Abstract
φYeO3-12 is a T3-related lytic bacteriophage of Yersinia enterocolitica serotype O:3. The nucleotide sequence of the 39,600-bp linear double-stranded DNA (dsDNA) genome was determined. The phage genome has direct terminal repeats of 232 bp, a GC content of 50.6%, and 54 putative genes, which are all transcribed from the same DNA strand. Functions were assigned to 30 genes based on the similarity of the predicted products to known proteins. A striking feature of the φYeO3-12 genome is its extensive similarity to the coliphage T3 and T7 genomes; most of the predicted φYeO3-12 gene products were >70% identical to those of T3, and the overall organizations of the genomes were similar. In addition to an identical promoter specificity, φYeO3-12 shares several common features with T3, nonsubjectibility to F exclusion and growth on Shigella sonnei D2371-48 (M. Pajunen, S. Kiljunen, and M. Skurnik, J. Bacteriol. 182:5114–5120, 2000). These findings indicate that φYeO3-12 is a T3-like phage that has adapted to Y. enterocolitica O:3 or vice versa. This is the first dsDNA yersiniophage genome sequence to be reported.
Yersinia enterocolitica is a gram-negative species which has ∼70 serotypes, some of which are pathogenic to humans. The major pathogens in Europe, Canada, Japan, and South Africa belong to serotypes O:3 and O:9, and those in the United States belong to serotype O:8 (10) The main reservoir in nature for Y. enterocolitica is pigs (13), and infection usually occurs by ingestion of contaminated foodstuffs.
Several yersiniophages have been described in the literature, but only a few have been characterized in detail. In our laboratory, a number of Yersinia-specific bacteriophages have been isolated, all originating from the raw incoming sewage of the Turku, Finland, city sewage treatment plant, and they have been used as genetic tools (44). One phage, φYeO3-12, was isolated as specific to Y. enterocolitica serotype O:3 (YeO3). It infects the Escherichia coli C600 strain expressing the cloned O-antigen of YeO3, and spontaneous phage-resistant YeO3 strains were missing the O-antigen, thus indicating that O-antigen is the phage receptor (1, 2). The serotype O:3 specificity makes the φYeO3-12 a potential biotechnological tool, and therefore we have initiated a detailed characterization; its biological and physical properties were reported previously (35). The dimensions of the icosahedral virion are ∼57 nm in diameter for the head and 15 by 8 nm for the tail, and thus φYeO3-12 belongs to the family Podoviridae. Based on an N-terminal sequence analysis of the major capsid protein and on its host requirements, nonsubjectibility to F exclusion, and growth on Shigella sonnei D2371-48, it was concluded that φYeO3-12 belongs to the T7 group and that it is the first described close relative of bacteriophage T3.
The T7 group comprises about 60 phages that have been divided into three subgroups on the basis of the promoter specificity of the phage-encoded RNA polymerase (RNAP). Bacteriophage T3 is the only member of one subgroup; BA14, BA127, and BA156 make a second; and the rest are all T7-like (24). Members of a given subgroup efficiently recombine with each other, but recombinants between phages in different subgroup are rare. In addition, T3 and T7 exhibit mutual exclusion in coinfections; only 2 to 5% of these cells produce a mixed burst while others produce exclusively either T3 or T7. From these findings it is evident that T3 and T7 do not belong to a phage population that efficiently interbreeds. Nevertheless, it has been suggested that bacteriophages T3 and T7 have recombined (7), both with each other and with other members of a pool of T7-like phages, during their coevolution, because the gene 17 promoter in T3 is actually of T7 specificity and is not utilized during a normal T3 infection (39). It seems likely that the theory of modular evolution of bacteriophages (9) is also applicable to the T7 group, as the varyious levels of homology that exist between the DNAs include regions of high sequence conservation immediately adjacent to regions that have no apparent homology (7, 17). It has also been suggested that many and probably all of the double-stranded DNA (dsDNA)-tailed bacteriophages share common ancestry in at least some of their genes and that they partake of a common gene pool through horizontal exchange of DNA (26).
In this study we report the complete genomic sequence of φYeO3-12. It has a linear DNA of 39,600 bp with 54 putative genes. The sequence data conclusively show the very close relationship between φYeO3-12 and T3.
MATERIALS AND METHODS
Bacterial strains, phages, and media.
Plasmid-cured Y. enterocolitica serotype O:3 strain 6471/76-c (YeO3-c) (43) was the usual host for propagation of phage φYeO3-12. Tryptic soy broth medium (Oxoid) was employed throughout, and incubations were done at room temperature unless specified otherwise. E. coli K-12 strains C600 (thi thr leuB tonA lacY supE) (4), JM109 {recA1 endA1 gyrA96 thi hsdR17 supE44 relA1 λ− Δ(lac-proAB) [F′ traD36 proAB lacIqZΔM15]} (47), and IJ719 [ara Δ(lac-proAB) thi hsdR-Tn10)] (from I. J. Molineux) were grown at 37°C in Luria broth; ampicillin (100 μg/ml) was added when required. Solid and soft agar media were those described above supplemented with 2 or 0.5% (wt/vol) agar, respectively. Bacteria were stored as frozen stocks at −70°C in culture medium supplemented with 20% (vol/vol) glycerol (5, 41).
Bacteriophage φYeO3-12 was isolated earlier (1) and characterized microbiologically elsewhere (35). The protein kinase deletion derivative of φYeO3-12 (designated ΔPK) and the large-scale preparations of bacteriophages were also described previously (35). Bacteriophage T3+ and the T3 gene 17 amber mutant amH26 (originally from R. Hausmann) were from I. J. Molineux.
DNA extraction and analysis.
Phage DNA was obtained from high-titer phage stock as described for bacteriophage lambda (41). Plasmid DNA was isolated from E. coli by the alkaline lysis procedure (8). All enzymatic treatments of DNA were performed as recommended by the suppliers. DNA electrophoresis was carried out in agarose gels using standard TAE buffer (41). DNA fragments were stained with ethidium bromide and photographed under UV illumination.
DNA sequencing and analysis.
Phage DNA was partially digested with HincII. DNA fragments of 1 to 5 kb were purified from agarose after electrophoretic separation and were ligated with pUC18 (47), previously digested with HincII and treated with calf intestinal phosphatase. The ligation mixture was transformed into E. coli strains C600 and JM109 according to the procedure described by Hanahan et al. (22). Clones carrying phage DNA were identified, and the insertions were sequenced using an Applied Biosystems 377 automated DNA sequencer. Synthetic oligonucleotide primers (MedProbe Inc., Oslo, Norway) as well as universal forward and reverse primers were used for sequencing reactions. The phage genome sequence was completed by primer walking using the total phage genome as a template. The sequence of the direct terminal repeats was confirmed by analyzing an 876-bp PCR fragment obtained by using a phosporylated and circulized-ligated phage DNA as a template and the oligonucleotides mp78for2 (5′-CTTACCTAAAGTGGATGCC-3′) and mp27back (5′-GGCTGTCTACTTATCCGG-3′) (positions 39280 to 39298 and 555 to 538, respectively) as primers. Sequences were assembled and the consensus sequence was edited using the GelAssemble sequence assembly program and other programs included in the Genetics Computer Group (GCG) suite of programs, (versions 9.0 and 10.0; GCG, Madison, Wis.). Open reading frames of more than 15 codons were searched for amino acid sequence similarities in the databases (GenBank, EMBL, SwissProt, PIR, PDB, and DDBJ) using BLAST version 2.0.10 (3) and FASTA version 3.28 (SwissProt) (36).
Cloning of gene 17 into pUC18 for in vivo recombination experiments.
Genomic φYeO3-12 DNA between nucleotides 33155 and 36558 carrying gene 17 and ca. 800 bp of flanking DNA on both sides was amplified by PCR using primers pfk3for2 (5′-TGTGGCGACTGGCTGAC-3′) and mp78bck5 (5′-CCAAGGGAGTGCGCTGCG-3′). The resulting 3,403-bp PCR fragment was electrophoretically purified, phosphorylated, and then ligated with HincII-digested pUC18 (47) vector digested with HincII. The resulting plasmid (pgp17u) was transformed into E. coli strains C600 and IJ719. C600/pgp17u was infected with T3+, and IJ719/pgp17u was infected with either T3+ or 17amH26, all at a multiplicity of 1. Lysates were then plated on Y. enterocolitica serotype O:3 to test for viable recombinant phages. DNA was isolated from purified phage particles and total cell lysates and was checked by PCR for the presence of φYeO3-12 DNA in the T3 genome using φYeO3-12 specific primers.
