Abstract
Two tandem cellulosome-associated genes were identified in the cellulolytic rumen bacterium, Ruminococcus flavefaciens. The deduced gene products represent multimodular scaffoldin-related proteins (termed ScaA and ScaB), both of which include several copies of explicit cellulosome signature sequences. The scaB gene was completely sequenced, and its upstream neighbor scaA was partially sequenced. The sequenced portion of scaA contains repeating cohesin modules and a C-terminal dockerin domain. ScaB contains seven relatively divergent cohesin modules, two extremely long T-rich linkers, and a C-terminal domain of unknown function. Collectively, the cohesins of ScaA and ScaB are phylogenetically distinct from the previously described type I and type II cohesins, and we propose that they define a new group, which we designated here type III cohesins. Selected modules from both genes were overexpressed in Escherichia coli, and the recombinant proteins were used as probes in affinity-blotting experiments. The results strongly indicate that ScaA serves as a cellulosomal scaffoldin-like protein for several R. flavefaciens enzymes. The data are supported by the direct interaction of a recombinant ScaA cohesin with an expressed dockerin-containing enzyme construct from the same bacterium. The evidence also demonstrates that the ScaA dockerin binds to a specialized cohesin(s) on ScaB, suggesting that ScaB may act as an anchoring protein, linked either directly or indirectly to the bacterial cell surface. This study is the first direct demonstration in a cellulolytic rumen bacterium of a cellulosome system, mediated by distinctive cohesin-dockerin interactions.
The cellulosome is believed to be one of the principle mechanisms by which cellulolytic microorganisms achieve efficient breakdown of the recalcitrant polysaccharides present in plant cell walls, particularly among anaerobic microorganisms (2, 7). The cellulosome has been described as a multiprotein complex, existing in cell-associated and/or extracellular forms in a wide range of cellulolytic organisms (5, 45). A typical cellulosome was first described in Clostridium thermocellum (6, 27, 29) which includes a noncatalytic subunit, called scaffoldin, and dozens of different enzymes.
The scaffoldin subunit plays a critical role in cellulosome assembly by its repetitive cohesin domains, each of which interacts with the dockerin domain of the individual cellulosomal enzymes. A cellulose-binding domain (CBD) of scaffoldin is responsible for mediating the binding of the cellulosome to the substrate. In C. thermocellum, the scaffoldin is believed to bind to another type of cellulosome-related protein (i.e., cell surface anchoring proteins) that carry C-terminal surface-layer homology domains (19, 32). The binding is mediated via a second type (type II) of cohesin-dockerin interaction, involving a C-terminal dockerin of scaffoldin and one or more cohesin-like domains in the anchoring proteins (30, 31).
Until recently, detailed molecular support for cellulosome organization was available only for the cellulolytic Clostridium species, C. thermocellum, Clostridium cellulovorans, Clostridium cellulolyticum, and Clostridium josui. In addition, a number of cellulosome-related signature sequences (i.e., cohesins and dockerins) have been discovered in different bacteria and fungi, although most of them are dockerin-tagged enzymes. The four clostridial scaffoldin genes that have been fully sequenced (20, 24, 37, 46) all contained a single CBD and a multiplicity of cohesin modules. Of these scaffoldins, only that of C. thermocellum exhibited a dockerin domain. Recently a scaffoldin from Acetivibrio cellulolyticus was described that also exhibited a C-terminal dockerin domain, similar to that of C. thermocellum, but differed from all known clostridial scaffoldins in that the gene included a catalytic module at its N terminus (13). Finally, another scaffoldin has been described from Bacteroides cellulosolvens that contains a similar arrangement of CBD, multiple cohesin modules (but of type II), and a single C-terminal dockerin domain (12). The growing evidence suggests a broad species-specific diversity in the sequence content and modular arrangement of the cellulosomal scaffoldins among the cellulolytic bacteria.
The rumen is a highly anaerobic environment that represents one of the most active sites for breakdown of plant cell wall material in nature (22). Genes encoding glycoside hydrolases have been characterized from the major cellulolytic ruminal bacteria, ruminal fungi (15, 16, 44), and ruminal protozoa (11). Nevertheless, the organization of enzyme systems that allow ruminal microorganisms to degrade lignocellulosic material is not well understood. In Ruminococcus species, which are among the most important ruminal cellulolytic bacteria in the rumen, previous biochemical and ultrastructural evidence indicated that protuberances are present on the cell surface that resemble those identified with cellulosomes in C. thermocellum (28). More recently dockerin sequences resembling those in Clostridium spp. were found in enzymes from ruminococci. These include xylanases, cellulases, and esterases from Ruminococcus flavefaciens 17 (1, 25) and cellulases from Ruminococcus albus (35, 36). The detection of dockerins in such enzymes is strongly suggestive of cellulosome organization in Ruminococcus spp., implying the presence of a scaffoldin-like protein and/or anchoring proteins that may organize enzyme subunits into such complexes. We report here the identification of two linked genes in R. flavefaciens 17 that appear to encode key structural components of a cellulosome complex in this species and present evidence for specific dockerin-cohesin interactions between the gene products.
MATERIALS AND METHODS
Strains and growth conditions.
R. flavefaciens 17 was grown anaerobically at 39°C overnight according to the method of Bryant (8) in medium M2GSC (21) as modified by Miyazaki et al. (33). The culture was then used to inoculate Hungate-Stack medium (23), containing 0.2% Avicel (E. Merck, Darmstadt, Germany) as an energy source, and allowed to grow for up to 10 days in this medium. Escherichia coli strains used for cloning (XL10-gold and XL1-blue) and for subsequent expression of pET constructs [BL21(DE3) pLySs gold] were obtained from Stratagene. The vector pET28a was obtained from Novagen (Madison, Wis.). E. coli strains were routinely grown on LB medium with appropriate antibiotic selection at 37°C; conditions used for expression of cloned products are given below.
Preparation of protein samples from R. flavefaciens 17.
R. flavefaciens cells and culture supernatant fluids were harvested after 10 days of incubation, by centrifuging (13,000 × g, for 10 min) the medium at room temperature. The pellet, containing residual Avicel plus cells, was resuspended in 1/40 of the original culture volume in 50 mM sodium phosphate buffer (pH 6.5) and 2 mM dithiothreitol and freeze-thawed twice, and the suspension was aliquoted and recentrifuged. The pellet from a 1-ml aliquot was resuspended in 250 μl of sample buffer containing 2% sodium dodecyl sulfate (SDS) (at 100°C for 5 min), and the proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE). Proteins were either visualized using Coomassie brilliant blue R250 or studied further by blot transfer or specific staining methods as described below.
PCR sequencing.
