Abstract
High mobility group (HMG) proteins are chromatin regulators with essential functions in development, cell differentiation and cell proliferation. The protein HMG20A is predicted by the AlphaFold2 software to contain three distinct structural elements, which we have functionally characterized: i) an amino-terminal, intrinsically disordered domain with transactivation activity; ii) an HMG box with higher binding affinity for double-stranded, four-way-junction DNA than for linear DNA; and iii) a long coiled-coil domain. Our proteomic study followed by a deletion analysis and structural modeling demonstrates that HMG20A forms a complex with the histone reader PHF14, via the establishment of a two-stranded alpha-helical coiled-coil structure. siRNA-mediated knockdown of either PHF14 or HMG20A in MDA-MB-231 cells causes similar defects in cell migration, invasion and homotypic cell–cell adhesion ability, but neither affects proliferation. Transcriptomic analyses demonstrate that PHF14 and HMG20A share a large subset of targets. We show that the PHF14-HMG20A complex modulates the Hippo pathway through a direct interaction with the TEAD1 transcription factor. PHF14 or HMG20A deficiency increases epithelial markers, including E-cadherin and the epithelial master regulator TP63 and impaired normal TGFβ-trigged epithelial-to-mesenchymal transition. Taken together, these data indicate that PHF14 and HMG20A cooperate in regulating several pathways involved in epithelial–mesenchymal plasticity.
INTRODUCTION
The HMG protein HMG20A (also called iBRAF) was originally described as an antagonist of the histone H3 lysine 4 monomethyl and dimethyl (H3K4m2/1) demethylase LSD1/Co-REST complex during neuronal differentiation (1). HMG20A is a paralogue of HMG20B (also called BRAF35), a core subunit of the LSD1/Co-REST complex (2,3). We have previously shown that HMG20A can form homodimers as well as heterodimers with HMG20B through coiled-coil domains (CCDs) present in the carboxy (C)-terminus of both proteins (4,5). HMG20A can also replace HMG20B in the LSD1/Co-REST complex by interacting with PHF21A (BHC80). We also discovered that HMG20B sumoylation is required for its activity, and that HMG20A can impair this sumoylation by forming HMG20A–HMG20B heterodimers. Therefore, HMG20A seems to antagonize the role of HMG20B in the LSD1/Co-REST complex in at least three ways: (i) replacing it, (ii) titrating it, resulting in its decreased ability to reintegrate into the complex and (iii) inhibiting its sumoylation (4,5). REST is a master repressor of neuronal genes (6). As the LSD1/Co-REST complex is essential for REST-dependent repression, an antagonist of LSD1/Co-REST should improve neuronal differentiation. Consistent with this, HMG20A is highly expressed in the brain (1,7) and is required for human neuronal differentiation in vitro and neuronal differentiation in ovo in the chicken neural tube (5).
The HMG20A gene has also been associated with both gestational and type 2 diabetes mellitus in GWAS studies performed in Asian and European populations (8–11). In agreement with this, we reported that HMG20A is important for pancreatic islet beta-cell functional maturation and adaptation to stress conditions, such as hyperglycemia and pregnancy (12). Furthermore, we have shown that HMG20A potentiates astrocyte survival and reactivates astroglyosis, thereby promoting the survival of hypothalamic neurons (7). We proposed that this function of HMG20A has an impact on the brain/islet axis and its role in glucose homeostasis (13). We have also shown that HMG20A is required for the epithelial–to–mesenchymal transition (EMT) (4). Our data suggested that HMG20A regulates EMT within the context of the LSD1-CoREST complex. However, some phenotypes of HMG20A-depleted cells differ from LSD1-depleted cells, suggesting a more complex scenario (4). Inspection of high-throughput proteomic studies also suggested that HMG20A may have additional partners that could explain additional functions of the protein (14,15).
Here, we report an additional interactor of HMG20A, the histone reader PHF14, which has also been associated with mesenchymal growth (16–18). Knockout of PHF14 in mice results in neonatal lethality due to defects in several organs including fibrosis in kidney and lung (16,19). We have now characterized the HMG20A-PHF14 complex molecularly and in silico and show that HMG20A and PHF14 share common functions in controlling the transcriptome, cell migration and invasion and cell-cell adhesion. Specifically, they modulate essential epithelial signaling pathways, including the Hippo and TGFβ pathways and the epithelial development regulator TP63.
MATERIALS AND METHODS
Cell culture and treatments
Human triple-negative breast cancer cell line MDA-MB-231, human embryonic kidney cell line HEK293T and human osteosarcoma cell line U2OS were grown in Dulbecco's Modified Eagle's Medium (DMEM). Normal murine mammary gland NMuMG cells (provided by José Antonio Pintor-Toro, CABIMER) were cultured in DMEM supplemented with 10 μg/ml insulin. All the cell culture media were supplemented with 10% fetal bovine serum, penicillin (100 U/ml) and streptomycin (100 μg/ml). Cells were maintained at 37°C in 5% CO2 and cultured until reaching about 80%-90% confluence. EMT was induced by the addition of TGFβ (5 ng/ml; R&D Systems, Minneapolis, MN, USA) diluted in 4 mM HCl and 1 mg/ml BSA (240-B, R&D Systems) for the indicated time. In order to analyze AKT signaling pathway activation, MDA-MB-231 cells were cultured in the absence of fetal bovine serum for 3 h followed by stimulation with 100 nM insulin (Sigma) for 3 min.
Cloning and DNA constructs
Full-length wild-type mouse HA-HMG20A and HA-HMG20B, and mutant HA-HMG20A-Δcc1 expression vectors were previously described (4,5). For the HA-HMG20A-Δcc2 expression vector, 45 bp corresponding to aa 292–278 were deleted using standard PCR techniques to generate the plasmid pRSV-Hmg20aΔcc2. The plasmid for expression of HA-HMG20A-ΔIDR was generated deleting nucleotides corresponding the first 78 amino acids of HMG20A by standard PCR techniques, to generate pRSV-Hmg20a-ΔIDR. HA-HMG20A-ΔIDR maintains the nuclear localization signal of HMG20A (81-RSKRGGWSKGRKRKKPLR-98). The A/B chimera (HMG20A1–217:HMG20B187–317) was generated by replacing nucleotides 1–558 of HMG20B with nucleotides 1–651 of HMG20A. The B/A chimera (HMG20B1–186:HMG20A217–346) was generated by replacing nucleotides 1–651 of HMG20A with nucleotides 1–558 of HMG20B. Chimeras constructs maintain HA-tags in their N-terminal regions and were generated by standard PCR techniques. Full-length human PHF14 cDNA was obtained from pMSCV-IRES-EGFP-PHF14 that was kindly provided by Dr Michinori Kitagawa and Dr Takumi Era (16). To generate pRSV-2Flag-PHF14, an EcoRI/ XhoI fragment of pMSCV-IRES-EGFP-PHF14 containing the Flag-PHF14 cDNA was cloned into the SnaBI site of pAdRSV-Sp (20). The luciferase reporter plasmid for the SCN2A gene (pNa-Luc) was constructed by cloning an EcoRI fragment from pMB4 (kindly provided by Dr G. Mandel) into SmaI-digested pGL2. To generate the pET24a(+)-HMG20A plasmid that was used for the synthesis and purification of the 6xHis-tagged HMG20A protein, the cDNA of the gene was produced by standard PCR techniques and subsequently cloned into the NdeI and XhoI restriction sites of the pET24a(+) vector (Novagen).
Transfections, siRNA and luciferase assay
Transient transfections of HEK293T and U2OS cells with expression vectors were performed with FuGENE (Roche) or Lipofectamine 2000 (ThermoFisher), respectively, except for proteomic experiments and virus production (see below). The siRNAs were transiently transfected in MDA-MB-231 cells for knockdown experiments using Lipofectamine RNAiMAX Reagent (ThermoFisher) according to the manufacturer′s instructions. The following siRNA sequences were used: siPHF14: 5′-GAUGGAACCAAACGAUCAA-3′; siHMG20A: 5′-CCCUAUAUUUACAGAGGAA-3′; siPHF14-2: 5′-GAUUAAGGAACCAGUGAAA-3′; siHMG20A-2: 5′- AGGCAAAUCUCAUAGGCAA-3′. siTP63: 5′-CCACUGAACUGAAGAAACU-3′. Cells were collected 48 or 72 h after transfection. Luciferase assays were performed in HEK293T cells as previously described (21). The total amount of DNA in each individual well was kept constant by adding empty vector (Flag or HA) as appropriate.
Retrovirus production and transduction
The pQCXIH-Myc-YAP-5SA retroviral vector for expression of a non-phosphorylable mutant of YAP has been previously described (22). pQCXIH-Myc-YAP-5SA was a gift from Kunliang Guan (Addgene #33093, https://www.addgene.org/33093/; RRID:Addgene_33093). Retroviruses were produced by transfection of HEK293T cells by the calcium phosphate method with the corresponding packaging vectors. Retroviruses-containing supernatants were collected 48 h after transfection and concentrated by ultracentrifugation at 22 000 rpm for 90 min at 4°C. For ectopic expression of YAP-5SA, MDA-MB-231 cells were first transfected with siControl, siPHF14 or siHMG20A. Twenty-four h later cells were infected with pQCXIH-Myc-YAP-5SA retroviruses and cultured during 48 h. After that, cells were processed for RNA isolation.
Expression and purification of His-HMG20A
HMG20A was purified by cobalt-affinity chromatography through its fusion to an oligopeptide consisting of six consecutive histidines. In brief, the pET24a(+)-HMG20A vector (see Cloning and DNA constructs) was first electroporated into the Escherichia coli BL21 (DE3) strain and 1 liter of the resulting culture was induced at an OD600 of 0.6 with 1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) for 2 h. Bacteria were then lysed with 10 ultrasound pulses from a Digital Sonifier S-450D device (Branson) at 4°C and in 30 ml of PBS 1× supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF). Recombinant HMG20A was finally purified by immobilized metal ion affinity chromatography using Econo-Column Standard columns (Bio-Rad) and Talon® resin charged with Co2+ (BD Biosciences), following the manufacturers’ instructions.
Electrophoretic mobility shift assays (EMSA)
To investigate the ability of HMG20A to interact with linear or cruciform DNA, different amounts of the purified protein were incubated at room temperature in 20 μl of EMSA buffer (100 mM Tris–HCl (pH 7.5), 2 mM MgCl2, 1 mM dithiothreitol (DTT), 35 mM NaCl, 10% glycerol and 0.01% bovine serum albumin (BSA)) with a fixed amount of radioactive DNA probe equivalent to 10 000 counts per min (cpm). Assays with linear DNA that were carried out in the presence of competitor molecules also included from 5 ng to 1 μg of poly(dA-dT) or poly(dC-dG) (Amersham). Cruciform DNA assays were always performed in the presence of 1 μg of poly(dI-dC) (Amersham). After 15 min incubation, samples were run at 180 V and 4°C on native polyacrylamide gels (6–10% acrylamide/bis, 29:1, 10% glycerol, 0.25× TBE) that were subsequently dried onto 3MM Chr chromatography paper (Whatman) using a Drygel SE1160 device (Hoefer Scientific Instruments). Radioactivity was revealed using an Instantimager (Packard).
To generate the linear DNA probe, we used the XhoI–XbaI fragment (125 bp) of the pwt(25bp)5 vector, kindly donated by Terumi Kohwi-Shigematsu, containing five repeats of the sequence tctttaatttctaatatatatttagaa. This probe was radioactively labeled at its 5' overhangs by incubating for 15 min, and at 30°C, 100–500 ng of the DNA fragment with 20–30 μCi of 5' [α-32P] dCTP (3000 mCi/mmol), 0.5 mM of the other three dNTP and 6.5 U of SequenaseTM 2.0 enzyme (USB) in its reaction buffer. Unincorporated nucleotides were finally removed using MicroSpinTM S-200 columns (Amersham).
The cruciform DNA structure and its two control linear sequences were obtained by pairing the primers described in (23), and the T4 polynucleotide kinase (PNK) enzyme (USB) was used for their radioactive labeling. Specifically, for the cruciform DNA probe 100 pmol of one of the four oligonucleotides required for the structure were labeled for 1 h at 37°C with 10 μCi of [γ-32P] dATP (3000 mCi/mmol), using 30 U of PNK and its reaction buffer. After removing the unincorporated nucleotides with MicroSpinTM G-25 columns (Amersham), the entire eluate was combined with 100 pmol of each of the other three oligonucleotides in annealing buffer (50 mM Tris–HCl (pH 7.5) and 10 mM MgCl2). This mixture was then heated to 90–100°C and subsequently cooled down slowly to room temperature, thus allowing the formation of the cruciform structure. For each of the control linear probes, one of the oligonucleotides was labeled according to the same protocol and then paired with the other oligonucleotide by the same heating and cooling procedure.
Protein lysates, co-immunoprecipitation and immunoblot analysis
Protein lysates were performed using IP buffer (50 mM Tris·HCl, pH 8, 150 mM NaCl, 1 mM EDTA, pH 8, 1% (vol/vol) Triton X-100, 1 mM PMSF) for co-immunoprecipitation, and urea buffer (8 M urea, 50 mM Tris–HCl, 1 mM EDTA) or RIPA buffer (50 mM Tris–HCl pH 7.8, 150 mM NaCl, 0.5% sodium deoxycholate, 0.1% SDS, 1% NP40), for whole cell extract. In all the cases, buffers were supplemented with a protease inhibitor cocktail.