Nucleotide sequence accession number.
Nucleotide sequence data for φYeO3-12 have been deposited in the GenBank/EMBL/DDBJ databases under accession no. AJ251805.
RESULTS AND DISCUSSION
Determination of the nucleotide sequence.
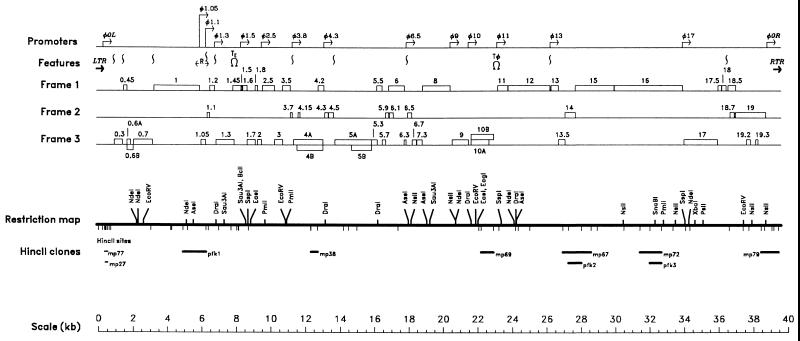
The nucleotide sequence was determined on both DNA strands from purified phage particles of wild-type φYeO3-12, using genomic HincII clones as a starting point (Fig. 1). Only a few of the possible HincII subclones were obtained, because some genes cannot be cloned either due to their lethality or due to site preferences in a partial restriction digest. Restriction sites predicted from the sequence confirmed the restriction map reported earlier (35). In the sections to follow the transcriptional, translational, and other features in φYeO3-12 DNA will be discussed in detail.
FIG. 1.
Genetic organization and analysis of the genome of bacteriophage φYeO3-12. In the upper part of the figure are shown the locations of the promoters, putative RNase III sites (represented by vertical wave lines), and the genes in the three forward (from left to right) reading frames of φYeO3-12. The genes are represented by open boxes, and the names of the genes are given above the boxes. LTR and RTR are the left and right TRs, respectively. R is the primary origin of replication. TE and Tø are transcription termination sites of host and φYeO3-12 RNAPs, respectively. In the lower part of the figure the restriction map with all the HincII sites indicated below the line, the obtained HincII-clones used for sequencing analysis, and a scale are shown.
Identification of φYeO3-12 genes.
The linear DNA of φYeO3-12 contains 39,600 bp and includes terminal repeats of 232 bp. The sequence was searched for functional genes, and all open reading frames (ORFs) longer than 15 codons were considered. As a first criterion, the CodonPreference program of the GCG package with the CodonFrequency table for highly expressed E. coli genes was used. Presence of an appropriately located potential Shine-Dalgarno (SD) sequence was used as the next criterion. Finally, similarity of the ORFs to the sequences in the databases was taken into account. Altogether 54 potential genes were identified (Fig. 1 and Table 1). Most genes showed highest similarity to coliphages T3 and T7; however, two genes were most similar to mycobacteriophage L5 gp59 and to lactococcal phage bIL170 e20 (Table 1). Based on amino acid similarities, putative functions were assigned to 30 gene products. Genes with predicted in-frame internal initiations or ribosomal frameshifts affording two related proteins were considered to be one gene. Because of the extensive similarity between φYeO3-12 and T3 or T7 we named φYeO3-12 genes according to the T3 or T7 nomenclature; the 19 genes on the original T7 genetic map have integral numbers, and genes added subsequently have decimal numbers. In those cases where in φYeO3-12 a location was occupied by an extra gene or a gene showing no similarity to the T3 or T7 counterparts, the gene was named according to its position on the genome by adding a new decimal number (genes 0.45, 1.45, 4.15, 6.1, and 13.5) (Table 1).
TABLE 1.
Predicted genes and gene products of bacteriophage φYeO3-12
| Gene
|
Gene product
|
SD sequenceb | Protein name | Identity with:
|
Function(s) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Name | Range | % G+C | Size (aaa) | Molecular mass (kDa) | pI | T3 (%) | T7 (%) | Other (accession no.) | |||
| 0.3 | 1035–1490 | 52.1 | 152 | 17.0 | 7.72 | ACATGAGGTAACACCAAATG | gp0.3 | 98.0 | 20.9 | SAMase (adometase) | |
| 0.45 | 1558–1755 | 44.4 | 66 | 7.5 | 7.36 | CTTATAGGACTAACACCATG | |||||
| 0.6a | 1758–1958 | 45.0 | 67 | 7.9 | 11.33 | GGTGGAATGACTAATG | gp0.6A | 34.0 | |||
| 0.6b | 1758–2115 | 46.2 | 120 | 14.0 | 11.70 | GGTGGAATGACTAATG | gp0.6B | 25.2 | |||
| 0.7 | 2133–3239 | 53.6 | 369 | 42.2 | 7.84 | ACAGGACACTGAACGATG | gp0.7 | 51.4 | Protein kinase | ||
| 1 | 3313–5964 | 53.6 | 884 | 98.8 | 7.32 | CAATGAGGTAAGCAATG | gp1 | 99.1 | 82.4 | RNA polymerase | |
| 1.05 | 6054–6323 | 45.7 | 90 | 10.4 | 10.04 | CTAAGAGATTAAATTTATG | gp1.05 | 97.8 | |||
| 1.1 | 6419–6556 | 53.3 | 46 | 5.9 | 11.57 | ACATGAGGTAAGATACTATG | gp1.1 | 97.8 | 45.0 | ||
| 1.2 | 6562–6834 | 51.5 | 91 | 10.5 | 8.45 | AGTGGAACTAATAATG | gp1.2 | 94.5 | 40.0 | Deoxyguanosine triphos-phohydrolase inhibitor | |
| 1.3 | 6933–7970 | 51.1 | 346 | 39.4 | 4.94 | CAATGAGGAACAACCGTATG | gp1.3 | 95.4 | 73.5 | DNA ligase | |
| 1.45 | 7903–8373 | 49.5 | 157 | 17.7 | 10.36 | TATGGAGGAAACACCTGATG | e20 | Phage bIL170 (AF009630)c | HNH endonuclease homologue | ||
| 1.5 | 8392–8466 | 46.7 | 25 | 2.8 | 3.33 | ACAGGAGACACACACCATG | gp1.5 | 96.0 | 37.5 | ||
| 1.6 | 8482–8736 | 53.7 | 85 | 9.9 | 11.78 | TAAGGAGACAACATCATG | gp1.6 | 98.8 | 57.6 | ||
| 1.7 | 8739–9206 | 52.1 | 156 | 17.7 | 10.08 | TAAGGAGGTGCTGTAATG | gp1.7 | 82.1 | 47.7 | ||
| 1.8 | 9196–9330 | 55.6 | 45 | 5.3 | 4.97 | ACCGGGGGCTGTGTTATG | gp1.8 | 93.0 | 32.4 | ||
| 2 | 9330–9563 | 52.7 | 78 | 8.9 | 4.69 | TAAGGAGGCCAATAAATG | gp2 | 85.2 | 39.1 | Bacterial RNAP inhibitor | |
| 2.5 | 9619–10314 | 52.2 | 232 | 26.0 | 4.62 | AAAGGAGAAACACTATG | gp2.5 | 98.3 | 85.5 | Single-stranded DNA- binding protein | |
| 3 | 10317–10776 | 49.1 | 153 | 17.6 | 10.28 | GGAAGAGGACTTCTAATG | gp3 | 93.4 | 84.6 | Endonuclease | |
| 3.5 | 10771–11223 | 53.0 | 151 | 16.9 | 9.29 | AAAGGAGTAAAGAAAAATG | gp3.5 | 96.7 | 91.4 | N-Acetylmuramoyl-l-alanine amidase (lysozyme) | |
| 3.7 | 11231–11335 | 48.6 | 35 | 4.2 | 8.18 | GACCGAGGGTGATACCATG | gp3.7 | 97.1 | |||