PCRs were performed using a Mastercycler Personal device (Eppendorf, Hamburg, Germany). Samples (50 μl) contained 100 ng of template DNA, 10 pmol of each primer, 2.5 U of Super Taq (HT Biotechnology Ltd., Cambridge, England), 10 mM Tris-HCl (pH 9.0), 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, and 0.01% (wt/vol) stabilizer. Cycling was programmed as follows: a 3-min predenaturation step at 95°C was followed by 30 cycles comprising a 30-s denaturation step at 94°C, 55°C for 45 s, and an extension step at 72°C for 10 min. PCR products were separated on a 1% agarose gel and purified using a Spin-X centrifuge tube filter (Corning Inc., Corning, N.Y.). When possible, purified PCR products were cloned into the pGEM-T easy vector system (Promega, Madison, Wis.) and sequenced. Otherwise, selected fragments were sequenced directly using PCR primers on a model 377 DNA sequencer (PE Biosystems Foster City, Calif.). For longer PCR fragments, additional primers were designed from previously sequenced portions of the fragment in order to further extend the sequence.
Cloning and overexpression of recombinant proteins.
Cohesin 2 and dockerin from ScaA and cohesin 4/5 from ScaB were amplified using primers designed to contain XhoI and NcoI sites (see Fig. 2 for details). The product was cloned into an NcoI/XhoI-treated pET28a(+) vector, such that a six-histidine stretch was fused at the C terminus of each recombinant module. Clones were expressed in E. coli Solopack Gold BL21 (DE3) pLysS (Stratagene, La Jolla, Calif.), at either 37 or 16°C as follows: single colonies were transferred into 50 ml of Luria-Bertani (LB) medium (30 μg of kanamycin per ml) and grown overnight at 37°C. A flask, containing 1 liter of LB medium, with added kanamycin (30 μg/ml), was inoculated with the 50-ml overnight culture and allowed to grow until an optical density at 600 nm of 0.8 to 1.0 was reached. Isopropyl-β-d-thiogalactopyranoside (IPTG) (1 mM) was then added, and the culture was incubated further for 4 h at 37°C. For expression at 16°C, 50 ml of an overnight culture was introduced into 1 liter of LB medium containing 1.2% glycerol and kanamycin (30 μg/ml), and the culture was incubated at 37°C until an optical density at 600 nm of 0.8 to 1.0 was reached. The culture was maintained at 4°C for 1 h, and 1 mM IPTG was added. The flask was incubated at 16°C without shaking for 1 h and then incubated with shaking (200 rpm) at 16°C overnight.
FIG. 2.
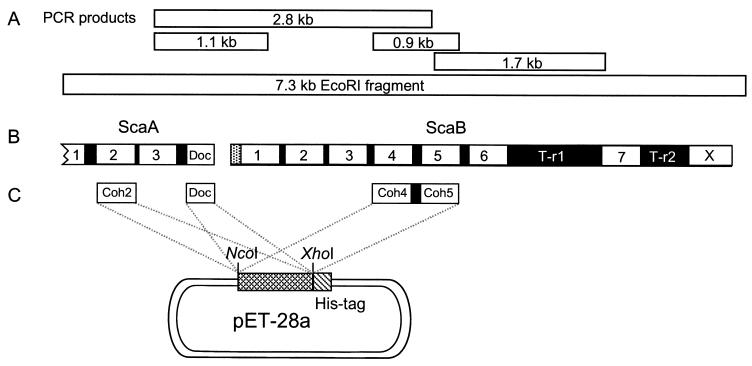
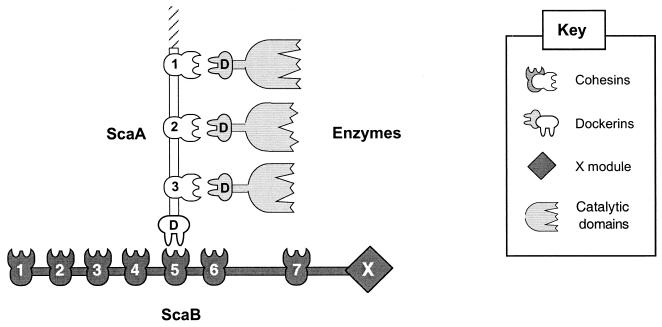
Overview of the 7.3-kb fragment selected from an R. flavefaciens EcoRI∗ library. (A) DNA fragments crucial for solving the sequence. (B) Domain organization of the C-terminal portion of scaA and the complete scaB gene. Both genes contain multiple copies of cohesin domains (numbered), and scaA contains a C-terminal dockerin domain (Doc). All of the domains are separated by short linker sequences (black); ScaB also includes two long T-rich linking segments (T-r1 and T-r2). The sequence of scaB shows a typical signal peptide at its N terminus and a C-terminal module (X) of unknown function. (C) Overexpression of selected modules. The C-terminal cohesin 3 and dockerin domains from ScaA were expressed separately, together with a His tag, in the pET-28a vector. A double-cohesin segment (cohesins 4 and 5) from ScaB was similarly expressed.
Cultures were centrifuged (5,000 × g at 4°C for 10 min), and the pellet was washed twice in 100 ml of lysis buffer (50 mM sodium phosphate buffer [pH 8.0], 0.3 M NaCl, and 10 mM imidazole) and resuspended in 10 ml of lysis buffer, to which 100 μl of Sigma protease inhibitor cocktail and lysosyme (4 mg/ml) were added. The suspension was incubated at 37°C for 30 min and the cells (cooled in an ice bath) were sonicated in a Soniprep 150 device (Sanyo, Gallenkamp PLC, Leicester, United Kingdom), using a 12-μm-diameter exponential microprobe with three strokes of 1 min each. The sonicated cell suspension was centrifuged (15,000 × g at 4°C for 20 min), and the supernatant fluids were collected and stored in the presence of 0.1% sodium azide.
Purification of His-tagged proteins.
The supernatant fluids from sonicated cell samples were incubated with Ni-NTA resin (Novagen) at a ratio of 1 ml of resin per 5 ml of supernatant fluids, and the suspension was incubated for 1 h at 4°C with gentle rocking. The resin was loaded in a plastic chromatographic column and washed with 10 volumes of wash buffer (50 mM sodium phosphate buffer [pH 8.0], 1 M NaCl, 20 mM imidazole, and 10% [vol/vol] glycerol). The His-tagged protein was then eluted using elution buffer (50 mM sodium phosphate buffer [pH 8.0], 0.3 M NaCl, 250 mM imidazole). The purified fraction was concentrated (10,000-Da molecular cutoff; VivaScience, Lincoln, United Kingdom) and stored at 4°C until further use.
