Abstract
Integration of the human papillomavirus (HPV) genome into host cell chromosomes has been observed in a majority of HPV-positive cervical cancers and a subset of oral HPV-associated cancers. HPV integration also occurs in long-term cell culture. Screening for HPV integration can be labor intensive and yield results that are difficult to interpret. Here we describe an assay based on exonuclease V (ExoV/RecBCD) and quantitative polymerase chain reaction (qPCR) to determine if samples from cell lines and tissues contain episomal or integrated HPV. This assay can be applied to screen other small DNA viruses with episomal/linear genome configurations in their viral lifecycle and has the potential to be used in clinical settings to define viral genomic conformations associated with disease.
Basic Protocol:
Exonuclease V genomic DNA digestion and qPCR for detection of HPV16 genome configuration in cells
Support Protocol:
Exonuclease V analysis of HPV16 genome configuration in tissues
Alternate Protocol:
Determining HPV integration type or integrity of HPV episome
Keywords: episomal, integrated, exonuclease V, human papillomavirus (HPV), RecBCD
INTRODUCTION
Papillomaviruses are small epitheliotropic viruses that carry a circular, extrachromosomal (episomal) genome ~8 kilobase pairs (kb) in length. There are over 200 different papillomaviruses that infect birds, reptiles, marsupials, and mammals (Egawa & Doorbar, 2017). Human papillomavirus (HPV) infections are common and typically result in benign hyperproliferative lesions that are cleared by the immune system (Doorbar, Egawa, Griffin, Kranjec, & Murakami, 2015). Persistent HPV infections are associated with severe pathologies that include recurrent respiratory papillomatosis, condylomas, and cancer. HPVs are divided into low- and high-risk types depending on their association with malignant carcinoma. HPV infection with high-risk types such as HPV16 and HPV18 have been associated with a majority of cervical cancers and an increasing number of oropharyngeal squamous cell carcinomas (Burd, 2003; de Martel, Plummer, Vignat, & Franceschi, 2017; Gooi, Chan, & Fakhry, 2016; Walboomers et al., 1999).
HPV persistence in the epithelia is a key step in oncogenesis, with persistent HPV infections resulting from integration of the HPV episomal genome into the host chromosome. HPV integration is an abortive infection event, which inhibits viral progeny production and regulation of viral oncogene (HPV E6 and E7) expression (Jeon, Allen-Hoffmann, & Lambert, 1995). HPV16 integration is frequently observed in HPV-positive anogenital carcinomas, in a subset of HPV-positive oral squamous cell carcinomas, and in vitro in HPV-infected cell lines (Olthof et al., 2014; Parfenov et al., 2014; Pett et al., 2006; Wentzensen, Vinokurova, & von Knebel Doeberitz, 2004). Although the mechanisms guiding HPV genome integration are not known, HPV16 can integrate as either a single copy integrant (type 1) or as multimeric/concatemeric copies of the complete viral genome (type 2; McBride & Warburton, 2017). Type 1 integration exhibits a frequent loss of viral genetic information involving loss or truncation of the HPV E2 gene, a viral negative transcriptional regulator of E6 and E7 expression.
Traditionally, Southern blots have been the standard method for determining the HPV genome status. However, Southern blots require large amounts of DNA from samples, which may not be achievable from small tissue biopsies, and require several days to complete (Hubbard, 2003; Southern, 1975). To circumvent this problem, several polymerase chain reaction (PCR)-based approaches have been developed to detect integrated HPV, each furthering our understanding of the prevalence of HPV integration in cancer. Analysis of the copy number ratio of HPV E2 to E6 in quantitative PCR (qPCR) can identify loss of E2 sequences that is associated with HPV type 1 integration but is unable to discern HPV type 2 integration (Yoshinouchi et al., 1999). DNA amplification of papillomavirus oncogene transcripts (APOT-PCR) uses reverse transcription PCR to detect loss of genetic information measured by differences in the length of viral transcripts between integrated and episomal HPV genomes (Klaes et al., 1999). APOT-PCR can also fail to detect type 2 HPV integrants or integrated HPV when present with episomal forms. Detection of integrated papillomavirus sequences (DIPS-PCR) uses ligation-mediated PCR to identify the cellular and viral junctions where HPV integration occurred (Luft et al., 2001). DIPS-PCR does not detect episomal forms that can be present in a background of integrated HPV genomes.