| 4a | 11406–13103 | 50.8 | 566 | 63.0 | 4.90 | TAAGGATTAACCACATG | gp4A | 95.6 | 82.4 | Primase-helicase | |
| 4b | 11592–13103 | 51.1 | 504 | 55.9 | 4.98 | ACAGGAGGCAGCAAGCCTATG | gp4B | 99.2 | 84.7 | Helicase | |
| 4.15 | 11672–11776 | 56.2 | 35 | 3.9 | 11.24 | CTCGAAGGAGACATG | |||||
| 4.2 | 12826–13152 | 49.8 | 109 | 12.0 | 7.58 | GGAGAAGGGAAAAGCACATG | gp4.2 | 83.5 | 37.6 | ||
| 4.3 | 13202–13411 | 48.1 | 70 | 7.7 | 10.80 | ATAGGAGACACACCATG | gp4.3 | 97.1 | 41.4 | ||
| 4.5 | 13427–13708 | 55.0 | 94 | 10.8 | 10.69 | TAAGGAGCGCACACTATG | gp4.5 | 96.8 | 56.2 | ||
| 5 | 13779–15890 | 51.0 | 704 | 79.8 | 6.77 | AAAGGAGGGCATTATG | gp5 | 87.1 | 85.6 | DNA polymerase | |
| 5b | 14739–15890 | 51.6 | 384 | 42.9 | 7.01 | ACACGAGGGATTACATG | gp5B | 89.8 | |||
| 5.3 | 15903–16232 | 50.3 | 110 | 13.0 | 10.05 | TAAGGAGGATTTATG | gp5.3 | 91.1 | 25.0 | ||
| 5.5 | 16213–16515 | 49.7 | 101 | 11.2 | 7.56 | AAAGGAGAAACATTATG | gp5.5 | 54.1 | 44.9 | Growth on lambda lysogens | |
| 5.5–5.7 | 16213–16721 | 50.9 | 171 | 18.6 | 9.72 | AAAGGAGAAACATTATG | gp5.5–5.7 | 67.9 | 61.9 | ||
| 5.7 | 16515–16721 | 52.7 | 69 | 7.3 | 10.53 | GCGAGAGGTGTTCAAATG | gp5.7 | 88.4 | 87.0 | ||
| 5.9 | 16721–16900 | 48.3 | 60 | 6.8 | 3.76 | ATGGGAGGTTGCGTATG | gp5.9 | 32.7 | 32.7 | ||
| 6 | 16900–17808 | 52.1 | 303 | 34.8 | 4.77 | CGGGGAGGATGACGAATG | gp6 | 79.5 | 73.0 | Exonuclease | |
| 6.1 | 16937–17164 | 52.2 | 76 | 9.1 | 12.23 | CGCGGAGATGCGTG | |||||
| 6.3 | 17793–17903 | 55.9 | 37 | 4.2 | 10.48 | CAAGGAGATTTACTTATG | gp6.3 | 97.3 | 26.5 | ||
| 6.5 | 17999–18241 | 47.7 | 81 | 9.4 | 6.26 | TTAAGAGGTGAAATTATG | gp6.5 | 98.8 | 55.6 | ||
| 6.7 | 18249–18497 | 52.2 | 83 | 8.9 | 9.79 | ACAGGAGTAATTATATG | gp6.7 | 98.8 | 63.6 | Adsorption | |
| 7.3 | 18528–18845 | 52.5 | 106 | 10.9 | 10.56 | TAGGGAGAAACATCATG | gp7.3 | 95.3 | 66.0 | Host specificity | |
| 8 | 18859–20463 | 51.3 | 535 | 58.6 | 4.37 | TGAGGAGGACTGAATG | gp8 | 99.1 | 84.6 | Head-tail connector | |
| 9 | 20568–21497 | 51.7 | 310 | 33.7 | 4.13 | TTAGGAGATTTAACAATG | gp9 | 94.8 | 64.3 | Scaffolding protein | |
| 10a | 21657–22697 | 55.9 | 347 | 36.9 | 6.74 | TAAGGAGATTCAACATG | gp10A | 99.1 | 80.5 | Major capsid protein | |
| 10b | 21657–22954 | 55.2 | 433 | 45.4 | 6.74 | TAAGGAGATTCAACATG | gp10B | 98.4 | 73.7 | Minor capsid protein | |
| 11 | 23191–23778 | 50.5 | 196 | 22.2 | 4.31 | ACAGGAGGTAACATCATG | gp11 | 80.1 | Tail protein | ||
| 12 | 23797–26199 | 50.2 | 801 | 89.9 | 6.35 | CAAGGAGGCTCTATG | gp12 | 69.7 | Tail protein | ||
| 13 | 26275–26688 | 43.5 | 138 | 16.1 | 5.73 | ACGAGGGGTTAAAGCATTATG | gp13 | 55.1 | Internal head protein | ||
| 13.5 | 26688–27074 | 50.8 | 129 | 14.6 | 8.72 | CAAGGAGGAACCCTTG | gp59 | Phage L5 (Q05272)d | T4 endonuclease VII homologue | ||
| 14 | 27080–27670 | 53.3 | 197 | 21.3 | 10.16 | AGAGGAGAATAATTATG | gp14 | 68.9 | Internal core protein | ||
| 15 | 27676–29916 | 51.2 | 747 | 85.3 | 5.51 | CGGGGAGGTAATAATG | gp15 | 66.7 | Internal core protein | ||
| 16 | 29938–33897 | 52.0 | 1320 | 143.6 | 9.10 | TAAGGAGGCTCCATG | gp16 | 67.2 | Internal core protein | ||
| 17 | 33972–35906 | 50.2 | 645 | 69.4 | 6.33 | AAAGGAGGTCACATG | gp171–645 | 37.7 | 40.0 | Tail fiber protein (adsorption) | |
| gp171–150 | 78.7 | 78.7 | |||||||||
| gp17150–645 | 22.5 | 24.7 | |||||||||
| 17.5 | 35962–36162 | 47.3 | 67 | 7.4 | 6.75 | ATAGGAGGACACAATG | gp17.5 | 85.1 | 91.0 | Holin for cell lysis | |
| 18 | 36169–36432 | 52.3 | 88 | 9.86 | 4.54 | TAAGGAGTAACCTATG | gp18 | 71.6 | 71.6 | DNA packaging, small subunit | |
| 18.5 | 36526–36975 | 49.8 | 150 | 17.0 | 10.16 | ATGGGAGGTGTTATG | gp18.5 | 53.8 | 56.0 | Endopeptidase, λ Rz homologue | |
| 18.7 | 36641–36892 | 48.0 | 84 | 9.3 | 10.35 | GAAGGAGGTAATCCAAAATG | gp18.7 | 45.8 | 47.0 | Cell lysis, λ RzI homologue | |
| 19 | 36953–38713 | 52.6 | 587 | 66.7 | 5.26 | TAAGGAGATGCAGAATG | gp19 | 95.9 | 84.5 | DNA packaging, large subunit | |
| 19.2 | 37602–37832 | 56.7 | 77 | 8.3 | 11.48 | CTCGAAGATAACCGTG | gp19.2 | 81.8 | 42.9 | ||
| 19.3 | 38139–38264 | 50.8 | 42 | 4.8 | 12.39 | CTGGCGGGTTCCGTGATG | gp19.3 | 92.8 | 59.5 | ||
| 19.5 | 38959–39105 | 50.3 | 49 | 5.5 | 8.11 | AAAGGAGGTGGCTCAATG | gp19.5 | 98.0 | 65.3 | ||
aa, amino acid.
SD sequence is indicated by underlining, and the initiation codon is indicated by boldface type.
Identity, 43% (26 of 60 amino acids).
Identity, 39% (41 of 105 amino acids).
Similar to the T3 and T7 genomes, φYeO3-12 genes could be divided into three classes: I (early), II (middle) and III (late). During a T3 or T7 infection class I genes are transcribed by the host RNAP and include functions to overcome host restriction and to convert the metabolism of the host cell to the production of phage proteins, e.g., synthesis of phage promoter-specific RNAP (gene 1). The phage RNAP transcribes the class II genes, mainly involved in phage DNA metabolism, and the class III genes, whose functions are predominantly morphogenetic (34).
Organization of genes.
Genes are all transcribed from the same DNA strand and occupy almost 92% of the nucleotide sequence (Fig. 1), the same value found for T7. This efficiency is presumably the result of evolutionary packaging of the maximum amount of useful information into a DNA molecule whose length is limited by a virion of fixed size (19). Where large noncoding sequences occur, recognizable genetic signals are almost always found; the longest noncoding stretches are the terminal repeats. Comparison φYeO3-12 genes with those of T3 and T7 shows that some are only present in one or two of the three phages.
Transcriptional features of φYeO3-12. (i) Promoters for host RNAP.