GST-XynD fusion construct.
A HindIII fragment of 1,423 kb that specifies residues 337 to 802 of the R. flavefaciens xynD product (i.e., a bifunctional xylanase-lichenase enzyme) was excised from the clone L956 (17). This fragment encodes a dockerin domain, a T-rich region, and the C-terminal catalytic domain, the product of which exhibits lichenase activity. The fragment was first cloned into the Pinpoint AX3 vector (Promega Corp.) and then excised using the enzymes NruI and SmaI, whose cleavage sites flank the insert. The resulting blunt-ended fragment was ligated into the vector pGEX2TK that had been cut with SmaI and phosphatase treated (Amersham Pharmacia Biotech Ltd., Little Chalfont, Bucks, United Kingdom), so as to create an in-frame fusion with a 235-amino-acid vector-determined polypeptide fragment, carrying glutathione-S-transferase (GST) at the amino terminus. Ampicillin-resistant transformant colonies of E. coli XL1-blue were isolated that expressed lichenase activity, which was detected by Congo red staining of agar overlays containing lichenan (17). The isolated transformants were shown to carry the predicted insert by sequencing across the fusion site and by PCR and restriction enzyme mapping.
Purification of GST-tagged proteins.
The clarified supernatant fluids from sonicated host cells were incubated with shaking for 1 h at 4°C with glutathione-Sepharose 4B (Pharmacia Biotech, Uppsala, Sweden), previously equilibrated with phosphate-buffered saline (140 mM NaCl, 2.7 mM KCl, 10 mM N2HPO4, 1.8 mM KH2PO4 [pH 7.2]) at a ratio of 1 ml of slurry per 5 ml of clarified supernatant fluids. The resin was then loaded into a plastic chromatographic column and washed with 15 volumes of the same buffer. The bound protein was then eluted using 5 bed volumes of elution buffer (10 mM reduced glutathione in 50 mM Tris-HCl [pH 8.0]). The purified fraction was concentrated (molecular cutoff weight 10,000; Vivaspin 4), 0.05% (wt/vol) sodium azide was added, and the preparation was stored at 4°C until further use.
Immunoblotting.
Concentrated, cell R. flavefaciens 17 supernatant fluids, cell-associated proteins, or the partially purified GST-XynD construct was subjected to SDS-PAGE (7% polyacrylamide). Separated proteins were then transferred onto a polyvinylidene difluoride (PVDF) transfer membrane (Immobilon-P; Millipore, Bedford, United Kingdom) and rinsed with wash buffer (50 mM Tris-HCl buffer [pH 7.5] containing 150 mM NaCl and 0.01% Tween 20). The membrane was then incubated for 1 h with blocking buffer (3% bovine serum albumin in wash buffer) and rinsed three times with wash buffer. The membrane was then incubated with the recombinant His-tagged protein (5 μg of protein/ml in blocking buffer, containing 5 mM CaCl2) for 1 h at room temperature and washed three times with wash buffer. The membrane was incubated for 1 h at room temperature with peroxidase-conjugated antibody [Anti-His(C-term)-HRP mouse antibody; (Invitrogen, Groningen, The Netherlands), 0.24 μg/ml in blocking buffer] and washed five times with wash buffer. The bands were visualized using an appropriate chemiluminescent substrate (SuperSignal Substrate, Western Blotting; Pierce, Rockford, Ill.) following the manufacturer's instructions.
Glycoprotein staining.
SDS-PAGE-separated proteins were transferred to a PVDF membrane in a semidry system (LKB 2117 Multiphor II electrophoresis unit). The membrane was stained for glycoproteins by periodate-induced biotinylation and visualization using streptavidin-peroxidase, according to the manufacturer's instructions (Glycotrack carbohydrate detection kit; Oxford GlycoSystems, Abingdon, United Kingdom).
Phylogenetic analysis.
Phylogenetic trees were generated using the ClustalW program (http://www2.ebi.ac.uk/clustalw/). Protein sequences were obtained from the GenBank website (http://www.ncbi.nlm.nih.gov/) or via the Carbohydrate-Active Enzymes server (CAZy and CAZyModO websites, http://afmb.cnrs-mrs.fr/∼pedro/CAZY/db.html and http://afmb.cnrs-mrs.fr/∼pedro/DB/db.html, respectively), designed by Coutinho and Henrissat (10). See also the phylogenetic treatment of cellulosomal components in previous publications (4, 12, 13).
Nucleotide sequence accession number.
The DNA sequence for the fragment and the complete scaB gene has been deposited in the GenBank database under accession number AJ278969.
RESULTS
Identification of a putative R. flavefaciens cellulosomal protein.
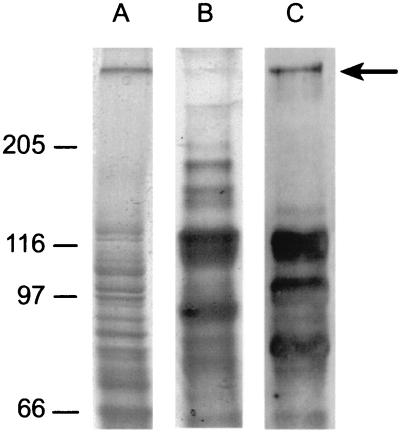
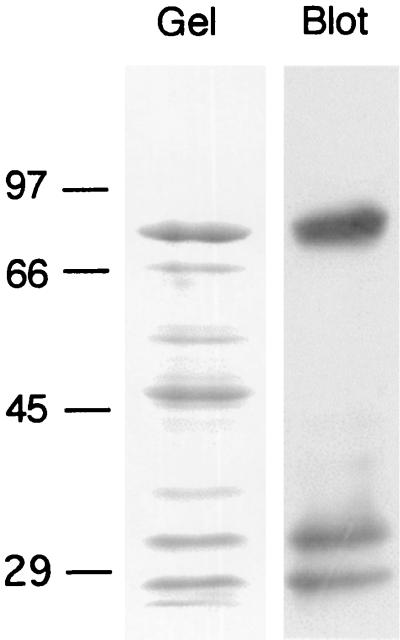
Cell-bound proteins from the culture fluids of R. flavefaciens 17 were separated by SDS-PAGE. A well-separated high-molecular-mass (∼350-kDa) band was identified as a glycosylated protein (Fig. 1). The designated band was extracted from the gel and subjected to proteolysis, and the amino acid sequence of selected peptides was determined. Four degenerate primers (1F, 1B, 2F, and 2B [Table 1]) were designed based on two peptide sequences (Seq-1, DSIAWVVDTVAAYPGDEVTL; Seq-2, VVDLDNSQLPIAGAQFNIV [amino acids used for primer design are underlined]).
