Abstract
Neurospora crassa protein QDE-1, a member of the two-barrel polymerase superfamily, possesses both DNA- and RNA-dependent RNA polymerase (DdRP and RdRP) activities. The dual activities are essential for the production of double-stranded RNAs (dsRNAs), the precursors of small interfering RNAs (siRNAs) in N. crassa. Here, we report five complex structures of N-terminal truncated QDE-1 (QDE-1ΔN), representing four different reaction states: DNA/RNA-templated elongation, the de novo initiation of RNA synthesis, the first step of nucleotide condensation during de novo initiation and initial NTP loading. The template strand is aligned by a bridge-helix and double-psi beta-barrels 2 (DPBB2), the RNA product is held by DPBB1 and the slab domain. The DNA template unpairs with the RNA product at position –7, but the RNA template remains paired. The NTP analog coordinates with cations and is precisely positioned at the addition site by a rigid trigger loop and a proline-containing loop in the active center. The unique C-terminal tail from the QDE-1 dimer partner inserts into the substrate-binding cleft and plays regulatory roles in RNA synthesis. Collectively, this work elucidates the conserved mechanisms for DNA/RNA-dependent dual activities by QDE-1 and other two-barrel polymerase superfamily members.
INTRODUCTION
RNA interference (RNAi), triggered by double-stranded (dsRNA), is a highly conserved gene regulation mechanism in eukaryotes and plays important roles in various biological processes, including heterochromatin formation, the regulation of repeated sequences, transcriptional gene silencing and post-transcriptional gene silencing. RNAi functions through base-pairing of siRNAs with target mRNAs (1–3). In protozoa, fungi, plants and nematodes, siRNA biogenesis depends on multiple cellular proteins, including the RNA-dependent RNA polymerase (RdRP) which produces dsRNAs (4–9), and Dicer, an RNase III class ribonuclease that cleaves dsRNAs and generates 21–25 nt siRNAs (10).
QDE-1 of the filamentous fungus Neurospora crassa (4,11,12), the first discovered RdRP protein in the RNAi pathway, is key in transgene-induced gene silencing and small RNA (known as qiRNA) production (13,14). In addition to RdRP activity, QDE-1 also possesses strong DNA-dependent RNA polymerase (DdRP) activity that is critical for the in vivo production of DNA damage-induced aberrant RNAs (aRNAs) (14). Consistently, in vitro studies confirmed that QDE-1 can use either single-stranded DNA (ssDNA) as a template to produce aRNAs or aRNAs as a template to generate dsRNAs (14–16). The proteins homologous to QDE-1 participate in diverse gene silencing pathways. In Arabidopsis, RdRP 2 (RDR2) partners with RNA polymerase IV (Pol IV) to convert Pol IV transcripts into dsRNAs, which are cleaved by Dicer-like 3 (DCL3) protein into 24 nt siRNAs (17–19). RdRP 6 (RDR6) participates in trans-acting secondary siRNA production and RNAi response amplification (20). EGO-1 and two other RdRPs (RRF1 and RRF3) are involved in the production of secondary siRNAs and silencing signal amplification in Caenorhabditis elegans RNAi pathways (7,21–23). In the fission yeast Schizosaccharomyces pombe, RdRP 1 (RDR1) produces dsRNA from non-coding transcripts in centromeric repeats, triggering RNAi and RNA-mediated heterochromatin formation (24–26).
QDE-1 is a two-barrel polymerase superfamily member (27–29); as confirmed by the apo-form structure, the catalytic domain of QDE-1 consists of two characteristic double-psi beta-barrels (DPBBs) (30). The two-barrel polymerase superfamily also includes single-subunit Cellulophage baltica crAss-like phage 14:2 DdRP protein gp66 (31), archaeal replicative DNA polymerase PolD (32,33) and many multisubunit cellular DdRPs (34,35). Among these polymerases, Arabidopsis RDR6 has strong DdRP activities (36), and Saccharomyces cerevisiae RNA polymerase II (Pol II) possesses RdRP activity (37). These observations, combined with the dual DdRP and RdRP activities of QDE-1, suggested that the two-barrel polymerase superfamily intrinsically uses DNA and/or RNA as templates. However, due to the lack of high-resolution substrate-bound complex structures, the molecular basis of DNA/RNA template binding and RNA synthesis by the two-barrel polymerase superfamily is not fully understood.
Here, we report five structures of QDE-1ΔN (residues 377–1402) in complex with DNA/RNA-templated substrates, revealing the detailed molecular basis of the dual DdRP and RdRP activities of QDE-1. Both the DNA and RNA templates are bound by a bridge-helix (BH) and DPBB2, and form A-form-like duplexes with the RNA products that are recognized by DPBB1 and the slab domain. The DNA template unpairs with the RNA product at position –7, whereas the corresponding nucleotide of the RNA template forms regular Watson–Crick base pairing. Structural analysis suggests that QDE-1 follows a conserved cation-dependent mechanism in catalysis, and the incoming NTP is positioned at the activation site (A site) by the trigger loop (TL) and a proline-containing loop (Pro-Gate loop). QDE-1 contains a unique C-terminal tail (C-tail), which plays a regulatory role in RNA synthesis catalyzed by QDE-1. This work provides structural insights into the molecular mechanism of QDE-1 and related cellular RdRPs in RNAi, and expands our understanding of the two-barrel polymerase superfamily.
MATERIALS AND METHODS
Cloning and expression
cDNA of QDE-1 (XM_953954) was amplified from the plasmid pEM41-QDE-1 and inserted into a modified pFastBac vector under the BamHI site, expressing full-length QDE-1 with a His-GST (glutathione S-transferase) tag at the N-terminus. cDNAs of N- and/or C-terminal truncated QDE-1 protein were amplified by polymerase chain reaction (PCR), and plasmid pEM41-QDE-1 was used as a template. All QDE-1ΔN mutants were constructed by overlap PCR, using cDNA of wild-type (WT) QDE-1ΔN as a template. All target sequences were treated with EcoRI and HindIII, and inserted into the pET28-Sumo vector, expressing proteins with a His-Sumo tag at the N-termini. Sequences of all constructed QDE-1 plasmids were verified by DNA sequencing. The primers used in plasmid construction are listed in Supplementary Table S1.
To produce QDE-1ΔNΔC heterodimers, the cDNA of QDE-1ΔNm3 (in which the three catalytic residues D1007, D1009 and D1011 were substituted with an Ala residue) and QDE-1ΔNΔC-m3 (variant with both a C-terminal 30 residue deletion and a catalytic residue mutation) were first cloned into the pGEX-6P-1 vector to express inactive GST-tagged QDE-1 protomer. The cDNAs of the GST-tagged QDE-1ΔNm3 or QDE-1ΔNΔC30-m3 mutant and His-SUMO-tagged QDE-1ΔNΔC were then cloned into the pETDuet-1 vector.
Full-length QDE-1 protein was expressed in insect cells using a Bac-to-Bac baculovirus expression system. Briefly, the recombinant pFastBac plasmid was transformed into DH10Bac Escherichia coli competent cells to produce recombinant bacmid DNA. P1 and P2 viruses were produced in Sf9 cells according to the manufacturer's instructions. After growing to a density of ∼1.5 × 106 cells/ml in suspension culture at 27°C, 1 l of Sf9 cells were infected by 10 ml of P2 virus. Twenty hours after infection, the growth temperature was decreased to 20°C to produce more soluble protein. Cells were collected 80 h after infection by centrifugation for further purification. Escherichia coli BL21 (DE3) cells transfected with the above plasmids derived from the pET28-Sumo or the pETDuet-1 fusion vector were grown at 37 °C in LB medium containing antibiotic to an optical density of 0.6–0.8 at 600 nm. Protein expression was induced by the addition of 0.1 mM isopropyl β-d-1-thiogalactopyranoside at 16 °C. After overnight growth, the cells were collected by centrifugation at 5000 rpm for 15 min at 4 °C and stored at −80°C.
Protein purification and oligonucleotide preparation
The collected cells were resuspended in lysis buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl and 25 mM imidazole) and lysed with an ultrahigh-pressure homogenizer. Cellular debris was removed by centrifugation using 38 758 × g for 1 h at 4°C. The supernatant was loaded onto a HisTrap HP column (GE Healthcare), and the target protein was washed with lysis buffer and eluted with a linear gradient of elution buffer (20 mM Tris–HCl pH 8.0, 500 mM NaCl and 500 mM imidazole). The proteins were treated with Ulp1 to remove the N-terminal His-SUMO tag and dialyzed against buffer consisting of 20 mM Tris–HCl pH 8.0 and 500 mM NaCl. The samples were loaded onto the HisTrap HP column again, and the flow-through sample was collected and diluted to 100 mM NaCl. The diluted sample was applied to a HisTrap Heparin HP column (GE Healthcare) pre-equilibrated with buffer containing 20 mM Tris–HCl pH 8.0, 100 mM NaCl and 2 mM dithiothreitol (DTT). The target protein was eluted with a linear gradient of elution buffer (20 mM Tris–HCl pH 8.0, 1 M NaCl and 2 mM DTT), pooled and applied to a HiLoad 16/600 Superdex 200 size-exclusion column (GE Healthcare) pre-equilibrated with SEC buffer (10 mM Tris–HCl pH 8.0, 100 mM NaCl and 2 mM DTT). Peak fractions were pooled and concentrated. The purity of the protein was analyzed using 12% sodium dodecylsulfate–polyacryamide gel electrophoresis (SDS–PAGE), and the concentration was measured using a UV-spectrophotometer at 280 nm. Purified proteins were concentrated to ∼15 mg/ml for crystallization or flash-frozen in liquid nitrogen before storage at –80 °C.
For purification of QDE-1ΔNΔC heterodimers, the flow-through collected from the second HisTrap HP column was pooled and applied to the a GSTrap HP column (GE Healthcare) pre-equilibrated in binding buffer (20 mM Tris–HCl pH 8.0, 200 mM NaCl, 2 mM DTT). The target protein was washed with binding buffer and eluted with buffer containing 20 mM Tris–HCl pH 8.0, 200 mM NaCl, 2 mM DTT and 10 mM glutathione. The GST tag was removed by PreScission protease. The collected sample was purified by Heparin and Superdex 200 columns as described above. Full-length QDE-1 was purified using a similar procedure. To inhibit degradation, protease inhibitors were included in the lysis buffer, and all purification buffers were supplemented with 5% glycerol.
The sequences of DNAs or RNAs used in this study are listed in Supplementary Table S1. All unmodified DNA oligonucleotides were synthesized and purified by PAGE or high-performance liquid chromatography (HPLC) by Sangon Biotech. All RNA oligonucleotides for crystallization were synthesized in the laboratory, deprotected and purified by denaturing PAGE. All 5′-FAM-labeled RNA oligonucleotides and 3′-OH-phosphorylated DNA oligonucleotides were purchased from Takara. The concentrations of the DNAs and RNAs were measured by UV spectrometry at 260 nm. DNA/RNA hybrids or RNA/RNA duplexes used for crystallization (template and primer molar ratio, 1:1) and primer-extension (template and primer molar ratio, 1.5:1) assays were annealed in 50 mM NaCl, 10 mM Tris–HCl pH 7.5, and 10 mM MgCl2 buffer. Annealing was performed by heating the mixture for 2 min at 95°C, followed by slow cooling to room temperature and incubation on ice.