There are no experimental data on how φYeO3-12 DNA is transcribed during infection. By analogy with the T7 and T3 genomes three major early promoters for host RNAP, A1 (488), A2 (618), and A3 (664), were identified in the noncoding region near the left end of φYeO3-12 DNA. The numbers in parentheses are the positions in the φYeO3-12 genome corresponding to the predicted first nucleotide of the RNA chain initiated at that promoter. In addition, a minor promoter, leftward promoter A0 (111), was identified. We emphasize that these are putative promoters and their functions needs to be confirmed by transcription analysis.
(ii) Promoters for φYeO3-12 RNAP.
One characteristic of T7 group phages is that they code for an RNAP with a strict template specificity for its own genome. Upstream of φYeO3-12 gene 10 a sequence identical to the T3 gene 10 promoter was identified, and this was assumed to be a φYeO3-12 promoter. Altogether 15 putative φYeO3-12 promoters (Fig. 1) were identified in the phage genome (Fig. 2); all are located in the same relative positions as phage promoters on the T3 genome. Only ø10 and ø13 are 100% identical with the consensus sequence.
FIG. 2.
φYeO3-12 promoters. In the upper part of the figure the 15 putative promoter sequences of φYeO3-12 are aligned and a consensus sequence is presented. The positions of the first nucleotides of the promoter sequences in the phage genome are given as well as the number of mismatches (indicated by uppercase letters) to the consensus sequence and the name of the promoter. Residues likely to determine the promoter specificity are in boldface type. In the lower part of the figure is an alignment of the consensus promoter sequences of φYeO3-12, T3, T7, K11, and SP6. Sequences of the nontemplate strand are presented; the transcription start site is at +1. Conserved nucleotides, when compared against the φYeO3-12 promoter are indicated by vertical bars.
A T3 promoter consists of a highly conserved 23-bp sequence that runs from −17 to +6 relative to the transcription start. It is known that positions −10 and −11 determine the specificity of the phage RNAP and position −2 determines the strength of the promoter (29). A T at position −2 gives the highest relative utilization of the T3 promoter. The φYeO3-12 promoter positions −2, −10, and −11 are identical with those of a T3 promoter, suggesting comparable promoter strengths and specificities. Furthermore, similar to the situation in T3 (7), five class II promoters (ø1.05, ø1.1, ø1.3, ø1.5, ø4.3) and the ø11 promoter have a C residue rather than an A residue at position −1 (Fig. 2). All these T3 promoters having a −1C are weaker than the salt-insensitive class III promoters (6), suggesting that the −1 position is also important in determining the promoter strength. Similar results on the effect of position −1A on promoter strength were obtained for T7 RNAP using a collection of T7 promoter variants (28). Also, analogous to T3 (7), ø3.8 differed from consensus at only one position (−10), but one that is thought to be important in determining promoter strength and/or specificity. The ability of the putative φYeO3-12 ø10 promoter to direct transcription was shown using the TnT T3 Coupled Reticulocyte Lysate System, Promega, Madison, Wis. (Söderholm et al., unpublished).
(iii) Transcription termination sites.
A putative rho-independent early transcriptional terminator TE (calculated −ΔG = 10.8 kcal/mol) (Fig. 1) for the host RNAP was identified at position 7986 to 8001 using the Terminator and StemLoop programs of GCG. The putative φYeO3-12 TE is located immediately downstream of gene 1.3, in an analogous position to TE of T3 and T7. A putative major terminator Tø for the φYeO3-12 RNAP was identified just downstream of gene 10 at position 23131 to 23153, and the stem-loop structure (−ΔG = 8.8 kcal/mol) is followed by a strech of six Ts; both the structure and location of this terminator are also similar to those found in T3 and T7.
(iv) φYeO3-12 RNase III cleavage sites and mRNAs.
Cleavage of T3 and T7 RNAs at specific sites by the host enzyme RNase III is a prominent feature of both early and late transcription. The φYeO3-12 sequence was also checked for RNA secondary structure (MFold program of the GCG package), as the RNA around these RNase III sites can be arranged in a characteristic pattern of base pairing within which lies the point of cleavage. Altogether 10 putative RNase III sites (R0.3 through R18.5), in positions analogous to those found in T3 and T7, were identified in φYeO3-12 (Fig. 1 and Table 2). Each site is referred to by R followed by the number of the first gene to the right of the putative cleavage site. It is reasonable to assume that φYeO3-12 transcripts are also processed by RNase III and that the Y. enterocolitica serotype O:3 RNase III has the same specificity as the E. coli enzyme. Like T7 (19), φYeO3-12 is thus predicted to produce a rather large set of overlapping late transcripts that originate from multiple promoters. Also like T7, these primary transcripts are likely to include readthough products across Tø and may also be incompletely processed by RNase III (19).
TABLE 2.
The predicted RNase III sites of bacteriophage φYeO3-12
| Putative RNaseIII site
|
Sequence of predicted stem-loopa | |||
|---|---|---|---|---|
| Name | Range | Putative cleavage site | ΔG (kcal/mol) | |
| R0.3 | 956–1009 | 997–998 | −14.9 | UAAGCGAAUAACUCAAGGUCGCACUGAAAGCGUGGCCUUUAU↓GAUAUUCACUUA |
| R0.45 | 1490–1545 | 1532–1533 | −20.8 | GUAAGUGUUAAACUCAAGGUCGCUCCAUGCGAGUGGCCUUUAU↓GAUUAUCACUUAU |
| R1 | 3245–3293 | 3282–3283 | −19.6 | GAGUCUUUUCUUACAGGUCAUCAUGUGGUGGCCUGAAU↓AGGAACGAUUU |
| R1.1 | 6340–6391 | 6379–6380 | −20.9 | GAGAGUUAAACUUAAGGUCAUCACCGACGGUGGCCUUUGU↓GAUUAACUUUC |
| R1.3 | 6856–6896 | 6891–6892 | −18.8 | GAAUCCU(↓)UAAGGUCACUUAACAUGAGUGGCCUUUGU↓GAUUC |
| R3.8 | 11350–11377 | Not clear | −15.0 | UAAAGGGAGACUUAACGGUUUCCCUUUG |
| R4.7 | 13706–13754 | 13743–13744 | −18.4 | AAGUGAUAAACUCAAGGUCGCCCAAGGGUGGCCUUUAU↓GAUUAUCAUUU |
| R6.5 | 17901–17971 | 17960–17961 | −23.3 | AAGUGAUAAACUCAAGGCUCUCUGUAUUAACCCUCACUAAAGGGAAGAGGGAGCCUUU AU↓GAUUAUUACUU |
| R13 | 26209–26246 | Not clear | −21.8 | GUCUCCCUGUGGUGAAUUAACCCUCACUAAAGGGAGAC |
| R18.5 | 36433–36486 | 36474–36475 | −22.0 | UAAGUGACAUACUCAAGGUUCUCCACUCGGGGGAGCCUUUAU↓GGAUGUUAUUUG |
↓, cleavage site; (↓), secondary cleavage site.
Translational features of φYeO3-12. (i) Protein initiation sites and termination codons of φYeO3-12.
The 54 close-packed genes of φYeO3-12 may actually specify 56 independent proteins, as both genes 4 and 5 are predicted to contain an in-frame internal initiation site that would produce truncated proteins with the same sequence as the C terminus of the full-length gene products. The nucleotide sequences in the mRNAs around the start sites for each of these 56 proteins, as well as those around the start sites for the six potential overlapping, but out-of-frame, proteins discussed in the next section, are given in Table 1.
The initiation codon for all but 3 of the 56 proteins specified by the φYeO3-12 genes is AUG: that for gp6.1 and gp19.2 is GUG, and gp13.5 is initiated at a UUG codon. All predicted genes are preceeded by a potential SD sequence of 3 to 10 nucleotides capable of uninterrupted pairing with nucleotides near the 3′ end of 16S rRNA. The distance from the A (or its equivalent) in the GGAGG ribosome binding sequence to the first nucleotide of the initiation codon ranges between 7 and 13 nucleotides. The shortest interval between the last paired nucleotide of the ribosome binding sequence and the first nucleotide of the initiation codon is 2 nucleotides, and the longest is ten. These features are the same as those found in the T7 genome (19). Also as found in the analysis of the T7 genome, the use of GCU (alanine) as the second codon correlates well with high expression of φYeO3-12 genes (19). Nine actively syntesized proteins in the cell during the time they are expressed—gp2.5, gp3, gp3.5, gp8, gp9, gp10, gp12, gp15, and gp17—have GCU as the second codon. All three stop codons, UAA, UGA, and UAG, are used in φYeO3-12, with UAA being the most frequent. There are also instances in which a termination codon overlaps the initiation codon of the next protein. The sequence UAAUG includes the termination codon for genes 0.45, 1.6, and 2.5 and the initiation codon for genes 0.6A, 1.7, and 3, respectively. The sequence AUGA includes the termination codon for genes 5.5, 5.7, and 5.9 and the initiation codon for genes 5.7, 5.9, and 6, respectively, and UUGA terminates gene 13 and initiates gene 13.5. Similar overlapping is found for the homologous T7 genes.