FIG. 1.
Identification of putative cellulosome-related glycoproteins from R. flavefaciens 17. A cellulose-grown culture was centrifuged, and both pellet (lane A) and supernatant fluids (lane B) were analyzed by SDS-PAGE on the same gel (7.5% polyacrylamide separating gel). A duplicate sample of the supernatant fluids was blotted onto a PVDF membrane and stained for the presence of glycosylated proteins (lane C). Molecular mass markers (in kilodaltons) are shown to the left of the gel.
TABLE 1.
Primers used in this study
| Name | Nucleotide sequencea | Location | Comments |
|---|---|---|---|
| BAN | ATATCCATGGCTGCCGGTGGTTTA | Coh-1, ScaB | Cohesin expression |
| BAX | CGGTCTCGAGATCCTTCTTAACAAT | Coh-1, ScaB | |
| A2N | ACAACCATGGTTACAGGCGGCCAGACATCAAA | Coh-2, ScaA | Cohesin expression |
| A2X | CTGTCTCGAGTGTACCAGGATTAGGTGTACC | Coh-2, ScaA | |
| ADN | ACTACCATGGTAACAACTCCTCAGCCCGGCA | Doc, ScaA | Dockerin expression |
| ADX | CCGTCTCGAGCTGAGGAAGTGTGATGAGTT | Doc, ScaA | |
| DEN | GGTACCATGGAGTGGGTTATTCCTAAG | Coh-4, ScaB | Cohesin expression, PCR sequencing |
| DEC | CAACCTCGAGCTTAACGATAATAGCACC | Coh-5, ScaB | |
| 1F | GCATAYCCNGGNGAYGARGT | Initially unidentified cohesin | Degenerate primer, PCR sequencing |
| 1B | ACYTCRTCNCCNGGRTATGC | Degenerate primer | |
| 2F | GGNGCNCARTTYAAYATHGT | Degenerate primer | |
| 2B | ACDATRTTRAAYTGNGCNCC | Degenerate primer | |
| 16C-5 | TAGTTCCCAAGGATTCTATCGCA | Coh-7, ScaB | |
| 20N-5 | TTGACGGCAGCATTATCG | Coh-7, ScaB | |
| 20C-3 | GTCAAGAGCTGACGAAACCTG | X, ScaB | |
| 16-5C | CAGCAACACTCAATATCAAG | Coh-6, ScaB | |
| 28-3N | GTCTGTAAATGTCACCTTATC | Coh-4, ScaB | |
| 11-C | TTCAAGTCTTGCAGTTGC | Coh-1, ScaB | |
| M13/F | GTTTTCCCAGTCACGACGTTG | Lambda vector | |
| M13/R | AGCGGATAACAATTTCACACAGGA |
Abbreviation for degenerate nucleotides: K, G or T; Y, C or T; W, A or T; R, A or G; S, C or G; N, A, C, G, or T. Restriction sites are underlined in primers used for protein expression.
Gene cloning and sequencing.
Several fragments were amplified from the R. flavefaciens 17 genomic DNA, using the degenerate primers given above. Three major products (1.1, 1.7, and 2.8 kb) were purified. The 1.7-kb fragment was amplified, using primers 2F and 1B, and sequenced by PCR sequencing using primers 1F, 2B, and 16-5C. The deduced polypeptide contained three cohesin-like domains and a T-rich linker. The other two fragments were both amplified in the same PCR, using primers 1F and 2B. The 1.1-kb fragment was cloned and sequenced. The 2.8-kb fragment was sequenced using primers 1F, 2B, 28-3N, DEN, and 11-C. The resultant sequences indicated that the 1.1-kb fragment was part of the 2.8-kb fragment. The sequence of the 2.8-kb PCR fragment revealed the presence of two tandem open reading frames (ORFs). The first ORF, later termed scaA, represented an incomplete portion of a gene, coding for a protein that contains several cohesin-like domains and a dockerin-like domain at its C terminus. The remaining downstream portion of the 2.8-kb fragment included a second ORF, later termed scaB, which contained a series of cohesin-like domains. Two primers were designed from the two larger fragments (DEN from the 2.8-kb fragment and DEC from the 1.7-kb fragment) and used to determine whether these fragments are linked. Indeed, the resultant 0.9-kb PCR product was found to contain two complete cohesin-like domains and the linker segment between them. This short fragment, in fact, proved to be of added importance, since it verified the actual sequence of the site masked by the degenerate primer. The remainder of the scaB gene and an additional portion of scaA were obtained by library screening. For this purpose, the 0.9-kb PCR product was labeled with 32P and employed as a probe to screen a Lambda-ZAPII EcoRI∗ R. flavefaciens 17 genomic library. Five positive clones were selected from 20,000 phage plaques. The positive clones were verified by PCR using primers DEN and DEC. Primers M13/F, M13/R, 16C-5, 20N-5, and 20C-3 were applied to amplify and sequence the inserted EcoRI fragment from the positive phage clones. In this manner, a 7.3-kb EcoRI fragment from R. flavefaciens was completely sequenced (Fig. 2). All portions of the 7.3-kb fragment were sequenced at least twice from different PCR products, except for the first T-rich linker of scaB, for which unique primers could not be designed in view of the numerous internal repeats. ScaB contains the originally sequenced peptide (Seq-1 and Seq-2), which indicated that ScaB indeed represents the ∼350-kDa glycosylated protein initially extracted and isolated from the R. flavefaciens cells.
Domain structure of cellulosomal proteins from R. flavefaciens.
The overall modular architecture of the C-terminal portion of ScaA and the complete ScaB is shown in Fig. 2B. ScaA harbors three successive cohesins and a dockerin at its C terminus. The linkers separating the ScaA modules are about 20 residues in length, most of which are threonines. The first four cohesins of ScaB are very closely linked. The two segments linking cohesin 4 to 5 and cohesin 5 to 6 are very similar to the threonine-rich linkers of ScaA. However, the two linking segments that flank cohesin 7 are special. They are particularly long (275 and 156 residues, respectively) and also contain a high content of threonine. Finally, ScaB includes at its C terminus an X module of unknown function.
Expression and function of selected ScaA and ScaB modules.