In vitro primer-extension assays
Primer-extension assays were carried out using annealed DNA/RNA or RNA/RNA duplexes. Duplexes labeled with 5′-FAM (0.2 μM) were incubated with 0.025 μM QDE-1ΔN (either WT or mutated) and 0.8 U/μl RNase inhibitor at 20°C for 20 min in reaction buffer (10 mM Tris–HCl pH 8.0, 70 mM NaCl, 2 mM DTT and 5 mM MgCl2). The reactions were initialized by adding 2 μM UTP at 25°C and quenched by adding 2× loading buffer (20 mM EDTA pH 8.0, 0.05% bromophenol blue, 0.05% xylene blue and 90% formamide) at various time points. The reaction products were separated in 20% 19:1 acrylamide:bis-acrylamide, 7 M urea and 1× TBE gels (1 pmol of RNA primer per lane). The gel was visualized using a Typhoon FLA 9000 (GE Healthcare), and the intensities of the substrate and product bands were quantified by ImageQuantTL. Data were then fitted to a one-phase exponential association Y = Ymax[1–e(–Kobst)] using non-linear regression in GraphPad Prism 7. The observed rate constant (Kobs) and maximum polymerization yield (Ymax) were determined from the regression curve.
De novo RNA synthesis assays
The de novo RNA synthesis reactions were carried out at 25°C for 60 min using 0.1 μM QDE-1ΔN (WT or C-tail truncated mutants), 0.2 μM 15 nt ssDNA (Supplementary Table S1), 20 μM each of ATP, GTP and 3′-dCTP, and 0.4 μCi of [γ-32P]ATP in reaction buffer. The reactions were quenched by adding 2× loading buffer. Samples were heated at 95°C for 3 min and chilled on ice, and the RNA products were resolved on 18% polyacrylamide–7 M urea gels. The gels were exposed to a phosphor screen overnight and scanned on a Typhoon FLA 9000.
Crystallization and data collection
For crystallization, QDE-1ΔN protein was mixed with the DNA or RNA template strand or the template/primer duplex (Supplementary Table S2) at a molar ratio of 1:1.2 in buffer containing 5 mM MgCl2. NTPs and non-hydrolyzable nucleotides, such as 3′-dGTP or AMPNPP, were then added to form quaternary complexes. The mixture was incubated at 20°C for 30 min and then diluted to a protein concentration of ∼6 mg/ml. All crystals were grown by the hanging-drop vapor diffusion method. The optimized reservoir buffer contained 6–10% polyethylene glycol (PEG) 8000, 0.2 M KCl, 10 mM magnesium acetate, 50 mM sodium cacodylate pH 6.3–6.5 for the DdRP-no primer crystal. For all other crystals, the optimized reservoir buffer contained 6% PEG 4000, 0.2 M KCl, 10 mM CaCl2, 50 mM sodium cacodylate pH 6.3. Crystals were cryo-protected by mother liquid supplemented with 25% (v/v) glycerol and flash-frozen in liquid nitrogen. Diffraction data were collected on beamlines BL17U1 and BL19U1 at Shanghai Synchrotron Radiation Facility (SSRF) at cryogenic temperatures maintained with a cryogenic system. Data processing was carried out using HKL2000 or HKL3000 programs (38,39). The data collection statistics are summarized in Supplementary Table S3.
Structure determination and refinement
The phase of the QDE-1ΔN complex structures was determined by molecular replacement using the phaser program of the CCP4 suite (40,41); the apo-form QDE-1ΔN structure (PDB code: 2J7N) was used as the search model (30). Iterative cycles of crystallographic refinement were performed using PHENIX and COOT (42,43). The 2Fo–Fc and Fo–Fc electron density maps were calculated by standard methods and used as guides for the building of nucleic acids. The structural refinement statistics are summarized in Supplementary Table S3. All structural figures were prepared with PyMOL (http://www.pymol.org/).
Quantification and statistical analysis
The primer-extension experiments were repeated three times, and representative results are shown. The double helical parameters of the template–product captured by QDE-1ΔN were calculated with the w3DNA 2.0 server (http://web.x3dna.org) (Supplementary Table S4).
Size-exclusion chromatography coupled with the multiangle light scattering (SEC-MALS) assay
The absolute molar mass of the target protein was determined by SEC-MALS. The experiments were performed by an Agilent system connected to a Wyatt DAWN HELEOS-II multiangle light scattering instrument and a Wyatt Optilab T-rEX differential refractometer. A Superdex 200 Increase 10/300 GL column (GE Healthcare) was used with a 100 μl sample loop at a flow rate of 0.4 ml/min in the running buffer containing 10 mM Tris pH 8.0, 200 mM NaCl and 2 mM DTT. The purified and filtrated samples were injected at a concentration of 1 mg/ml in running buffer. The molar mass was calculated by light scattering intensity and differential refractive index using Wyatt ASTRA 6 software.
RESULTS
Overall structures of QDE-1ΔN complexes
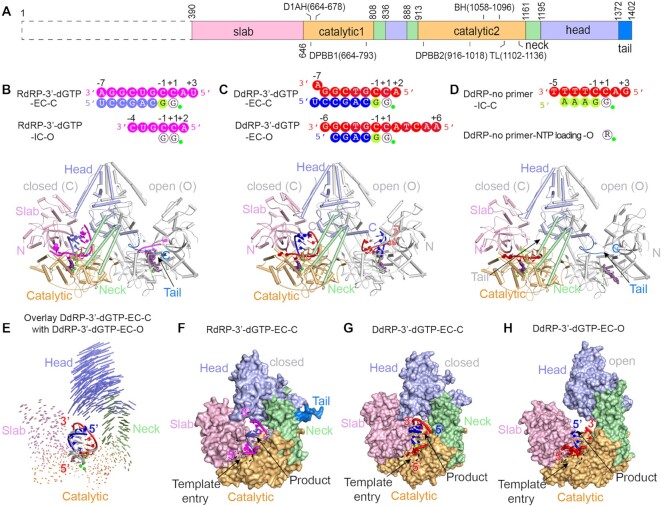
QDE-1 contains five interlaced domains: slab, catalytic, neck, head and C-tail domains (Figure 1A). To unravel the molecular basis of the dual DdRP and RdRP activities of QDE-1, we purified full-length QDE-1 and one QDE-1 variant with the N-terminus removed (QDE-1ΔN, amino acids 377–1402) (Supplementary Figure S1A). Although both proteins are active in vitro, the RNA polymerization activity of full-length QDE-1 is weaker than that of QDE-1ΔN (Supplementary Figure S1B). The reason for the lower catalytic activity of full-length QDE-1 is not clear at present. Unlike QDE-1ΔN, full-length QDE-1 is unstable during purification. Even when protected by protease inhibitors, degradation of full-length QDE-1 occurred (Supplementary Figure S1A). No crystal of full-length QDE-1 was obtained, but we successfully obtained and solved five complex structures of QDE-1ΔN (Supplementary Table S2). The crystals of RdRP-3′-dGTP and DdRP-3′-dGTP were grown in the presence of QDE-1ΔN protein with the 14 nt RNA or DNA template strand and a 6 nt primer in the presence of 3′-dGTP (opposite template CC). Both RdRP-3′-dGTP and DdRP-3′-dGTP structures (Figure 1B, C) had one 3′-dGMP incorporated at the primer 3′ end, indicating that QDE-1ΔN protein is catalytically active. The crystals of RdRP-AMPNPP and DdRP-AMPNPP structures were grown in the presence of QDE-1ΔN, 14 nt RNA or DNA template, 7 nt primer and non-hydrolyzable AMPNPP. To obtain the crystal structures of the complex in de novo initiation, the crystals of the DdRP-no primer (Figure 1D) were grown with QDE-1ΔN with a 12 nt DNA template in the presence of ATP and 3′-dGTP, which paired with TTTT and CC in the template, respectively. In addition to crystallization, we demonstrated the de novo RNA synthesis activity of QDE-1ΔN in vitro using an identical template and NTPs (Supplementary Figure S1C).
Figure 1.
Co-crystal structures of QDE-1ΔN in complex with substrates. (A) Domain architecture of QDE-1, N-terminus (black dashed box), slab (light pink), catalytic (light orange), neck (pale green), head (light blue) and C-tail (aquamarine) domains. (B) Co-crystal structure of QDE-1ΔN with a 14 nt RNA template, 6 nt RNA primer, 3′-dGTP and cations, designated QDE-1ΔN RdRP-3′-dGTP. (C) Co-crystal structure of QDE-1ΔN with a 14 nt DNA template, 6 nt RNA primer, 3′-dGTP and cations, designated QDE-1ΔN DdRP-3′-dGTP. (D) Co-crystal structure of QDE-1ΔN with a 12 nt DNA template, ATP, 3′-dGTP and cations, designated QDE-1ΔN DdRP-no primer. (E) Structural comparison between the open (O) and closed (C) molecules of the QDE-1ΔN dimer in the DdRP-3′-dGTP co-crystal structure. The DNA–RNA hybrid bound by the closed QDE-1ΔN molecule is colored in red and blue, whereas it is colored in gray in the open QDE-1ΔN molecule. Vector length correlates with the domain translation scale. (F) The closed QDE-1ΔN molecule and the bound RNA–RNA duplex in the RdRP-3′-dGTP co-crystal structure. The QDE-1ΔN molecule is represented as a surface, and the RNA–RNA duplex is represented as a cartoon. (G) The closed QDE-1ΔN molecule and the bound DNA–RNA hybrid in the DdRP-3′-dGTP co-crystal structure. The QDE-1ΔN molecule is represented as a surface, and the DNA–RNA hybrid is represented as a cartoon. (H) The open QDE-1ΔN molecule and the bound DNA–RNA hybrid in the DdRP-3′-dGTP co-crystal structure. The QDE-1ΔN molecule is represented as a surface, and the DNA–RNA hybrid is represented as a cartoon. In (B–D), sequences of the visible template strand, product strand and NTP analog are illustrated at the top of the structure. The nucleotide at the addition site is denoted +1, and the upstream and downstream nucleotides are labeled with negative and positive numbers, respectively. The RNA and DNA template strands are colored in magenta and red, respectively; the primer strands are colored in slate and blue in the RdRP and DdRP complex structures, respectively. The newly incorporated nucleotides and the cations are colored in lemon and green, respectively. The NTP analog is violet–purple. These colors were used throughout the paper unless specifically annotated.
All complex structures were solved by the molecular replacement method using the apo-form QDE-1ΔN structure (30) as a search model and refined at high resolution, from 2.05 to 2.70 Å (Supplementary Table S3). The root mean square deviation (RMSD) values between the apo- and complex structures were <0.9 Å (Supplementary Figure S2A–E), suggesting that complex formation did not cause an overall conformational change in the QDE-1ΔN protein. QDE-1ΔN forms an asymmetric homodimer in both apo- and complexed structures. Similar to the apo-form structure, the overall conformation of one QDE-1ΔN molecule is more open than that of the partner molecule. Superposition showed that the structural difference between the monomers is mainly caused by the swing of the head domain, whereas the catalytic domains have similar conformations (Figure 1E). The complex structures were originally designed to capture the elongation state (RdRP-3′-dGTP, DdRP-3′-dGTP, RdRP-AMPNPP and DdRP-AMPNPP) and the de novo initiation state of RNA synthesis (DdRP-no primer). Unlike the other structures, the RdRP-3′-dGTP and DdRP-no primer structures belong to the P1 space group (Supplementary Table S3). Due to their different packing in the crystal lattices, they captured two additional reaction states (the first step of nucleotide condensation during de novo initiation and initial NTP loading).
