. Effect of selected wastewater characteristics on estimation of SARS-CoV-2 viral load in wastewater. Environ. Res. 2022;203 doi: 10.1016/j.envres.2021.111877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabzadeh R., Grunbacher D.M., Insam H., Kreuzinger N., Markt R., Rauch W. Data filtering methods for SARS-CoV-2 wastewater surveillance. Water Sci. Technol. 2021;84:1324–1339. doi: 10.2166/wst.2021.343. [DOI] [PubMed] [Google Scholar]
- Been F., Rossi L., Ort C., Rudaz S., Delemont O., Esseiva P. Population normalization with ammonium in wastewater-based epidemiology: application to illicit drug monitoring. Environ. Sci. Technol. 2014;48:8162–8169. doi: 10.1021/es5008388. [DOI] [PubMed] [Google Scholar]
- Bibby K., Bivins A., Wu Z., North D. Making waves: plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021;202 doi: 10.1016/j.watres.2021.117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bivins A., Greaves J., Fischer R., Yinda K., Ahmed W., Kitajima M., et al. Persistence of SARS-CoV-2 in water and wastewater. Environ.Sci.Technol.Lett. 2020;7:937–942. doi: 10.1021/acs.estlett.0c00730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castiglioni S., Bijlsma L., Covaci A., Emke E., Hernández F., Reid M., et al. Evaluation of uncertainties associated with the determination of community drug use through the measurement of sewage drug biomarkers. Environ.Sci.Technol. 2013;47:1452–1460. doi: 10.1021/es302722f. [DOI] [PubMed] [Google Scholar]
- Centre for Disease Control and Prevention . 2022. Wastewater Surveillance Data Reporting and Analytics. 2022. [Google Scholar]
- Choi P.M., Tscharke B., Samanipour S., Hall W.D., Gartner C.E., Mueller J.F., et al. Social, demographic, and economic correlates of food and chemical consumption measured by wastewater-based epidemiology. Proc. Natl. Acad. Sci. U. S. A. 2019;116:21864–21873. doi: 10.1073/pnas.1910242116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluzel N., Courbariaux M., Wang S., Moulin L., Wurtzer S., Bertrand I., et al. A nationwide indicator to smooth and normalize heterogeneous SARS-CoV-2 RNA data in wastewater. Environ. Int. 2022;158 doi: 10.1016/j.envint.2021.106998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crank K., Chen W., Bivins A., Lowry S., Bibby K. Contribution of SARS-CoV-2 RNA shedding routes to RNA loads in wastewater. Sci. Total Environ. 2022;806 doi: 10.1016/j.scitotenv.2021.150376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Graber T.E., Mercier E., Montpetit D., Alexandrov I., Neault N., et al. Catching a resurgence: increase in SARS-CoV-2 viral RNA identified in wastewater 48 h before COVID-19 clinical tests and 96 h before hospitalizations. Sci. Total Environ. 2021;770 doi: 10.1016/j.scitotenv.2021.145319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Mercier E., Montpetit D., Jia J.-J., Alexandrov I., Neault N., et al. Quantitative analysis of SARS-CoV-2 RNA from wastewater solids in communities with low COVID-19 incidence and prevalence. Water Res. 2021;188 doi: 10.1016/j.watres.2020.116560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Aoust P.M., Towhid S.T., Mercier É., Hegazy N., Tian X., Bhatnagar K., et al. COVID-19 wastewater surveillance in rural communities: comparison of lagoon and pumping station samples. Sci. Total Environ. 2021;801 doi: 10.1016/j.scitotenv.2021.149618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C. The international imperative to rapidly and inexpensively monitor community-wide Covid-19 infection status and trends. Sci. Total Environ. 2020;726 doi: 10.1016/j.scitotenv.2020.138149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughton C.G. Wastewater surveillance for population-wide Covid-19: the present and future. Sci. Total Environ. 2020;736 doi: 10.1016/j.scitotenv.2020.139631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvallet C., Wu F., KA McElroy, Imakaev M., Endo N., Xiao A., et al. Nationwide trends in COVID-19 cases and SARS-CoV-2 RNA wastewater concentrations in the United States. ACS ES&T Water. 2022 doi: 10.1021/acsestwater.1c00434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Roguet A., McClary-Gutierrez J.S., Newton R.J., Kloczko N., Meiman J.G., et al. Evaluation of sampling, analysis, and normalization methods for SARS-CoV-2 concentrations in wastewater to assess COVID-19 burdens in Wisconsin communities. ACS Es&T Water. 2021;1:1955–1965. [Google Scholar]