For fractionation experiments cells were incubated during 10 min in IP buffer. Then, lysates were centrifuged at 12 000 rpm for 10 min at 4°C. The supernatant was considered the soluble fraction and the pellets were dissociated in Laemmli buffer and were considered the insoluble fraction.
Co-immunoprecipitation was performed as previously described (5), using 1 mg of protein lysates using IP buffer and 1 μg of the indicated antibodies, or 500 ng in the case of anti-HMG20A. Antibodies used for immunoprecipitation were as followed: anti-PHF14 (ProteinTech, Cat# 24787-1-AP), anti-HMG20A (Sigma, Cat# HPA008126) and anti-HMG20B (Abnova, H00010362-M01) and anti-TEAD1 (Santa Cruz, sc-376113). For immunoblotting, protein extracts were denatured and separated by SDS-PAGE on 10% polyacrylamide gels. After transfer to 0.45 μm nitrocellulose membranes (Bio-Rad), membranes were blocked with a solution containing 0.5% Tween-20, 5% non-fat milk powder in 1× PBS for 1 h at room temperature and incubated with primary antibodies overnight at 4°C. After washing several times with 1× PBS, 0.1% Tween-20, membranes were incubated with secondary antibody for 1 h at room temperature. Finally, membranes were visualized using the Clarity Western ECL Substrate on a ChemiDoc Gel Imaging System. Primary antibodies used for immunoblotting were anti-PHF14 (ProteinTech, Cat# 24787-1-AP, 1:1000), anti-HMG20A (Sigma, Cat# HPA008126, 1:500), anti-HMG20B (Abnova, H00010362-M01, 1:2000), anti-CDH1 (BD, Cat# 610182, 1:2000), anti-HSP70 (Abcam, Cat# ab5439, 1:10000), anti-FlagM2 (Sigma, Cat# F1804, 1:500), anti-HA (Sigma, H9658, 1:2000), anti-phospho-AKT(Thr308) (Cell Signaling #13038), anti-phospho-AKT(Ser473) (Cell Signaling #4060), anti-phospho-S6K(Thr389) (Cell Signaling #9234), anti-phospho-S6K(Ser235/236) (Cell Signaling #4856), anti-AKT (Cell Signaling #4691), anti-S6K (Cell Signaling #2708), anti-S6 (Cell Signaling #2217), anti-actin (Cell Signaling #4970), anti-TEAD1 (Santa Cruz, sc-376113), anti-phospho-LATS1 (Thr1079) D57D3 (Cell signaling) and anti-YAP (63.7) (Santa Cruz, sc-101199). For secondary antibodies, anti-mouse IgG-H + L HRP Conjugated (Bethyl, Cat# A90-516P, 1:10 000) and anti-rabbit IgG– H + L HRP Conjugated, (Bethyl, Cat# A120-101P, 1:10000) were used.
Wound-healing, invasion and adhesion assays
Wound-healing assays for determination of migration were performed as previously described (24). Following transfection with either control, PHF14 or HMG20A siRNAs, an equal number of cells was plated into culture inserts (Ibidi, Cat# 80209) and incubated at 37°C for 24 h. The next day, culture inserts were removed and the wound was imaged at 0 h, and again after 24 h.
Invasion assays were carried out as previously described with Boyden chambers (25). For that, 24-well-plate cell culture insert (8 μm filters, Corning, Cat# 353097) were coated with 0.6 μg growth factor-reduced Matrigel (Trevigen, Cat# 3431-005-02). Cells suspension of transfected cells was plated at a density of 5 × 104 cells/0.5 ml DMEM, 0.2% BSA into the upper compartment of the chamber, while complete medium was used as chemoattractant. After 24 h, the filter containing invaded cells was placed on a slide with Vectashield mounting medium-DAPI (Vectorlabs), and was imaged to count invading cells.
For adhesion assays, 5 × 105 cells were plated in 24-well plates, transfected with control, PHF14 or HMG20A siRNAs, and grown during 48 h until forming a monolayer. Then, 3 × 105 siControl-, siHMG20A- and siPHF14-treated cells were labelled with 5 μM Calcein during 30 min at 37°C and seeded over monolayers of cells transfected with the same siRNA. After 45 min, non-adherent cells were washed away and the percentage of attached Calcein-positive cells was determined by flow cytometry.
RNA extraction and RT-qPCR
Total RNA was prepared using the RNeasy Kit (Qiagen), as described in the manufacturer's instructions. One microgram of RNA extract was treated with 1 μl of RQ1 RNase-free DNase (Promega) in a final volume of 10 μl for 30 min and then inactivated with Stop Solution for 10 min at 65°C to avoid potential DNA contamination. Complementary DNA (cDNA) was generated from 8 μl of the DNase-digested RNA by using the Maxima H Minus First Strand cDNA Synthesis Kit (ThermoFisher), according to the manufacturer's instruction in a final volume of 20 μl. Two microliters of the generated cDNA solution were used as a template for real-time PCR (qPCR) in a final volume of 20 μl, using 10 μl of Applied Biosystems Power SYBR Green Master Mix (Applied Biosystems) and oligonucleotides at 0.75 μM. The run protocol has four stages: two holding stages of 50°C 2 min and 95°C 10 min, a cycling stage of 95°C 15 s and 60°C 1 min of 40 cycles, and melt curve stage of 95°C 15 s, 60°C 1 min, 95°C 30 s and 60°C 15 s. Quantification of gene products was performed by qPCR with a 7500 FAST Real-Time PCR System (Applied Biosystems). Values were normalized to the expression of the GAPDH reference gene. Invariant expression of this gene under the experimental conditions described was verified using other normalization gene (CHD8) and RNA-seq data. Three to five independent replicates of each experiment were performed. Two technical replicates of each sample were performed. Sequences of all oligonucleotides used are listed in Supplementary Table S1. Sequence-specificity of the designed oligonucleotides was checked by BLAST and In-Silico PCR tool of UCSC and the amplification efficiency was determined from the slope of the calibration curve.
RNA-seq analysis
Total RNA from MDA-MB-231 cells was extracted using the RNeasy kit (74106, QIAGEN). Libraries were prepared with the TruSeq Stranded mRNA kit (Illumina) and sequencing was performed with a Novaseq system (Illumina) with 75 bp single-end reads at the Genomic Unit of CABIMER (Sevilla, Spain). Two independent biological replicates for each condition were sequenced.
Data were aligned using the subjunc function from the Rsubread package, and reads were mapped to the hg19 human reference genome using TH1 = 2 and unique = TRUE parameters. The downstream analysis was performed on .bam files with duplicates removed using the samtools (v0.1.19) rmdup command. The FeatureCounts() function from the Rsubread package was used to assign reads to the UCSC hg19 KnownGenes (miRNAs were discarded from the analysis) using GTF.featureType = ‘exon’, GTF.attrType = ‘gene_id’ and strandSpecific = 2 parameters with the duplicate-removed .bam files. Then, differential gene expression analysis and statistics were performed using the DESeq2 Bioconductor. Differentially expressed genes with adjusted P value <0.05 and Log2FC >0.5 (upregulated genes) or log2FC < –0.5 (downregulated genes) were selected for further analysis.
Gene ontology (GO) functional categories were analyzed using DAVID (26). Bonferroni-adjusted P values of the Fisher exact test were used to determine enrichment significance. GSEA was performed using the GSEA v2.0.14 software (GSEA, Broad Institute, Cambridge, MA, USA) with 1000 phenotype permutations (27). The TGFβ-, EMT- and Hippo pathway-related gene sets were download from MSigDB (https://www.gsea-msigdb.org/gsea/msigdb/). Venn diagrams were performed with Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/).
Confocal microscopy and co-localization analyses
U2OS transiently cotransfected with wild-type flag-PHF14 and either wild-type HA-HMG20A or mutant HA-HMG20A-Δcc1 expression vectors were cultured on coverslips for immunofluorescence staining. Cells were fixed with 4% paraformaldehyde for 10 min at room temperature. After fixation, cells were permeabilized using 0.05% Triton-X100 for 10 min followed by a 30 min incubation with PBS-5% BSA to block unspecific sites, on a rocking platform at room temperature. Cells were incubated with the primary antibodies mouse anti-FlagM2 (Sigma, Cat# F1804 1:1000) and rat anti-HA (Sigma, Cat# H9658, 1:30) for 1 h at room temperature. Secondary antibodies conjugated to fluorochromes 488 and Cy3 were used at 1:500 and 1:200, respectively (Invitrogen), for 1h at room temperature. Antibodies were diluted in the blocking solution PBS–5% BSA. Finally, the coverslips were mounted on slides with Prolong containing DAPI (Invitrogen) and the samples were imaged with a LEICA TSC SP5 confocal microscope and analysed with Imaris image software (Oxford Instruments).
JACoP analysis (ImageJ) on images of the nuclei was used to study colocalization of the fluorochromes. Co-localization was represented by the Pearson correlation coefficient.
Generation of inducible CRISPR/Cas9 stable cell lines
For the inducible mutation of Phf14 and Hmg20a in NMuMG cells we generated stable cell lines by using the CRISPR technology described in (28). This system involves a Cas9 constitutively expressed from the FUCas9Cherry expression vector, and a doxycycline-dependent inducible sgRNA encode in the FgH1tUTG expression vector. Both plasmids were a gift from Marco Herold (Addgene plasmid #70182; http://n2t.net/addgene:70182; RRID:Addgene_70182 and #70183; http://n2t.net/addgene:70183; RRID:Addgene_70183). To design sgRNAs IDT (https://eu.idtdna.com/site/order/designtool/index/CRISPR_CUSTOM) and CRIS-POR (http://crispor.tefor.net/) tools were used. Designed sgRNAs were inserted into the expression vector FgH1tUTG. The sequences of the sgRNA guides were: sgPhf14: 5′-TCCCCCAAGAAATGGAATCTTCGT-3′ and sgHmg20a: 5′ TCCCGCACTGCCATACGGACCCTC-3′. These vectors were packaged on lentiviral particles as described in (29). NMuMG cells were first infected using 8 μg/ml polybrene with lentiviral particles containing the FUCas9Cherry vector. Single Cherry-positive cells were then sorted and grown in 96-well plates. One clone was chosen to be subsequently infected with lentiviral particles containing FgH1tUTG-sgHmg20a or FgH1tUTG-sgPhf14. After that, single Cherry- and GFP-positive cells were grown on 96-well plates for further analysis of the system efficiency. Clones were validated by RT-qPCR and western blot by treating cells with different concentrations of doxycycline (Sigma-Aldrich D9891) (0.1–1 μg/ml) for several periods of time.
Flag-HMG20A expression and affinity purification for proteomic analysis
Eight million HEK293T cells were seeded per 10-cm plate and transfected 16 h later with 1 μg of pCMV-FLAG-HMG20A (human coding sequence of HMG20A into BamHI/XhoI of pCMV-Tag-2B) or empty control vector with the calcium-phosphate method. After 48 h, cells were washed with PBS, collected and stored at −80°C as pellets.
Nuclei were purified from cell pellets corresponding to 24 10-cm plates of transfected HEK293T cells per sample using a sucrose cushion at 4°C. Briefly, cells were resuspended in sucrose buffer (0.32 M sucrose, 10 mM Tris pH 8, 3 mM CaCl2, 2 mM MgOAc, 0.1 mM EDTA) freshly added 1 mM PMSF and 50 μg/ml aprotinin. A 10% Triton X-100 solution was added to a final concentration of 0.1%. Cell suspensions were incubated on ice and nuclei were pelleted at 2800 rpm. Pelleted nuclei were washed once in sucrose buffer. For the preparation of a high salt extract, nuclei were first resuspended in 1.2 ml of low salt buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 20 mM KCl, 0.2 mM EDTA, 25% glycerol (v/v), 1 mM PMSF). Then, one volume of high salt buffer (20 mM HEPES pH 7.9, 1.5 mM MgCl2, 800 mM KCl, 0.2 mM EDTA, 25% glycerol (v/v), 1% NP-40, 1 mM PMSF) was added while vortexing. Nuclei suspensions were incubated for 1 h while rotating. Soluble and insoluble fractions were separated by ultracentrifugation at 50 000 rpm for 45 min. The supernatant was dialyzed to binding buffer (10 mM Tris pH 7.5, 135 mM NaCl, 10% glycerol (w/v), 0.05% Triton X-100) overnight. Aggregates were removed by centrifugation. Extracts were cleared by incubating with 200 μl of BSA-blocked Sepharose beads for 1 h. For affinity binding, extracts were incubated with 90 μl BSA-blocked anti-FLAG M2 beads (Sigma) overnight, while rotating. Beads were transferred to vertical columns and washed with 75 ml of binding buffer by gravity flow. Specifically bound proteins were eluted by incubating beads with 200 μM FLAG peptide in PBS (three times with 200 ul). Eluate fractions were checked by anti-FLAG immunoblotting and those of interest were pooled. Proteins were precipitated using acetone precipitation and dried with and SpeedVac vacuum concentrator.
Mass spectrometry
Protein samples were dissolved in 6 M urea, 200 mM ammonium bicarbonate, reduced with dithiothreitol (10 mM, 1 h, 37°C) and alkylated in the dark with iodoacetamide (20 mM, 30 min, 25°C). The protein mixture was then diluted 10 times with 200 mM ammonium bicarbonate before being digested at 37°C overnight with trypsin (ration protein:enzyme 10:1). Peptides generated in the digestion were desalted with a C18 column (Empore 3M, St. Paul, MN, USA), evaporated to dryness and resuspended in 0.1% formic acid.