Elongation state of QDE-1ΔN in RdRP and DdRP complexes (EC)
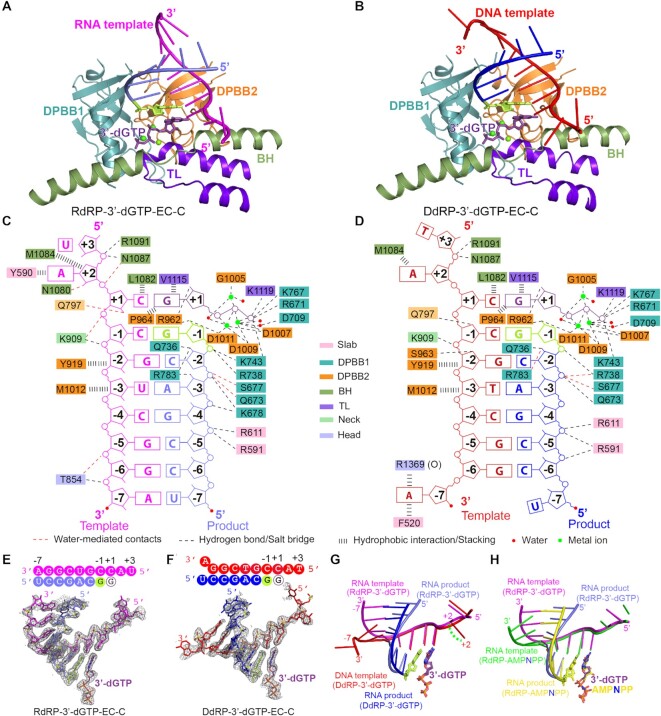
The overall conformations of RdRP-3′-dGTP, DdRP-3′-dGTP, RdRP-AMPNPP and DdRP-AMPNPP complexes are exactly the same in captured elongation states (EC). Compared with the two latter structures, RdRP-3′-dGTP and DdRP-3′-dGTP had higher resolution (Figure 1B, C; Supplementary Table S3); therefore, they were selected and presented in detail. The closed QDE-1ΔN captured the reaction in the EC state in both RdRP-3′-dGTP and DdRP-3′-dGTP complexes (Figure 1F, G). The EC state was also captured by the open QDE-1ΔN in the DdRP-3′-dGTP structure (Figure 1H). In all the EC state structures, the product strand forms at least five base pairs with the template strand (Figure 1B, C; Supplementary Figure S3A–E). The incoming 3′-dGTP or AMPNPP resides at the A site and pairs with the nucleotide from the template strand.
The template–product duplexes were captured in the cleft mainly formed by two barrels (DPBB1 and DPBB2) in the catalytic domain. The RNA and DNA templates enter the active center in a similar manner (Figure 1F–H) and pair with the product strands, following the path along the surface of DPBB1 (Figure 2A, B). The 5′ end of the product strand leaves the cleft from the distal end. For the EC state captured by the RdRP-3′-dGTP structure (Figure 1F; Supplementary Figure S2F), the 3′ end of the template pairs with the product (Figure 2A, C), suggesting that they probably leave the cleft together as a duplex. In the closed QDE-1ΔN of the DdRP-3′-dGTP structure (Figure 1G), the template 3′ end nucleotide is separated from the product strand at position –7 (Figure 2B, D) and probably exits through a narrow channel between the head and the slab domains (Supplementary Figure S2G). The conformations of the captured RNA–RNA duplexes or DNA–RNA hybrids are well supported by their composite omit electron density maps (Figure 2E, F; Supplementary Figure S3A–E).
Figure 2.
Structures of the catalytic center of QDE-1ΔN-bound RdRP and DdRP substrates in elongation states. (A and B) The core architecture of the catalytic center of QDE-1ΔN-bound RdRP and DdRP substrates in the RdRP-3′-dGTP-EC-C and DdRP-3′-dGTP-EC-C structures, respectively. The core architecture consists of four subdomains: DPBB1 (light teal), DPBB2 (dark orange), BH (sage green) and TL (purple–blue). (C and D) Schematic representation of nucleic acid and cation recognition by the closed QDE-1ΔN molecule in the RdRP-3′-dGTP and DdRP-3′-dGTP structures, respectively. (E and F) The slow annealing composite omit electron density maps of the RNA–RNA duplexes or DNA–RNA hybrids captured in the QDE-1ΔN complex structures. The maps are all contoured at the 1.0 σ level. Sequences of the DNA–RNA hybrid and RNA–RNA duplex are provided at the top of the panels. (G) Superposition of the DNA–RNA hybrid and RNA–RNA duplex captured by the DdRP-3′-dGTP-EC-C and RdRP-3′-dGTP-EC-C structures. (H) Superposition of RNA–RNA duplexes captured by the RdRP-3′-dGTP-EC-C (magenta and light blue) and RdRP-AMPNPP-EC-C (green and yellow) structures.
Although both RNA–RNA duplexes in the RdRP complexes and DNA–RNA hybrids in the DdRP complexes adopt an A-form-like conformation, superimposition suggested that the duplexes and hybrids aligned well from +1 to –4, but diverged from –5 to –7 (Figure 2G). In particular, the –7 nucleotide of the DdRP template unpaired with the product and turned in the opposite direction (Figure 2G). Compared with other duplexes, the RNA–RNA duplex in the RdRP-3′-dGTP structure is closer to the ideal A-form conformation (Figure 2H; Supplementary Figure S3F, G). Analysis of the helical parameters suggested that the base–base distance of the RNA–RNA duplex is shorter than that of the DNA–RNA hybrid (Supplementary Table S4), allowing the substrate-binding cleft of QDE-1 to accommodate eight base pairs of the RNA–RNA duplex or seven base pairs of the DNA–RNA hybrid, including the base pair between the incoming NTP and the template.
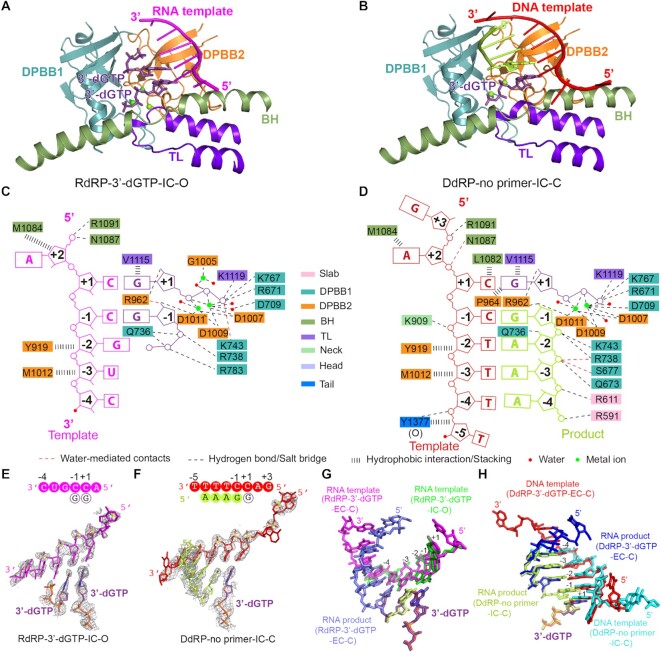
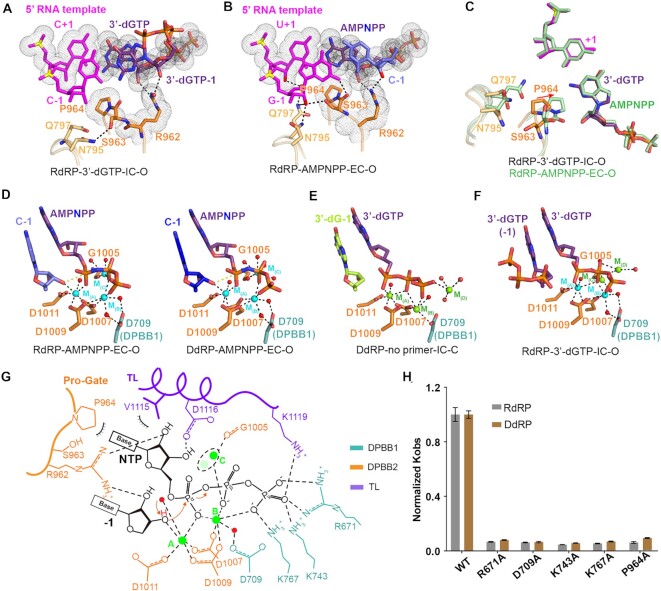
De novo initiation states of QDE-1ΔN in RdRP and DdRP complexes (IC)
Although the closed molecule RdRP-3′-dGTP captured a complex in the elongation state, unexpectedly the open molecule RdRP-3′-dGTP captured a complex in the early de novo initiation state, namely the first step of nucleotide condensation (Figure 3A, C, E). Both NTPs were identified as 3′-dGTP, pairing with C nucleotides at positions +1 and –1 of the RNA template, respectively. The de novo initiation state was also captured by the closed QDE-1ΔN molecule of the DdRP-no primer complex (Figure 3B, D, F). In the structure, the DNA template forms a hybrid with one 4 nt RNA product (5′-AAAdG-3′), which was synthesized during the crystallization process. The three A nucleotides at the 5′ end of the RNA product pair with the T nucleotides at positions –4 to –2 of the DNA template, and the 3′-dG pairs with the C nucleotide at position –1 of the template. Similar to the RdRP-3′-dGTP structure, one individual 3′-dGTP resides at the A site and pairs with the C nucleotide at position +1 of the template. Both the RNA and DNA templates in the IC state are well superimposed with templates in the EC state from positions +1 to –4 (Figure 3G, H), suggesting that either DNA or RNA templates adopt similar A-form conformations during the initial RNA synthesis process.
Figure 3.
Structures of the catalytic center of QDE-1ΔN-bound RdRP and DdRP substrates in de novo initiation states. (A and B) The core architecture of the catalytic center of QDE-1ΔN-bound RNA template paired with two nucleotides (3′-dGTP) for the first step of condensation and DNA template paired with a 4 nt product (5′-AAAdG-3′) and 3′-dGTP in the RdRP-3′-dGTP-IC-O and DdRP-no primer-IC-C structures, respectively. Colors are the same as in Figure 2A and B. (C and D) Schematic representation of nucleic acid and cation recognition by the open QDE-1ΔN molecule in the RdRP-3′-dGTP and the closed QDE-1ΔN molecule in the DdRP-no primer structures, respectively. (E) The slow annealing composite omit electron density maps of RNA-3′-dGTP captured in the open QDE-1ΔN molecule in the RdRP-3′-dGTP structure. The map is contoured at the 1.0 σ level. The sequence is provided at the top of the panel. (F) The slow annealing composite omit electron density maps of the DNA–RNA hybrid captured in the closed QDE-1ΔN molecule in the DdRP-no primer structure. The map is contoured at the 1.0 σ level. The sequence is provided at the top of the panel. (G) Superposition of RNA-3′-dGTP (green and violet–purple) and RNA–RNA (magenta and light blue) duplexes captured by the open and closed QDE-1ΔN molecules in the RdRP-3′-dGTP structure. (H) Superposition of DNA–RNA hybrids captured by the closed QDE-1ΔN molecules in the DdRP-3′-dGTP (red and blue) and DdRP-no primer (cyan and lemon) structures.