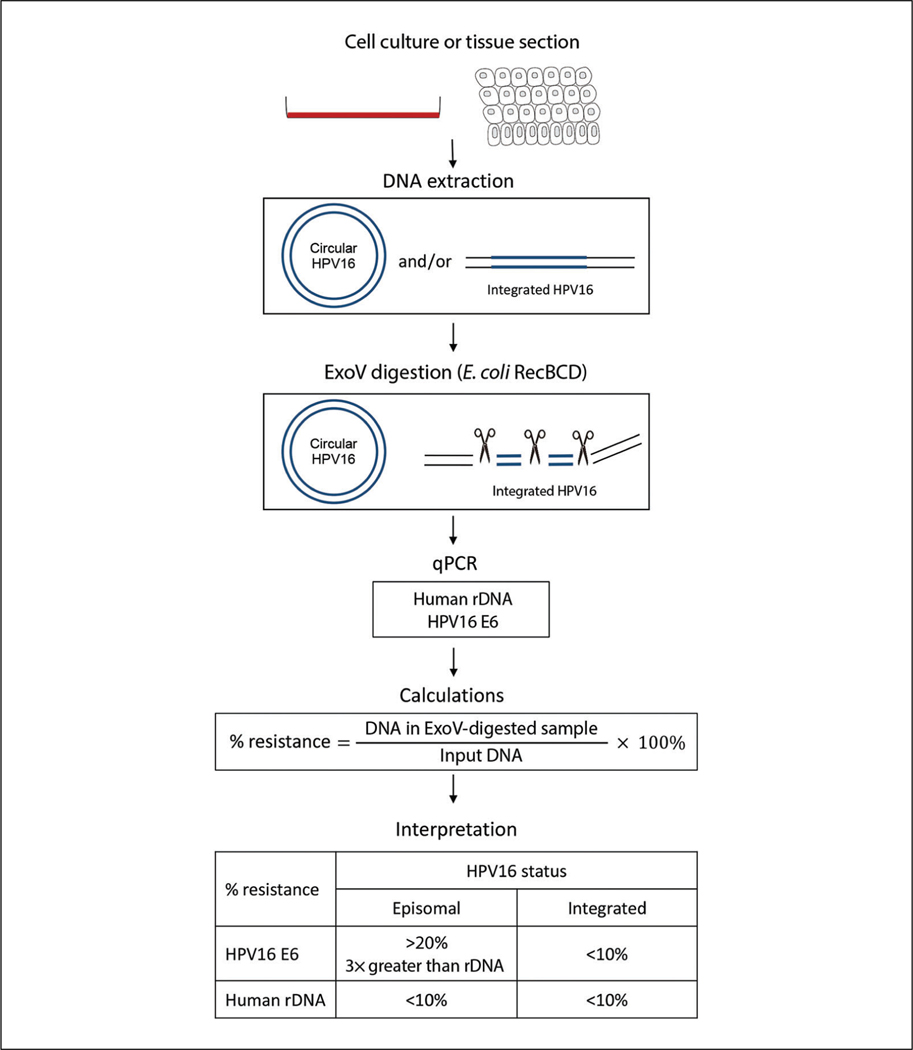
In this article, we provide a step-by-step protocol for a sensitive assay based on exonuclease V (ExoV) digestion of linear DNA and qPCR that can be used to rapidly screen HPV16 genome configurations in cell lines and tissues (Fig. 1). Genomic DNA is digested with ExoV, an enzyme that selectively degrades double-stranded and single-stranded linear DNA while preserving nicked and supercoiled circular DNA. After digestion, qPCR is used to determine the relative DNA concentration for each sample relative to an undigested sample. In the Basic Protocol, the fraction of HPV DNA resistant to ExoV digestion is used as a readout for the presence of episomal or integrated HPV. Human 18S ribosomal DNA (rDNA) is used as an internal control for linear DNA and to monitor the efficiency of ExoV nuclease activity. In the Alternate Protocol, the integration type and integrity of the viral episome can be assessed using a copy number ratio of various viral genomic regions relative to E6. The protocol is specific for HPV16 but can be easily adapted for assessment of the genome configuration of other papillomavirus strains or extended to other circular DNA viruses (polyomavirus) by changing the primers used in qPCR (Myers et al., 2019).
Figure 1.
Graphical overview of the exonuclease V (ExoV) assay to determine the conformation of the human papillomavirus (HPV) genome in cells and tissues. ExoV selectively degrades linear DNA, leaving circular DNA intact. Genomic DNA from HPV-positive cell lines and tissues are digested with ExoV. HPV episomes are preserved, while integrated HPV and 18S ribosomal DNA (rDNA) are degraded. rDNA is a cellular internal control to monitor the efficiency of ExoV digestion. The DNA remaining after ExoV digestion is analyzed by qPCR using specific primers. The relative DNA amount from ExoV-digested DNA divided by input DNA from no-enzyme controls determines the fraction of DNA resistant to ExoV digestion. The percentage of DNA resistant to ExoV digestion is used as a readout to determine the HPV genome status. Samples with episomal HPV are identified by having a percent resistance to ExoV above 20% and at least three times greater than the percent resistance observed for rDNA.
CAUTION: HPV16 is a biosafety level 2 (BSL-2) pathogen. Follow all appropriate guidelines and regulations for the use and handling of BSL-2 pathogens (see Current Protocols article: Burnett, Lunn, & Coico, 2009). Follow proper use of personal protective equipment, and handle cells and tissues in a BSL-2 cabinet. Liquid waste can be disinfected by treatment with 10% (v/v) bleach for 10 min, and solid waste can be sterilized by autoclaving prior to disposal.
NOTE: Disposable gloves should be worn at all times when handling samples to avoid nuclease contamination. All cell culture incubations should be performed in a humidified 37°C, 5% CO2 incubator unless otherwise specified.