Samples were analyzed using an LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific) coupled to an Agilent Technologies 1200 Series Gradient HPLC System. Peptides were loaded onto a C18 Zorbax precolumn (Agilent Technologies), and were separated by reversed-phase chromatography using a 12-cm column with an inner diameter of 75 μm, packed with 5 μm C18 particles (Nikkyo Technos Co.) using a 60 minute chromatographic gradients (3–35% acetonitrile, 0.1% formic acid). The mass spectrometer was operated in positive ionization mode with nanospray voltage set at 2.5 kV and source temperature at 200°C. Ultramark 1621 was used for the external calibration of the FT mass analyzer prior the analyses. An internal calibration was also performed using the background polysiloxane ion signal at m/z 445.1200. The instrument was operated in data-dependent acquisition (DDA) mode and full MS scans, with 1 micro scans at resolution of 60 000, over a mass range of m/z 350–2000 with detection in the Orbitrap. Auto gain control (AGC) was set to 100 000, dynamic exclusion (60 s) and charge state filtering disqualifying singly charged peptides were activated. In each cycle of DDA analysis, following each survey scan, the top ten most intense ions, with multiple charged ions, above a threshold ion count of 5000 were selected for fragmentation at normalized collision energy of 35%. Fragment ion spectra produced via collision-induced dissociation (CID) were acquired in the Ion Trap, AGC was set to 50 000 and isolation window to 2.0 m/z. All data were acquired with the Xcalibur software.
Mass spectrometry data analysis
Proteome Discoverer software suite (Thermo Fisher Scientific) and Mascot search engine (Matrix Science) were used for peptide identification and quantitation. Data were searched against the Swiss-Prot human database. A precursor ion mass tolerance of 7 ppm at the MS1 level was used, and up to three miscleavages for trypsin were allowed. The fragment ion mass tolerance was set to 0.5 Da. Methionine oxidation and N-terminal protein acetylation were used as variable modifications, whereas carbamidomethylation on cysteines was set as a fixed modification. False discovery rate (FDR) in peptide identification was set to a maximum of 5%. Raw proteomics data are currently being deposited in the PRIDE repository and will be available in the revised version of the manuscript.
ChIP assays
Chromatin immunoprecipitation (ChIP) assays were performed as previously described (29). Quantification of immunoprecipitated DNA was performed by qPCR. Provided data are the result of two-three independent biological replicates with three technical replicates each. For HMG20A ChIPs, anti-HMG20A antibodies (Sigma, Cat# HPA008126) were used. For PHF14 ChIPs, two different anti-PHF14 antibodies (ProteinTech, Cat# 24787-1-AP and Sigma, SAB1304812) were used, however, no specific PHF14 ChIP signal was obtained. All oligonucleotide sequences used are listed in Supplementary Table S1.
Alphafold2 modeling
AlphaFold2 was downloaded from https://github.com/deepmind/alphafold/releases. To perform PHF14-HMG20A complex models we used Alphafold_multimers (v2.1.1) (30) with –db_preset=reduced_dbs, –model_preset=multimer –max_template_date=2021-11-01, and default parameters. Full length amino acid sequences of human PHF14 and HMG20A were used. Computation was performed in the CESGA Supercomputing Center. RCSB PDB (https://www.rcsb.org/3d-view) or Swiss PDB viewer (DeepView) were used for structure viewing.
Statistical analysis
Statistical and graphical data analyses were performed using either Prism 5, Prism 9 (Graphpad) software or R package. To determine the significance of the differences between groups, two-tailed Student’s t-tests with confidence interval of 95% were computed. For correlations, the parametric Pearson coefficients or the non-parametric Spearman coefficients  were computed and significance was calculated using Fisher exact test. Significance of enrichments was calculated with the Fisher exact test using the fisher.test() function from R. Probabilities of overlapping were calculated using the hypergeometric distribution using dhyper() function from the stats R package or the Keisan Online Calculator (https://keisan.casio.com/exec/system/1180573201). Data were judged to be statistically significant when P value <0.05 in applied statistical analyses, unless otherwise noted. Other statistical methods are described above.
were computed and significance was calculated using Fisher exact test. Significance of enrichments was calculated with the Fisher exact test using the fisher.test() function from R. Probabilities of overlapping were calculated using the hypergeometric distribution using dhyper() function from the stats R package or the Keisan Online Calculator (https://keisan.casio.com/exec/system/1180573201). Data were judged to be statistically significant when P value <0.05 in applied statistical analyses, unless otherwise noted. Other statistical methods are described above.
RESULTS
Structure–function relationships in HMG20A
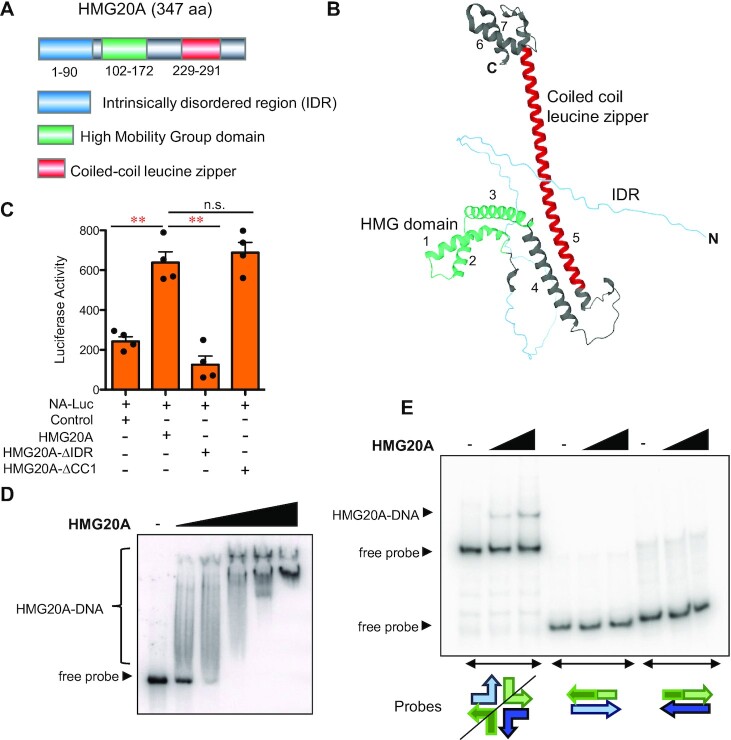
To gain further insight into the structural elements that determine the function of HMG20A, we took advantage of Alphafold2, a recently developed deep learning–based tool for protein structure prediction (31). The predicted structure of human HMG20A presents an amino (N)-terminal intrinsically disordered region (IDR) of 90 amino acids (aa), followed by seven alpha helices (labeled 1 to 7) that expand over the rest of the protein (Figure 1A, B). Mouse, rat and zebrafish HMG20A orthologues revealed a very similar structure (although the unstructured N-terminal region is shorter in zebrafish, from aa 1 to 42). Prediction of IDRs by the IUPred3 web server (32) also identified a region with low probability of secondary structure in the amino terminus of HMG20A (Supplementary Figure S1A). The first 80 amino acids are enriched for serines and acidic residues (isoelectric point 3.87 vs. 9.79 for the rest of the protein). As acidic domains are well-known transactivation domains (33,34), we assessed whether HMG20A can transactivate the SCN2A gene promoter (a previously identified target of HMG20A (5)) fused to a luciferase reporter gene in transient transfections. Of note, expression of the full-length HMG20A increased luciferase activity but a mutant lacking the N-terminal IDR (HMG20A-ΔIDR) did not (Figure 1C), revealing that this region has a necessary role in the transactivation activity of HMG20A.
Figure 1.
Structural elements of the HMG20A protein. (A) Schematic depiction of the domain architecture of human HMG20A. (B) HMG20A structure as predicted by AlphaFold2. Structural elements depicted are named and represented with the same colors as in A. Alpha helices are numbered from 1 to 7. (C) Reporter assays showing activation of the SCNA2 promoter by HMG20A in HEK293T cells. Cells were transfected with empty vector (control) or expression vectors encoding the indicated proteins. Data are the average of four determinations from two independent experiments ± SE. Significance of the difference between distributions was determined using two-tailed Student’s t-tests. **P < 0.01. (D) Electrophoretic mobility shift assays (EMSA) of a double-stranded synthetic probe with increasing quantities (0.06, 0.3, 0.6, 1.5 and 3 μg) of purified His-HMG20A. (E) EMSA of four-way junction (4WJ) DNA structures with His-HMG20A (0.6 and 1.5 μg), in the presence of an excess of linear competitor DNA (poly(dI-dC)). As a control, EMSA was also performed with linear double-stranded probes containing the same sequence than the 4WJ structure. Schematics of the probes structure are shown.
Alpha helices 1, 2 and 3 display the typical ‘L’-shaped fold of the HMG box (35) (Figure 1B, Supplementary Figure S1B, C). The HMG box domains mediate either non–sequence-specific (e.g. HMGB-type proteins) or sequence-specific (e.g. SOX family) DNA binding activity. Amino acid sequence determinants that define sequence-specific or non- sequence-specific binding have yet to be clearly defined (36). As HMG20A can interact with several specific promoters (4,12), we opted to characterize its double-stranded DNA binding activity, using a purified recombinant 6× His-tagged version of HMG20A (His-HMG20A) (Supplementary Figure S1D) in electrophoretic mobility shift assays (EMSA). Increasing amounts of His-HMG20A retarded a synthetic probe containing five tandem repeats of a 25-bp, AT-rich sequence (Figure 1D). His-HMG20A generated a smear pattern, rather than a specific retarded complex, indicative of multiple, non-specific binding sites along the probe. Binding activity was also efficiently competed by poly(dA-dT), poly(dI-dC) and (to a lesser degree) poly(dC-dG) double-stranded DNA (Supplementary Figure S1E). Taken together, these data suggest that HMG20A binds DNA with either no or a very low sequence specificity. Several HMG box proteins are able to bind structured or bent DNA, such as 4-way junction structures (4WJ) (23,37). Notably, HMG20A was able to bind to a synthetic 4WJ DNA molecule in the presence of excess of poly(dI-dC) competitor DNA, but not to linear double-stranded oligonucleotides with the same sequence (Figure 1E). This indicates that HMG20A displays higher affinity for bent DNA than for linear DNA.
The long alpha helix 5 of HMG20A contains a CCD with a leucine zipper motif (Figure 1B, Supplementary Figure S1F). A 15-aa deletion (residues 252–266) of part of this motif (HMG20A-Δcc1) had no effect on the transactivation activity of the protein in transient transfection experiments (Figure 1C). However, we have previously shown that this deletion abolishes i) HMG20A homodimerization, ii) HMG20A’s heterodimerization with HMG20B, and iii) HMG20A’s interaction with the LSD1-CoREST complex (4,5). As HMG20A binds DNA with little sequence specificity, we postulate that the interactions between HMG20A and other proteins dictate its chromatin targeting.
HMG20A and PHF14 interact through their coiled-coil leucine zipper motifs
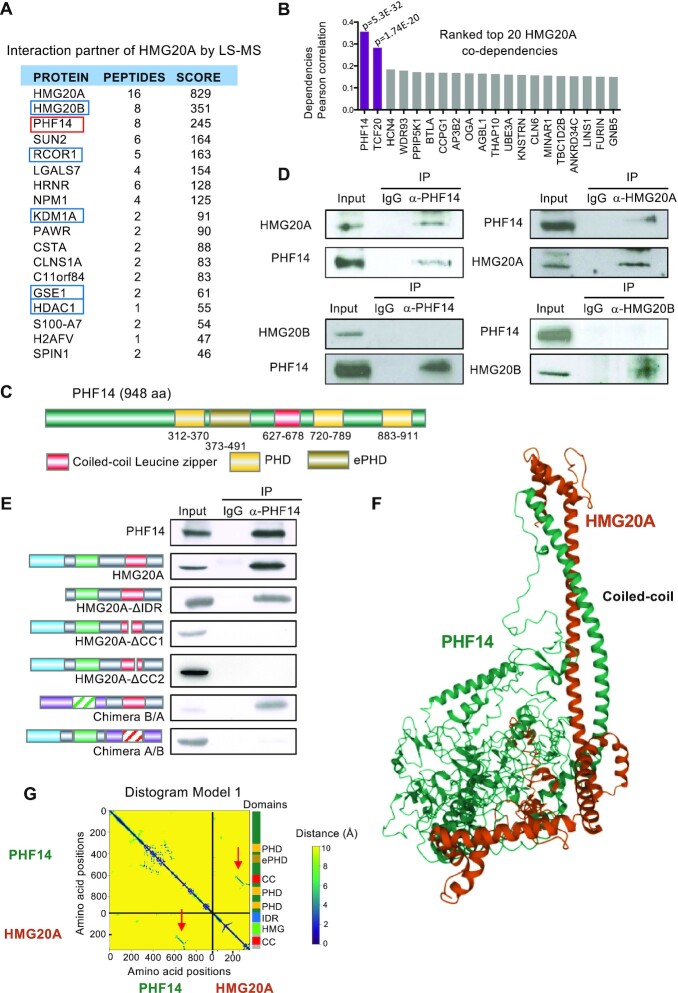
To identify additional putative interactors of HMG20A, we performed a proteomic analysis. After expressing a Flag-tagged version of HMG20A in HEK293T cells, we purified the Flag-HMG20A and its associated factors from nuclear extracts using anti-Flag antibodies (Supplementary Figure S2A, B). The same purification protocol was used with control cells that do not express Flag-HMG20A. Eluted proteins were subjected to liquid chromatography associated to mass spectrometry (LC-MS). Consistent with our previous work (4,5), five subunits of the LSD1-CoREST complex were identified as interactive partners: LSD1, RCOR1, HMG20B, GSE1, and HDAC1 (Figure 2A, Supplementary Table S2). In addition, novel interactors were also identified (including the proteins PHF14, SUN2, LGALS7, and NPM1), with comparable score to those of the previously identified interactors. Of particular interest is PHF14, for which we found a strong co-dependency (Pearson coefficient 0.36) with HMG20A in an analysis of the publicly available DepMap dependencies dataset (CRISPR(Avana) Public 21Q3) (38,39) (Figure 2B). Correlation between dependencies of HMG20A and PHF14 in 940 cell lines is shown in Supplementary Figure S2C. Furthermore, the mRNA levels of the HMG20A and PHF14 genes in the 1269 cell lines of the Cancer Cell Line Encyclopedia (CCLE) (40) are positively correlated (Pearson coefficient 0.31) (Supplementary Figure S2D). Taken together, these data suggest that PHF14 and HMG20A are functionally related.