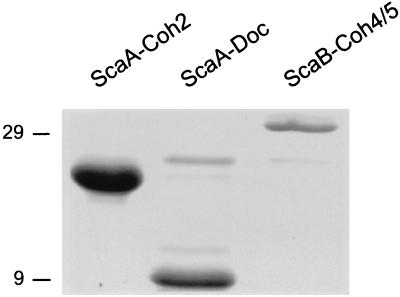
In order to test the possible interactions between ScaA and ScaB, several of their key modules were subcloned and expressed with a C-terminal His-Tag in E. coli (Fig. 2C). These include the second cohesin and C-terminal dockerin from ScaA (ScaA-Coh2 and ScaA-Doc, respectively) and the double cohesin from ScaB (ScaB-Coh4/5). The expressed proteins were isolated on a Ni-nitrilotriacetic acid column and each preparation exhibited a major SDS-PAGE band, consistent with the estimated molecular weight according to the deduced sequence (Fig. 3). The calculated molecular weights (including the His tag) of ScaA-Coh2, ScaA-Doc, and ScaB-Coh4/5 are 17,784, 10,712, and 32,047, respectively.
FIG. 3.
SDS-PAGE of purified recombinant proteins, derived from R. flavefaciens ScaA and ScaB. Cohesin 2, the dockerin from ScaA, and the double cohesin (cohesins 4 and 5) from ScaB were subcloned and expressed as indicated in Fig. 2C. The His-tagged proteins were isolated on an Ni-nitrilotriacetic acid column; the eluted proteins were subjected to SDS-PAGE and stained with Coomassie brilliant blue. Molecular mass markers (in kilodaltons) are shown to the left of the gel.
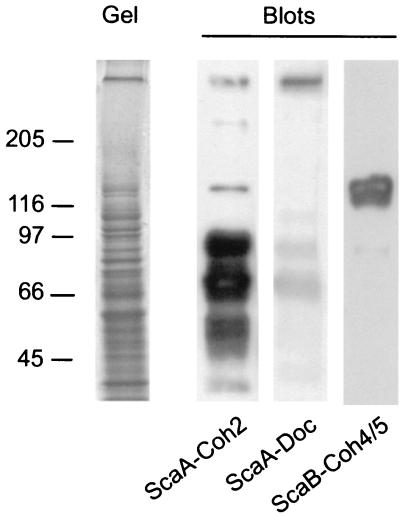
In order to examine whether the expressed modules would interact specifically with other R. flavefaciens proteins, a cell culture was grown on cellulose, the cells and residual substrate were centrifuged, and macromolecules from the pelleted samples were separated by SDS-PAGE. The proteins were transferred electrophoretically onto blots, and the purified recombinant proteins were used as probes. The blots shown in Fig. 4 show that the different probes recognize a different set of target proteins. The ScaA cohesin labels a series of bands, notably from 50 to 100 kDa, consistent with the molecular dimensions of several known enzymes from this bacterium, including xylanases, cellulases, and esterases, which carry dockerin domains (1, 18, 25). Other faint bands, including a very-high-molecular-mass band, were also observed. On the other hand, the ScaA dockerin mainly recognized the ∼350-kDa band, putatively identified as ScaB, with some low-level labeling of other bands. In contrast to the ScaA cohesin, the double cohesin from ScaB appeared to label a major band of ∼130 kDa, presumably representing ScaA.
FIG. 4.
Affinity blotting of cell-derived material from R. flavefaciens, using selected recombinant protein domains from ScaA and ScaB. Samples containing the pelleted, cellulose-grown culture (Fig. 1) were subjected to SDS-PAGE (Gel), and blotted onto PVDF membranes (Blots). The blots were probed with the indicated recombinant protein sample, and labeled bands (labeled at left [in kilodaltons]) were detected by chemiluminescence using peroxidase-conjugated, anti-His tag antibody.
Interaction of ScaA cohesin with an expressed R. flavefaciens glycanase.
In order to further investigate the possible role of ScaA as a scaffoldin that integrates relevant enzymes into a cellulosome complex, we examined the interaction of the ScaA cohesin construct with an expressed recombinant form of a known R. flavefaciens dockerin-containing enzyme, XynD. The enzyme construct represented a 701-residue GST-XynD fusion protein that comprised a 1,423-bp HindIII fragment of the XynD gene (encoding for the dockerin domain) and T-rich linker and C-terminal lichenase domains of XynD, fused to the C-terminal end of GST. The estimated molecular weight of the construct was 78,953. The expressed protein was partially purified on a glutathione-Sepharose column. SDS-PAGE of the preparation exhibited a major ∼80-kDa band (consistent with the predicted size of the intact construct) plus a series of lower-molecular-mass contaminating bands and breakdown products (Fig. 5). Affinity blotting of the contents of the gel, probed with the recombinant ScaA-Coh2, revealed a strong signal with the ∼80-kDa band and two additional low-molecular-mass bands, presumably representing dockerin-containing proteolytic fragments of the intact construct. Such fragmentation has been observed previously for expressed preparations of this enzyme (17).
FIG. 5.
Affinity blotting of a known R. flavefaciens enzyme construct with a recombinant ScaA cohesin. A GST-XynD fusion protein (molecular weight, 78,953) was subjected to SDS-PAGE (Gel), and the proteins were transferred to a PVDF membrane (Blot), and probed with ScaA-Coh2 as described in the legend to Fig. 4. Molecular mass markers (in kilodaltons) are shown to the left of the gel.
DISCUSSION
Although indirect biochemical evidence has suggested the presence of a cellulosome in R. flavefaciens (14), direct support has only recently been reported upon detection of cellulosome signature sequences, i.e., dockerin domains from different cloned enzymes. Dockerin-containing enzymes suggest the existence of a scaffoldin-like counterpart, namely, a multiple-cohesin-containing protein, which would organize the various glycosyl hydrolases into a cellulosome complex. This hypothesis prompted the quest for scaffoldins, described in the present communication.
Preliminary studies on the binding of cloned dockerin regions from two R. flavefaciens 17 polysaccharidases had indicated recognition of an extracellular proteins(s) of approximately 130 kDa from cellulose-grown R. flavefaciens cultures (unpublished results), but this protein could not easily be recovered in a pure form. Another potential candidate for a cellulosome component, however, was a high-molecular-mass (>300-kDa) glycosylated polypeptide that was readily separable by one-dimensional SDS-PAGE. Limited peptide sequencing followed by the design of degenerate oligonucleotide primers allowed the amplification by PCR of portions of the coding sequence, following the strategy recently adopted successfully for A. cellulolyticus and B. cellulosolvens (12, 13). However, many Ruminococcus sequences are notoriously unstable in standard plasmid cloning vectors. Likewise, a similar instability has been experienced with the cloned PCR products. Consequently, the new sequence described in this work was achieved mainly through limited direct sequencing of appropriate PCR products and through analysis of a 7.3-kb fragment cloned in a phage vector. Quite fortuitously, the sequencing revealed portions of two ORFs, separated by a short segment of only 70 bp. Both of these ORFs, later designated scaA and scaB, encode polypeptides that contain cohesin-like sequences. Both of these were therefore strong candidates for components of a cellulosome complex.