BASIC PROTOCOL
EXONUCLEASE V GENOMIC DNA DIGESTION AND qPCR FOR DETECTION OF HPV16 GENOME CONFIGURATION IN CELLS
By combining ExoV digestion with qPCR, we can detect the presence of HPV episomal DNA in a rapid and sensitive assay that may not be observable by Southern blot. Integrated HPV will be digested by ExoV and identified as having a significantly reduced signal in qPCR compared with undigested input DNA. The tandemly repeated rDNA, present on five separate chromosomes, is used as an internal control for the efficiency of enzyme digestion (Stults, Killen, Pierce, & Pierce, 2008). The ExoV assay can be easily completed in 1 day. An important consideration is that plasmid DNA containing HPV gene segments can easily contaminate samples to produce false episomal calls. To prevent such contamination, all work should be performed in a plasmid/DNA-free clean room environment. Negative controls need to be included to monitor potential contamination. HPV-positive samples can also be screened using primers to alternate HPV genes to identify the integration type and episome integrity (see Alternate Protocol). This assay provides a novel technique for screening cell lines and tissues grown in vitro for the HPV16 genome status and has the potential to be used in a clinical setting to determine HPV genome configurations.
Materials
UMSCC47 cell line (e.g., Millipore, SCC071)
1× Dulbecco’s modified Eagle medium (DMEM; e.g., Corning, 10–014-CV)
Fetal bovine serum (FBS; e.g., HyClone, SH30070.03)
1× Dulbecco’s phosphate-buffered saline (PBS; e.g., Corning, 21–031-CM)
Qiagen QIAamp Blood Mini Kit (e.g., Qiagen, 51106) or NucleoSpin Blood QuickPure kit (e.g., Macherey-Nagel, 740569)
20 mg/ml proteinase K
0.25% (w/v) trypsin/0.1% (w/v) EDTA (e.g., Corning 25–053-CI)
200 proof ethanol, molecular biology grade
10 mM Tris, pH 8.0 (see recipe)
pGL3 basic plasmid (e.g., Promega, E1751)
Nuclease-free water
Exonuclease V (recBCD, 10,00 units/mL; e.g., New England Biolabs, M0345S) containing:
10 mM ATP
10× buffer
Power SYBR® Green PCR Master Mix (e.g., Applied Biosystems, 4367659) or iQ™ SYBR® Green Supermix (e.g., Bio-Rad, 170–8882)
qPCR primers
HPV16 E6 forward: 5′-GAGAACTGCAATGTTTCAGGACC-3′
HPV16 E6 reverse: 5′-TGTATAGTTGTTTGCAGCTCTGTGC-3′
18S rDNA forward: 5′-GCAATTATTCCCCATGAACG-3′
18S rDNA reverse: 5′-GGGACTTAATCAACGCAAGC-3′
Luciferase forward: 5′-GAAAGGCCCGGCGCCATTCT-3′
Luciferase reverse: 5′-TTCATAGCTTCTGCCAACCG-3′
25-cm2 flask
Humidified 37°C, 5% CO2 incubator
Vortex mixer
Variable temperature incubator
Centrifuge with plate adaptor
Microvolume spectrophotometer (e.g., NanoDrop)
1.5-ml microcentrifuge tube, sterile, DNase-free
Thermal heating block
Microcentrifuge
Optical low-profile 96-well PCR plates compatible with real-time PCR system (e.g., Phenix, 3890; Bio-Rad, HSP9601)
Optical adhesive covers (e.g., Applied Biosystems, 4360954; Bio-Rad, MSB 1001)
Real-time PCR thermal cycler (e.g., Applied Biosystems 7500 Real-Time PCR System or Bio-Rad CFX96 Touch Real-Time PCR Detection System)
Computer running spreadsheet analysis software (e.g., Excel)
UMSCC47 cell culture
UMSCC47 is an HPV-positive head and neck carcinoma cell line carrying ~18 copies of type II integrated HPV (Akagi et al., 2014; Brenner et al., 2010; Olthof et al., 2015). UMSCC47 DNA is used in serial dilutions for the standard curve analysis during qPCR and as a control for integrated HPV DNA.
-
1
Seed ~1 × 105 UMSCC47 cells into a 25-cm2 flask, and grow in DMEM supplemented with 10% FBS.
UMSCC47 cells are typically passaged 1:5 twice a week with trypsin/EDTA. Heat inactivate serum for 30 min in a 56°C water bath before use.
-
2Once cells reach confluency, harvest cells for genomic DNA isolation:
- Remove cell culture medium using a sterile pipette, and discard into a waste container.
- Rinse with 4 ml of 1× PBS.
-
Harvest DNA directly from plate or from detached cells after trypsin/EDTA treatment as described in steps 3 through 6.Cell pellet may be stored at −20°C until ready for DNA isolation.
Genomic DNA isolation from cells
Genomic DNA from cell lines and/or tissues (Support Protocol) can be isolated using the Qiagen QIAamp Blood Mini Kit or the NucleoSpin Blood QuickPure kit. Follow the manufacturer’s protocol. The procedure to isolate DNA from samples of interest using the Qiagen kit is outlined below.