- Gerrity D., Papp K., Stoker M., Sims A., Frehner W. Early-pandemic wastewater surveillance of SARS-CoV-2 in Southern Nevada: methodology, occurrence, and incidence/prevalence considerations. Water Res.X. 2021;10 doi: 10.1016/j.wroa.2020.100086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez R., Curtis K., Bivins A., Bibby K., Weir M.H., Yetka K., et al. COVID-19 surveillance in Southeastern Virginia using wastewater-based epidemiology. Water Res. 2020;186 doi: 10.1016/j.watres.2020.116296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greaves J., Stone D., Wu Z., Bibby K. Persistence of emerging viral fecal indicators in large-scale freshwater mesocosms. Water Res. X. 2020;9 doi: 10.1016/j.wroa.2020.100067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald H.D., Kennedy L.C., Hinkle A., Whitney O.N., Fan V.B., Crits-Christoph A., et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Res.X. 2021;12 doi: 10.1016/j.wroa.2021.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald H.D., Kennedy L.C., Hinkle A., Whitney O.N., Fan V.B., Crits-Christoph A., et al. Tools for interpretation of wastewater SARS-CoV-2 temporal and spatial trends demonstrated with data collected in the San Francisco Bay Area. Water Res. X. 2021;12 doi: 10.1016/j.wroa.2021.100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudra D., Dejus S., Bartkevics V., Roga A., Kalnina I., Strods M., et al. Detection of SARS-CoV-2 RNA in wastewater and importance of population size assessment in smaller cities: an exploratory case study from two municipalities in Latvia. Sci. Total Environ. 2022;823 doi: 10.1016/j.scitotenv.2022.153775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez V., Hassard F., Vu M., Leitao R., Burczynska B., Wildeboer D., et al. medRxiv; 2021. Monitoring Occurrence of SARS-CoV-2 in School Populations: A Wastewater-based Approach. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamouda M., Mustafa F., Maraqa M., Rizvi T., Aly Hassan A. Wastewater surveillance for SARS-CoV-2: lessons learnt from recent studies to define future applications. Sci. Total Environ. 2021;759 doi: 10.1016/j.scitotenv.2020.143493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamza I.A., Bibby K. Critical issues in application of molecular methods to environmental virology. J. Virol. Methods. 2019;266:11–24. doi: 10.1016/j.jviromet.2019.01.008. [DOI] [PubMed] [Google Scholar]
- Holm R., Nagarkar M., Yeager R., Talley D., Chaney A., Rai J., et al. Surveillance of RNase P, PMMoV, and CrAssphage in wastewater as indicators of human fecal concentration across urban sewer neighborhoods, Kentucky. FEMS Microbes. 2022;3 doi: 10.1093/femsmc/xtac003. March 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard G., Bartram J., Brocklehurst C., Colford J.M., Costa F., Cunliffe D., et al. COVID-19: urgent actions, critical reflections and future relevance of ‘WaSH’: lessons for the current and future pandemics. J. Water Health. 2020;18:613–630. doi: 10.2166/wh.2020.162. [DOI] [PubMed] [Google Scholar]
- Isaksson F., Lundy L., Hedström A., Székely A.J., Mohamed N. Evaluating the use of alternative normalization approaches on SARS-CoV-2 concentrations in wastewater: experiences from two catchments in Northern Sweden. Environments. 2022;9:39. [Google Scholar]
- Johns Hopkins Coronavirus Resource Center . 2022. New COVID-19 Cases Worldwide. 2022. [Google Scholar]
- Kevill J.L., Pellett C., Farkas K., Brown M.R., Bassano I., Denise H., et al. A comparison of precipitation and filtration-based SARS-CoV-2 recovery methods and the influence of temperature, turbidity, and surfactant load in urban wastewater. Sci. Total Environ. 2022;808 doi: 10.1016/j.scitotenv.2021.151916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima M., Sassi H., Torrey J. Pepper mild mottle virus as a water quality indicator. NPJ Clean Water. 2018;1:1–9. [Google Scholar]