The template strand is mainly aligned by BH and DPBB2 subdomains
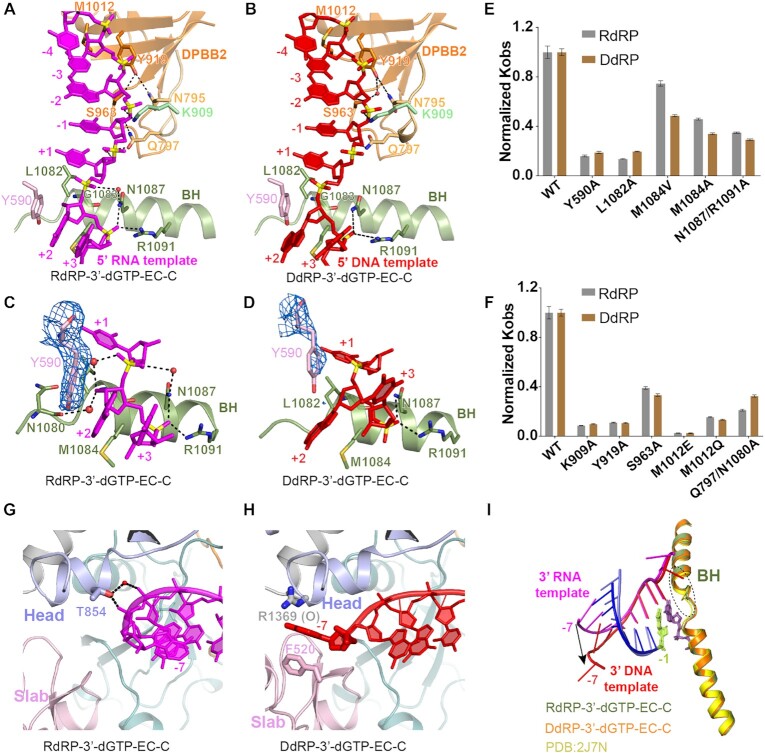
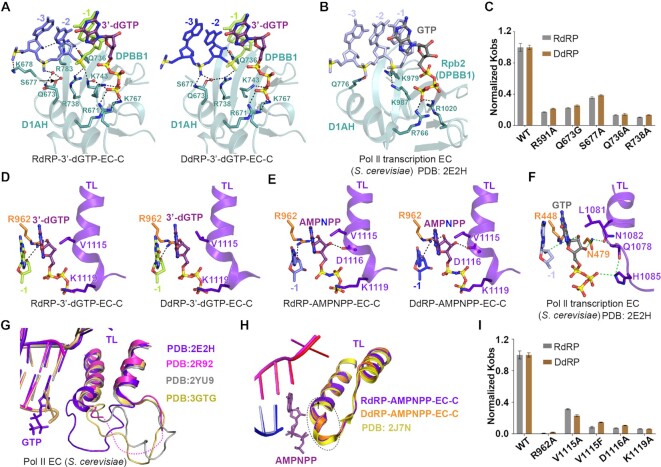
Similar to the BH in Pol II (Supplementary Figure S4A), the BH in QDE-1 also recognizes nucleotides near the entrance site (Figure 4A, B). N1087 and R1091 from the BH form H-bonds with the phosphate backbone of the +2 nucleotide, which are conserved in both RdRP and DdRP complexes. Also, the base of the +2 nucleotide is stabilized by π–π stacking with Y590 of the slab domain in the RdRP-3′-dGTP complex (Figure 4C). Compared with the RNA template, the nucleobase of the DNA template +2 nucleotide is rotated ∼80° and forms hydrophobic interactions with the side chain of M1084 (Figures 2G and 4D; Supplementary Figure S5A, B). Instead of 3′-endo, the sugar pucker of the template +2 nucleotide adopts a 2′-endo conformation in the DdRP-3′-dGTP complex (Supplementary Figure S5B). In addition, the relatively small residues (G1083 of QDE-1 and A832 of Pol II) allow the template to easily enter the active center alongside the BH (Figure 4A, B; Supplementary Figure S5C). Once the template entered the active center, L1082 of QDE-1 or T831 of Pol II stabilized the template via van der Waals interactions with the +1 nucleobase. M1084 mutation caused a more significant reduction in the DdRP activity of QDE-1ΔN than in the RdRP activity, and single or double Ala substitutions of Y590, L1082, N1087 and R1091 reduced both the DdRP and RdRP activities of QDE-1ΔN (Figure 4E; Supplementary Figure S5G).
Figure 4.
The detailed structures of template recognition by the DPBB2 subdomain and BH in DdRP and RdRP elongation states. (A and B) The RNA and DNA template strands are aligned by BH and DPBB2 subdomains in the closed QDE-1ΔN molecules in the RdRP-3′-dGTP and DdRP-3′-dGTP structures, respectively. (C and D) Interactions between the template entrance site nucleotides and the BH subdomain of QDE-1ΔN in the RdRP-3′-dGTP-EC-C and DdRP-3′-dGTP-EC-C structures. (E and F) The normalized Kobs showing the impacts of mutations at the template entrance site and the template-binding cleft of QDE-1ΔN. The data represent the mean of three independent experiments, with SD values indicated by error bars. (G and H) Interactions between QDE-1ΔN and the 3′-end nucleotides of the template in the RdRP-3′-dGTP-EC-C structure and the DdRP-3′-dGTP-EC-C structure. (I) Superposition showing the conformational change between the 3′-end nucleotides of the RNA template and DNA template and the rigid BH subdomain of QDE-1.
In addition to recognition by BH, nucleotides –1 to –4 of the DNA and RNA templates are also extensively contacted by the DPBB2 subdomain of QDE-1 from the minor groove side (Figure 4A, B). K909 of the neck domain in the closed QDE-1ΔN interacts with the phosphate backbone of the template between –1 and –2. Residues Y919 and M1012 of the DPBB2 subdomain contact the ribose groups of the template between –3 and –4 via van der Waals interactions. The mutations of K909, Y919 and M1012 dramatically reduced both RdRP and DdRP activities (Figure 4F; Supplementary Figure S5H). Interestingly, there was little recognition of ribose 2′-OH groups in the RdRP complex, including Q797, which forms weak or water-mediated H-bonds with the 2′-OH group of ribose at the +1 and –1 positions of the RNA template (Figures 2C and 4A; Supplementary Figure S4F, G), consistent with the observations that the Q797A-containing mutation caused a more significant reduction in the RdRP activity than in the DdRP activity (Figure 4F; Supplementary Figure S5H). We note that the conformations of RNA and DNA templates differ from –5 to –7 (Figures 2G and 4G–I). For the RNA template, the phosphate backbone between positions –5 and –7 forms H-bonds with T854 of the head domain directly or mediated by a water molecule (Figure 4G). As shown before, unlike the RNA templates that keep pairing with the RNA product at position –7, the nucleobase at position –7 of the DNA template flips out and stacks with the side chains of F520 from the slab domain and of R1369 from the head domain of the open molecule (Figure 4H).
The RNA product strand is mainly bound by DPBB1 and the slab domain
Similar to the template strands, we also observed some conformational differences in the nucleotides at positions –7 to –5 of the RNA products, but the conformation of nucleotides –4 to –1 is well conserved and plays important roles in interacting with QDE-1 (Figures 2G–H and 3H). The DPBB1 and one small α-helix prior to DPBB1 (residues 664–678 in QDE-1), termed DPBB1 auxiliary helix (D1AH), participate in product strand binding in QDE-1ΔN elongation complexes (Figure 5A). Q736 of DPBB1 forms an H-bond with the 2′-OH group of the –2 nucleotide. R671 of D1AH and R738, K743 and K767 of DPBB1 interact with the –1 and –2 nucleotides and 3′-dGTP phosphate groups, respectively. Q673 and S677 of D1AH form either direct or water-mediated H-bonds with the –2 nucleotide (Figure 5A). The Q673 residue is unique to QDE-1 (Supplementary Figure S6A). R738 of QDE-1 is conserved in many homologous proteins involved in RNAi, but the corresponding residue is substituted by Ala in Pol II (Supplementary Figure S6A). Other residues involved in the binding product strand are conserved and form similar interactions in the Pol II EC structure (Figure 5B). The less conserved residues R591 and R611 of the slab domain form H-bonds with the phosphate backbones of the –4 to –6 nucleotides of the product strand (Supplementary Figure S6B, C). QDE-1ΔN-derived mutagenesis and in vitro catalytic assays showed that R591, Q673, S677, Q736 and R738 are all important for the QDE-1ΔN activity (Figure 5C; Supplementary Figure S6D).
Figure 5.
Detailed structures of the NTP analog and product strand recognition by the DPBB1 and TL subdomains. (A) Interactions between the RNA product and the DPBB1 and D1AH subdomains of QDE-1 in the RdRP-3′-dGTP-EC-C structure (left) and the DdRP-3′-dGTP-EC-C structure (right). (B) Interactions between the RNA product and the DPBB1 and D1AH subdomains in the S. cerevisiae Pol II transcription EC structure. (C) The normalized Kobs showing the impacts of mutations at the DPBB1, D1AH and slab subdomains of QDE-1ΔN. The data represent the mean ± SD of three independent experiments. (D) Interactions between NTP and the TL subdomain of QDE-1ΔN in the RdRP-3′-dGTP-EC-C structure (left) and the DdRP-3′-dGTP-EC-C structure (right). (E) Interactions between the NTP analog and the TL subdomain of QDE-1ΔN in the RdRP-AMPNPP-EC-C structure (left) and the DdRP-AMPNPP-EC-C structure (right). (F) Interactions between the NTP analog and the Pol II TL subdomain in the S. cerevisiae Pol II transcription EC structure. (G) Superposition of four S. cerevisiae Pol II EC structures showing the multiple conformations of the TL subdomain of Pol II. (H) Superposition of apo-QDE-1ΔN and two QDE-1ΔN complex structures showing the relatively rigid conformation of the TL subdomain of QDE-1. (I) The normalized Kobs showing the impacts of mutations at the TL subdomain of QDE-1ΔN. The data represent the mean ± SD of three independent experiments.