-
3Harvest DNA directly from the plate or from trypsinized cells:
- To harvest DNA directly from adherent cells on a plate: Add 400 μl of a 1:1 dilution of AL lysis buffer (Qiagen) and 1× PBS. Add 20 μl of 20 mg/ml proteinase K to the lysate, and mix by pulse vortexing for 10 to 15 s. Incubate at 56°C for 10 min. Briefly centrifuge to remove any condensation from the lid.
-
To harvest DNA from trypsinized cells: Add 0.5 ml trypsin/EDTA. Collect detached cells with 1 to 2 ml growth medium, and centrifuge 5 min at 800 × g, room temperature. Rinse cells with 2 ml of 1× PBS, and centrifuge an additional 5 min at 800 × g, room temperature. Resuspend cell pellet with 200 μl of 1× PBS and 20 μl of 20 mg/ml proteinase K. Add 200 μl AL lysis buffer (Qiagen), and mix by pulse vortexing for 10 to 15 s. Incubate at 56°C for 10 min. Briefly centrifuge to remove any condensation from the lid.Do not use more than 5 × 106 cells per column. If isolating DNA from more than 5 × 106 cells, double the amount of 1× PBS, lysis buffer, and proteinase K; and separate lysate into two columns.
-
4
Add 200 μl of 200 proof ethanol to 400 μl DNA lysate, and vortex to mix. Add DNA lysate mixture to the supplied column. Follow recommended wash and centrifugation steps. Include the optional spin (1 min at 20,000 × g) after using buffer AW2 (Qiagen) to ensure no AW2 buffer remains in the column.
Any residual buffer in the eluate may disrupt downstream applications.
-
5
To elute DNA, add 50 μl of 10 mM Tris, pH 8.0, to the column. Incubate for 5 min at room temperature. Centrifuge 1 min at 6000 × g, room temperature.
-
6
Measure concentration of the eluate DNA using a microvolume spectrophotometer. Optional: Add purified pGL3 basic plasmid DNA to each sample to monitor DNA breakage. For every 5 μg genomic DNA, add 7 × 108 copies (3.71 ng) purified pGL3 plasmid.
DNA can be stored at −20°C for long-term storage or at 4°C for up to 1 week. Aliquot DNA to minimize freezing and thawing, which can cause breakage of episomal DNA.
Exonuclease V digestion
-
7
Prepare two separate master mix reactions: one master mix with ExoV enzyme(ExoV+) and one without ExoV enzyme (Exo–). Prepare master mixes by multiplying the number of total reactions by the values described below. Add an additional reaction as overage:
1 μl ATP (1 mM final)
1 μl 10× buffer (1× final)
6.34 μl nuclease-free water
0.66 μl ExoV (6.6 units final) or nuclease-free water (input/undigested control).
The ExoV– reaction will measure the amount of input DNA.
Store ATP, 10× buffer, and enzyme on ice when in use. Always add the enzyme last to the master mix. ExoV activity has been observed to decrease over time and may need to be replaced every 6 months to a year. Enzyme activity is monitored by loss of 18S rDNA signal in each sample following PCR. UMSCC47 DNA can also be included as a control for ExoV activity.
The 10× buffer contains 500 mM potassium acetate, 200 mM Tris-acetate, 100 mM magnesium acetate, and 10 mM dithiothreitol.
-
8
Add 9 μl master mix to a DNase-free, sterile 1.5-ml microcentrifuge tube. Place tubes on ice. For each sample, include one tube with enzyme (ExoV+) and one tube with no enzyme added (ExoV–).
For high-throughput analysis, reactions can be performed in a 96-well plate format instead of individual microcentrifuge tubes.
-
9
Dilute sample DNA to 100 ng/μl, and place on ice.
Include a mock reaction by adding nuclease-free water in place of DNA to monitor for potential DNA contamination. DNA from an HPV-negative cell line can also be included as a no-template control for the HPV primers. UMSCC47 genomic DNA should be included as an integrated HPV control. The minimum DNA quantity for this assay is 100 ng. If desired, the reaction can be scaled up to 1 μg of DNA.
-
10
For each sample DNA add 1 μl of 100 ng/μl DNA into one tube containing the ExoV+ reaction and 1 μl of 100 ng/μl DNA into a tube containing the no-enzyme reaction.
-
11
Incubate for 1 hr at 37°C.
-
12
Heat inactivate reactions for 10 min at 95°C.
If using a heating block, turn on the heating block before setting up the reactions to ensure it reaches temperature before the 1-hr incubation is over. The 1-hr incubation time is sufficient for DNA digestion; longer incubation times will not significantly improve digestion.
-
13
Cool samples on ice for 5 min.
-
14
If performing qPCR the same day, keep samples on ice. Otherwise, store samples at −20°C until ready for qPCR analysis.
qPCR analysis
-
15
Centrifuge samples to collect condensation on the lid of the microcentrifuge tube.
-
16
Dilute 2 μl of each sample DNA in 18 μl nuclease-free water in a 1.5-ml microcentrifuge tube for a 1:10 dilution. Place dilutions on ice.