(ii) Frameshifting during translation.
Frameshifting occurs naturally in bacteriophage T7 gene 10 at a frequency that depends on the nucleotide sequence in the region of the frameshift and also on the 3′ noncoding region (15). The φYeO3-12 sequence was also analyzed for putative sites of frameshifting, and we found three sites that are analogous to those predicted or shown to occur in T3 and T7. Frameshifting to the +1 frame during translation of gene 0.6 and to the −1 frame during translation of gene 5.5 and gene 10 affords, respectively, gp0.6B, a 171-residue 5.5-5.7 fusion product, and gp10B. The presence of gp10B has been already confirmed (35) by analysis of two proteins with identical N termini. The predicted sizes of gp10A and gp10B correspond well with the eletrophoretic mobilities of these two proteins and were thus concluded to be the major and minor capsid proteins.
DNA sequences around the putative gene 10 frameshifting site were compared, and the φYeO3-12 sequence showed 100% identity to that of T3 (14). Thus, the gene 10 shift is proposed to happen at the sequence C-CCA-AAG when tRNA Thr3 sometimes recognizes the two-base codon CC (18). Consequently, the ribosome would only translocate two bases down the mRNA, thereby shifting into the −1 reading frame.
In T7 it is not known whether gp0.6B results from a +1 frameshift or from a readthrough of gene 0.6A stop codon, allowing a longer protein to be made (19). We believe that one mechanism or another is also used in φYeO3-12 to produce gp0.6B, although there is limited sequence similarity between the genomes at the predicted site of frameshifting in T7. The gene 0.6B reading frame is the only one that is open, but no good ribosome binding sequence is present. If no gp0.6B is made there would be a noncoding region of almost 200 bp in the genome, which would be anomalous in the otherwise close packing of φYeO3-12 genes.
T7-infected cells contain a protein that is a fusion product of genes 5.5 and 5.7 (19). In T3 it is not known whether this gene 5.5-5.7 fusion product is made because the sequence of the predicted site of frameshifting differs in two positions between T3 and T7. Even greater differences are found in this region when φYeO3-12 is compared to T7, and the existence of a gene 5.5-5.7 fusion protein should thus be considered speculative.
Origins of DNA replication.
The T3 primary origin of replication, the preferred origin for the first replication of parental DNA, has been mapped downstream of gene 1 overlapping the 5′ end of gene 1.05 (42). The site is an AT-rich region of 142 nucleotides; using pairwise alignments an identical (100%) region was identified in φYeO3-12. Thus the putative primary origin of replication of φYeO3-12 DNA (R) (Fig. 1) is located at position 5995 to 6136, also between genes 1 and 1.05. The φYeO3-12 and T3 primary origins of replication do not have the same location as that of T7, which is located at the noncoding region between genes 1 and 1.1 (40). The T7 øOL and øOR promoters were proposed to be secondary origins of replication (19). Counterparts to both promoters are found in the φYeO3-12 genome, but their role, if any, in DNA replication needs to be determined.
Ends of φYeO3-12 DNA and the concatamer junction.
The longest stretches of φYeO3-12 DNA that do not code for any proteins are at the ends of the molecule. Redundant direct terminal repeats (TRs) (indicated in Fig. 1) of 232 bp were identified from left and right genome ends of the linear dsDNA molecule, and they were 87% identical to the 230-bp TR of T3 when compared at nucleotide level. The 160-bp TR of T7 showed low similarity compared with φYeO3-12 (56%) and T3 (59%). It is noteworthy that the sequences at the beginning and end of the TRs of all three phages are significantly similar to those in the middle of the TRs, indicating that the mechanisms of maturation of the DNA ends might be similar. Otherwise the organization of the noncoding regions of mature φYeO3-12 DNA is essentially identical to that of T7 (19). The left end of φYeO3-12 DNA contains the terminal repetition; a regular array of short repeated sequences (CCTAAAG, or variants); an AT-rich region that contains the øL replication origin; the A1, A2, and A3 promoters for host RNAP; the R0.3 RNase III cleavage site; and finally, the start of the coding sequence of gene 0.3. The right end of φYeO3-12 DNA contains the terminal repetition, an array of short repeated sequences similar to that found near the left end, the coding sequence of gene 19.5, an AT-rich region that contains the ØR replication origin, and finally, the end of the coding sequence of gene 19.
Other features of the nucleotide sequence.
The genomic DNA of φYeO3-12 has an overall GC content of 50.6%, compared to 48.5 ± 1.5 mol% for its host (11). The GC contents of the common representatives of the T7 group, T7 (accession no. V01146 [complete sequence of 39,937 bp]) and T3 (accession no. X17255 [partial sequence of 19,680 bp]), are 48.4 and 50.6%, respectively. The sequences corresponding to several restriction enzyme recognition sites are grossly underrepresented in φYeO3-12 DNA (data not shown). Specific cases include GATC and CC(A/T)GG, which are present only three and zero times in φYeO3-12 DNA, although those sequences are expected statistically 155 and 77 times, respectively. The biological significance of this feature is not known, but it may reflect the desire of the phage to avoid the methylation activities present in the host; Dam (DNA adenine methyltransferase) methylase modifies GATC, and Dcm (DNA cytosine methyltransferase) methylase modifies CC(A/T)GG sequences (32). Although φYeO3-12 is predicted to code for S-adenosyl-l-methionine hydrolase (SAMase), which degrades the methyl group donor in the host (45). If the methylation sites were abundant in the phage sequence, the host methylation activity might modify the translocated phage DNA before it is shut off by the SAMase activity of the phage. The SAMase activity is the reason why T3 DNA is not methylated, a situation likely the same in φYeO3-12. No modified nucleotides were detected in φYeO3-12 DNA by high-performance liquid chromatography analysis (S. J. Kiljunen et al., unpublished data).
φYeO3-12 genes are all very closely packed, and in six cases genes occupy overlapping reading frames (Fig. 1). Genes 4.15 and 4.2 lie almost entirely within the gene 4 coding sequence, gene 6.1 overlaps gene 6, gene 18.7 lies within gene 18.5, and both genes 19.2 and 19.3 overlap the gene 19 sequence. All the overlapping genes are in the exact same positions as in T3 and T7, except for gene 6.1.
Mutations.
We previously identified a gene 0.7 deletion derivative of φYeO3-12 (designated ΔPK) (35), and the nature of the deletion was analyzed in the present study. ΔPK was shown to carry a 705-bp deletion at position 2181 to 2886, deleting the majority of the gene 0.7 coding region. The deleted region was flanked by 12-bp direct repeats (CGATTGACCGCT) that most probably were targets for short-range recombination. Similar deletions have been identified in both T3 and T7. Various models have been proposed to account for deletion mechanisms, but it seems possible that enzyme components capable of promoting break rejoining play a crucial role (30). The deletion results in an in-frame deletion within gene 0.7 that allows synthesis of a protein containing the 16 N-terminal residues of gp0.7 fused to its 115 C-terminal residues. The N-terminal domain of T7 gp0.7 acts a serine-threonine kinase that phophorylates several host proteins, including RNase III (38). T7 gp0.7 also shuts off host-catalyzed transcription by an unknown mechanism that is distinct from protein kinase activity (33). It is also known that 0.7 deletion mutants grow better than the wild type in rich medium, but gene 0.7 is important for growth in poor media and at elevated temperatures (27, 34). Thus, the ΔPK derivative of φYeO3-12 most likely arose from serial growth of new phage stocks in rich medium from the remains of the previous stock.
Lysis genes.
All known dsDNA phages produce a soluble, muralytic enzyme known as endolysin. Endolysins require access to the cell wall, which is provided by a second lysis factor, a small membrane protein designated holin. In addition many phages of gram-negative hosts also bear two overlapping genes that specify auxiliary lysis proteins (for reviews, see references 48 and 49). All four genes involved in host cell lysis were identified in the φYeO3-12 genome. Based on amino acid homology with T3 and T7 φYeO3-12 gp3.5 is proposed to be the endolysin that would have N-acetylmuramoyl-l-alanine amidase activity and gp17.5 is proposed to be the holin. Using the TMHMM program (version 1.0) (Department of Biotechnology, The Technical University of Denmark) to predict transmembrane helices, φYeO3-12 gp17.5 contains two transmembrane domains and is thus a class II holin. φYeO3-12 gp18.5 and gp18.7 are proposed to be homologues of λ Rz and Rz1, respectively. The exact functions of Rz and Rz1 are unknown; they are required for λ-induced cell lysis only when the outer membrane is stabilized in medium containing millimolar concentrations of divalent cations (50, 51).