Our evidence points to a specific interaction between the C-terminal dockerin region of ScaA and cohesins 4 or 5 (or both) of ScaB. This conclusion is supported by the interaction between these two domains. Moreover, the cloned ScaA dockerin recognizes selectively the large ∼350-kDa polypeptide derived from R. flavefaciens cells, and the cloned cohesin 4/5 from ScaB binds specifically to the 130-kDa polypeptide from this strain. These data imply that the ScaA product is around 130 kDa, while the apparent size of ScaB is very roughly estimated at about 350 kDa. The two lengthy T-rich linkers in ScaB are probable sites for the glycosylation observed for the ∼350 kDa polypeptide. Their sequences display a considerable internal homology within and between the two linker segments (Fig. 6). Secondary structure analysis (41–43) of the linker regions indicates a near-complete β-sheet structure (data not shown), interspersed with distinctive loops at or near the glycine residues (notably the Gly-Pro dyads).
FIG. 6.
Alignment of internal segments within the T-rich linkers of ScaB. Note the high degree of internal repetition, the marked similarity among segments of these two linker regions, and the presence of distinctive Gly-Pro dyads (shown in boldface type). T-r1 comprises residues 939 to 1213, and T-r2 comprises residues 1356 to 1511.
The molecular mass predicted for the ScaB polypeptide is around 200 kDa, which suggests that anomalous migration and/or glycosylation would contribute to the observed low mobility in SDS gels. At this stage we can propose tentatively that the ScaA product may bind to the cell surface via the larger ScaB product. The C-terminal domain of unknown function is a possible candidate for involvement in cell surface attachment. In this context, preliminary evidence (Rinchon et al., unpublished) has shown that this domain is not involved in cellulose binding.
The cloned ScaA-Coh2 was able to interact with multiple R. flavefaciens polypeptides ranging from 60 to 100 kDa. The sizes of the polypeptides recognized by ScaA-Coh2 are consistent with the contention that they include plant cell wall-degrading enzymes, since known dockerin-containing enzymes from R. flavefaciens 17 range in size from 80 to 95 kDa (1, 18). This implies recognition of other dockerin domains present in a variety of R. flavefaciens proteins via another type of dockerin-cohesin interaction. Indeed, a recombinant dockerin-bearing construct, based on a known R. flavefaciens enzyme (17), was selectively recognized by ScaA-Coh2. If we assume that the R. flavefaciens system is analogous to those found in cellulolytic Clostridium spp., ScaA appears the more likely candidate to act as a scaffolding protein for the assembly of plant cell wall-degrading enzymes, while ScaB might play a cell wall anchoring role (Fig. 7).
FIG. 7.
Schematic representation of the proposed binding specificity of cellulosomal components from R. flavefaciens.
The mechanism for attachment of the complex to the substrate has yet to be established. As for other known cellulosomes, the process might involve an appropriate carbohydrate-binding module(s) within the unsequenced portion of the scaffolding protein or within an enzyme or enzymes that belong to the complex. Another possibility would be that substrate binding would be mediated by a cellulose-binding pilus-like surface protein, reported recently in a related ruminococcal species (34, 38). A. cellulolyticus, B. cellulosolvens, and now R. flavefaciens, which synthesize cellulosomes resembling those in clostridia, all belong to the Bacillus/Clostridium subphylum of the gram-positive bacteria based on 16S rDNA sequences (9, 26, 40). However, given the special nature of the rumen environment and the genetic distance between R. flavefaciens and cellulolytic clostridia (39), it is reasonable that significant differences would exist in cellulosome organization between these bacteria.
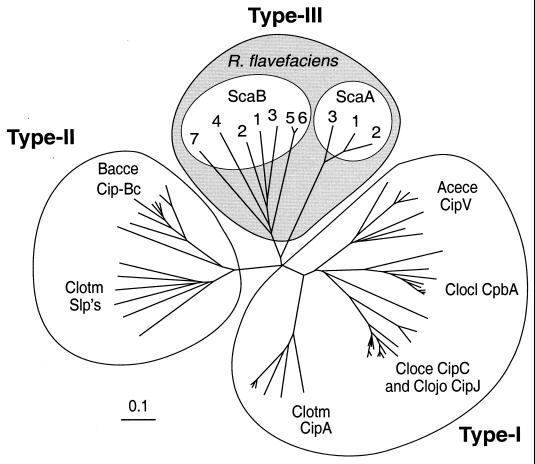
While ScaA and ScaB from R. flavefaciens both contain cohesin domains that have been identified by sequence similarity and by binding studies, the sequence relationships indicate a high degree of divergence from previously described cohesins. The closest percent identity between the R. flavefaciens ScaA and ScaB cohesins and those in Clostridium spp. is less than 27% for certain ScaB cohesins. The ScaA cohesin sequences are very similar to each other (85% identity) but show little overall similarity (less than 25%) to the clostridial cohesins. In fact, phylogenetic comparison of the known cohesins (Fig. 8) reveals that the R. flavefaciens cohesins represent a group distinct from those previously identified either in the clostridia or more recently in A. cellulolyticus and B. cellulosolvens. The ruminococcal cohesins emanate from a separate branch of the phylogenetic tree and are therefore herein designated type III cohesins. Finally, it can be noted that the seven cohesins described here from ScaB include two (cohesins 5 and 6) that are very similar to each other, while the remaining five are relatively divergent in sequence (Fig. 8). We have yet to investigate the binding specificity of all ScaB cohesins and so cannot rule out the possibility that other ScaB cohesins differ in their binding specificity from the cohesin 4/5 doublet studied here. The possible functional significance of cohesin and dockerin (1) divergence in R. flavefaciens cellulosome components is currently under investigation.
FIG. 8.
Phylogenetic relationship of R. flavefaciens cohesin domains. The individual ScaA and ScaB cohesins are numbered as they appear in sequence from the N terminus of the gene. The type I cohesins include those from the known scaffoldins (CipA from C. thermocellum, C. cellulolyticum CipC, C. josui CipJ, C. cellulovorans CpbA, and A. cellulolyticus CipV). Type II cohesins include those from the B. cellulosolvens CipBc scaffoldin and the surface anchoring proteins (Slp's) of C. thermocellum. The sources of the sequences used in this figure are given in reference 13. The scale bar indicates percentage (0.1) of amino acid substitutions.