Figure 2.
HMG20A interacts with PHF14 through the coiled-coil domain. (A) Mass spectrometry analysis of proteins co-purified with HMG20A. Peptides from these proteins were exclusively identified in HMG20A-immunoprecipitation but not in mock control. Chaperones and common proteomic contaminants are excluded from the list. Subunits of the LSD1/CoREST complex are in blue boxes. (B) Top 20 co-dependencies of HMG20A. Depicted values are Pearson coefficients of the correlation between dependencies of HMG20A gene in 940 cancer cell lines, versus those of the indicated genes (data from Depmap Portal). Pearson coefficients with P value ≤ 10–10 are shown in purple. (C) Schematic showing the domain architecture of human PHF14. (D) Co-immunoprecipitation of endogenous HMG20A or HMG20B with PHF14. Cell extracts were subjected to immunoprecipitation with the indicated antibody or with IgG bulk antibodies as control. (E) PHF14 interacts with the coiled-coil domain of HMG20A. HEK293T cells were transfected with expression vectors encoding wild-type or the indicated mutant versions of HMG20A. Cell extracts were subjected to immunoprecipitation with anti-PHF14 antibody or with IgG bulk antibodies as control. (F) Alphafold2_multimers prediction of the PHF14-HMG20A complex through the formation of a two-stranded alpha-helical coiled-coil. (G) Distogram showing predicted relative distances between the amino acids of HMG20A and PHF14 for the structural model shown in F. A stretch of short distances corresponding to the contacts predicted in the two-stranded alpha-helical coiled-coil are visible (indicated with red arrows). A schematic showing the domain architecture of HMG20A and PHF14 is represented on the right of the plot.
Consequently, we elected to study in detail the putative interactions between HMG20A and PHF14. PHF14 is a histone reader with three PHD domains and an extended PHD (ePHD) domain (Figure 2C) (16–18). We first confirmed that HMG20A, but not its close homolog HMG20B, interacted with PHF14 by co-immunoprecipitation of the endogenous proteins (Figure 2D). Next, we mapped the interaction domain of HMG20A using different mutants. Deletion of the N-terminal IDR of HMG20A did not impede the interaction (Figure 2E), while two different partial deletions of the coiled-coil leucine zipper (HMG20A-Δcc1 and HMG20A-Δcc2) (see Supplementary Figure S1F) completely suppressed its co-immunoprecipitation with PHF14 (Figure 2E). We also constructed chimeric proteins by replacing the N-terminal half of HMG20A with the same region of HMG20B (B/A chimera), or the C-terminal half of HMG20A by the same region of HMG20B (A/B chimera). Only the B/A chimera, which keeps the CCD of HMG20A, interacted with PHF14 (Figure 2E).
The Alphafold2 software has successfully been used to predict protein-protein complex formation and interaction domains (30,41). As such, we have modeled the complex formed by HMG20A and PHF14 using Alphafold2_multimers (30) (Figure 2F). In all the five predicted models, PHF14 interacted with HMG20A through a two-stranded alpha-helical coiled-coil leucine zipper structure formed between the CCDs of both proteins. Predicted relative distances between the amino acids in the five models, represented as binned distance distributions (distograms), are shown in Figure 2G and Supplementary Figure S2E. The leucine zipper motif of PHF14 is shown in Supplementary Figure S2F.
Finally, we also studied the nuclear distribution of co-expressed HA-HMG20A and Flag-PHF14 by immunofluorescence, using confocal microscopy. Both factors displayed a finely punctuated staining pattern inside the nuclei (Supplementary Figure S3A). Most puncta were stained with both antibodies, as revealed by colocalization analysis (Pearson's correlation coefficient of approximately 0.8) (Supplementary Figure S3B). However, a very significant drop of colocalization was observed when HA-HMG20A-Δcc1 was used. Taken together, these data strongly suggest that HMG20A and PHF14 form a stable nuclear complex by interacting through their CCDs.
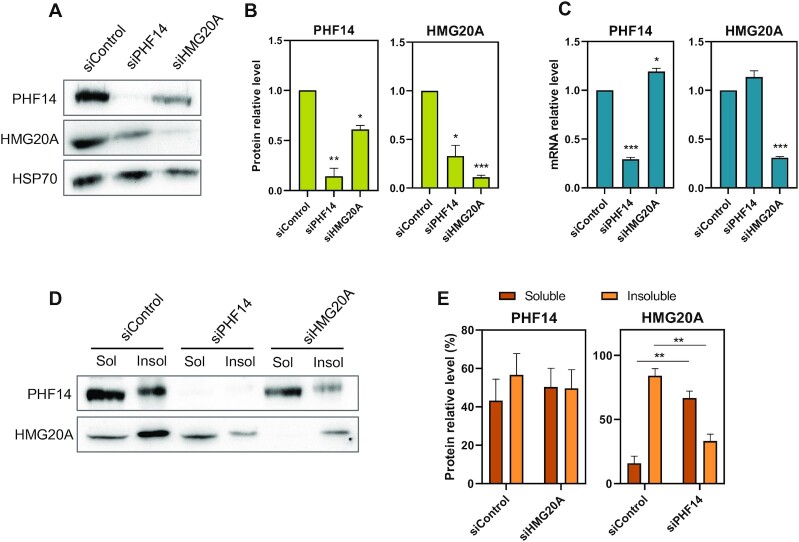
HMG20A levels and localization are impaired in PHF14-depleted cells
Protein levels of complexes subunits are often coordinated (42,43). Consistent with this idea, we observed that MDA-MB-231 cells with an siRNA-mediated PHF14 knockdown (KD) had decreased HMG20A protein levels but slightly increased HMG20A mRNA levels (Figure 3A–C). HMG20A KD also slightly decreased protein levels of PHF14 but not of its mRNA. These results indicate that the control is exerted at the protein, not at the mRNA level. Further, fractionation experiments demonstrated that PHF14 KD strongly altered the solubilization of HMG20A, leading to its enrichment in the soluble fraction; however, PHF14 solubilization was not modified after HMG20A KD (Figure 3D, E). These results suggest that HMG20A is associated with the nuclear compartment, at least in part, through its interaction with PHF14.
Figure 3.
Protein levels of HMG20A decreased in the absence of PHF14. (A) Determination by immunoblotting of PHF14 and HMG20A protein levels upon treatment of MDA-MB-231 cells with siPHF14, siHMG20A or a scrambled siRNA (siControl) for 48 h. HSP70 protein levels were also determined as a control. (B) Quantification of PHF14 and HMG20A protein levels from three independent immunoblotting experiments. Data were normalized with respect to HSP70. (C) Determination of the HMG20A and PHF14 mRNA by RT-qPCR in cells treated with siControl, siHMG20A or siPHF14. Data were normalized with respect to GAPDH mRNA levels. (D) MDA-MB-231 cells were treated with siPHF14, siHMG20A or siControl during 48 h. After fractionation, the presence of PHF14 and HMG20A proteins in the soluble (Sol) and insoluble (Insol) fractions was detected by immunoblotting. (E) Quantification of the PHF14 or HMG20A levels present in the soluble or insoluble fractions, as obtained in three independent experiment. (B, C, E) Data are the average of three independent experiments ± SE. Significance of the difference between distributions was determined using two-tailed Student’s t-tests. *P < 0.05; **P < 0.01, ***P < 0.001.
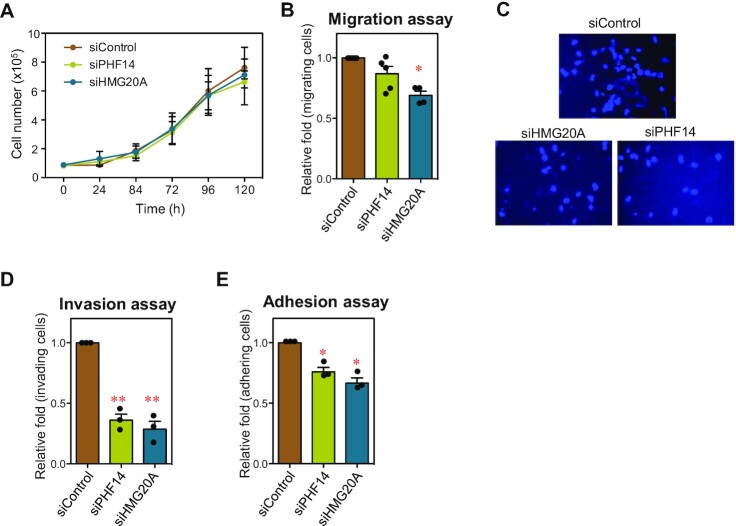
PHF14 and HMG20A control migration, invasion and cell–cell adhesion
Given the tight physical interaction between PHF14 and HMG20A, we speculated that they might perform common functions. Therefore, we functionally characterized the effects of the PHF14 KD or HMG20A KD in MDA-MB-231 cells, a human triple-negative breast cancer (TNBC) cell line. Cell proliferation was not significantly affected in either case (Figure 4A). Analysis of the Depmap database (39,40) revealed that, in general, mutation of PHF14 had little effect or even a positive effect on cell proliferation, while the effect of HMG20A mutation is cell type–specific (Supplementary Figure S4A, B). We have previously shown that HMG20A deficiency causes defects in cell migration (4). Either PHF14 KD or HMG20A KD caused a weak reduction in motility in wound-healing assays in MDA-MB-231 cells (Figure 4B). However, a strong decrease of invasiveness was observed in Boyden chamber migration assays in cells depleted of any of the two factors (Figure 4C, D). Finally, homotypic adhesion assays demonstrated that HMG20A KD or PHF14 KD cells showed a reduction in cell–cell adhesion (Figure 4E). These analyses confirmed that deficiency of PHF14 or HMG20A cause similar cellular phenotypic alterations.
Figure 4.
HMG20A and PHF14-depleted cells display common phenotypes. (A) Growth curve of MDA-MB-231 cells treated with siControl, siHMG20A or siPHF14. (B) Wound-healing assays were performed with MDA-MB-231 cells treated for 48 h with the indicated siRNAs. Quantification of the assays is represented. (C) Transwell invasion analyses of cells treated during 48 h with the indicated siRNAs. Micrographs of DAPI-stained cells migrated to the opposite side of the transwell membrane are shown. (D) Quantification of the invasion assays. (E) Quantification of the cell-cell adhesion assays under the indicated conditions. (A, B, D, E) Values are the average of three to five independent experiments ± SE. Significance of the differences with respect to the control were determined using two-tailed Student’s t-tests. *P ≤ 0.05; **P ≤ 0.01.
The AKT-mTOR pathway is not altered following PHF14 or HMG20A depletion
The AKT-mTOR pathway has been shown to be modulated by PHF14 (44,45). As the AKT pathway is involved in cell migration and invasion (46), we next investigated if it was also altered in HMG20A KD and/or PHF14 KD cells by analyzing its level of activation. Thus, we monitored by Western blot AKT activation via the phosphorylation of residues T308 and S473, as well as the phosphorylation of the well-known downstream targets of mTOR, S6K (at T389) and S6 (at S235/236). As shown in Supplementary Figure S5A, knockdown of PHF14, HMG20A or both genes did not alter signaling though the AKT-mTOR pathway under steady state conditions. We therefore assessed whether an acute stimulus would alter this pathway. To this end, MDA-MB-231 cells were subjected to serum starvation during 3 h and then treated with 100 nM of insulin during 3 min) which strongly induced phosphorylation of AKT and S6K. Depletion of PHF14, HMG20A or both genes did not change significantly the level of phosphorylation of the indicated proteins (Supplementary Figure S5B). In sum, our data indicate that HMG20A and PHF14 are not involved in controlling the AKT-mTOR pathway in MDA-MB-231 cells.