Tail fiber.
The tail fibers are assembled directly onto the preformed head-tail complex and are required to give an infectious phage particle. Gene 17 of φYeO3-12 encodes a protein of 645 amino acid residues, more than 100 residues larger than its T3 and T7 homologues. The N-terminal 150 amino acids of φYeO3-12 gp17 show marked (78.7%) homology to the N-terminal part of gp17 of both T3 and T7, whereas the C-terminal portion shows only about 23% identity (Table 1 and Fig. 3). The N-terminal part of T7 gp17 attaches the fiber to the tail just below its junction with the head-tail connector protein (46). By analogy this description likely applies to φYeO3-12 also; the high level of identity between the N-terminal region of gp17 with that of T7 is partly mirrored in a higher-than-average level of homology between the φYeO3-12 and T7 tail proteins gp11 (80.1%) and gp12 (69.7%).
FIG. 3.
Alignment of the N-terminal parts of tail fiber proteins (gp17) of T3, T7, φYeO3-12, and the endosialidase of bacteriophage K1F. Identical residues are shaded with black and similar residues are shaded with grey. Gaps (indicated by horizontal lines) were introduced into the sequences to maximize the alignments. Numbering is from the N-terminal methionine. Alignment of the C termini of the proteins was not included in the figure due to the very low similarity.
Sequences of five tail fiber proteins of the family Podoviridae are now known. Figure 3 shows an alignment over the significantly similar N-terminal parts of gp17 proteins of T3, T7, and φYeO3-12 and of the endosialidase of E. coli phage K1F. The endosialidase of bacteriophage K1E was not included in the alignment due to very low similarity between the other sequences (31). The first 113 N-terminal amino acid residues of K1F endosialidase are 40% identical with T7 gp17 and 60% similar when conservative changes are considered (37). In addition, the 113 N-terminal amino acids of K1F endo-N-endosialidase show 42.6 and 43.5% identity to the φYeO3-12 and T3 gp17 proteins (Fig. 3), respectively.
However, the C-terminal two-thirds of the tail fiber proteins are much more different (Table 1). The C-terminal two-thirds of φYeO3-12 gp17 exhibits ca. 31% similarity compared with T3 and T7, whereas T3 and T7 are 75.8% similar. The C-terminal portion of T7 gp17 forms the distal part of the tail fiber that is involved in the binding to the host cell receptor (46). T7 tail fibers attach to the lipopolysaccharide outer core of the E. coli to initiate adsorption. The absence of similarity between the C-terminal regions of φYeO3-12 gp17 and T3 or T7 gp17 may thus be a reflection of host specificity. In addition, bacteriophages K1F (37) and K1E (31), which recognize and infect strains of E. coli displaying the α-2,8-linked polysialic acid K1 capsule, show a high degree of identity (unbroken over 532 residues) towards the endo-N-endosialidase C termini. The absence of homology in the C-terminal parts of φYeO3-12 gp17 and T3 or T7 gp17 is consistent with the finding that in many dsDNA phages it has been demonstrated that the C-terminal parts of the tail fiber proteins, responsible for binding to host receptors, evolve faster than other phage genes as a result of intense host range selection (21).
We tried to isolate T3 recombinants carrying the φYeO3-12 gene 17 to ask the question whether gp17 is sufficient to turn a coliphage into a yersiniophage (see Materials and Methods). No viable YeO3-c specific T3 recombinants were obtained (data not shown), a result that could reflect a requirement for a φYeO3-12 adsorption protein in addition to gp17, a failure of φYeO3-12 gp17 to attach to a T3 tail (gp11 and gp12), or the fact that recombination did not take place. The level of homology between φYeO3-12 and T3 tail proteins gp11 and gp12 could not be analyzed as the T3 sequence is not available. The efficiency of recombination might have been below levels of detection, because the regions of φYeO3-12 DNA flanking gene 17 have only 60 to 80% DNA identity to the corresponding T3 sequence. Further work on the specificity determinants is needed to understand these host-phage interactions.
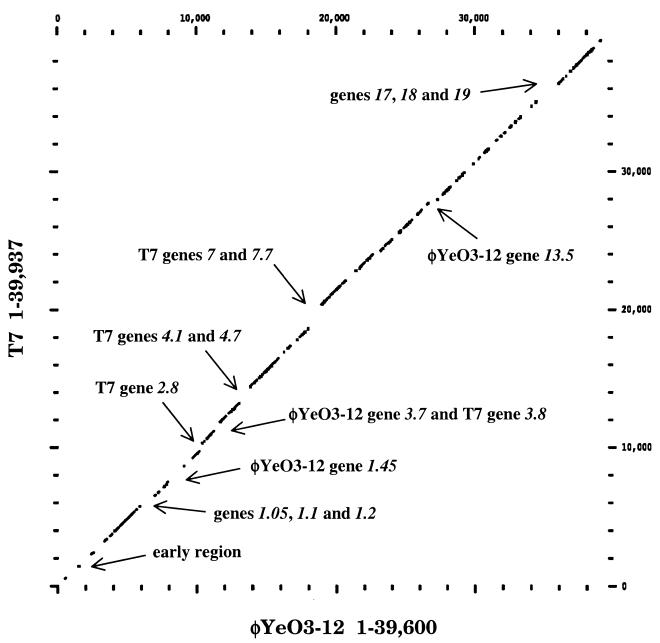
Comparison to T7 and T3 genomes.
Comparison of the nucleotide sequences of φYeO3-12 and T7 was performed, and the results were plotted using the DOTPLOT program of the GCG package (Fig. 4). The genomes can be aligned over their entire lengths as indicated by the identity line. In a few places the line was broken by upward or downward shifts, indicating the presence of insertions and deletions. A whole genome analysis could not be performed with T3; sequences upstream of gene 1 are incompletely known, and those from gene 11 through gene 16 have not been reported. Analysis of the available sequence of T3 by Dotplot was performed, but no differences, beyond those seen with T7, were noted. The major differences between the φYeO3-12 and T7 genomes, and the known parts of T3, are summarized in Table 3.
FIG. 4.
Comparison of the φYeO3-12 and T7 genomes using the COMPARE and DOTPLOT programs. Sequences were scanned using a 50-nucleotide window. Each data point represents ≥80% identity between the sequences. The φYeO3-12 genome is plotted on the x axis, and the T7 genome is plotted on the y axis. Regions with gross sequence differences between genomes are indicated (see text and Table 3 for details).
TABLE 3.
Major differences between bacteriophages φYeO3-12, T3, and T7
| φYeO3-12 sequence range | Expressed gene product(s) (size [aa]) in phage:
|
Pairwise comparison of gene products (% identity)
|
||||
|---|---|---|---|---|---|---|
| φYeO3-12 | T3 | T7 | φYeO3-12 vs T3 | φYeO3-12 vs T7 | T3 vs T7 | |
| 1035–1490 | gp0.3 (152) | gp0.3 (152) | gp0.3 (117) | 98.0 | 20.9 | 22.6 |
| 1558–1755 | gp0.45 | SNAa | gp0.4, gp0.5 | ANAb | <20c | ANA |
| 1758–2115 | gp0.6A, gp0.6B | SNA | gp0.6A, gp0.6B | ANA | 34.0 and 25.2, respectively | ANA |
| 6054–6323 | gp1.05 (90) | gp1.05 (90) | —d | 97.8 | ||
| 6419–6556 | gp1.1 | gp1.1 | gp1.1 | 97.8 | 45.0 | 45.0 |
| 6562–6834 | gp1.2 | gp1.2 | gp1.2 | 94.5 | 40.0 | 41.2 |
| 7903–8373 | gp1.45 (157) | — | gp1.4 (51) | <20 | ||
| 9330–9563 | gp2 (78) | gp2 (54) | gp2 (64) | 85.2 | 39.1 | 50.0 |
| — | — | — | gp2.8 (139) | |||
| 11231–11335 | gp3.7 (35) | gp3.7 (35) | gp3.8 (121) | 97.1 | <20 | <20 |
| 11672–11776e | gp4.15 (35) | Putative ORF (35) | gp4.1 (40) | 100.0 | <20 | <20 |
| — | — | — | gp4.7 (135) | |||
| — | No putative ORF | gp5.1 (29)f | No putative ORF | |||
| 16721–16900 | gp5.9 (60) | gp5.9 (52) | gp5.9 (52) | 32.7 | 32.7 | 98.1 |
| — | — | — | gp7 (133) | |||
| — | — | — | gp7.7 (130) | |||
| 16937–17164g | gp6.1 (76) | No putative ORF | No putative ORF | |||
| 26688–27074 | gp13.5 (129) | SNA | — | ANA | ANA | |
| 33972–35906 | gp17 (645) | gp17 (557) | gp17 (553) | 37.7 | 40.0 | 79.4 |
SNA, sequence not available.