- La Rosa G., Mancini P., Ferraro G.B., Veneri C., Iaconelli M., Bonadonna L., et al. SARS-CoV-2 has been circulating in northern Italy since December 2019: evidence from environmental monitoring. Sci. Total Environ. 2021;750 doi: 10.1016/j.scitotenv.2020.141711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaTurner Z.W., Zong D.M., Kalvapalle P., Gamas K.R., Terwilliger A., Crosby T., et al. Evaluating recovery, cost, and throughput of different concentration methods for SARS-CoV-2 wastewater-based epidemiology. Water Res. 2021;197 doi: 10.1016/j.watres.2021.117043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Mazurowski L., Dewan A., Carine M., Haak L., Guarin T.C., et al. Longitudinal monitoring of SARS-CoV-2 in wastewater using viral genetic markers and the estimation of unconfirmed COVID-19 cases. Sci. Total Environ. 2022;817 doi: 10.1016/j.scitotenv.2022.152958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazumder P., Dash S., Honda R., Sonne C., Kumar M. Sewage surveillance for SARS-CoV-2: molecular detection, quantification and normalization factors. Curr.Opin.Environ.Sci.Health. 2022;100363 doi: 10.1016/j.coesh.2022.100363. Apr 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClary-Gutierrez J.S., Aanderud Z.T., Al-Faliti M., Duvallet C., Gonzalez R., Guzman J., et al. Standardizing data reporting in the research community to enhance the utility of open data for SARS-CoV-2 wastewater surveillance. Environ. Sci. (Camb.) 2021;9 doi: 10.1039/d1ew00235j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClary-Gutierrez J.S., Mattioli M.C., Marcenac P., Silverman A.I., Boehm A.B., Bibby K., et al. SARS-CoV-2 wastewater surveillance for public health action. Emerg. Infect. Dis. 2021;27:1–8. doi: 10.3201/eid2709.210753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf Eddy, Abu-Orf M., Bowden G., Burton F.L., Pfrang W., et al. McGraw Hill Education; 2014. Wastewater Engineering: Treatment and Resource Recovery. [Google Scholar]
- Nagarkar M., Keely S., Jahne M., Wheaton E., Hart C., Smith B., et al. SARS-CoV-2 monitoring at three sewersheds of different scales and complexity demonstrates distinctive relationships between wastewater measurements and COVID-19 case data. Sci. Total Environ. 2022;816 doi: 10.1016/j.scitotenv.2021.151534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabbaraju K., Wong A.A., Douesnard M., Ma R., Gill K., Dieu P., et al. Development and validation of RT-PCR assays for testing for SARS-CoV-2. Off.J.Assoc.Med.Microbiol.Infect.Dis.Can. 2021;6:16–22. doi: 10.3138/jammi-2020-0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philo S.E., Keim E.K., Swanstrom R., Ong A.Q.W., Burnor E.A., Kossik A.L., et al. A comparison of SARS-CoV-2 wastewater concentration methods for environmental surveillance. Sci. Total Environ. 2021;760 doi: 10.1016/j.scitotenv.2020.144215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y., Yu J., Pabbaraju K., Lee B.E., Gao T., Ashbolt N.J., et al. Validating and optimizing the method for molecular detection and quantification of SARS-CoV-2 in wastewater. Sci. Total Environ. 2022;812 doi: 10.1016/j.scitotenv.2021.151434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randazzo W., Truchado P., Ferrando E.C., Simon P., Allende A., Sanchez G. SARS-CoV-2 RNA titers in wastewater anticipated COVID-19 occurrence in a low prevalence area. Water Res. 2020;181 doi: 10.1016/j.watres.2020.115942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusiñol M., Zammit I., Itarte M., Forés E., Martínez-Puchol S., Girones R., et al. Monitoring waves of the COVID-19 pandemic: inferences from WWTPs of different sizes. Sci. Total Environ. 2021;787 doi: 10.1016/j.scitotenv.2021.147463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakarovitch C., Schlosser O., Courtois S., Proust-Lima C., Couallier J., Petrau A., et al. Monitoring of SARS-CoV-2 in wastewater: what normalisation for improved understanding of epidemic trends? J. Water Health. 2022;20:712–726. doi: 10.2166/wh.2022.012. [DOI] [PubMed] [Google Scholar]