PHF14 KD or HMG20A KD causes defects of the Hippo pathway
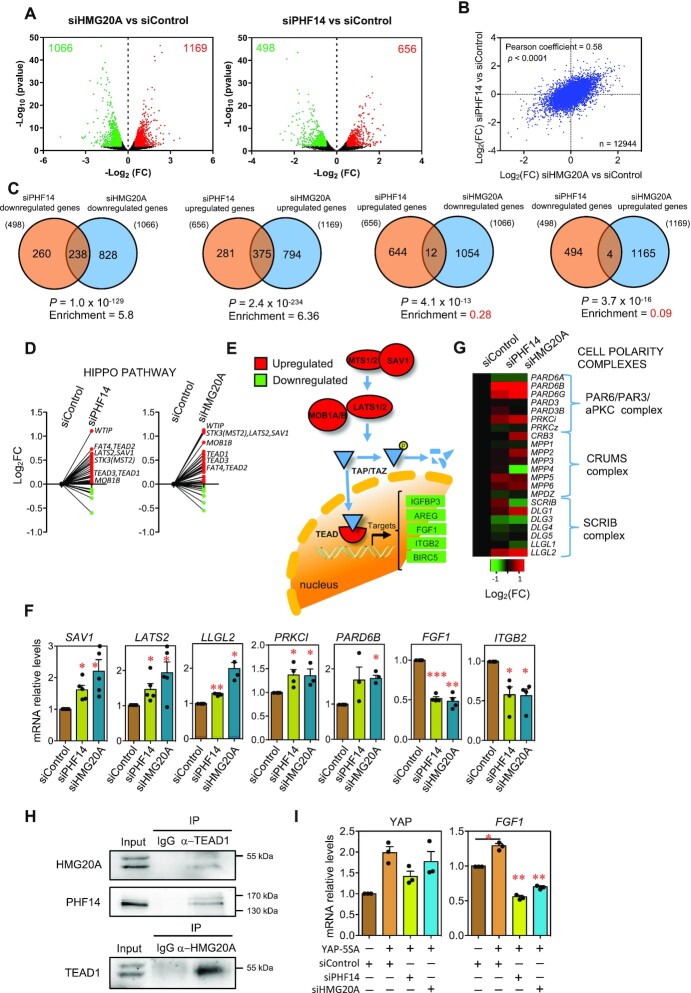
Next, we analyzed changes in the transcriptome in HMG20A KD or PHF14 KD MDA-MB-231 cells by RNA sequencing (RNA-seq). Principal component analysis (PCA) of the data demonstrated that both replicates of each condition (siHMG20A, siPHF14 or siControl) displayed a similar behavior (Supplementary Figure S6A). Differential expression analysis demonstrated that the HMG20A KD deregulated 2,235 genes (adjusted P value < 0.05 and |log2(fold change, FC)| > 0.5), with 1169 upregulated and 1066 downregulated; in turn, PHF14 KD deregulated 1154 genes (adjusted P value < 0.05 and |log2(FC)| > 0.5), with 656 upregulated and 498 downregulated (Figure 5A, Supplementary Table S3). Interestingly, changes in gene expression between the two KD conditions were highly correlated with each other (Pearson correlation coefficient = 0.56) (Figure 5B). Consistently, we observed a very significant enrichment of genes simultaneously downregulated (238 genes, 5.8-fold enrichment, P = 1.0 × 10–129) or upregulated (375 genes, 6.36-fold enrichment, P = 2.4 × 10–234) upon depletion of either HMG20A or PHF14 (Figure 5C). In contrast, we also observed that the number of conversely regulated genes was much lower than randomly expected (Figure 5C). These data indicate that a large subset of genes is commonly regulated by PHF14 and HMG20A, in agreement with the physical and genetic interactions described above. Gene ontology (GO) analysis and gene set enrichment analysis (GSEA) (27) of differentially expressed genes indicated that PHF14 and HMG20A are involved in the regulation of several epithelial-mesenchymal plasticity signaling pathways (Supplementary Figure S6, Supplementary Figure S7). For instance, upregulated genes were enriched in regulation of signaling, epithelium development, Hippo signaling and regulation of transcription GO categories, while downregulated genes were enriched in extracellular matrix organization, cell migration, cell adhesion and localization of cell categories (Supplementary Figure S6B, C). Many of the genes that were upregulated in HMG20A KD cells, or up- or downregulated in PHF14 KD cells, were also found concomitantly deregulated in the osteosarcoma U2OS cell line upon HDAC1/2 KD (47) (Supplementary Figure S7A, B).
Figure 5.
PHF14 or HMG20A depletion activates the Hippo pathway. MDA-MB-231 cells treated with siPHF14, siHMG20A or siControl for 48 h were analyzed by RNA-seq. (A) Volcano plots of the RNA-seq data at the indicated conditions. Significant deregulated genes (adjusted p value < 0.05 and |log2(FC)| > 0.5) are depicted in red (upregulated) or green (downregulated). Numbers inside the plots indicate downregulated or upregulated genes. (B) Correlation between gene expression changes (log2(FC)) after HMG20A or PHF14 KD. (C) Venn diagrams representing overlapping of deregulated genes after knocking down the indicated genes. Enrichment with respect to the randomly expected values, and probability of the result (P) assuming an hypergeometric distribution, are also indicated for the different comparisons. (D) Before–after plots showing changes of mRNA levels (log2(FC)) in Hippo pathway genes after KD of PHF14 (left panel) or HMG20A (right panel). (E) Schematic depiction of the Hippo pathway. Proteins in red or green are upregulated or downregulated, respectively, after HMG20A and PHF14 KD. (F) Determination of SAV1, LATS2, LLGL2, PRKCI, PARD6B, FGF1 and ITGB2 mRNA levels by RT-qPCR in MDA-MB-231 cells at 48 h after transfection with the indicated siRNAs. (G) Heatmap showing changes of mRNA levels (log2(FC)) of genes encoding proteins of cell polarity complexes. (H) Co-immunoprecipitation of endogenous HMG20A and PHF14 with TEAD1. Cell extracts were subjected to immunoprecipitation with the indicated antibody or with IgG bulk antibodies as control. (I) MDA-MB-231 were transfected with the indicated siRNAs and 24 h later infected with retrovirus expressing YAP-5SA protein. 48h later samples were processed for RNA isolation and YAP and FGF1 mRNA levels were determines by RT-qPCR. (F, I) Values are the average of three to five independent experiments ± SE. Significance of the differences with respect to the control were determined using two-tailed Student’s t-tests. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
The evolutionarily conserved Hippo pathway integrates diverse signals, such as cell–cell and cell–matrix contacts, mechanical cues, cellular energy status, and various stresses, to control cell growth, differentiation, tissue/organ development, and homeostasis (48,49). A low signaling through the pathway is associated with cancer initiation, progression, or metastasis (50). Depletion of HMG20A or PHF14 in MDA-MB-231 cells increased the mRNA levels of most members of the Hippo pathway, including mRNAs for WTIP, LATS2, SAV1, MOB1B, MST2(STK3) and several TEAD proteins (Figure 5D, E, Supplementary Table S3). Upregulation of SAV1 and LATS2 was confirmed by RT-qPCR (Figure 5F, Supplementary Figure S6D for confirmation of SAV1 mRNA levels with a different siRNA). Cell polarity is fundamental for the functionality of epithelial cells (51). It has recently been shown that the LLGL, PRKCI, and CRB proteins, members of the three described cell polarity complexes, regulate the Hippo pathway (49). Our RNA-seq analyses indicated that 10–12 of the 22 genes encoding subunits of the cell polarity complexes were also upregulated upon HMG20A KD or PHF14 KD (Figure 5G). Increased levels of PARD6B, PRKCI and LLGL2 mRNAs in siHMG20A- and siPHF14-treated cells were confirmed by RT-qPCR (Figure 5F). These results indicate that HMG20A or PHF14-depleted cells presented an alteration of the Hippo pathway.
The Hippo pathway effectors YAP and TAZ are paralogous transcription co-activators that cooperate with TEAD transcription factors to activate genes involved in proliferation and organ growth (Figure 5E) (48,49). When the Hippo pathway is activated, YAP/TAZ are phosphorylated by the LATS1/2 kinases, resulting in their proteasomal degradation. We found that several well-known YAP/TAZ-TEAD target genes, such as IGFBP3, ITGB2, FGF1, and BIRC5, were downregulated in HMG20A KD or PHF14 KD cells (Figure 5E, Supplementary Table S3). Decreased levels of FGF1 and ITGB2 mRNAs in HMG20A KD or PHF14 KD cells were confirmed by RT-qPCR (Figure 5F). However, despite decreased expression of YAP/TAZ-TEAD target genes, we found that levels of phospho-LATS1 and YAP proteins were not significantly affected by HMG20A KD or PHF14 KD (Supplementary Figure S7C), suggesting that signaling through the cytoplasmic part of the Hippo pathway was not altered in the absence of normal levels of these chromatin factors. Next, we reasoned that if signaling through the pathway is normal, defects in gene activation should be due to a direct interaction of HMG20A and/or PHF14 with the YAP/TAZ-TEAD complex. In fact, we were able to demonstrate by co-immunoprecipitation that endogenous HMG20A and PHF14 interact with TEAD1 protein (Figure 5H), suggesting that the HMG20A-PHF14 complex cooperates with the YAP/TAZ-TEAD complexes for gene activation. Consistent with this, downregulation of FGF1 caused by HMG20A or PHF14 KD, was not suppressed by expression of a YAP mutant (YAP-5SA) which is not under the negative control of the Hippo pathway due to mutation of key phosphorylation sites (22) (Figure 5I).
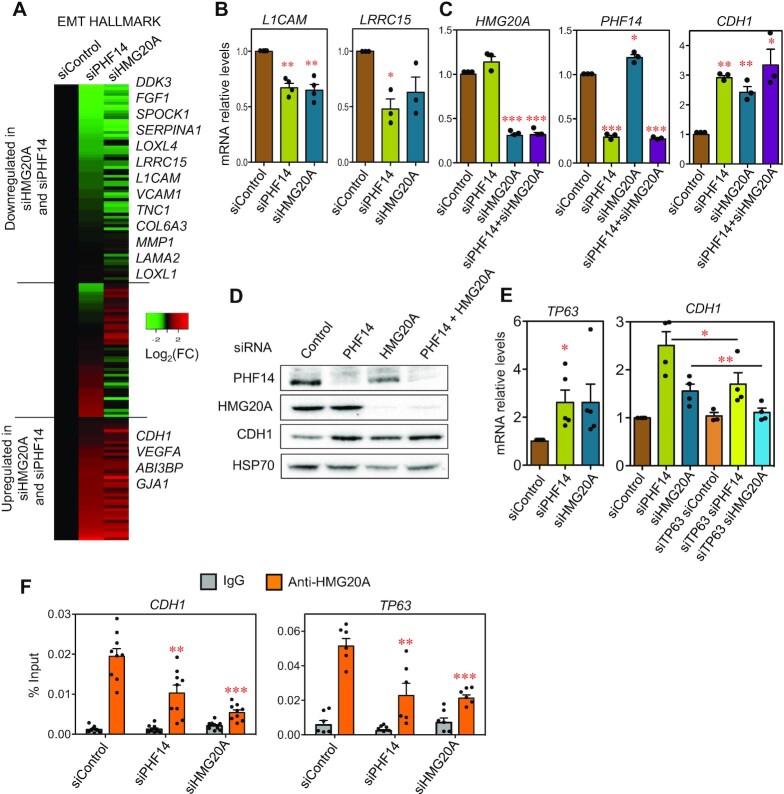
HMG20A KD or PHF14 KD MDA-MB-231 cells have increased epithelial markers
GSEA analysis also indicated that the Epithelial to Mesenchymal Transition Hallmark was enriched among the genes downregulated in both HMG20A KD and PHF14 KD cells (Figure 6A, Supplementary Figure S7A, B). Of note, HMG20A KD and PHF14 KD cells displayed downregulation of two well-known mesenchymal and pro-metastatic adhesion molecule genes, L1CAM and LRRC15 (52,53) (Figure 6B, Supplementary Figure S6D for confirmation of L1CAM mRNA levels with a different siRNA), consistent with the observed decrease in cell-cell adhesion in the depleted cells (Figure 4E). Concomitantly, a strong upregulation of E-cadherin (CDH1 gene), one of the best known cell–cell adhesion epithelial markers was observed (Figure 6C, D). Depletion of both HMG20A and PHF14 did not have greater effect than single KDs (Figure 6B, C). These results are consistent with an increase of the epithelial phenotypes upon depletion of PHF14 or HMG20A, as described above.
Figure 6.
HMG20A KD and PHF14 KD MDA-MB-231 cells display decreased mesenchymal and increased epithelial markers. (A) Heatmap showing changes of mRNA levels (log2(FC)) in genes encoding EMT hallmark proteins. (B, C) Determination of L1CAM, and LRR15 mRNAs (B), or PHF14, HMG20A, and CDH1 mRNAs (C) by RT-qPCR in MDA-MB-231 cells 48 h after transfection with the indicated siRNAs. (D) Immunoblot analysis of PHF14, HMG20A and E-cadherin (CDH1) proteins in MDA-MB-231 cells treated during 48 h with the indicated siRNAs. HSP70 levels were also determined as control. (E) Determination of TP63 or CHD1 mRNAs by RT-qPCR in MDA-MB-231 cells at 48 h after transfection with the indicated siRNAs. (F) HMG20A binds to the CDH1 and TP63 promoters. MDA-MD-231 cells transfected with siControl, siHMG20A or siPHF14 were subjected to chromatin immunoprecipitation (ChIP) assays with anti-HMG20A and bulk IgG (control) antibodies. All data points of two (for TP63) or three (for CDH1) biological replicates with three technical replicate each are shown. (B, C, E) Values are the average of three to five independent experiments ± SE. (B, C, E, F) Significance of the differences with respect to the control were determined using two-tailed Student’s t-tests. *P ≤ 0.05; **P ≤ 0.01; ***P < 0.001.
Also consistent with an increase of epithelial features, the TP63 tumor-suppressor gene was upregulated in HMG20A KD or PHF14 KD cells (Figure 6E, Supplementary Figure S6D for confirmation with a different siRNA); TP63 is a master transcription regulator of the p53 family involved in mammary gland and skin epithelial development (54–56). Furthermore, TP63 KD partially suppressed the upregulation of CDH1 caused by HMG20A or PHF14 KD (Figure 6D, Supplementary Figure S7D). Taken together, our data indicate that HMG20A and PHF14 are important regulators of several pathways that affect the mesenchymal–epithelial plasticity in MDA-MB-231 cells.