-
17Dilute stock UMSCC47 DNA to 100 ng/μl. Then from the 100 ng/μl UMSCC47 dilution, prepare a 10-fold dilution series of UMSCC47 DNA at 25 ng/μl, 2.5 ng/μl, 0.25 ng/μl, 0.025 ng/μl, and 0.0025 ng/μl. To prepare the dilution series:
- Add 37.5 μl nuclease-free water to a microcentrifuge tube. Add 12.5 μl of 100 ng/μl UMSCC47 DNA to make the 25 ng/μl dilution. Mix by vortexing, and briefly centrifuge to remove any drops from the lid of the microcentrifuge tube.
- To the next four tubes, add 45 μl nuclease-free water. Add 5 μl of 25 ng/μl UMSCC47 DNA for a final concentration of 2.5 ng/μl. Mix by vortexing and briefly centrifuge.
-
Repeat by adding 5 μl of the latter dilution to the subsequent tube in the series. Mix by vortexing and then centrifuge. Change pipette tips between each dilution.These dilutions will have a final concentration of 0.25 ng/μl, 0.025 ng/μl, and 0.0025 ng/μl.For qPCR, 4 μl will be added as template. These quantities will allow analysis of the standard DNA in duplicate with five sets of primers and can be adjusted as needed. If using the pGL3 plasmid control, include the pGL3 plasmid in the standard curve. The standard curve is prepared by adding 7 × 108 copies (3.71 ng) purified pGL3 plasmid to 5 μg UMSCC genomic DNA. A 10-fold dilution series is prepared as outlined in step 17.
-
18
Prepare qPCR master mix. Multiply volumes below by the number of reactions. Add at least two additional reactions as overage.
7.5 μl of 2× Power SYBR Green Master Mix or 2× iQ SYBR Green Supermix
Forward primer (300 nM final)
Reverse primer (300 nM final)
Bring volume to 11 μl with nuclease-free water.
Each sample and DNA standard should be analyzed in at least duplicate.
Separate master mix reactions will need to be prepared for each primer set being analyzed. If using the pGL3 plasmid control, a separate master mix containing primers to luciferase should also be included. PCR master mixes with primers to other HPV16 genes can also be included to monitor for episome integrity or plasmid contamination. See the Alternate Protocol for instructions.
-
19
Add 11 μl qPCR master mix to each well in a 96-well PCR plate.
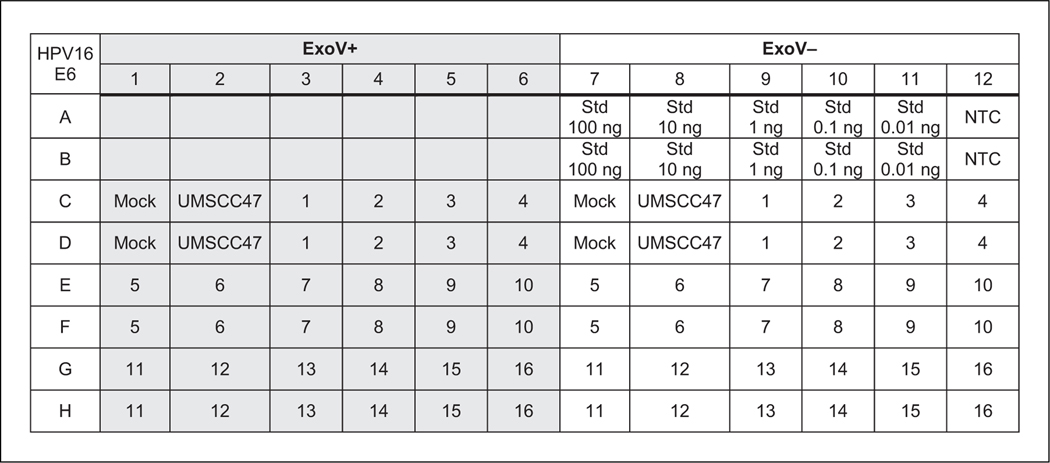
For an example 96-well plate setup, see Figure 2.
-
20
Add 4 μl standard DNA dilution series or 1:10 diluted sample DNA to the appropriate wells of the 96-well PCR plate.
-
21
Seal top of plate with adhesive optical film. Press firmly to seal around the edges of each well.
-
22
Centrifuge plate 1 min at 300 × g, room temperature, to remove bubbles from the bottom of the wells.
-
23Add qPCR plate to a thermal cycler with the following cycling conditions:
-
If using the Applied Biosystems Fast 7500 thermal cycler:
1 cycle 2 min 50°C 1 cycle 10 min 95°C 40 cycles 15 s 95°C 1 cycle 15 s 95°C (dissociation stage) 1 min 60°C 15 s 95°C 15 s 60°C. The dissociation/melt curve will monitor primer specificity in the assay.Run time is 2.5 hr. -
If using the Bio-Rad CFX96 Touch thermal cycler:
1 cycle 3 min 95°C 40 cycles 20 s 95°C 1 min 58°C Melting curve 10 s 95°C 5 s 0.5°C increments from 65°C to 95°C. Run time is 1.75 hr.
-
-
24
After the cycles have completed, export data as an Excel spreadsheet for further analysis.
Figure 2.