Recognition of the NTP analog at the A site by the TL subdomain
In addition to the BH subdomain, QDE-1 also shares one TL subdomain (residues 1102–1136) with Pol II (Figure 5D–F; Supplementary Figure S4A). In the complex structures, V1115 and K1119 of the QDE-1 TL subdomain interact with NTP analogs at the A site (Figure 5D, E). Residue V1115 forms hydrophobic interactions with the nucleobase, and K1119 forms H-bonds with the γ-phosphate group of NTP analogs. The 3′-OH group is missing in the 3′-dGTP utilized in the RdRP-3′-dGTP, DdRP-3′-dGTP and DdRP-no primer structures, whereas it is present in the AMPNPP captured in the RdRP-AMPNPP and DdRP-AMPNPP structures. The side chain of D1116 forms direct H-bonds with the 3′-OH group of AMPNPP at the A site (Figure 5E). Residues V1115 and K1119 of QDE-1 correspond to L1081 and H1085 of Pol II (Figure 5D–F; Supplementary Figure S6G), respectively; they form similar interactions with NTP in the QDE-1 and Pol II structures. Residue D1116 is highly conserved in QDE-1 and many homologous proteins, but is replaced by N1082 in Pol II (Supplementary Figure S6G). Upon NTP binding, the TL subdomain of Pol II undergoes obvious conformational changes (Figure 5G), but the TL subdomain of QDE-1 bound to NTP has no obvious change in comparison with the apo-form QDE-1ΔN structure (Figure 5H). QDE-1ΔN-derived mutagenesis and in vitro polymerization assays confirmed the functional importance of NTP-binding residues (Figure 5I; Supplementary Figure S6E).
Minor groove recognition of NTP at the A site by the Pro-Gate loop
The open QDE-1ΔN molecule in the RdRP-3′-dGTP structure captured the reaction in the early initiation state, and the two individual 3′-dGTPs formed base pairs with C nucleotides at positions +1 and –1 of the template (Figure 6A). It was noted that a steric loop (residues 962–964), composed of a conserved R–X–P (where X is variable) motif from the DPBB2 subdomain (Supplementary Figure S7A), directly interacts with the two 3′-dGTPs at the active center (Figure 6A; Supplementary Figure S7B). The middle residue, X, and the last residue, proline, are linked by a characteristic cis-peptide bond. The highly conserved residue R962 forms H-bonds with the 2′-OH groups of the two 3′-dGTPs, and the side chain of P964 contacts the minor groove of 3′-dGTP at the A site (Figure 6A). Similar recognition was also observed in the elongation state structure, such as RdRP-AMPNPP-EC-O (Figure 6B), but the distance between the side chain of P964 and the minor groove of AMPNPP at the A site was much less. Superposition of the above QDE-1ΔN structures revealed that P964 shifted ∼2 Å from the initiation state to the elongation state (Figure 6C). In addition, the side chain of S963 points away from the –1 site base pair (C-1:3′-dGTP) in the de novo initiation state of the RdRP-3′-dGTP structure (Figure 6A), whereas the side chain of S963 in the EC state forms one stable H-bond with the 2′-OH group of the template –1 site nucleotide that interacts with N795 and Q797 (Figure 6B), allowing P964 to move closer to the minor groove of NTP at the A site (Figure 6C). As defined in the Pol II structure (Supplementary Figure S7C) (44,45), the steric R–X–P loop from DPBB2 is also termed the Pro-Gate loop in QDE-1. Ala substitution of P964 dramatically reduced the catalytic efficiency of QDE-1ΔN (Figure 6H; Supplementary Figure S6F), indicating the functional importance of the Pro-Gate loop.
Figure 6.
The recognition of NTP at the A site by the Pro-Gate loop and multiple cation binding in the catalytic center of QDE-1ΔN. (A) Stick–dot–dash representation showing the conformation, relative orientation and interactions between 3′-dGTP and the Pro-Gate loop in the initiation state structure. (B) Stick–dot representation showing the shape-complementarity of the Pro-Gate loop and AMPNPP in the elongation state structure. (C) Superposition showing the conformational changes of the Pro-Gate loop during the initiation to elongation state transition. The elongation state complex is colored pale green. (D) Conformation of the catalytic residues and their coordination with cations (Mg2+ in green, Ca2+ in cyan) at the active sites of the RdRP-AMPNPP-EC-C and DdRP-AMPNPP-EC-C structures, representing the elongation state of the reaction. (E) Cation coordination at the active sites of the closed QDE-1ΔN molecule in the DdRP-no primer structure, representing the de novo initiation state of the reaction. (F) Cation (Mg2+ in green, Ca2+ in cyan) coordination at the active sites of the open QDE-1ΔN molecule in the RdRP-3′-dGTP structure, representing the de novo initiation state of the reaction. (G) Summarized catalytic site assembly of QDE-1ΔN and the proposed catalytic mechanism for QDE-1. (H) The normalized Kobs showing the impacts of mutations of the NTP-interacting residues of QDE-1ΔN. The data represent the mean ± SD of three independent experiments.
Multiple metal ions in the catalytic center
All the crystals of QDE-1ΔN elongation complexes in this study were grown in the buffer containing Mg2+ and Ca2+ cations. In both the RdRP-AMPNPP and DdRP-AMPNPP structures, two cations, designated cation A and cation B, coordinate with four conserved aspartic acids. Cation A coordinates with D1007, D1009, D1011, the 3′-OH group at the growing end of the product strand, AMPNPP α-phosphate and one water molecule, and cation B coordinates with D1007, D1009, two non-bridging phosphate oxygens of AMPNPP and two water molecules (Figure 6D; Supplementary Figure S7B). D709 of DPBB1 forms a water-mediated coordination with cation B (Figure 6D). It is likely that due to the absence of the OH group at the 3′ position of 3′-dG or 3′-dGTP, the coordination of cation A and cation B in the DdRP-3′-dGTP and RdRP-3′-dGTP structures (Figure 6E, F) is slightly different from that in the RdRP-AMPNPP and DdRP-AMPNPP structures; in the DdRP-3′-dGTP and RdRP-3′-dGTP structures, both cation A and cation B coordinate directly with the β-phosphate groups. D709, D1007, D1009 and D1011 are highly conserved (Supplementary Figures S6A and S7A). QDE-1ΔN-derived mutagenesis and in vitro catalytic assays supported the functional importance of the conserved aspartate triad. The catalytic activity of the QDE-1ΔND709A mutant was much weaker than that of WT QDE-1ΔN, and no detectable activity could be observed for the QDE-1ΔND1007A, QDE-1ΔND1009A or QDE-1ΔND1011A mutants (Figure 6H; Supplementary Figures S6F and S7I).
In addition to cations A and B, most QDE-1ΔN complex structures in this study captured a third cation (designated cation C), coordinating with the main-chain carboxyl oxygen atom of G1005 and interacting with the non-bridging oxygen of β-phosphate either directly or mediated by water molecules (Figure 6D, F; Supplementary Figure S7J). Interestingly, in the closed DdRP-no primer-IC structure and the open RdRP-3′-dGTP-IC structure, one additional cation (designated cation D) was observed, interacting with the non-bridging oxygen of γ-phosphate (Figure 6E), the bridging oxygen between β- and γ-phosphates and the non-bridging oxygen of β-phosphate (Figure 6F), respectively.
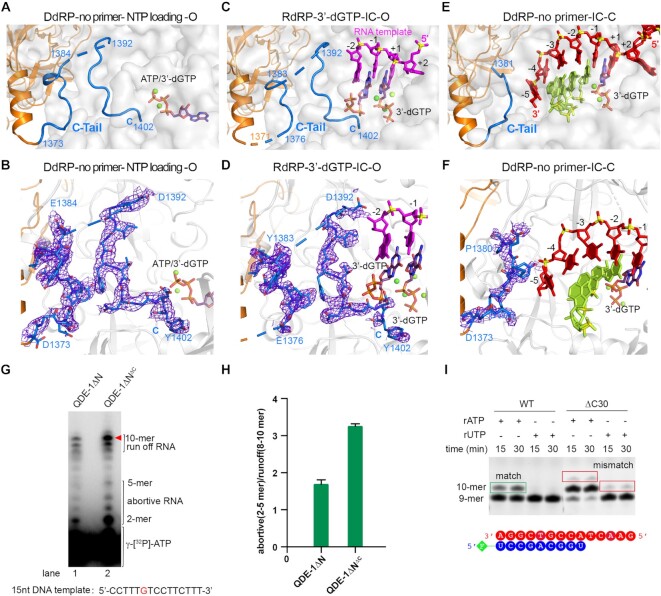
The C-tail occupied the substrate-binding cleft in de novo initiation states
In four complex structures, QDE-1ΔN molecules are bound to a DNA or RNA template, but in the open QDE-1ΔN molecule in the DdRP-no primer structure that only captured one NTP at the entry site (E site) (Figure 7A; Supplementary Figure S8A, B) the NTP resides in a narrow tunnel identified in the QDE-1ΔN structures (Supplementary Figure S8A, C). The phosphate groups coordinate with two cations and form conserved interactions with QDE-1ΔN at both the A and E site (Supplementary Figure S8B, D, E); a similar phenomenon has also been observed in the Pol II structure (46). Instead of the template or product strand, the long C-tail (residues 1373–1402) from the closed QDE-1ΔN molecule inserts into the substrate-binding cleft of the open QDE-1ΔN molecule in the DdRP-no primer structure (Figure 7A). The 1385–1391 region is disordered, whereas all other C-tail residues are well defined, as supported by the clear electron density maps (Figure 7B). The C-tail forms extensive interactions with the substrate-binding cleft residues. For example, Y1377 forms hydrophobic interactions with L726, L728 and V785, and I1394 forms hydrophobic interactions with Y919. The last C-terminal residue, Y1402, forms H-bonds with S677 and R783 and contacts with Q673, F584 and H613 through hydrophobic interactions (Supplementary Figure S9A).
Figure 7.
The C-tail occupied the substrate-binding cleft of QDE-1ΔN in de novo initiation states. (A and B) Cartoon presentation and slow annealing composite omit maps of the QDE-1 C-tail observed in the DdRP-no primer-NTP loading-O structure. (C) Cartoon-stick presentations of the C-tail, RNA template and 3′-dGTP observed in the RdRP-3′-dGTP-IC-O structure. (D) The slow annealing composite omit maps of the QDE-1 C-tail observed in the RdRP-3′-dGTP-IC-O structure. (E) Cartoon and stick representations of the C-tail, DNA template and RNA product observed in the DdRP-no primer-IC-C structure. (F) The slow annealing composite omit maps of the QDE-1 C-tail observed in the DdRP-no primer-IC-C structure. (G) De novo RNA synthesis using the 15 nt DNA as a template (0.2 μM). The reaction was carried out in the presence of 20 μM ATP, GTP and 3′-dCTP spiked with γ-[32P]ATP. The reaction was quenched at 60 min. The gels were exposed to a phosphor screen overnight and scanned on a Typhoon FLA 9000. (H) Abortive to runoff ratio on the 15 nt DNA. Error bars are from three measurements. (I) Analysis of primer-extension products by a 20% polyacrylamide–7 M urea denaturing gel. The target products are indicated by green boxes, whereas the misincorporated products are indicated by red boxes. All electron density maps are contoured at the 1.0 σ level.
The C-tail was also ordered in the early initiation state in the RdRP-3′-dGTP structure (Figure 7C, D; Supplementary Figure S9B), sharing an almost identical conformation with the NTP loading state in the DdRP-no primer structure (Figure 7A, B). Probably due to the clash with the residual –3 and –4 nucleotides of the template, the C-tail 1392–1396 region has an estimated occupancy of 0.6 (Supplementary Figure S9B). Together with Q736, R738 and R783, the C-tail residues 1397–1402 form one well-fitted binding pocket for 3′-dGTP, which pairs with the C-1 nucleotide of the template strand (Supplementary Figure S9C). In the initiation state in the DdRP-no primer structure (Figure 7E), residues 1382–1402 are disordered (Figure 7F), probably due to the growth of the product strand and translocation of the template strand. Residues 1376–1380 of the C-tail are ordered in the NTP-loading state (Figure 7A, B), the early initiation state (Figure 7C, D) and the initiation state (Figure 7E, F). Interestingly, Y1377 forms an H-bond with the non-bridging oxygen of phosphate between –4 and –5 in the initiation state (Supplementary Figure S9D).