To test whether HMG20A and PHF14 regulate CDH1 and TP63 genes directly or indirectly, we performed chromatin immunoprecipitation (ChIP) experiments using anti-HMG20A and anti-PHF14 antibodies in the CDH1 and TP63 promoters. We showed that HMG20A binds both promoters. As a control, we verified that HMG20A KD decreased HMG20A occupancy, confirming the specificity of the ChIP signals (Figure 6F). Interestingly, PHF14 KD also decreased the occupancy of HMG20A at both the CDH1 and TP63 promoters, consistent with the cooperation between these two factors. In contrast, we were unable to obtain specific ChIP signal using two different commercial anti-PHF14 antibodies (see Materials and Methods). Others have also reported that specific ChIP experiments cannot be done using either anti-PHF14 antibodies or anti-Flag antibodies with an endogenous Flag-tagged PHF14 (57). This fact is probably a consequence of the lack of contact between PHF14 and DNA.
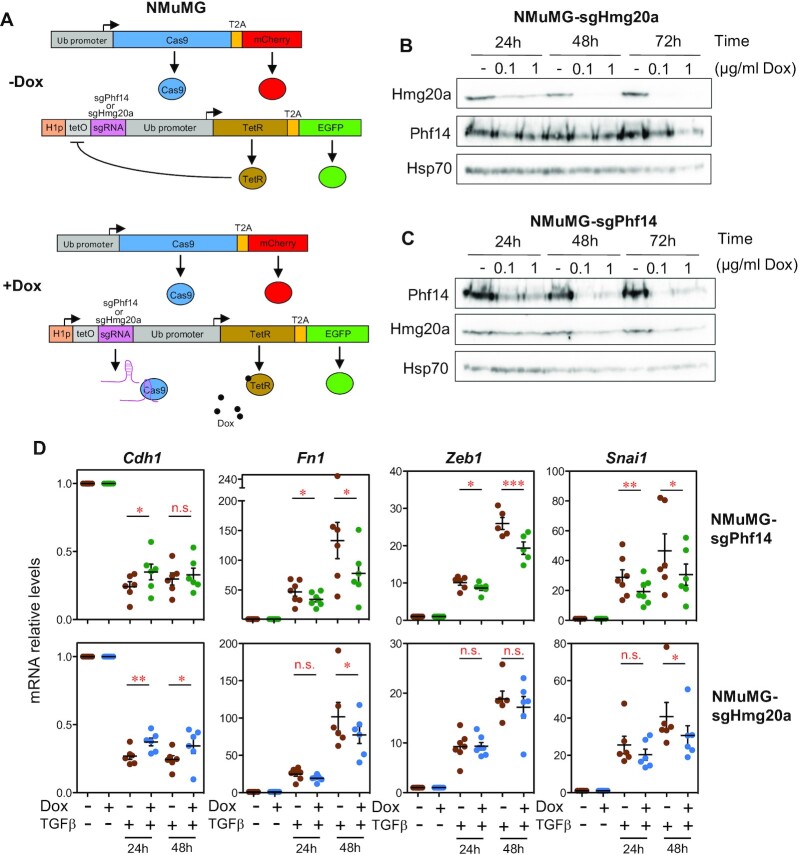
HMG20A and PHF14 are required for TGFβ-induced EMT
In PHF14 KD cells, we observed that TGFβ-upregulated and TGFβ-downregulated gene sets were enriched among downregulated and upregulated genes, respectively (Supplementary Figure S7B). To investigate whether HMG20A and PHF14 are required for TGFβ signaling, we selected the murine epithelial mammary non-tumoral NMuMG cell line, in which EMT can be induced by TGFβ (58). We used a lentiviral doxycycline (Dox)-inducible CRISPR/Cas9 system (28) to knock out the Hmg20a or Phf14 genes (Figure 7A). Efficiency of the CRISPR/Cas9-mediated inactivation was evaluated at different Dox concentrations and days of treatment. A treatment with Dox for 48 h was enough to strongly decrease the Hmg20a or Phf14 protein levels at the two concentrations used (Figure 7B, C). We also observed a strong decrease in the level of Hmg20a upon inactivation of Phf14, confirming the observation performed in the human MDA-MB-231 cancer cells. After 48 h of Dox treatment, depletion of Phf14 or Hmg20a was stable for at least the following four passages, even when Dox was removed from the culture media (Supplementary Figure S8A). Proliferation of Phf14 or Hmg20a mutant cells was not significantly affected with respect to the parental cell line even after 6 passages in the presence of Dox (Supplementary Figure S8B). Next, we triggered EMT by TGFβ addition to 48 h Dox-treated cells or to control cells. EMT-dependent repression of Cdh1 and induction of Fn1 and Snai1 genes were impaired in the Hmg20a or Phf14 mutated cells (Figure 7D). Induction of Zeb1 was also impaired in Hmg20a or Phf14 mutated cells, although it was not significant in Hmg20a mutants. These results indicate that both proteins are required for the correct TGFβ−dependent regulation of expression of typical EMT markers.
Figure 7.
Inducible CRISPR/Cas9 Phf14 or Hmg20a mutants in mouse NMuMG cells display impaired EMT. (A) Diagram depicting the Dox-inducible CRISPR/Cas9 system used to generate Phf14 and Hmg20a mutant NMuMG cell lines (28). (B, C) Immunoblotting showing Phf14 and Hmg20a protein levels, at the indicated times, after induction of CRISPR/Cas9-dependent mutagenesis of Phf14 and Hmg20a genes by addition of doxycycline (Dox). (D) Evolution of EMT markers upon induction of EMT by TGFβ addition to wild type (-Dox) or Phf14 and Hmg20a induced mutants (+Dox). Levels of Cdh1, Fn1, Zeb1 and Snai1 mRNA were determined by RT-qPCR at the indicated times. Values are the average of six to seven independent experiments ± SE. Significance of the differences with respect to the control were determined using two-tailed Student’s t-tests. *P ≤ 0.05; **P ≤ 0.01; ***P < 0.001.
DISCUSSION
Here we describe that the HMG protein HMG20A forms a nuclear complex with the histone reader PHF14 through the establishment of a two-stranded alpha-helical coiled-coil. We have previously shown that HMG20A also forms homodimers as well as heterodimers with HMG20B through this CCD, and interacts with the LSD1-CoREST complex through PHF21A. Taken together, these data reveal HMG20A as an important chromatin regulator that displays a complex choreography of interactions (Supplementary Figure S9). We also demonstrate that the PHF14–HMG20A complex plays important roles in modulating several pathways related to the epithelial-mesenchymal plasticity.
Early mass spectrometry analysis of proteins associated with nucleosomes whose histones presented specific posttranslational modifications demonstrated that PHF14 and HMG20A were among the few proteins excluded from nucleosomes containing the H3K4me3 modification, but associated with H3K4-unmodified nucleosomes (59,60). This is consistent with very recent structural studies demonstrating that a module of PHF14, denoted as the PZP domain (formed by the first PHD and the ePHD domains), reads unmodified histone H3 (18). Co-purification of PHF14 with nucleosomes containing H2AX (61), H2A.Z (57,62–64) or macroH2A (65) has also been reported. Our data indicate that HMG20A binds DNA either with no, or with very little, sequence specificity. However, HMG20A binds 4WJ DNA with higher affinity than linear double-stranded DNA, suggesting that HMG20A displays significant affinity for bent DNA. Which chromatin substrate is being bound by the PHF14-HMG20A complex? HMG box-containing proteins do not normally bind intact nucleosomal DNA (66,67). In fact, they have been characterized by their ability to destabilize the nucleosomal structure (68–71). Of note, the SSRP1 protein (a subunit of the FACT complex, with an HMG box in its C-terminus and an H3/H4 tetramer interaction domain in its central part) cannot bind intact nucleosomes in vitro but does stably bind partially assembled nucleosome structures, such as the hexasome (H2A·H2B)·(H3·H4)2, or the tetrasome (H3·H4)2 (72–75). Similar to SSRP1, the PHF14-HMG20A complex displays both a histone interaction domain in PHF14 and an HMG box in HMG20A. Therefore, a potential scenario is that the PHF14-HMG20A complex binds partially disassembled nucleosomes. This type of structures is common in regulatory regions such as enhancers and promoters (76,77). This is consistent with our ChIP data that identify HMG20A at the CDH1 and TP63 promoters.
How the PHF14-HMG20A complex controls transcription is currently unknown. Zheng et al. have shown that the PZP module (PHD1 + ePHD) of PHF14 binds unmodified histone H3, and that this interaction is abolished by some active histone marks, such as H3K4me3, H3K18ac and H3K23ac, but not by repressive marks, such as H3K9me3 and H3K27me3 (18). Further, they have proposed that PHF14 may associate to a chromatin ‘ground state’ that defines a repressive or poised state. Interestingly, our GSEA analyses indicated that PHF14 and HMG20A partially share targets with HDACs (Supplementary Figure S7). Therefore, it is possible that the PHF14-HMG20A complex contributes to maintaining chromatin in that ‘ground state’ by preserving the deacetylated state of specific nucleosomes. In the absence of the PHF14-HMG20A complex, or upon the appropriated signaling, histone acetyltransferases would gain access to these nucleosomes, causing transcription activation. Interestingly, in transient transfection experiments, in which the template is not assembled into normal chromatin, and HMG20A is probably not acting in a complex with PHF14, the IDR of HMG20A is able to activate transcription. IDRs of transcription factors and co-factors are involved in the formation of condensates by liquid-liquid phase separation, which have been associated with transcription activation (78,79). This fact suggests a possible dual activity of HMG20A that could serve as a bridge between repression and activation. Furthermore, H2A.Z has often been implicated in poised chromatin states (80). PHF14 has been found to co-purify with H2A.Z in four different proteomic studies (57,62–64). In addition, our proteomic analysis also identified H2A.Z.2 (H2AFV) as a HMG20A partner. These data are also in agreement with a role for the PHF14–HMG20A complex in maintaining poised configurations.
During the revision of this manuscript a paper describing the existence of a complex between PHF14 and the MECP2 protein, which also contains the TCF20 protein, has been published. They report that PHF14 interacts with MECP2 through the PZP region of PHF14. While they also describe that PHF14 co-immunoprecipitates with HMG20A, it is unclear whether a complex containing MECP2, PHF14, TCF20 and HMG20A exists (81).
We show here that HMG20A and PHF14 share an important number of targets related to the mesenchymal phenotype of the TNBC cell line MDA-MB-231. Defects in migration and invasion due to PHF14 KD have been previously reported in glioblastoma multiforme, lung, bladder and gastric cancer cell lines (44,45,82,83). However, the signaling pathways involved in this regulation are unclear. Zhao et al. and Park et al. have proposed that PHF14 modulates the AKT-mTOR pathway (44,45), while Wu et al. reported that the Wnt signaling pathway is affected by PHF14 silencing (83). However, our results in MDA-MB-231 cells revealed that the AKT-mTOR signaling pathway was not affected by either HMG20A or PHF14 KD, and our RNA-seq transcriptomic analysis does not support the downregulation of Wnt signaling in either KD condition. In contrast, we find that PHF14 and HMG20A modulate the Hippo pathway, the epithelial development master regulator TP63 and the TGFβ pathway. As interactions between these three pathways have been described (84,85), we cannot rule out that PHF14 and/or HMG20A are implicated in one of the pathways and thereby indirectly affect the others. Nevertheless, the data here presented, together with data previously reported, suggest direct roles of PHF14/HMG20A in these three pathways. First, we found that PHF14/HMG20A deficiency causes increased mRNA levels of genes encoding several members of the Hippo cytoplasmic kinases cascade, and decreased expression of YAP/TAZ-TEAD gene targets. We were unable to demonstrate an increased activation of the Hippo pathway, suggesting a defect at the level of the final transcription factor effectors, the YAP/TAZ-TEAD complexes. Consistent with this, we observed a direct interaction of TEAD1 with both HMG20A and PHF14. The mechanism by which the PHF14/HMG20A complex helps to activate the YAP/TAZ-TEAD targets is unknown. Indeed, as discussed above, the histone binding preferences of PHF14 suggest that the PHF14/HMG20A complex might be associated with poised regions (18). It is commonly accepted that TEAD factors bind to their DNA targets regardless of the presence or absence of YAP/TAZ coactivators—and therefore also under non-activation conditions (48,49,85). One possibility is that PHF14-HMG20A complex helps to stabilize TEAD factors at regulatory elements with a poised chromatin configuration. Thus, under conditions of PHF14/HMG20A deficiency, TEAD might not be properly positioned/stabilized in the chromatin, which in turn would lead to poor activation of YAP/TAZ-TEAD targets. Increased mRNA levels of genes encoding elements of the Hippo signaling cascade may be a consequence of the feedback regulatory mechanisms previously described (84).
Depletion of PHF14/HMG20A complex also affected TGFβ signaling. Previous results demonstrated interaction of HMG20A with the transcription factor SMAD4 (4,86), which acts as a mediator of TGFβ signal transduction. We have also shown that a Hmg20a knockdown in mouse astrocytes downregulates the EMT hallmark gene set (7). Furthermore, a very recent preprint shows that Hmg20a depletion in Xenopus laevis causes defects in neural crest cell migration, a developmental example of EMT (87). Finally, we also demonstrated direct binding of HMG20A to the promoter of the TP63 gene, a regulator of epithelial development (54–56). Taken together, these results seem to suggest that—during evolution and in different cellular systems—the PHF14/HMG20A complex plays important roles in several pathways related to the epithelial–mesenchymal plasticity.