ANA, analysis not applicable
<20, gene product(s) considered nonhomologous.
—, gene not present.
Within gene 4 coding region.
Within gene 5 coding region.
Within gene 6 coding region.
The early region genes 0.45, 0.6a, and 0.6b are very different from T7 genes 0.4, 0.5, and 0.6; the corresponding region in T3 is not known. No information of the functions of these genes is available. Elsewhere in the early region, φYeO3-12 contains genes 0.3, 1.05, 1.1, and 1.2; these are highly homologous to those of T3 but not to those of T7. Like T3, counterparts (or homologues) of T7 genes 2.8, 3.8, 4.7, 7, and 7.7 are missing from φYeO3-12. In contrast, T3 and φYeO3-12 both contain gene 3.7, which encodes a protein similar to a number of trypsin inhibitors (7) and is not in the T7 genome. A putative gene 4.1 (overlapping gene 4) has been identified in T7 but was not recognized in T3. At the DNA level T3 and φYeO3-12 sequences are similar to T7, but a putative ribosome binding sequence for gp4.1 synthesis is missing. In φYeO3-12 gene 4.15 was identified as a gene overlapping gene 4; it showed no significant similarity to T7 gp4.1 or to any sequences in the databases and may be a novel gene. A putative ORF that is identical to gene 4.15 (except for a GUG initiation codon), can also be found in the T3 sequence. In contrast to T3, but analogous to T7, φYeO3-12 seems to be missing the putative gene 5.1 (overlaps gene 5). Gp5.9 of T3 and T7 are extremely similar (98.1% identity), but surprisingly φYeO3-12 gp5.9 shows only 32.7% identity to the T3 and T7 gp5.9 sequences. Whether φYeO3-12 gp5.9 has the anti-RecBCD (exonuclease V) activity of T7 gp5.9 (34) is not known. φYeO3-12 gene 6.1 (overlaps gene 6) is preceeded by a fairly good SD, but the predicted gene product has no significant similarities to those in the protein databases. φYeO3-12 gene 6.1 may be a novel gene.
In the location corresponding to gene 1.4 of T7, φYeO3-12 has gene 1.45, which shows no similarity either to gene 1.4 or to any gene in T3 (in T3 there is no gene in this location). Gene 1.45 encodes a putative novel protein with similarity to lactococcal phage bIL170 e20 protein. E20 is a member of the HNH endonuclease protein family that are found in bacteria and viruses, e.g., T7 gp5.3 (16). The HNH endonucleases are thought to be derived from group I introns.
Gene 13.5, located between genes 13 and 14, encodes a putative novel protein with similarity (ca. 40% identity) to mycobacteriophage L5 gp59; this gene is missing from T7, and the corresponding T3 sequences are not available. However, heteroduplex analysis suggests that there are no genetic differences between T3 and T7 from gene 11 through gene 16 (17), suggesting that T3 may be missing gene 13.5. L5 gp59 is homologous to bacteriophage T4 endonuclease VII. An alignment of φYeO3-12 gp13.5, L5 gp59, and T4 endoVII amino acid sequences shows conservation of cysteine residues, including three CXXC sequences, (data not shown). This conservation may imply that these residues are important for function. Although the CXXC zinc finger motif is often associated with DNA-binding proteins, no homology between gp13.5 and proteins involved in DNA binding was detected using the Pfam protein motif search routine. Interestingly, the initiation codon of gene 13.5 is UUG, which is a common initiation codon in mycobacteriophages (e.g., L5 and D29) as well as in mycobacteria (20, 23), but one that is only rarely used in E. coli. Altogether these data suggest a common origin for φYeO3-12 gene 13.5 and L5 gene 59.
In comparisons of closely related phage genomes, occasionally an “extra” gene is present in one of the phages, inserted between two genes that are adjacent in the related set of phages. Hendrix et al. recently named these extra genes “morons” because they apparently add more DNA to the genome (25). Typically, the nucleotide and dinucleotide compositions of the moron genes are substantially different from those of adjacent genes, indicating that the moron has recently entered the genome from an outside source. The AT content of gene 13.5 is slightly higher than that of the rest of the phage, suggesting that gene 13.5 is a moron. What biological relevance if any this gene has to φYeO3-12 remains to be elucidated. φYeO3-12 showed, in addition to gene 13.5, no regions with markedly higher-than-average AT content, giving no indication of additional morons originating from an outside source of different AT content.
Final conclusions and evolutionary aspects.
Phage φYeO3-12 is the first Y. enterocolitica phage characterized at the molecular level. This lytic phage belongs to the T7 group of phages. The genome of φYeO3-12 shows striking homology to the genome of bacteriophage T7 and even more so to the partially sequenced genome of bacteriophage T3. The similarity between φYeO3-12 and T3 convincingly shows that they belong to the same lineage of phages that have developed different host specificities. This very close relationship was a surprising finding since φYeO3-12 and T3 have been isolated far apart both geographically and temporally. The order of genes is identical in T3 and φYeO3-12, and all essential genes exhibit very high identities, indicating that the two phages share a common ancestor. In comparison, although all of the lambdoid phages studied to date have maintained essentially identical orders of genes and regulatory elements, they can encode radically different proteins to perform analogous functions (12). Very few genetic differences were noticed between φYeO3-12 and the known sequence of T3; therefore, completion of the T3 sequence will allow detailed mapping of the differences and will make a basis for further work to elucidate the roles the genetic differences play in the different host specificities of these phages.
ACKNOWLEDGMENTS
We express our thanks to the personnel of the Sequencing Laboratory of the Department of Genetics (University of Turku, Turku, Finland). Heli Kaukoranta and Anne Peippo are thanked for technical assistance, and Jyri Kurkinen is thanked for the digital art images. Ian Molineux is greatly appreciated for strains, phages, and critical reviewing of the manuscript.
The Turku Graduate School for Biomedical Sciences (for M.I.P.) and the Technology Development Centre of Finland and Academy of Finland are thanked for financial support.