To understand the functional role of the QDE-1 C-tail, we constructed a QDE-1ΔN variant with 30 amino acids deleted from the C-terminus, QDE-1ΔNΔC. The in vitro catalytic assay results showed that the de novo RNA synthesis activity of QDE-1ΔNΔC was higher than that of QDE-1ΔN (Figure 7G), suggesting that the QDE-1 C-tail has an inhibitory effect on RNA synthesis. The ratio between the abortive (2–5 mer) and runoff (8–10 mer) products was higher for QDE-1ΔNΔC than for QDE-1ΔN (Figure 7H). In addition to de novo synthesis assays, we also performed primer-extension assays. As depicted in Figure 7I, QDE-1ΔNΔC showed higher primer-extension activity than QDE-1ΔN. Interestingly, in addition to matching NTPs, QDE-1ΔNΔC can also catalyze the incorporation of mismatched NTPs. In contrast, no clear misincorporated product was observed for QDE-1ΔN, suggesting that it has higher fidelity.
Dimerization is not required for the catalytic activity of QDE-1ΔN in vitro
QDE-1ΔN exists as a homodimer (Figure 1B–D). To investigate whether dimerization is strictly required for the function of QDE-1, we first constructed two QDE-1ΔN heterodimers, composed of one active His-SUMO-tagged QDE-1ΔNΔC protomer and one inactive GST-tagged QDE-1ΔNm3 or QDE-1ΔNΔC-m3 protomer (Supplementary Figure S10A, B). In vitro catalytic assay results showed that both heterodimers are active (Supplementary Figure S10C). Compared with that of QDE-1ΔNΔC/QDE-1ΔNm3, the de novo RNA synthesis activity of QDE-1ΔNΔC/QDE-1ΔNΔC-m3 was higher, which further confirmed the inhibitory effect of the QDE-1 C-tail. To obtain monomeric QDE-1ΔN protein, we then performed systematic truncation on QDE-1ΔN (Supplementary Figure S11A, B). Similar to QDE-1ΔN, the QDE-1377–1339 variant exists as a homodimer. However, the SEC-MALS assay results showed that the QDE-1377–1192 variant exists as a monomer in solution (Supplementary Figure S11C; Supplementary Table S5). In vitro primer-extension assay results showed that both QDE-1377–1339 and QDE-1377–1192 have RdRP and DdRP activities. Compared with those of QDE-1377–1339, the catalytic activities of QDE-1377–1192 were higher (Supplementary Figure S11D). These observations suggested that dimerization is not strictly required for the catalytic activity of QDE-1ΔN in vitro.
DISCUSSION
RNAi is a highly conserved gene expression regulation mechanism. During the vegetative stage of N. crassa, both repetitive transgenes and damaged rDNA loci stimulate the production of aRNAs by QDE-1 and QDE-3. QDE-1 subsequently converts the aRNAs to dsRNA precursors (2). Both aRNA and dsRNA precursor production indicates the functional importance of the dual polymerization activities of QDE-1. In this work, we report structures of QDE-1–nucleic acid complexes, in which QDE-1 could bind both the A-form-like DNA–RNA hybrid and RNA–RNA duplex in the duplex-binding cleft (Figure 2; Supplementary Figure S3), revealing the structural basis of the dual DdRP and RdRP activities of QDE-1.
Conserved BH for template recognition
The two-barrel polymerase superfamily contains various classes of polymerases. In addition to QDE-1 or homologous proteins involved in RNAi, multisubunit DdRPs (including eukaryotic Pol II and prokaryotic RNA polymerase), single subunit DdRP (such as phage pg66) and archaeal DNA polymerase PolD also belong to the two-barrel polymerase superfamily. In addition to the characteristic DPBBs, the QDE-1ΔN complex structures in this study revealed that the D1AH, BH and TL subdomains also constitute the core architecture of the two-barrel polymerase superfamily (Figure 2A, B and 3A, B; Supplementary Figure S4D, E) (31,47–49). Recently reported structures confirmed that PolD has TL- and BH-related motifs, but the motifs fulfill different roles (50,51).
In QDE-1 and many multisubunit DdRPs, the template is mainly recognized by the BH subdomain near the entrance site (Figure 4A, B; Supplementary Figures S4A, B and S5C). The QDE-1 BH subdomain interacts with both DNA and RNA templates and does not strictly distinguish between DNA and RNA templates. Pol II also has RdRP activity (37); it can recognize certain RNAs, such as the non-coding B2 RNA (52). The Pol II BH subdomain could undergo conformational change, selecting the right NTP that pairs with the complementary template at the +1 site. In Pol II, T831 of the BH subdomain serves as a sensor for the 3′-terminal base pairing of the DNA–RNA hybrid, and Y836 promotes continuous BH bending motions (Supplementary Figure S5C, D). The conformational bending of BH is critical for Pol II backtracking (53). However, the QDE-1 BH subdomain is relatively rigid and changes slightly in QDE-1ΔN EC structures compared with the apo- or IC structures (Figure 4I; Supplementary Figure S7H).
DNA and RNA templates exit from the substrate-binding cleft differently
During the synthesis of the product strand, the upstream template–product duplex will exit the binding cleft. The RNA template probably exits the cleft together with the paired product strand (Figure 1F). However, the 3′-end nucleotide of the DNA template is separated from the product strand in all DdRP-EC-C structures and probably exits through a narrow channel between the head and slab domains (Figure 4H; Supplementary Figure 2G). The 5′-end nucleotide of the RNA product leaves the channel from the distal end. Previously, Liu and co-workers reported that the ssDNA-binding protein RPA strongly promotes dsRNA production from ssDNA by QDE-1 (16). These results are consistent with the structural observations in this study. While the DNA template separates from the RNA product, the RNA product 3′ end reenters the binding cleft and serves as an RNA template for dsRNA synthesis.
In all the apo- and complexed structures, the two molecules of the QDE-1ΔN dimer adopt different conformations (Figure 1; Supplementary Figure S2). The open molecules captured the reaction in various states, whereas the closed molecules were mostly in the elongation state (Figure 1B, C). Most probably due to the extrusion caused by the head domain from the partner QDE-1ΔN molecule, the template–product duplexes exit differently from the distal end of the active center (Figure 4G–I; Supplementary Figure 2F, G). For the RNA-templated elongation complex structures, the RNA–RNA duplex always pairs at position –7 in the closed molecule (Figure 2E; Supplementary Figure S3A, B). For the DNA-templated elongation complex structures, the DNA–RNA hybrid forms only 5–6 base pairs (Figure 2F; Supplementary Figure S3C–E). Although the template nucleotides at position –7 could be observed, they were separated from the product strands in all DdRP-EC-C structures, probably due to the lower stability of the DNA–RNA hybrid compared with the RNA–RNA duplex (54). The separated nucleotide at the 3′ end of the DNA template has a 2′-endo conformation, which is favorable for DNA (Supplementary Figure S5F).
The RNA product is mainly recognized by the DPBB1 subdomain
In many two-barrel polymerases, two DPBB1 lysine residues play important roles in product binding. For example, in Pol II EC, two DPBB1 lysine residues (K979 and K987 of the Rpb2 subunit) recognize RNA products (Figure 5B). However, the Pol II K979 is substituted by Q736 in the QDE-1 DPBB1 subdomain, and an arginine residue (R738) specifically evolved in RNAi-related RdRPs (Figure 5A, B; Supplementary Figure S6A). Q736 and R738 are conserved in many QDE-1 homologous proteins (Supplementary Figure S6A). Q736 forms one H-bond with the 2′-OH group of the –2 nucleotide. R738 forms H-bonds with the phosphate backbone of the –1 nucleotide, which may help product strand stabilization (Figure 5A). The DPBB1 and D1AH subdomains both participate in product binding (Figure 5A, B), which is conserved in the two-barrel polymerase superfamily (Figures 2A, B and 3A, B; Supplementary Figure S4) (29).
NTP at the A site is stabilized by the conserved TL subdomain
The TL subdomain of QDE-1 and Pol II shares similar interactions, such as hydrophobic interactions, with the nucleobase of NTP at the A site by V1115 from the QDE-1 TL and by L1081 from the Pol II TL, respectively. However, the interaction between QDE-1 D1116 and the 3′-OH group of NTP may be more stable than the indirect interaction with N1082 of Pol II EC (Figure 5E, F). Structural superposition also indicated that the Pol II TL subdomain undergoes conformational changes upon NTP binding, playing an important role in catalysis in Pol II (Figure 5G) (48). However, the QDE-1 TL subdomain is relatively rigid (Figure 5H) and changed slightly in the elongation state compared with the apo- or early initiation state structure (Supplementary Figure S7H). The QDE-1 TL always adopts one closed conformation, which may be due to the extensive hydrophobic interactions between TL and BH subdomains (Supplementary Figure S6H).
NTP at the A site is selected by the Pro-Gate loop
The A site NTP is recognized by the QDE-1 Pro-Gate loop from the minor groove side (Figure 6A–C; Supplementary Figure S7H); the minor-groove-sensing Pro-Gate loop is also present in Pol II (44,45). R446 from the Pro-Gate loop of Pol II and the corresponding R425 residue of the multisubunit E. coli RNA polymerase affect the ssRNA product synthesis and incoming NTP selection (55). The highly conserved P448 from Rpb1 of Pol II has been demonstrated to play an essential role during translocation and to sense the environment of the minor groove after nucleotide incorporation (45). In addition to stabilization, we speculated that the QDE-1 Pro-Gate loop also enhances NTP selection. Residue R962 of the DPBB2 subdomain forms H-bonds with the 2′-OH groups at the 3′ end of the product strand and the NTP analog at the A site (Figure 5D, E). The interactions with R962 of the Pro-Gate loop and D1116 of the TL subdomain in QDE-1 keep the sugar moieties of the 3′-end nucleotide and the NTP analog in the 3′-endo conformation, which is favorable for RNA (55).
Conserved catalytic mechanism of the two-barrel polymerase superfamily
The polymerization activities of QDE-1 and the two-barrel polymerases are cation dependent (29). The QDE-1ΔN complex structures show that the four catalytic aspartate residues (one in DPBB1 and three in DPBB2) coordinate with two cations, similar to other two-barrel polymerases (Figure 6D–F; Supplementary Figure S7B–D). Therefore, the two-barrel polymerase superfamily shares the conserved two-cation catalytic mechanism (Figure 6G). In addition to cations A and B, most QDE-1ΔN complexes here have cation C (Figure 6D; Supplementary Figure S7J) or a third cation at different positions (Figure 6E), and some even have a fourth cation (Figure 6F). The third cations with different orientations and coordination have been reported in DNA polymerase η and β structures and have been proposed to stabilize the transition state or deter the reverse reaction in the catalysis (56,57). Unlike the third cations in DNA polymerase η and β that are located between α and β phosphates, the third or fourth cations observed in QDE-1ΔN complexes interact with β and γ phosphates, which is similar to the behavior of cation C observed in the transcribing complex of S. cerevisiae Pol II (48). However, cations C and D in the QDE-1ΔN complexes were observed at lower metal ion concentrations, and cations A and B were stable in all structures, suggesting that cations C and D may not be inhibitory but propitious to the reaction by stabilizing the leaving pyrophosphate group.