In summary, we have uncovered the existence of a PHF14–HMG20A complex and characterized its functions in essential pathways. The fact that HMG20A has multiple partners increases the complexity of its function and raises important questions. For example, is PHF14 implicated in the well-known HMG20A functions in neuronal development or in islet beta-cell functions? Which molecular factors determine the association of HMG20A with its different partners (Supplementary Figure S9)? What is the genomic distribution of HMG20A and its different partners? These questions require further investigation.
DATA AVAILABILITY
Publicly available databases used in our analysis included the DepMap portal (21Q3 data release; https://depmap.org/), the CCLE portal (https://portals.broadinstitute.org/ccle), the TCGA Research Network (https://www.cancer.gov/tcga), the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb/), the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/). The structural model is available in ModelArchive at https://modelarchive.org/doi/10.5452/ma-1us57. These data can be accessed through the Token: asTzxrLlCj. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030769. All NGS data have been deposited to Gene Expression Omnibus (GEO) under the accesión number GSE193476.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Benoit Gauthier for critical reading of the manuscript and continuous discussion. We thank Michinori Kitagawa, Takumi Era, Terumi Kohwi-Shigematsu, Gail Mandel, Jose Antonio Pintor-Toro Kunliang Guan and Abelardo López-Rivas for providing different plasmids, antibodies and cell lines. We thank Cristina Chivas and Eva Borras from UPF-CRG Proteomics facility lead by Eduard Sabidó for MS sample preparation and MS raw data analysis, E. Andújar and M. Pérez from the CABIMER Genomics Unit for NGS and P. Domínguez-Giménez from the CABIMER Microscopy Unit.
Contributor Information
Elena Gómez-Marín, Genome Biology Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
Melanija Posavec-Marjanović, Program for Predictive and Personalized Medicine of Cancer, Germans Trias i Pujol Research Institute (PMPPC-IGTP), Badalona, Spain.
Laura Zarzuela, Cell Dynamics and Signaling Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
Laura Basurto-Cayuela, Genome Biology Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
José A Guerrero-Martínez, Genome Biology Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
Gonzalo Arribas, Genome Biology Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
Rosario Yerbes, Cell Dynamics and Signaling Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
María Ceballos-Chávez, Genome Biology Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
Manuel Rodríguez-Paredes, Institute of Toxicology, University Medical Center Mainz, Johannes Gutenberg University, 55131 Mainz, Germany.
Mercedes Tomé, Cell Dynamics and Signaling Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
Raúl V Durán, Cell Dynamics and Signaling Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
Marcus Buschbeck, Program for Predictive and Personalized Medicine of Cancer, Germans Trias i Pujol Research Institute (PMPPC-IGTP), Badalona, Spain; Cancer and Leukaemia Epigenetics and Biology Program, Josep Carreras Leukaemia Research Institute (IJC), 08916 Badalona, Spain.
José C Reyes, Genome Biology Department. Centro Andaluz de Biología Molecular y Medicina Regenerativa-CABIMER, Consejo Superior de Investigaciones Científicas-Universidad de Sevilla-Universidad Pablo de Olavide (CSIC-USE-UPO), Av. Americo Vespucio, 41092 Seville, Spain.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Research in the J.C. Reyes lab was funded by the Spanish Ministry of Economy and Competitiveness MCIN/AEI/10.13039/501100011033/ [PID2020-118516GB-I00]; Junta de Andalucía [P18-FR-1962 and BIO-321]; Fundación Vencer El Cancer (VEC); European Union FEDER ‘A way to build Europe’ program; research at Buschbeck lab is further supported by FEDER/Ministerio de Ciencia e Innovación - Agencia Estatal de Investigación [RTI2018-094005-B-I00]; Marie Skłodowska Curie Training network ‘INTERCEPT-MDS’ [H2020-MSCA-ITN-2020–953407]; Deutsche José Carreras Leukämie Stiftung [DJCLS 14R/2018, AGAUR 2017-SGR-305]; Fundació La Marató de TV3 257/C/2019; CABIMER is a Center partially funded by the Junta de Andalucía; E.G.-M. is the recipient of an FPI fellowship from the Spanish Ministry of Science and Innovation; L.B.-C. is the recipient of an FPU fellowship from the Spanish Ministry of Education; M.P.M. was funded by an AGAUR FI-2010 fellowship. Funding for open access charge: Spanish Ministry of Economy and Competitiveness MCIN/AEI/10.13039/501100011033/ [PID2020-118516GB-I00].
Conflict of interest statement. None declared.
REFERENCES
- 1. Wynder C., Hakimi M.A., Epstein J.A., Shilatifard A., Shiekhattar R.. Recruitment of MLL by HMG-domain protein iBRAF promotes neural differentiation. Nat. Cell Biol. 2005; 7:1113–1117. [DOI] [PubMed] [Google Scholar]
- 2. Hakimi M.A., Bochar D.A., Chenoweth J., Lane W.S., Mandel G., Shiekhattar R.. A core-BRAF35 complex containing histone deacetylase mediates repression of neuronal-specific genes. Proc. Natl. Acad. Sci. U.S.A. 2002; 99:7420–7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lee M.G., Wynder C., Cooch N., Shiekhattar R.. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005; 437:432–435. [DOI] [PubMed] [Google Scholar]
- 4. Rivero S., Ceballos-Chavez M., Bhattacharya S.S., Reyes J.C.. HMG20A is required for SNAI1-mediated epithelial to mesenchymal transition. Oncogene. 2015; 34:5264–5276. [DOI] [PubMed] [Google Scholar]
- 5. Ceballos-Chavez M., Rivero S., Garcia-Gutierrez P., Rodriguez-Paredes M., Garcia-Dominguez M., Bhattacharya S., Reyes J.C.. Control of neuronal differentiation by sumoylation of BRAF35, a subunit of the LSD1-CoREST histone demethylase complex. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:8085–8090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ballas N., Mandel G.. The many faces of REST oversee epigenetic programming of neuronal genes. Curr. Opin. Neurobiol. 2005; 15:500–506. [DOI] [PubMed] [Google Scholar]
- 7. Lorenzo P.I., Martin Vazquez E., Lopez-Noriega L., Fuente-Martin E., Mellado-Gil J.M., Franco J.M., Cobo-Vuilleumier N., Guerrero Martinez J.A., Romero-Zerbo S.Y., Perez-Cabello J.A.et al.. The metabesity factor HMG20A potentiates astrocyte survival and reactive astrogliosis preserving neuronal integrity. Theranostics. 2021; 11:6983–7004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fukuda H., Imamura M., Tanaka Y., Iwata M., Hirose H., Kaku K., Maegawa H., Watada H., Tobe K., Kashiwagi A.et al.. A single nucleotide polymorphism within DUSP9 is associated with susceptibility to type 2 diabetes in a japanese population. PLoS One. 2012; 7:e46263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kooner J.S., Saleheen D., Sim X., Sehmi J., Zhang W., Frossard P., Been L.F., Chia K.S., Dimas A.S., Hassanali N.et al.. Genome-wide association study in individuals of south asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 2011; 43:984–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Perry J.R., Voight B.F., Yengo L., Amin N., Dupuis J., Ganser M., Grallert H., Navarro P., Li M., Qi L.et al.. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. PLos Genet. 2012; 8:e1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sim X., Ong R.T., Suo C., Tay W.T., Liu J., Ng D.P., Boehnke M., Chia K.S., Wong T.Y., Seielstad M.et al.. Transferability of type 2 diabetes implicated loci in multi-ethnic cohorts from southeast asia. PLoS Genet. 2011; 7:e1001363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mellado-Gil J.M., Fuente-Martin E., Lorenzo P.I., Cobo-Vuilleumier N., Lopez-Noriega L., Martin-Montalvo A., Gomez I.G.H., Ceballos-Chavez M., Gomez-Jaramillo L., Campos-Caro A.et al.. The type 2 diabetes-associated HMG20A gene is mandatory for islet beta cell functional maturity. Cell Death. Dis. 2018; 9:279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fuente-Martin E., Mellado-Gil J.M., Cobo-Vuilleumier N., Martin-Montalvo A., Romero-Zerbo S.Y., Diaz Contreras I., Hmadcha A., Soria B., Martin Bermudo F., Reyes J.C.et al.. Dissecting the brain/islet axis in metabesity. Genes (Basel). 2019; 10:350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F.et al.. A human interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015; 163:712–723. [DOI] [PubMed] [Google Scholar]
- 15. Huttlin E.L., Bruckner R.J., Paulo J.A., Cannon J.R., Ting L., Baltier K., Colby G., Gebreab F., Gygi M.P., Parzen H.et al.. Architecture of the human interactome defines protein communities and disease networks. Nature. 2017; 545:505–509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kitagawa M., Takebe A., Ono Y., Imai T., Nakao K., Nishikawa S., Era T.. Phf14, a novel regulator of mesenchyme growth via platelet-derived growth factor (PDGF) receptor-alpha. J. Biol. Chem. 2012; 287:27983–27996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Huang Q., Zhang L., Wang Y., Zhang C., Zhou S., Yang G., Li Z., Gao X., Chen Z., Zhang Z.. Depletion of PHF14, a novel histone-binding protein gene, causes neonatal lethality in mice due to respiratory failure. Acta Biochim. Biophys. Sin. (Shanghai). 2013; 45:622–633. [DOI] [PubMed] [Google Scholar]
- 18. Zheng S., Bi Y., Chen H., Gong B., Jia S., Li H.. Molecular basis for bipartite recognition of histone H3 by the PZP domain of PHF14. Nucleic Acids Res. 2021; 49:8961–8973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yang B., Chen S., Wu M., Zhang L., Ruan M., Chen X., Chen Z., Mei C., Mao Z.. PHF14: an innate inhibitor against the progression of renal fibrosis following folic acid-induced kidney injury. Sci. Rep. 2017; 7:39888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Giudicelli F., Gilardi-Hebenstreit P., Mechta-Grigoriou F., Poquet C., Charnay P.. Novel activities of mafb underlie its dual role in hindbrain segmentation and regional specification. Dev. Biol. 2003; 253:150–162. [DOI] [PubMed] [Google Scholar]
- 21. Subtil-Rodriguez A., Reyes J.C.. BRG1 helps RNA polymerase II to overcome a nucleosomal barrier during elongation, in vivo. EMBO Rep. 2010; 11:751–757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao B., Wei X., Li W., Udan R.S., Yang Q., Kim J., Xie J., Ikenoue T., Yu J., Li L.et al.. Inactivation of YAP oncoprotein by the hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev. 2007; 21:2747–2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bianchi M.E., Beltrame M., Paonessa G.. Specific recognition of cruciform DNA by nuclear protein HMG1. Science. 1989; 243:1056–1059. [DOI] [PubMed] [Google Scholar]
- 24. Liang C.C., Park A.Y., Guan J.L.. In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat. Protoc. 2007; 2:329–333. [DOI] [PubMed] [Google Scholar]
- 25. Hazan R.B., Phillips G.R., Qiao R.F., Norton L., Aaronson S.A.. Exogenous expression of N-cadherin in breast cancer cells induces cell migration, invasion, and metastasis. J. Cell Biol. 2000; 148:779–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huang da W., Sherman B.T., Lempicki R.A.. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009; 4:44–57. [DOI] [PubMed] [Google Scholar]
- 27. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S.et al.. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005; 102:15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Aubrey B.J., Kelly G.L., Kueh A.J., Brennan M.S., O’Connor L., Milla L., Wilcox S., Tai L., Strasser A., Herold M.J.. An inducible lentiviral guide RNA platform enables the identification of tumor-essential genes and tumor-promoting mutations in vivo. Cell Rep. 2015; 10:1422–1432. [DOI] [PubMed] [Google Scholar]
- 29. Guerrero-Martinez J.A., Ceballos-Chavez M., Koehler F., Peiro S., Reyes J.C.. TGFbeta promotes widespread enhancer chromatin opening and operates on genomic regulatory domains. Nat. Commun. 2020; 11:6196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Evans R., O’Neill M., Pritzel A., Antropova N., Senior A., Green T., Žídek A., Bates R., Blackwell S., Yim J.et al.. Protein complex prediction with alphafold-Multimer. 2022; biorxiv doi:10 March 2022, preprint: not peer reviewed 10.1101/2021.10.04.463034. [DOI]
- 31. Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., Tunyasuvunakool K., Bates R., Zidek A., Potapenko A.et al.. Highly accurate protein structure prediction with alphafold. Nature. 2021; 596:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Erdos G., Pajkos M., Dosztanyi Z.. IUPred3: prediction of protein disorder enhanced with unambiguous experimental annotation and visualization of evolutionary conservation. Nucleic Acids Res. 2021; 49:W297–W303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown S.A., Weirich C.S., Newton E.M., Kingston R.E.. Transcriptional activation domains stimulate initiation and elongation at different times and via different residues. EMBO J. 1998; 17:3146–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sadowski I., Ma J., Triezenberg S., Ptashne M.. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988; 335:563–564. [DOI] [PubMed] [Google Scholar]