REFERENCES
- 1.Al-Hendy A, Toivanen P, Skurnik M. Expression cloning of the Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb Pathog. 1991;10:47–59. doi: 10.1016/0882-4010(91)90065-i. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hendy A, Toivanen P, Skurnik M. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect Immun. 1992;60:870–875. doi: 10.1128/iai.60.3.870-875.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Appleyard R K. Segregation of new lysogenic types during growth of doubly lysogenic strain derived from Escherichia coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons; 1987. [Google Scholar]
- 6.Bailey J N, McAllister W T. Mapping of promoter sites utilized by T3 RNA polymerase on T3 DNA. Nucleic Acids Res. 1980;8:5071–5088. doi: 10.1093/nar/8.21.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck P J, Gonzalez S, Ward C L, Molineux I J. Sequence of bacteriophage T3 DNA from gene 2.5 through gene 9. J Mol Biol. 1989;210:687–701. doi: 10.1016/0022-2836(89)90102-2. [DOI] [PubMed] [Google Scholar]
- 8.Birnboim H C, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Botstein D. A theory of modular evolution for bacteriophages. Ann NY Acad Sci. 1980;354:484–490. doi: 10.1111/j.1749-6632.1980.tb27987.x. [DOI] [PubMed] [Google Scholar]
- 10.Bottone E J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997;10:257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brenner D J, Ursing J, Bercovier H, Steigerwalt A G, Fanning G R, Alonso J M, Mollaret H H. Deoxyribonucleic acid relatedness in Yersinia enterocolitica and Yersinia enterocolitica-like organisms. Curr Microbiol. 1980;4:195–200. [Google Scholar]
- 12.Casjens S, Hatfull G, Hendrix R. Evolution of dsDNA tailed-bacteriophage genomes. Semin Virol. 1992;3:383–397. [Google Scholar]
- 13.Christensen S G. Yersinia enterocolitica in Danish pigs. J Appl Bacteriol. 1980;48:377–382. doi: 10.1111/j.1365-2672.1980.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 14.Condreay J P, Wright S E, Molineux I J. Nucleotide sequence and complementation studies of the gene 10 region of bacteriophage T3. J Mol Biol. 1989;207:555–561. doi: 10.1016/0022-2836(89)90464-6. [DOI] [PubMed] [Google Scholar]
- 15.Condron B G, Gesteland R F, Atkins J F. An analysis of sequences stimulating frameshifting in the decoding of gene 10 of bacteriophage T7. Nucleic Acids Res. 1991;19:5607–5612. doi: 10.1093/nar/19.20.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalgaard J Z, Klar A J, Moser M J, Holley W R, Chatterjee A, Mian I S. Statistical modeling and analysis of the LAGLIDADG family of site-specific endonucleases and identification of an intein that encodes a site-specific endonuclease of the HNH family. Nucleic Acids Res. 1997;25:4626–4638. doi: 10.1093/nar/25.22.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Davis R W, Hyman R W. A study in evolution: the DNA base sequence homology between coliphages T7 and T3. J Mol Biol. 1971;62:287–301. doi: 10.1016/0022-2836(71)90428-1. [DOI] [PubMed] [Google Scholar]
- 18.Dayhuff T J, Atkins J F, Gesteland R F. Characterization of ribosomal frameshift events by protein sequence analysis. J Biol Chem. 1986;261:7491–500. [PubMed] [Google Scholar]
- 19.Dunn J J, Studier F W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983;166:477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- 20.Ford M E, Sarkis G J, Belanger A E, Hendrix R W, Hatfull G F. Genome structure of mycobacteriophage D29: implications for phage evolution. J Mol Biol. 1998;279:143–164. doi: 10.1006/jmbi.1997.1610. [DOI] [PubMed] [Google Scholar]
- 21.Haggård-Ljungquist E, Halling C, Calendar R. DNA sequences of the tail fiber genes of bacteriophage P2: evidence for horizontal transfer of tail fiber genes among unrelated bacteriophages. J Bacteriol. 1992;174:1462–1477. doi: 10.1128/jb.174.5.1462-1477.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 23.Hatfull G F, Sarkis G J. DNA sequence, structure and gene expression of mycobacteriophage L5: a phage system for mycobacterial genetics. Mol Microbiol. 1993;7:395–405. doi: 10.1111/j.1365-2958.1993.tb01131.x. [DOI] [PubMed] [Google Scholar]
- 24.Hausmann R. The T7 group. In: Calendar R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Press; 1988. pp. 259–289. [Google Scholar]
- 25.Hendrix R W, Lawrence J G, Hatfull G F, Casjens S. The origins and ongoing evolution of viruses. Trends Microbiol. 2000;8:504–508. doi: 10.1016/s0966-842x(00)01863-1. [DOI] [PubMed] [Google Scholar]
- 26.Hendrix R W, Smith M C, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hirsch-Kauffmann M, Herrlich P, Ponta H, Schweiger M. Helper function of T7 protein kinase in virus propagation. Nature. 1975;255:508–510. doi: 10.1038/255508a0. [DOI] [PubMed] [Google Scholar]
- 28.Imburgio D, Rong M, Ma K, McAllister W T. Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry. 2000;39:10419–10430. doi: 10.1021/bi000365w. [DOI] [PubMed] [Google Scholar]
- 29.Klement J F, Moorefield M B, Jorgensen E, Brown J E, Risman S, McAllister W T. Discrimination between bacteriophage T3 and T7 promoters by the T3 and T7 RNA polymerases depends primarily upon a three base-pair region located 10 to 12 base-pairs upstream from the start site. J Mol Biol. 1990;215:21–29. doi: 10.1016/s0022-2836(05)80091-9. [DOI] [PubMed] [Google Scholar]
- 30.Kong D, Masker W. Deletion between direct repeats in T7 DNA stimulated by double-strand breaks. J Bacteriol. 1994;176:5904–5911. doi: 10.1128/jb.176.19.5904-5911.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long G S, Bryant J M, Taylor P W, Luzio J P. Complete nucleotide sequence of the gene encoding bacteriophage E endosialidase: implications for K1E endosialidase structure and function. Biochem J. 1995;309:543–550. doi: 10.1042/bj3090543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marinus M G. Methylation of DNA. In: Neidhart F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. I. Washington, D.C.: ASM Press; 1996. pp. 782–791. [Google Scholar]
- 33.Michalewicz J, Nicholson A W. Molecular cloning and expression of the bacteriophage T7 0.7 (protein kinase) gene. Virology. 1992;186:452–462. doi: 10.1016/0042-6822(92)90010-m. [DOI] [PubMed] [Google Scholar]
- 34.Molineux I J. The T7 family of bacteriophages. In: Creighton T E, editor. Encyclopedia of molecular biology. New York, N.Y: John Wiley and Co.; 1999. pp. 2495–2507. [Google Scholar]
- 35.Pajunen M, Kiljunen S, Skurnik M. Bacteriophage φYeO3–12 specific for Yersinia enterocolitica serotype O:3 is related to coliphages T3 and T7. J Bacteriol. 2000;182:5114–5120. doi: 10.1128/jb.182.18.5114-5120.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petter J G, Vimr E R. Complete nucleotide sequence of the bacteriophage K1F tail gene encoding endo-N-acylneuraminidase (endo-N) and comparison to an endo-N homolog in bacteriophage PK1E. J Bacteriol. 1993;175:4354–4363. doi: 10.1128/jb.175.14.4354-4363.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robertson E S, Aggison L A, Nicholson A W. Phosphorylation of elongation factor G and ribosomal protein S6 in bacteriophage T7-infected Escherichia coli. Mol Microbiol. 1994;11:1045–1057. doi: 10.1111/j.1365-2958.1994.tb00382.x. [DOI] [PubMed] [Google Scholar]
- 39.Rosa M D, Andrews N C. Phage T3 DNA contains an exact copy of the 23 base-pair phage T7 RNA polymerase promoter sequence. J Mol Biol. 1981;147:41–53. doi: 10.1016/0022-2836(81)90078-4. [DOI] [PubMed] [Google Scholar]
- 40.Saito H, Tabor S, Tamanoi F, Richardson C C. Nucleotide sequence of the primary origin of bacteriophage T7 DNA replication: relationship to adjacent genes and regulatory elements. Proc Natl Acad Sci USA. 1980;77:3917–3921. doi: 10.1073/pnas.77.7.3917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 42.Schmitt M P, Beck P J, Kearney C A, Spence J L, DiGiovanni D, Condreay J P, Molineux I J. Sequence of a conditionally essential region of bacteriophage T3, including the primary origin of DNA replication. J Mol Biol. 1987;193:479–495. doi: 10.1016/0022-2836(87)90261-0. [DOI] [PubMed] [Google Scholar]
- 43.Skurnik M. Lack of correlation between the presence of plasmids and fimbriae in Yersinia enterocolitica and Yersinia pseudotuberculosis. J Appl Bacteriol. 1984;56:355–363. doi: 10.1111/j.1365-2672.1984.tb01362.x. [DOI] [PubMed] [Google Scholar]
- 44.Skurnik M. Molecular genetics of Yersinia lipopolysaccharide. In: Goldberg J, editor. Genetics of bacterial polysaccharides. Boca Raton, Fla: CRC Press; 1999. pp. 23–51. [Google Scholar]
- 45.Spoerel N, Herrlich P, Bickle T A. A novel bacteriophage defence mechanism: the anti-restriction protein. Nature. 1979;278:30–34. doi: 10.1038/278030a0. [DOI] [PubMed] [Google Scholar]
- 46.Steven A C, Trus B L, Maizel J V, Unser M, Parry D A D, Wall J S, Hainfeld J F, Studier F W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 47.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 48.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Young R, Wang I-N, Roof W D. Phages will out: strategies of host cell lysis. Trends Microbiol. 2000;8:120–128. doi: 10.1016/s0966-842x(00)01705-4. [DOI] [PubMed] [Google Scholar]
- 50.Young R, Way J, Way S, Yin J, Syvänen M. Transposition mutagenesis of bacteriophage lambda: a new gene affecting cell lysis. J Mol Biol. 1979;132:307–322. doi: 10.1016/0022-2836(79)90262-6. [DOI] [PubMed] [Google Scholar]
- 51.Zagotta M T, Wilson D B. Oligomerization of the bacteriophage lambda S protein in the inner membrane of Escherichia coli. J Bacteriol. 1990;172:912–921. doi: 10.1128/jb.172.2.912-921.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]