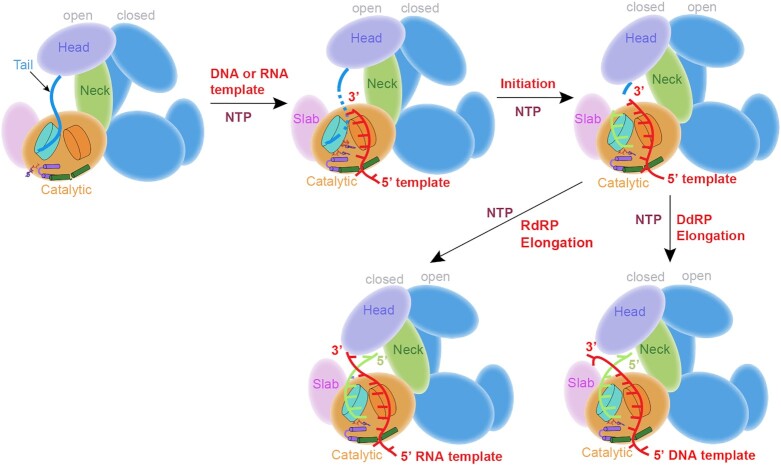
Regulatory roles of the QDE-1 C-tail
Unlike AtRDR2, ZmRDR2 or AtRDR6, which can function as monomers (Supplementary Figure S11C; Supplementary Table S5) (58,59), QDE-1ΔN exists as a homodimer (Figure 1B-D; Supplementary Figure S2). The C-tail observed in NTP loading and the two initiation states comes from the dimer partner. Dimerization placed the C-tail of one QDE-1 monomer in the substrate-binding cleft of the partner monomer (Figure 7A–F). Based on QDE-1ΔN complex structures, this study proposes one plausible working model for the dimerization and reaction state transition of QDE-1ΔN (Figure 8). In the early initiation state, the C-tail of QDE-1 enhances the first triphosphate nucleotide binding next to the active site and probably facilitates the de novo initiation reaction. When the RNA product extends in the initiation states of the QDE-1 dimer, the C-tail coordinates with extension of the RNA products and the alignment of DNA or RNA templates in the substrate-binding cleft through space occupation and direct interaction. The C-tail moves out of the cleft in the elongation state, then bounces back and waits for the next round of polymerization when the reaction is completed and releases the template–product duplex.
Figure 8.
Cartoon model summarizing the dynamic conformational change of the QDE-1 C-tail in different reaction states. Both QDE-1ΔN protomers are functional and undergo identical conformational changes. For clarity, conformational changes are only shown for one protomer in the figure.
Although dimerization is not strictly required for the catalytic activity of QDE-1 in vitro (Supplementary Figure S11; Supplementary Table S5), our in vitro catalytic assays suggested that dimerization may enable the QDE-1 C-tail to play a regulatory role in RNA synthesis. As depicted in Figure 7G, the de novo RNA synthesis activity of QDE-1ΔNΔC was higher than that of WT QDE-1ΔN and the two heterodimers (QDE-1ΔNΔC/QDE-1ΔN m3 and QDE-1ΔNΔC/QDE-1ΔNΔC-m3) (Supplementary Figure S10C). However, the percentage of abortive products of QDE-1ΔNΔC was also higher (Figure 7H). The primer-extension assays suggested that the fidelity of QDE-1ΔNΔC was lower than that of WT QDE-1ΔN (Figure 7I). The C-tail in the QDE-1ΔN dimer is similar to that of the mitochondrial transcription factor (Mtf1) C-tail observed in mitochondrial RNA polymerase complex structures (60,61) and may play regulatory roles in stabilizing the initiation state, coordinating template strand alignment, making the transition from initiation to elongation and maintaining fidelity during the RNA synthesis process. To better understand the functional roles of the dimerization and the unique C-tail of QDE-1, more mutagenesis and in vivo studies are needed in the future.
In plants, Pol IV functions as a DdRP, and RdRP activity is performed by RDR2 which is homologous to QDE-1 (62). Both Pol IV and RDR2 are two-barrel polymerase superfamily members. Genetic and genomic evidence indicates that the production of double-stranded siRNA precursors requires both Pol IV and RDR2. However, single pol IV or rdr2 mutants had undetectable siRNA precursors (63–65). Pikaard and co-workers recently demonstrated the co-dependence of Pol IV and RDR2 in vitro. Starting from Pol IV, RDR2 and ssDNA, the team successfully generated dsRNA products in an in vitro reaction system. Recent studies also demonstrated that Pol IV interacts directly with RDR2, forming the Pol IV–RDR2 heterodimer (66,67). No Pol IV homologous proteins exist in N. crassa, and the QDE-1 homodimer may mimic the Pol IV/RDR2 heterodimer in action.
In conclusion, these results provide structural and mechanistic insights into the dual DdRP and RdRP activities of QDE-1 and particularly shed light on the conserved recognition of the template, product and NTP by QDE-1, Pol II and other two-barrel polymerase superfamily members.
DATA AVAILABILITY
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 7Y7Q, 7Y7R, 7Y7P, 7Y7S and 7Y7T.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Prof. Yi Liu for plasmid pEM41 encoding QDE-1, Dr. Hehua Liu for guidance with RNA synthesis, Dr. Yiqing Chen for providing some ideas on the polymerase activity assay, Dr. Baixing Wu for helpful discussions and the initial manuscript revision, and the staff of BL17U1 beamline at the Shanghai Synchrotron Radiation Facility (SSRF) and BL19U1 beamlines of the National Faciltiy for Protein Science Shanghai for help during data collection.
Contributor Information
Ruixue Cui, Huashan Hospital affiliated to Fudan University, State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Multiscale Research Institute of Complex Systems, Department of Biochemistry and Biophysics, School of Life Sciences, Fudan University, Shanghai 200438, China.
Hao Li, Huashan Hospital affiliated to Fudan University, State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Multiscale Research Institute of Complex Systems, Department of Biochemistry and Biophysics, School of Life Sciences, Fudan University, Shanghai 200438, China.
Jin Zhao, Huashan Hospital affiliated to Fudan University, State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Multiscale Research Institute of Complex Systems, Department of Biochemistry and Biophysics, School of Life Sciences, Fudan University, Shanghai 200438, China.
Xuhang Li, Huashan Hospital affiliated to Fudan University, State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Multiscale Research Institute of Complex Systems, Department of Biochemistry and Biophysics, School of Life Sciences, Fudan University, Shanghai 200438, China.
Jianhua Gan, Shanghai Public Health Clinical Center, State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Department of Biochemistry and Biophysics, School of Life Sciences, Fudan University, Shanghai 200438, China.
Jinbiao Ma, Huashan Hospital affiliated to Fudan University, State Key Laboratory of Genetic Engineering, Collaborative Innovation Center of Genetics and Development, Multiscale Research Institute of Complex Systems, Department of Biochemistry and Biophysics, School of Life Sciences, Fudan University, Shanghai 200438, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China [31971130 to J.M.]. Funding for open access charge: National Natural Science Foundation of China [31971130].
Conflict of interest statement. None declared.
REFERENCES
- 1. Martienssen R., Moazed D. RNAi and heterochromatin assembly. Cold Spring Harb. Perspect. Biol. 2015; 7:a019323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chang S.S., Zhang Z., Liu Y.. RNA interference pathways in fungi: mechanisms and functions. Annu. Rev. Microbiol. 2012; 66:305–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wendte J.M., Pikaard C.S.. The RNAs of RNA-directed DNA methylation. Biochim. Biophys. Acta. 2017; 1860:140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cogoni C., Macino G.. Gene silencing in Neurospora crassa requires a protein homologous to RNA-dependent RNA polymerase. Nature. 1999; 399:166–169. [DOI] [PubMed] [Google Scholar]
- 5. Dalmay T., Hamilton A., Rudd S., Angell S., Baulcombe D.C.. An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell. 2000; 101:543–553. [DOI] [PubMed] [Google Scholar]
- 6. Mourrain P., Beclin C., Elmayan T., Feuerbach F., Godon C., Morel J.B., Jouette D., Lacombe A.M., Nikic S., Picault N.et al.. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000; 101:533–542. [DOI] [PubMed] [Google Scholar]
- 7. Smardon A., Spoerke J.M., Stacey S.C., Klein M.E., Mackin N., Maine E.M.. EGO-1 is related to RNA-directed RNA polymerase and functions in germ-line development and RNA interference in C. elegans. Curr. Biol. 2000; 10:169–178. [DOI] [PubMed] [Google Scholar]
- 8. Cogoni C., Macino G.. Isolation of quelling-defective (qde) mutants impaired in posttranscriptional transgene-induced gene silencing in Neurospora crassa. Proc. Natl Acad. Sci. USA. 1997; 94:10233–10238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forrest E.C., Cogoni C., Macino G.. The RNA-dependent RNA polymerase, QDE-1, is a rate-limiting factor in post-transcriptional gene silencing in Neurospora crassa. Nucleic Acids. Res. 2004; 32:2123–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bernstein E., Caudy A.A., Hammond S.M., Hannon G.J.. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001; 409:363–366. [DOI] [PubMed] [Google Scholar]
- 11. Cogoni C., Macino G.. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science. 1999; 286:2342–2344. [DOI] [PubMed] [Google Scholar]
- 12. Catalanotto C., Azzalin G., Macino G., Cogoni C.. Involvement of small RNAs and role of the qde genes in the gene silencing pathway in Neurospora. Genes Dev. 2002; 16:790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Makeyev E.V., Bamford D.H.. Cellular RNA-dependent RNA polymerase involved in posttranscriptional gene silencing has two distinct activity modes. Mol. Cell. 2002; 10:1417–1427. [DOI] [PubMed] [Google Scholar]
- 14. Lee H.C., Chang S.S., Choudhary S., Aalto A.P., Maiti M., Bamford D.H., Liu Y.. qiRNA is a new type of small interfering RNA induced by DNA damage. Nature. 2009; 459:274–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aalto A.P., Poranen M.M., Grimes J.M., Stuart D.I., Bamford D.H.. In vitro activities of the multifunctional RNA silencing polymerase QDE-1 of Neurospora crassa. J. Biol. Chem. 2010; 285:29367–29374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lee H.C., Aalto A.P., Yang Q., Chang S.S., Huang G., Fisher D., Cha J., Poranen M.M., Bamford D.H., Liu Y.. The DNA/RNA-dependent RNA polymerase QDE-1 generates aberrant RNA and dsRNA for RNAi in a process requiring replication protein A and a DNA helicase. PLoS Biol. 2010; 8:e1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xie Z., Johansen L.K., Gustafson A.M., Kasschau K.D., Lellis A.D., Zilberman D., Jacobsen S.E., Carrington J.C.. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004; 2:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Herr A.J., Jensen M.B., Dalmay T., Baulcombe D.C.. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005; 308:118–120. [DOI] [PubMed] [Google Scholar]
- 19. Onodera Y., Haag J.R., Ream T., Nunes P.C., Pontes O., Pikaard C.S.. Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell. 2005; 120:613–622. [DOI] [PubMed] [Google Scholar]
- 20. Axtell M.J., Jan C., Rajagopalan R., Bartel D.P.. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006; 127:565–577. [DOI] [PubMed] [Google Scholar]
- 21. Sijen T., Fleenor J., Simmer F., Thijssen K.L., Parrish S., Timmons L., Plasterk R.H.A., Fire A.. On the role of RNA amplification in dsRNA-triggered gene silencing. Cell. 2001; 107:465–476. [DOI] [PubMed] [Google Scholar]