- 35. Thomas J.O., Travers A.A.. HMG1 and 2, and related ‘architectural’ DNA-binding proteins. Trends Biochem. Sci. 2001; 26:167–174. [DOI] [PubMed] [Google Scholar]
- 36. Stros M., Launholt D., Grasser K.D.. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell. Mol. Life Sci. 2007; 64:2590–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ferrari S., Harley V.R., Pontiggia A., Goodfellow P.N., Lovell-Badge R., Bianchi M.E.. SRY, like HMG1, recognizes sharp angles in DNA. EMBO J. 1992; 11:4497–4506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wainberg M., Kamber R.A., Balsubramani A., Meyers R.M., Sinnott-Armstrong N., Hornburg D., Jiang L., Chan J., Jian R., Gu M.et al.. A genome-wide atlas of co-essential modules assigns function to uncharacterized genes. Nat. Genet. 2021; 53:638–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsherniak A., Vazquez F., Montgomery P.G., Weir B.A., Kryukov G., Cowley G.S., Gill S., Harrington W.F., Pantel S., Krill-Burger J.M.et al.. Defining a cancer dependency map. Cell. 2017; 170:564–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ghandi M., Huang F.W., Jane-Valbuena J., Kryukov G.V., Lo C.C., McDonald E.R. 3rd, Barretina J., Gelfand E.T., Bielski C.M., Li H.et al.. Next-generation characterization of the cancer cell line encyclopedia. Nature. 2019; 569:503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gao M., Nakajima An D., Parks J.M., Skolnick J.. AF2Complex predicts direct physical interactions in multimeric proteins with deep learning. Nat. Commun. 2022; 13:1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen J., Archer T.K.. Regulating SWI/SNF subunit levels via protein-protein interactions and proteasomal degradation: BAF155 and BAF170 limit expression of BAF57. Mol. Cell. Biol. 2005; 25:9016–9027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Mueller S., Wahlander A., Selevsek N., Otto C., Ngwa E.M., Poljak K., Frey A.D., Aebi M., Gauss R.. Protein degradation corrects for imbalanced subunit stoichiometry in OST complex assembly. Mol. Biol. Cell. 2015; 26:2596–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhao Y., He J., Li Y., Xu M., Peng X., Mao J., Xu B., Cui H.. PHF14 promotes cell proliferation and migration through the AKT and ERK1/2 pathways in gastric cancer cells. Biomed. Res. Int. 2020; 2020:6507510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park J.E., Tse S.W., Xue G., Assisi C., Maqueda A.S., Ramon G.P.X., Low J.K., Kon O.L., Tay C.Y., Tam J.P.et al.. Pulsed SILAC-based proteomic analysis unveils hypoxia- and serum starvation-induced de novo protein synthesis with PHD finger protein 14 (PHF14) as a hypoxia sensitive epigenetic regulator in cell cycle progression. Oncotarget. 2019; 10:2136–2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chin Y.R., Toker A.. Function of Akt/PKB signaling to cell motility, invasion and the tumor stroma in cancer. Cell. Signal. 2009; 21:470–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Senese S., Zaragoza K., Minardi S., Muradore I., Ronzoni S., Passafaro A., Bernard L., Draetta G.F., Alcalay M., Seiser C.et al.. Role for histone deacetylase 1 in human tumor cell proliferation. Mol. Cell. Biol. 2007; 27:4784–4795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Meng Z., Moroishi T., Guan K.L.. Mechanisms of hippo pathway regulation. Genes Dev. 2016; 30:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng Y., Pan D.. The hippo signaling pathway in development and disease. Dev. Cell. 2019; 50:264–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zanconato F., Cordenonsi M., Piccolo S.. YAP/TAZ at the roots of cancer. Cancer Cell. 2016; 29:783–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Martin-Belmonte F., Perez-Moreno M.. Epithelial cell polarity, stem cells and cancer. Nat. Rev. Cancer. 2011; 12:23–38. [DOI] [PubMed] [Google Scholar]
- 52. Ganesh K., Basnet H., Kaygusuz Y., Laughney A.M., He L., Sharma R., O’Rourke K.P., Reuter V.P., Huang Y.H., Turkekul M.et al.. L1CAM defines the regenerative origin of metastasis-initiating cells in colorectal cancer. Nat Cancer. 2020; 1:28–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Purcell J.W., Tanlimco S.G., Hickson J., Fox M., Sho M., Durkin L., Uziel T., Powers R., Foster K., McGonigal T.et al.. LRRC15 is a novel mesenchymal protein and stromal target for antibody-drug conjugates. Cancer Res. 2018; 78:4059–4072. [DOI] [PubMed] [Google Scholar]
- 54. Yang A., Schweitzer R., Sun D., Kaghad M., Walker N., Bronson R.T., Tabin C., Sharpe A., Caput D., Crum C.et al.. p63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 1999; 398:714–718. [DOI] [PubMed] [Google Scholar]
- 55. Forster N., Saladi S.V., van Bragt M., Sfondouris M.E., Jones F.E., Li Z., Ellisen L.W.. Basal cell signaling by p63 controls luminal progenitor function and lactation via NRG1. Dev. Cell. 2014; 28:147–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Deyoung M.P., Ellisen L.W.. p63 and p73 in human cancer: defining the network. Oncogene. 2007; 26:5169–5183. [DOI] [PubMed] [Google Scholar]
- 57. Lamaa A., Humbert J., Aguirrebengoa M., Cheng X., Nicolas E., Côté J., Trouche D.. Integrated analysis of H2A.Z isoforms function reveals a complex interplay in gene regulation. Elife. 2020; 9:e53375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miettinen P.J., Ebner R., Lopez A.R., Derynck R.. TGF-beta induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type i receptors. J. Cell Biol. 1994; 127:2021–2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Eberl H.C., Spruijt C.G., Kelstrup C.D., Vermeulen M., Mann M.. A map of general and specialized chromatin readers in mouse tissues generated by label-free interaction proteomics. Mol. Cell. 2013; 49:368–378. [DOI] [PubMed] [Google Scholar]
- 60. Bartke T., Vermeulen M., Xhemalce B., Robson S.C., Mann M., Kouzarides T.. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell. 2010; 143:470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Marcon E., Ni Z., Pu S., Turinsky A.L., Trimble S.S., Olsen J.B., Silverman-Gavrila R., Silverman-Gavrila L., Phanse S., Guo H.et al.. Human-chromatin-related protein interactions identify a demethylase complex required for chromosome segregation. Cell Rep. 2014; 8:297–310. [DOI] [PubMed] [Google Scholar]
- 62. Draker R., Ng M.K., Sarcinella E., Ignatchenko V., Kislinger T., Cheung P.. A combination of H2A.Z and H4 acetylation recruits brd2 to chromatin during transcriptional activation. PLoS Genet. 2012; 8:e1003047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zhang Y., Ku W.L., Liu S., Cui K., Jin W., Tang Q., Lu W., Ni B., Zhao K.. Genome-wide identification of histone H2A and histone variant H2A.Z-interacting proteins by bPPI-seq. Cell Res. 2017; 27:1258–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Punzeler S., Link S., Wagner G., Keilhauer E.C., Kronbeck N., Spitzer R.M., Leidescher S., Markaki Y., Mentele E., Regnard C.et al.. Multivalent binding of PWWP2A to H2A.Z regulates mitosis and neural crest differentiation. EMBO J. 2017; 36:2263–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ouararhni K., Hadj-Slimane R., Ait-Si-Ali S., Robin P., Mietton F., Harel-Bellan A., Dimitrov S., Hamiche A.. The histone variant mH2A1.1 interferes with transcription by down-regulating PARP-1 enzymatic activity. Genes Dev. 2006; 20:3324–3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nightingale K., Dimitrov S., Reeves R., Wolffe A.P.. Evidence for a shared structural role for HMG1 and linker histones B4 and H1 in organizing chromatin. EMBO J. 1996; 15:548–561. [PMC free article] [PubMed] [Google Scholar]
- 67. Gerlitz G., Hock R., Ueda T., Bustin M.. The dynamics of HMG protein-chromatin interactions in living cells. Biochem. Cell. Biol. 2009; 87:127–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bonaldi T., Langst G., Strohner R., Becker P.B., Bianchi M.E.. The DNA chaperone HMGB1 facilitates ACF/CHRAC-dependent nucleosome sliding. EMBO J. 2002; 21:6865–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yamada M., Ueda T., Sato K., Yoshida M.. ATP-dependent chromatin structural modulation by multiprotein complex including HMGB1. J. Biochem. 2004; 135:149–153. [DOI] [PubMed] [Google Scholar]
- 70. Ueda T., Chou H., Kawase T., Shirakawa H., Yoshida M.. Acidic C-tail of HMGB1 is required for its target binding to nucleosome linker DNA and transcription stimulation. Biochemistry. 2004; 43:9901–9908. [DOI] [PubMed] [Google Scholar]
- 71. Hepp M.I., Alarcon V., Dutta A., Workman J.L., Gutierrez J.L.. Nucleosome remodeling by the SWI/SNF complex is enhanced by yeast high mobility group box (HMGB) proteins. Biochim. Biophys. Acta. 2014; 1839:764–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Belotserkovskaya R., Oh S., Bondarenko V.A., Orphanides G., Studitsky V.M., Reinberg D.. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003; 301:1090–1093. [DOI] [PubMed] [Google Scholar]
- 73. Chen P., Dong L., Hu M., Wang Y.Z., Xiao X., Zhao Z., Yan J., Wang P.Y., Reinberg D., Li M.et al.. Functions of FACT in breaking the nucleosome and maintaining its integrity at the single-nucleosome level. Mol. Cell. 2018; 71:284–293. [DOI] [PubMed] [Google Scholar]
- 74. Zhou K., Liu Y., Luger K.. Histone chaperone FACT FAcilitates chromatin transcription: mechanistic and structural insights. Curr. Opin. Struct. Biol. 2020; 65:26–32. [DOI] [PubMed] [Google Scholar]
- 75. Liu Y., Zhou K., Zhang N., Wei H., Tan Y.Z., Zhang Z., Carragher B., Potter C.S., D’Arcy S., Luger K.. FACT caught in the act of manipulating the nucleosome. Nature. 2020; 577:426–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jin C., Felsenfeld G.. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 2007; 21:1519–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huang C., Zhang Z., Xu M., Li Y., Li Z., Ma Y., Cai T., Zhu B.. H3.3-H4 tetramer splitting events feature cell-type specific enhancers. PLoS Genet. 2013; 9:e1003558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Shrinivas K., Sabari B.R., Coffey E.L., Klein I.A., Boija A., Zamudio A.V., Schuijers J., Hannett N.M., Sharp P.A., Young R.A.et al.. Enhancer features that drive formation of transcriptional condensates. Mol. Cell. 2019; 75:549–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Boija A., Klein I.A., Sabari B.R., Dall’Agnese A., Coffey E.L., Zamudio A.V., Li C.H., Shrinivas K., Manteiga J.C., Hannett N.M.et al.. Transcription factors activate genes through the phase-separation capacity of their activation domains. Cell. 2018; 175:1842–1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Giaimo B.D., Ferrante F., Herchenrother A., Hake S.B., Borggrefe T.. The histone variant H2A.Z in gene regulation. Epigenetics Chromatin. 2019; 12:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Zhou J., Hamdan H., Yalamanchili H.K., Pang K., Pohodich A.E., Lopez J., Shao Y., Oses-Prieto J.A., Li L., Kim W.et al.. Disruption of mecp2-TCF20 complex underlies distinct neurodevelopmental disorders. Proc. Natl. Acad. Sci. U.S.A. 2022; 119:e2119078119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Miao L., Liu H.Y., Zhou C., He X.. LINC00612 enhances the proliferation and invasion ability of bladder cancer cells as ceRNA by sponging miR-590 to elevate expression of PHF14. J. Exp. Clin. Cancer Res. 2019; 38:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wu S., Luo C, Li F, Hameed NUF, Jin Q., Zhang J.. Silencing expression of PHF14 in glioblastoma promotes apoptosis, mitigates proliferation and invasiveness via wnt signal pathway. Cancer Cell Int. 2019; 19:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Huh H.D., Kim D.H., Jeong H.S., Park H.W.. Regulation of TEAD transcription factors in cancer biology. Cells. 2019; 8:600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Currey L., Thor S., Piper M.. TEAD family transcription factors in development and disease. Development. 2021; 148:dev196675. [DOI] [PubMed] [Google Scholar]
- 86. Bertero A., Brown S., Madrigal P., Osnato A., Ortmann D., Yiangou L., Kadiwala J., Hubner N.C., de Los Mozos I.R., Sadee C.et al.. The SMAD2/3 interactome reveals that TGFbeta controls m(6)A mRNA methylation in pluripotency. Nature. 2018; 555:256–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Herchenröther A., Gossen S., Friedrich T., Reim A., Daus N., Diegmüller F., Leers J., Sani H.M., Gerstner S., Schwarz L.et al.. The H2A.Z.1/PWWP2A/NuRD-associated protein HMG20A controls early head and heart developmental transcription programs. 2022; bioRxiv doi:04 May 2022, preprint: not peer reviewed 10.1101/2022.05.04.490592. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available databases used in our analysis included the DepMap portal (21Q3 data release; https://depmap.org/), the CCLE portal (https://portals.broadinstitute.org/ccle), the TCGA Research Network (https://www.cancer.gov/tcga), the MSigDB database (https://www.gsea-msigdb.org/gsea/msigdb/), the AlphaFold Protein Structure Database (https://alphafold.ebi.ac.uk/). The structural model is available in ModelArchive at https://modelarchive.org/doi/10.5452/ma-1us57. These data can be accessed through the Token: asTzxrLlCj. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030769. All NGS data have been deposited to Gene Expression Omnibus (GEO) under the accesión number GSE193476.