An example 96-well plate layout using one primer pair in qPCR. This example shows samples and standard curve dilutions in duplicate. ExoV+ labeled wells are samples digested with exonuclease V (ExoV). ExoV– labeled wells are no-enzyme controls to measure the starting input DNA. A total of 16 samples can be analyzed in this setup. Wells labeled Std (standards) represent wells loaded with the UMSCC47 10-fold dilution series. Mock refers to wells with no DNA added to the ExoV+ or Exo– reactions. UMSCC47 DNA monitors ExoV digestion of integrated human papillomavirus (HPV) genomes. NTC represents water as template (a no-template control) for qPCR.
Calculating relative DNA resistance to ExoV digestion
-
25Obtain the linear equation generated from the standard curve:
-
Average cycle threshold (Ct) values for each DNA dilution replicate used to generate the standard curve.Replicate wells should be within 1 Ct value of each other to ensure pipetting accuracy. Ct values should fall between 15 and 30.
- Calculate log concentration of the UMSCC47 standard curve DNA by first plotting the average Ct values against the log concentration of the standard DNA, using DNA input of 100, 10, 1, 0.1, and 0.01 ng. Obtain linear equation from the line by adding a trend line. Ensure correlation coefficient (R2) is above 0.98 and that the slope from the linear equation is between −3.1 and −3.6 to confirm PCR amplification efficiency.
- Repeat for each primer set (e.g., HPV16 E6 and rDNA primers).
-
-
26Calculate relative amount of HPV E6 or rDNA in each DNA sample:
- Average Ct values for each replicate DNA sample.
- Calculate relative log amount of HPV E6 or rDNA using the linear equation generated from the standard curve in step 25:
- Calculate relative amount of HPV E6 DNA and rDNA in each sample:
-
27Calculate percentage of DNA resistant to ExoV digestion:
-
28Interpret results as follows:
- Percent resistance for rDNA <10% indicates optimal enzyme digestion of the template DNA. Percent resistance for rDNA >10% indicates a failed digestion (see Troubleshooting for additional information).
- Integrated HPV genomes will have a percent resistance of <10% and will be similar to the percent resistance obtained for rDNA.
-
Samples with episomal HPV DNA are identified by two parameters: (1) HPV E6 percent resistance >20% and (2) HPV E6 percent resistance at least three-fold higher than the percent resistance observed for rDNAHPV-positive human keratinocyte cell lines with episomal HPV genomes showed a percent resistance to ExoV between 15% and 85% (Myers et al., 2019). The cut-off value of 20% is arbitrary with the confidence for calling a sample episomal increasing with a greater percent resistance to ExoV.
- Percent resistance for HPV-positive samples between 10% and 20% suggests mixed forms carrying both integrated and episomal HPV DNA.
SUPPORT PROTOCOL
EXONUCLEASE V ANALYSIS OF HPV16 GENOME CONFIGURATION IN TISSUES
The Basic Protocol describes how ExoV can be applied to determine the HPV16 genome status in cell lines. The Support Protocol can be used to analyze HPV16 genome configurations in HPV-positive tissues. This protocol has been successfully used to analyze the HPV16 genome in tissues grown using organotypic raft culture but has the potential to be applied to study HPV-positive clinical tissue samples (Bienkowska-Haba et al., 2018; Guidry et al., 2019; Myers et al., 2019).
Additional Materials (also see the Basic Protocol)
Tissue sample of interest
Qiagen QIAamp Blood Mini Kit with ALT buffer or NucleoSpin Blood QuickPure kit
Genomic DNA isolation from tissues
If using the Qiagen QIAamp Blood Mini Kit start with step 1. If using the NucleoSpin Blood mini columns, start with step 11.
QIAamp Blood Mini Kit
-
1
Add 180 μl ATL buffer (Qiagen) and 20 μl of 20 mg/ml proteinase K to each sample. Mix by vortexing.
-
2
Incubate samples overnight at 56°C.
-
3
Briefly centrifuge samples to remove any condensation from the lid, and add 200 μl AL buffer (Qiagen).
-
4
Mix by vortexing.
-
5
Incubate at 70°C for 10 min.
-
6
Briefly centrifuge to remove condensation from microcentrifuge tubes.
-
7
Add 200 μl ethanol and vortex. Briefly centrifuge and add to supplied column.
-
8
Continue with recommended wash and centrifugation steps according to the manufacturer’s protocol.
-
9
For the elution step: Add 50 μl of 10 mM Tris, pH 8.0, to the column. Incubate for 5 min at room temperature. Centrifuge 1 min at 6000 × g, room temperature.
Store the DNA at 4°C for up to 1 week or long term at −20°C.
-
10
Proceed to step 14.
NucleoSpin Blood mini columns
-
11
Add 200 μl of 1× PBS, 30 μl of 20 mg/ml proteinase K, and 200 μl BQ buffer (NucleoSpin).
-
12
Incubate at 70°C for 1 hr.
-
13
Centrifuge samples 5 min at 11,000 × g, room temperature. Use only the supernatant for subsequent steps as recommended by the manufacturer.
Do not use more than 25 mg tissue per column when using either kit. If isolating more than 25 mg, double the lysis buffer amount, and separate into two separate columns.