Initially, the cellulosome was described on the basis of biochemical characterization as a cellulose-binding, multienzyme complex. Later molecular biological data and recognition of dockerin-containing enzymes and scaffoldin-borne CBDs and cohesins appeared to support the biochemical information. The recent findings (3) of noncellulosomal cohesin- and dockerin-like sequences in the genome of the archeon Archeaeoglobus fulgidus suggested that such signature sequences cannot be used alone to identify a cellulosome in a given organism. However, the A. fulgidus genome contains no known polysaccharidase, and the arrangement of the two cohesins and single dockerin precludes the formation of a multienzyme complex. In contrast, R. flavefaciens ScaA and ScaB, together with the demonstrated intermolecular connectivities, clearly allow the formation of cellulosome-like multienzyme complex. It is hard to predict whether in the future the term cellulosome will adequately categorize all types of multiprotein polysaccharide-degrading complexes. Nevertheless, the cellulosome concept should continue to provide a framework by means of which future discoveries in this field can be compared.
ACKNOWLEDGMENTS
M.T.R. was supported by a Ph.D. studentship from Conicit/British Council. This work was partly supported by the Scottish Office Executive Rural Affairs Department. A contract from the European Commission (Fourth Framework, Biotechnology Programme, BIO4-97-2303) is gratefully acknowledged. Additional support was provided by grants from the Israel Science Foundation (administered by the Israel Academy of Sciences and Humanities, Jerusalem). Parts of this research were performed in the Otto Meyerhof Center for Biotechnology, established by the Minerva Foundation (Munich, Germany), with funding from the Technion-Niedersachsen Cooperation (Hannover, Germany).
REFERENCES
- 1.Aurilia V, Martin J C, McCrae S I, Scott K P, Rincon M T, Flint H J. Three multidomain esterases from the cellulolytic rumen anaerobe Ruminococcus flavefaciens 17 that carry divergent dockerin sequences. Microbiology. 2000;146:1391–1397. doi: 10.1099/00221287-146-6-1391. [DOI] [PubMed] [Google Scholar]
- 2.Bayer E A, Chanzy H, Lamed R, Shoham Y. Cellulose, cellulases and cellulosomes. Curr Opin Struct Biol. 1998;8:548–557. doi: 10.1016/s0959-440x(98)80143-7. [DOI] [PubMed] [Google Scholar]
- 3.Bayer E A, Coutinho P M, Henrissat B. Cellulosome-like sequences in Archaeoglobus fulgidus: an enigmatic vestige of cohesin and dockerin domains. FEBS Lett. 1999;463:277–280. doi: 10.1016/s0014-5793(99)01634-8. [DOI] [PubMed] [Google Scholar]
- 4.Bayer E A, Ding S-Y, Mechaly A, Shoham Y, Lamed R. Emerging phylogenetics of cellulosome structure. In: Gilbert H J, Davies G J, Henrissat B, Svensson B, editors. Recent advances in carbohydrate bioengineering. Cambridge, United Kingdom: The Royal Society of Chemistry; 1999. pp. 189–201. [Google Scholar]
- 5.Bayer E A, Morag E, Lamed R. The cellulosome—a treasure-trove for biotechnology. Trends Biotechnol. 1994;12:378–386. doi: 10.1016/0167-7799(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 6.Bayer E A, Shimon L J W, Lamed R, Shoham Y. Cellulosomes: structure and ultrastructure. J Struct Biol. 1998;124:221–234. doi: 10.1006/jsbi.1998.4065. [DOI] [PubMed] [Google Scholar]
- 7.Béguin P, Lemaire M. The cellulosome: an exocellular, multiprotein complex specialized in cellulose degradation. Crit Rev Biochem Mol Biol. 1996;31:201–236. doi: 10.3109/10409239609106584. [DOI] [PubMed] [Google Scholar]
- 8.Bryant M P. Commentary on the Hungate technique for culture of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- 9.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 10.Coutinho P M, Henrissat B. Carbohydrate-active enzymes: an integrated database approach. In: Gilbert H J, Davies G J, Henrissat B, Svensson B, editors. Recent advances in carbohydrate bioengineering. Cambridge, United Kingdom: The Royal Society of Chemistry; 1999. pp. 3–12. [Google Scholar]
- 11.Devillard E, Newbold C J, Scott K P, Forano E, Wallace R J, Jouany J-P, Flint H J. A family 11 xylanase from the ruminal protozoan Polyplastron multivesiculatum. FEMS Microbiol Lett. 1999;181:6720–6729. doi: 10.1111/j.1574-6968.1999.tb08837.x. [DOI] [PubMed] [Google Scholar]
- 12.Ding S-Y, Bayer E A, Steiner D, Shoham Y, Lamed R. An atypical scaffoldin of the Bacteroides cellulosolvens cellulosome that contains eleven type-II cohesins. J Bacteriol. 2000;182:4915–4925. doi: 10.1128/jb.182.17.4915-4925.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding S-Y, Bayer E A, Steiner D, Shoham Y, Lamed R. A novel cellulosomal scaffoldin from Acetivibrio cellulolyticus that contains a family-9 glycosyl hydrolase. J Bacteriol. 1999;181:6720–6729. doi: 10.1128/jb.181.21.6720-6729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doerner K C, White B A. Assessment of the endo-1,4-β-glucanase components of Ruminococcus flavefaciens FD-1. Appl Environ Microbiol. 1990;56:1844–1850. doi: 10.1128/aem.56.6.1844-1850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flint H J. The rumen microbial ecosystem—some recent developments. Trends Microbiol. 1997;5:483–488. doi: 10.1016/S0966-842X(97)01159-1. [DOI] [PubMed] [Google Scholar]
- 16.Flint H J, Forsberg C W. Polysaccharide degradation in the rumen: biochemistry and genetics. In: Engelhardt W V, Leonard-Marek S, Breves G, Giesecke D, editors. Ruminant physiology, digestion, metabolism, growth and reproduction. Proceedings of the Eighth International Symposium on Ruminant Physiology. Stuttgart, Germany: Ferdinand Enke Verlag; 1995. pp. 43–70. [Google Scholar]
- 17.Flint H J, Martin J, McPherson C A, Daniel A S, Zhang J X. A bifunctional enzyme, with separate xylanase and β(1,3-1,4)-glucanase domains, encoded by the xynD gene of Ruminococcus flavefaciens. J Bacteriol. 1993;175:2943–2951. doi: 10.1128/jb.175.10.2943-2951.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flint H J, Zhang J-X, Martin J C. Multiplicity and expression of xylanases in the rumen cellulolytic bacterium Ruminococcus flavefaciens. Curr Microbiol. 1994;29:139–143. [Google Scholar]