- Sapula S.A., Whittall J.J., Pandopulos A.J., Gerber C., Venter H. An optimized and robust PEG precipitation method for detection of SARS-CoV-2 in wastewater. Sci. Total Environ. 2021;785 doi: 10.1016/j.scitotenv.2021.147270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah S., Gwee S.X.W., Ng J.Q.X., Lau N., Koh J., Pang J. Wastewater surveillance to infer COVID-19 transmission: a systematic review. Sci. Total Environ. 2022;804 doi: 10.1016/j.scitotenv.2021.150060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steel R.G. A multiple comparison rank sum test: treatments versus control. Biometrics. 1959:560–572. [Google Scholar]
- Sweetapple C., Melville-Shreeve P., Chen A.S., Grimsley J.M.S., Bunce J.T., Gaze W., et al. Building knowledge of university campus population dynamics to enhance near-to-source sewage surveillance for SARS-CoV-2 detection. Sci. Total Environ. 2021;806 doi: 10.1016/j.scitotenv.2021.150406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E.M., Nguyen K.H., Harwood V.J., Breitbart M. Pepper mild mottle virus: a plant pathogen with a greater purpose in (waste)water treatment development and public health management. Water Res. 2018;144:1–12. doi: 10.1016/j.watres.2018.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thai P.K., O'Brien J.W., Banks A.P.W., Jiang G., Gao J., Choi P.M., et al. Evaluating the in-sewer stability of three potential population biomarkers for application in wastewater-based epidemiology. Sci. Total Environ. 2019;671:248–253. doi: 10.1016/j.scitotenv.2019.03.231. [DOI] [PubMed] [Google Scholar]
- Vadde K.K., Al-Duroobi H., Phan D.C., Jafarzadeh A., Moghadam S.V., Matta A., et al. Assessment of concentration, recovery, and normalization of SARS-CoV-2 RNA from two wastewater treatment plants in Texas and correlation with COVID-19 cases in the community. ACS ES&T Water. 2022 doi: 10.1021/acsestwater.2c00054. [DOI] [PubMed] [Google Scholar]
- Von Sperling M., Verbyla M., Oliveira S. Assessment of Treatment Plant Performance and Water Quality Data: A Guide for Students, Researchers and Practitioners. IWA Publishing; 2020. Relationship between monitoring variables: correlation and regression analysis; pp. 397–477. [Google Scholar]
- Wade M., Jacomo A., Armenise E., Brown M., Bunce J., Cameron G., et al. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 2022;15 doi: 10.1016/j.jhazmat.2021.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade M.J., Lo Jacomo A., Armenise E., Brown M.R., Bunce J.T., Cameron G.J., et al. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard. Mater. 2022;424 doi: 10.1016/j.jhazmat.2021.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775 doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilder M.L., Middleton F., Larsen D.A., Du Q., Fenty A., Zeng T., et al. Co-quantification of crAssphage increases confidence in wastewater-based epidemiology for SARS-CoV-2 in low prevalence areas. Water Res. X. 2021;11 doi: 10.1016/j.wroa.2021.100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Xiao A., Zhang J., Moniz K., Endo N., Armas F., et al. SARS-CoV-2 RNA concentrations in wastewater foreshadow dynamics and clinical presentation of new COVID-19 cases. Sci. Total Environ. 2022;805 doi: 10.1016/j.scitotenv.2021.150121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan Q., Babler K.M., Sharkey M.E., Amirali A., Beaver C.C., Boone M.M., et al. Relationships between SARS-CoV-2 in wastewater and COVID-19 clinical cases and hospitalizations, with and without normalization against indicators of human waste. ACS ES&T Water. 2022 doi: 10.1021/acsestwater.2c00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T., Breitbart M., Lee W.H., Run J.-Q., Wei C.L., Soh S.W.L., et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4(1):e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.