- 22. Pak J., Fire A.. Distinct populations of primary and secondary effectors during RNAi in C. elegans. Science. 2007; 315:241–244. [DOI] [PubMed] [Google Scholar]
- 23. Sijen T., Steiner F.A., Thijssen K.L., Plasterk R.H.. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science. 2007; 315:244–247. [DOI] [PubMed] [Google Scholar]
- 24. Volpe T.A., Kidner C., Hall I.M., Teng G., Grewal S.I.S., Martienssen R.A.. Regulation of heterochromatic silencing and histone H3 lysine-9 methylation by RNAi. Science. 2002; 297:1833–1837. [DOI] [PubMed] [Google Scholar]
- 25. Motamedi M.R., Verdel A., Colmenares S.U., Gerber S.A., Gygi S.P., Moazed D. Two RNAi complexes, RITS and RDRC, physically interact and localize to noncoding centromeric RNAs. Cell. 2004; 119:789–802. [DOI] [PubMed] [Google Scholar]
- 26. Verdel A., Jia S., Gerber S., Sugiyama T., Gygi S., Grewal S.I., Moazed D. RNAi-mediated targeting of heterochromatin by the RITS complex. Science. 2004; 303:672–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Castillo R.M., Mizuguchi K., Dhanaraj V., Albert A., Blundell T.L., Murzin A.G.. A six-stranded double-psi beta barrel is shared by several protein superfamilies. Structure. 1999; 7:227–236. [DOI] [PubMed] [Google Scholar]
- 28. Iyer L.M., Koonin E.V., Aravind L.. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 2003; 3:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sauguet L. The extended ‘two-barrel’ polymerases superfamily: structure, function and evolution. J. Mol. Biol. 2019; 431:4167–4183. [DOI] [PubMed] [Google Scholar]
- 30. Salgado P.S., Koivunen M.R., Makeyev E.V., Bamford D.H., Stuart D.I., Grimes J.M.. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006; 4:e434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Drobysheva A.V., Panafidina S.A., Kolesnik M.V., Klimuk E.I., Minakhin L., Yakunina M.V., Borukhov S., Nilsson E., Holmfeldt K., Yutin N.et al.. Structure and function of virion RNA polymerase of a crAss-like phage. Nature. 2021; 589:306–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sauguet L., Raia P., Henneke G., Delarue M.. Shared active site architecture between archaeal PolD and multi-subunit RNA polymerases revealed by X-ray crystallography. Nat. Commun. 2016; 7:12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raia P., Carroni M., Henry E., Pehau-Arnaudet G., Brule S., Beguin P., Henneke G., Lindahl E., Delarue M., Sauguet L.. Structure of the DP1–DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases. PLoS Biol. 2019; 17:e3000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cramer P., Bushnell D.A., Kornberg R.D.. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001; 292:1863–1876. [DOI] [PubMed] [Google Scholar]
- 35. Vassylyev D.G., Sekine S., Laptenko O., Lee J., Vassylyeva M.N., Borukhov S., Yokoyama S.. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002; 417:712–719. [DOI] [PubMed] [Google Scholar]
- 36. Curaba J., Chen X.. Biochemical activities of Arabidopsis RNA-dependent RNA polymerase 6. J. Biol. Chem. 2008; 283:3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lehmann E., Brueckner F., Cramer P.. Molecular basis of RNA-dependent RNA polymerase II activity. Nature. 2007; 450:445–449. [DOI] [PubMed] [Google Scholar]
- 38. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M.. HKL-3000: the integration of data reduction and structure solution—from diffraction images to an initial model in minutes. Acta Crystallogr. D. 2006; 62:859–866. [DOI] [PubMed] [Google Scholar]
- 39. Otwinowski Z., Minor W.. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997; 276:307–326. [DOI] [PubMed] [Google Scholar]
- 40. McCoy A.J., Grosse-Kunstleve R.W., Adams P.D., Winn M.D., Storoni L.C., Read R.J.. Phaser crystallographic software. J. Appl. Crystallogr. 2007; 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Potterton E., Briggs P., Turkenburg M., Dodson E.. A graphical user interface to the CCP4 program suite. Acta Crystallogr. D. 2003; 59:1131–1137. [DOI] [PubMed] [Google Scholar]
- 42. Adams P.D., Grosse-Kunstleve R.W., Hung L.W., Ioerger T.R., McCoy A.J., Moriarty N.W., Read R.J., Sacchettini J.C., Sauter N.K., Terwilliger T.C.. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. 2002; 58:1948–1954. [DOI] [PubMed] [Google Scholar]
- 43. Emsley P., Cowtan K.. Coot: model-building tools for molecular graphics. Acta. Crystallogr. D. 2004; 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 44. Oh J., Fleming A.M., Xu J., Chong J., Burrows C.J., Wang D. RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair–pi and CH–pi interactions. Proc. Natl Acad. Sci. USA. 2020; 117:9338–9348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xu L., Wang W., Wu J.B., Shin J.H., Wang P.C., Unarta I.C., Chong J., Wang Y.S., Wang D. Mechanism of DNA alkylation-induced transcriptional stalling, lesion bypass, and mutagenesis. Proc. Natl Acad. Sci. USA. 2017; 114:E7082–E7091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Westover K.D., Bushnell D.A., Kornberg R.D.. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004; 119:481–489. [DOI] [PubMed] [Google Scholar]
- 47. Qian X., Hamid F.M., El Sahili A., Darwis D.A., Wong Y.H., Bhushan S., Makeyev E.V., Lescar J.. Functional evolution in orthologous cell-encoded RNA-dependent RNA polymerases. J. Biol. Chem. 2016; 291:9295–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wang D., Bushnell D.A., Westover K.D., Kaplan C.D., Kornberg R.D.. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006; 127:941–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Vassylyev D.G., Vassylyeva M.N., Zhang J., Palangat M., Artsimovitch I., Landick R.. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007; 448:163–168. [DOI] [PubMed] [Google Scholar]
- 50. Lei L., Burton Z.F.. Early evolution of transcription systems and divergence of archaea and bacteria. Front. Mol. Biosci. 2021; 8:651134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Madru C., Henneke G., Raia P., Hugonneau-Beaufet I., Pehau-Arnaudet G., England P., Lindahl E., Delarue M., Carroni M., Sauguet L.. Structural basis for the increased processivity of D-family DNA polymerases in complex with PCNA. Nat. Commun. 2020; 11:1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wagner S.D., Yakovchuk P., Gilman B., Ponicsan S.L., Drullinger L.F., Kugel J.F., Goodrich J.A.. RNA polymerase II acts as an RNA-dependent RNA polymerase to extend and destabilize a non-coding RNA. EMBO J. 2013; 32:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Da L.T., Pardo-Avila F., Xu L., Silva D.A., Zhang L., Gao X., Wang D., Huang X.. Bridge helix bending promotes RNA polymerase II backtracking through a critical and conserved threonine residue. Nat. Commun. 2016; 7:11244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Clark C.L., Cecil P.K., Singh D., Gray D.M.. CD, absorption and thermodynamic analysis of repeating dinucleotide DNA, RNA and hybrid duplexes [d/r(AC)]12·[d/r(GT/U)]12 and the influence of phosphorothioate substitution. Nucleic Acids Res. 1997; 25:4098–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Makinen J.J., Shin Y., Vieras E., Virta P., Metsa-Ketela M., Murakami K.S., Belogurov G.A.. The mechanism of the nucleo-sugar selection by multi-subunit RNA polymerases. Nat. Commun. 2021; 12:796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gao Y., Yang W.. Capture of a third Mg2+ is essential for catalyzing DNA synthesis. Science. 2016; 352:1334–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Perera L., Freudenthal B.D., Beard W.A., Pedersen L.G., Wilson S.H.. Revealing the role of the product metal in DNA polymerase beta catalysis. Nucleic Acids Res. 2017; 45:2736–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Fukudome A., Singh J., Mishra V., Reddem E., Martinez-Marquez F., Wenzel S., Yan R., Shiozaki M., Yu Z., Wang J.C.et al.. Structure and RNA template requirements of Arabidopsis RNA-DEPENDENT RNA POLYMERASE 2. Proc. Natl Acad. Sci. USA. 2021; 118:e2115899118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Du X., Yang Z., Ariza A.J.F., Wang Q., Xie G., Li S., Du J.. Structure of plant RNA-DEPENDENT RNA POLYMERASE 2, an enzyme involved in small interfering RNA production. Plant Cell. 2022; 34:2140–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Basu U., Lee S.W., Deshpande A., Shen J., Sohn B.K., Cho H., Kim H., Patel S.S.. The C-terminal tail of the yeast mitochondrial transcription factor Mtf1 coordinates template strand alignment, DNA scrunching and timely transition into elongation. Nucleic Acids Res. 2020; 48:2604–2620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. De Wijngaert B., Sultana S., Singh A., Dharia C., Vanbuel H., Shen J., Vasilchuk D., Martinez S.E., Kandiah E., Patel S.S.et al.. Cryo-EM structures reveal transcription initiation steps by yeast mitochondrial RNA polymerase. Mol. Cell. 2021; 81:268–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Singh J., Mishra V., Wang F., Huang H.Y., Pikaard C.S.. Reaction mechanisms of Pol IV, RDR2, and DCL3 drive RNA channeling in the siRNA-directed DNA methylation pathway. Mol. Cell. 2019; 75:576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Blevins T., Podicheti R., Mishra V., Marasco M., Wang J., Rusch D., Tang H., Pikaard C.S.. Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. Elife. 2015; 4:e09591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li S., Vandivier L.E., Tu B., Gao L., Won S.Y., Li S., Zheng B., Gregory B.D., Chen X.. Detection of Pol IV/RDR2-dependent transcripts at the genomic scale in Arabidopsis reveals features and regulation of siRNA biogenesis. Genome Res. 2015; 25:235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhai J., Bischof S., Wang H., Feng S., Lee T.F., Teng C., Chen X., Park S.Y., Liu L., Gallego-Bartolome J.et al.. A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell. 2015; 163:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mishra V., Singh J., Wang F., Zhang Y., Fukudome A., Trinidad J.C., Takagi Y., Pikaard C.S.. Assembly of a dsRNA synthesizing complex: RNA-DEPENDENT RNA POLYMERASE 2 contacts the largest subunit of NUCLEAR RNA POLYMERASE IV. Proc. Natl Acad. Sci. USA. 2021; 118:e2019276118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Huang K., Wu X.X., Fang C.L., Xu Z.G., Zhang H.W., Gao J., Zhou C.M., You L.L., Gu Z.X., Mu W.H.et al.. Pol IV and RDR2: a two-RNA-polymerase machine that produces double-stranded RNA. Science. 2021; 374:1579–1586. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 7Y7Q, 7Y7R, 7Y7P, 7Y7S and 7Y7T.