- 19.Fujino T, Béguin P, Aubert J-P. Organization of a Clostridium thermocellum gene cluster encoding the cellulosomal scaffolding protein CipA and a protein possibly involved in attachment of the cellulosome to the cell surface. J Bacteriol. 1993;175:1891–1899. doi: 10.1128/jb.175.7.1891-1899.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerngross U T, Romaniec M P M, Kobayashi T, Huskisson N S, Demain A L. Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol Microbiol. 1993;8:325–334. doi: 10.1111/j.1365-2958.1993.tb01576.x. [DOI] [PubMed] [Google Scholar]
- 21.Hobson P N. Rumen bacteria. Methods Microbiol. 1969;3B:133–149. [Google Scholar]
- 22.Hungate R E. The rumen and its microbes. New York, N.Y: Academic Press; 1966. [Google Scholar]
- 23.Hungate R E, Stack R J. Phenylpropanoic acid: growth factor for Ruminococcus albus. Appl Environ Microbiol. 1982;44:79–83. doi: 10.1128/aem.44.1.79-83.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kakiuchi M, Isui A, Suzuki K, Fujino T, Fujino E, Kimura T, Karita S, Sakka K, Ohmiya K. Cloning and DNA sequencing of the genes encoding Clostridium josui scaffolding protein CipA and cellulase CelD and identification of their gene products as major components of the cellulosome. J Bacteriol. 1998;180:4303–4308. doi: 10.1128/jb.180.16.4303-4308.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kirby J, Martin J C, Daniel A S, Flint H J. Dockerin-like sequences in cellulases and xylanases from the rumen cellulolytic bacterium Ruminococcus flavefaciens. FEMS Microbiol Lett. 1997;149:213–219. doi: 10.1111/j.1574-6968.1997.tb10331.x. [DOI] [PubMed] [Google Scholar]
- 26.Krause D O, Dalrymple B P, Smith W J, Mackie R I, McSweeney C S. 16S rDNA sequencing of Ruminococcus albus and Ruminococcus flavefaciens: design of a signature probe and its application in adult sheep. Microbiology. 1999;145:1797–1807. doi: 10.1099/13500872-145-7-1797. [DOI] [PubMed] [Google Scholar]
- 27.Lamed R, Bayer E A. The cellulosome of Clostridium thermocellum. Adv Appl Microbiol. 1988;33:1–46. [Google Scholar]
- 28.Lamed R, Naimark J, Morgenstern E, Bayer E A. Specialized cell surface structures in cellulolytic bacteria. J Bacteriol. 1987;169:3792–3800. doi: 10.1128/jb.169.8.3792-3800.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamed R, Setter E, Bayer E A. Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J Bacteriol. 1983;156:828–836. doi: 10.1128/jb.156.2.828-836.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leibovitz E, Béguin P. A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J Bacteriol. 1996;178:3077–3084. doi: 10.1128/jb.178.11.3077-3084.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leibovitz E, Ohayon H, Gounon P, Béguin P. Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J Bacteriol. 1997;179:2519–2523. doi: 10.1128/jb.179.8.2519-2523.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemaire M, Ohayon H, Gounon P, Fujino T, Béguin P. OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J Bacteriol. 1995;177:2451–2459. doi: 10.1128/jb.177.9.2451-2459.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miyazaki K, Martin J C, Marinsek-Logar R, Flint H J. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- 34.Morrison M, Miron J. Adhesion to cellulose by Ruminococcus albus: a combination of cellulosomes and Pil-proteins? FEMS Microbiol Lett. 2000;185:109–115. doi: 10.1111/j.1574-6968.2000.tb09047.x. [DOI] [PubMed] [Google Scholar]
- 35.Ohara H, Karita S, Kimura T, Sakka K, Ohmiya K. Characterization of the cellulolytic complex (cellulosome) from Ruminococcus albus. Biosci Biotechnol Biochem. 2000;64:254–260. doi: 10.1271/bbb.64.254. [DOI] [PubMed] [Google Scholar]
- 36.Ohara H, Noguchi J, Karita S, Kimura T, Sakka K, Ohmiya K. Sequence of egV and properties of EgV, a Ruminococcus albus endoglucanase containing a dockerin domain. Biosci Biotechnol Biochem. 2000;64:80–88. doi: 10.1271/bbb.64.80. [DOI] [PubMed] [Google Scholar]
- 37.Pagès S, Belaich A, Fierobe H-P, Tardif C, Gaudin C, Belaich J-P. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J Bacteriol. 1999;181:1801–1810. doi: 10.1128/jb.181.6.1801-1810.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pegden R S, Larson M A, Grant R J, Morrison M. Adherence of the gram-positive bacterium Ruminococcus albus to cellulose and identification of a novel form of cellulose-binding protein which belongs to the Pil family of proteins. J Bacteriol. 1998;180:5921–5927. doi: 10.1128/jb.180.22.5921-5927.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rainey F A, Janssen P H. Phylogenetic analysis by 16S ribosomal DNA sequence comparison reveals two unrelated groups of species within the genus Ruminococcus. FEMS Microbiol Lett. 1995;129:69–73. doi: 10.1016/0378-1097(95)00138-U. [DOI] [PubMed] [Google Scholar]
- 40.Rainey F A, Stackebrandt E. 16S rDNA analysis reveals phylogenetic diversity among the polysaccharolytic clostridia. FEMS Microbiol Lett. 1993;113:125–128. doi: 10.1111/j.1574-6968.1993.tb06501.x. [DOI] [PubMed] [Google Scholar]
- 41.Rost B. PHD: predicting one-dimensional protein structure by profile based neural networks. Methods Enzymol. 1996;266:525–539. doi: 10.1016/s0076-6879(96)66033-9. [DOI] [PubMed] [Google Scholar]
- 42.Rost B, Sander C. Combining evolutionary information and neural networks to predict protein secondary structure. Proteins. 1994;19:55–72. doi: 10.1002/prot.340190108. [DOI] [PubMed] [Google Scholar]
- 43.Rost B, Sander C. Prediction of protein secondary structure at better than 70% accuracy. J Mol Biol. 1993;232:584–599. doi: 10.1006/jmbi.1993.1413. [DOI] [PubMed] [Google Scholar]
- 44.Selinger L B, Forsberg C W, Cheng K J. The rumen: a unique source of enzymes for enhancing livestock production. Anaerobe. 1996;2:263–284. doi: 10.1006/anae.1996.0036. [DOI] [PubMed] [Google Scholar]
- 45.Shoham Y, Lamed R, Bayer E A. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol. 1999;7:275–281. doi: 10.1016/s0966-842x(99)01533-4. [DOI] [PubMed] [Google Scholar]
- 46.Shoseyov O, Takagi M, Goldstein M A, Doi R H. Primary sequence analysis of Clostridium cellulovorans cellulose binding protein A. Proc Natl Acad Sci USA. 1992;89:3483–3487. doi: 10.1073/pnas.89.8.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]