ExoV digestion and qPCR analysis
-
14
Follow steps 7 to 28 of the Basic Protocol for ExoV digestion and qPCR analysis.
ALTERNATE PROTOCOL
DETERMINING HPV INTEGRATION TYPE OR INTEGRITY OF HPV EPISOME
In the Basic Protocol, the percent resistance to ExoV digestion identifies DNA samples carrying integrated or episomal HPV in cells. However, the method cannot inform on the type of integration and is prone to false positive results from accidentally introduced plasmid contamination. If plasmids carrying the E6 gene are being used in the laboratory, episomal integrity of the HPV genome should be confirmed by comparing the copy number ratio of the HPV E6 region to other parts of the viral genome. This assay will not be able to discern contamination from plasmids carrying the entire HPV genome. If such plasmids are used in the laboratory, DNA extraction and PCR should be performed in a clean room environment. A PCR hood should be used to prevent introduction of potential plasmid contaminants. Copy number ratio analysis can also be applied to define the integration type in samples known to have integrated HPV DNA. qPCR of undigested DNA with alternate HPV16 primers can be used to identify HPV integrant types, confirm HPV16 episome integrity, and identify potential plasmid contaminants (Myers et al., 2019).
Additional Materials (also see the Basic Protocol)
Alternate HPV16 primers compatible with SYBR real-time PCR
HPV16 E2 forward: 5′-CCATATAGACTATTGGAAACACATGCGCC-3′
HPV16 E2 reverse: 5′-CTGTAGTTGCAGTTCAATTGCTTGTAATGC-3′
HPV16 E5 forward: 5′-TACGTCCGCTGCTTTTGTCT-3′
HPV16 E5 reverse: 5′-AACGCAGAGGCTGCTGTTAT-3′
HPV16 L2 forward: 5′-TGCATCGGCTACCCAACTTT-3′
HPV16 L2 reverse: 5′-ACCCGACCCTGTTCCAATTC-3′
Use serially diluted UMSCC47 DNA as described in step 17 of the Basic Protocol to generate a standard curve using primers to E2, E5, L2, and E6 in qPCR.
-
Perform qPCR analysis following steps 18 to 24 of the Basic Protocol with sample DNA that was not treated with ExoV using the E2, E5, L2, and E6 primers.
For each primer pair, the UMSCC47 standard DNA and sample DNA are analyzed on the same qPCR plate.
Calculate relative amount of DNA for each sample relative to its standard curve as described in step 25 of the Basic Protocol.
-
Divide relative amount of DNA for E2, E5, or L2 by the relative amount of DNA estimated for E6 to determine the copy number ratio.
For interpretation of results, see Table 1.
Table 1.
Interpretation for the Percent Resistance After ExoV Digestion and Copy Number Ratio Analyses
| HPV genome state | Percent resistance of HPV E6 DNA after ExoV digestion | Ratio of E2/E6, E5/E6, or L2/E6 (not treated with ExoV) |
|---|---|---|
| Defective episome (plasmid contaminant) | >20% | <0.5 for at least 1 ratio |
| Intact episome | >20% | ~1 for all ratios (range 0.8–1.2) |
| Integrated type I | <10% | <0.5 for at least 1 ratio |
| Integrated type II | <10% | ~1 for all ratios (range 0.8–1.2) |
ExoV, exonuclease V; HPV, human papillomavirus.
REAGENTS AND SOLUTIONS
10 mM Tris, pH 8.0
0.5 ml 1 M Tris, pH 8.0 (see recipe)
49.5 ml nuclease-free water
Store at room temperature for up to 12 months
1 M Tris, pH 8.0
6.05 g Tris base (e.g., Sigma-Aldrich, T-1503)
30 ml nuclease-free water
Adjust pH to 8.0 with 12.1 M HCl (e.g., Fisher Scientific, A144–500)
Bring to 50 ml with nuclease-free water
Store at room temperature for up to 12 months
COMMENTARY
Background Information
ExoV is the RecBCD heterotrimeric complex from Escherichia coli. ExoV (RecBCD) protects the bacterial genome from double-stranded breaks that can stall or collapse replication forks and is required for bacterial DNA replication and DNA repair. ExoV allows for strand resection and loading of RecA to single-stranded DNA at crossover hotspot instigator DNA elements for homologous recombination and DNA repair. ExoV possesses various enzymatic activities. ExoV is a highly processive DNA helicase, DNA-dependent ATPase, single-stranded DNA endonuclease, and single-stranded and double-stranded DNA exonuclease (Dillingham & Kowalczykowski, 2008). The RecBCD (ExoV) complex has a high affinity to blunt ends or short 5′or 3′ overhangs on double-stranded DNA. ExoV exonuclease activity is bidirectional with the helicase and the single-stranded DNA endonuclease activity working together to hydrolyze DNA into short fragments. The nuclease activity is dependent on the concentration of Mg2+ ions. Ca2+ ions inhibit the nuclease activity but allow helicase activity without DNA degradation. Both Mg2+ and ATP are required for ExoV helicase activity.
The selectivity of ExoV for linear DNA and not circular or nicked DNA has been used by different laboratories to study circular DNAs. A method using ExoV has recently been applied to study the covalently closed circular hepatitis B virus (3.2 kb) DNA (Gao, Yan, & Li, 2019). ExoV has also been used to enrich for human mitochondrial DNA (16 kb) prior to sequencing (Yao et al., 2019). The protocol outlined in this article has been used to determine the state of the HPV genome in established cell lines and organotypic raft tissues (Bienkowska-Haba et al., 2018; Guidry et al., 2019; Myers et al., 2019). While episomal HPV would be resistant to ExoV degradation, integrated HPV would be enzymatically hydrolyzed resulting in a loss of signal in qPCR relative to the no-enzyme control. The assay proved to be sensitive, requiring only nanogram quantities of DNA. Circular DNA—such as the HPV 8 kb genome, plasmids up to 10 kb, and 16 kb mitochondrial DNA—was compatible with the ExoV analysis procedure described (Myers et al., 2019). The HPV episomal genome was resistant to ExoV digestion after various cycles of freeze–thawing, which would allow retrospective analysis of stored samples (Myers et al., 2019). Application of the ExoV assay to the analysis of other small DNA viruses with circular genomes (polyomaviruses) is feasible. Whether the ExoV assay can be applied to larger DNA viral genomes, such as herpesviruses, needs further testing.
Critical Parameters
While this assay is rapid and sensitive in the detection of episomal HPV DNA, it is a relative analysis and does not attempt to quantify the absolute amount of episomal HPV DNA in a cell. Although the HPV genome can tolerate several rounds of freeze–thawing and vortexing (Myers et al., 2019), breakage of the viral episomes can result in loss of signal. Sample DNA thawing should be limited as the reliability of the assay is dependent upon DNA integrity. After DNA isolation, small aliquots of genomic DNA should be made and used to prevent multiple rounds of thawing. The high sensitivity of the ExoV assay for detecting circular DNA can result in false positive signals due to plasmid contaminants. To limit false positives, solutions should be prepared in a clean room and work should be done in a PCR hood. Negative controls should be included to ensure the absence of plasmid contaminants. Negative controls can be a no-template control or DNA from an HPV-negative cell line. Loss of ExoV activity is seen over time. Proper handling and aliquoting of the enzyme and ATP are required to ensure efficient nuclease activity.
Troubleshooting
rDNA percent resistance after ExoV digestion above 10% indicates suboptimal enzymatic activity that may be due to deterioration of the ATP or the enzyme over time or impurities in DNA preparation. More enzyme can be added or the DNA sample reisolated to remove any impurities prior to retesting. ExoV will need to be replaced with fresh enzyme periodically. See other potential issues and solutions in Table 2.
Table 2.
Troubleshooting Guide for the ExoV Assay
| Problem | Possible cause | Solution |
|---|---|---|
| rDNA resistant fraction >10% | ExoV activity may have deteriorated during storage | Replace enzyme and ATP |
| Presence of inhibitors that affect ExoV activity | Reisolate sample DNA | |
| UMSCC47 resistant fraction >10% | ExoV activity may have deteriorated during storage | Replace enzyme |
| Presence of inhibitors that affect ExoV activity | Reisolate sample DNA | |
| Presence of contaminating circular DNA | Reisolate sample DNA | |
| HPV DNA resistant fraction >100% | Contamination in PCR DNA may not be well purified | Reisolate sample DNA Reisolate sample DNA |
ExoV, exonuclease V; HPV, human papillomavirus.
Understanding Results
The ExoV assay is unable to determine the absolute amount of episomal DNA in a given sample due to variable loss of episomal DNA during isolation and storage. Instead, the ExoV assay uses the percent resistant fraction of HPV16 E6 DNA above that measured for rDNA to determine the presence of episomal HPV. Samples that exhibit an HPV16 E6 percent resistant fraction above 20% and at least three-fold higher than the percent resistance observed for rDNA carry episomal HPV DNA (Myers et al., 2019). A higher E6 percent resistance increases the confidence for the presence of episomal HPV DNA in a sample. HPV percent resistance between 10% and 20% are difficult to define and may be mixed forms containing both integrated and episomal HPV DNA. HPV percent resistance below 10% and comparable to that of the rDNA percent resistance indicates integrated HPV.
Time Considerations
The ExoV assay can be performed in a single day once samples are ready for DNA isolation. DNA purification using either of the DNA isolation kits described and ExoV digestion can take up to 4 hr. qPCR plate setup takes an hour and a maximum of 2.5 hr on the thermal cycler. After qPCR, data analysis can take 1 to 2 hr. From DNA isolation to data analysis, the total time required to complete the assay is ~8 hr but is dependent on the number of samples analyzed.
Acknowledgments
The authors would like to acknowledge the Louisiana State University Health Science Center-Shreveport Research Core Facility for their help in the development of this assay. This work was supported by National Institutes of Health grant awards from the NIDCR DE025565, NCI CA211576, and NIGMS GM110703. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Conflicts of Interest
The authors report no conflicts of interest.
Literature Cited
- Akagi K, Li J, Broutian TR, Padilla-Nash H, Xiao W, Jiang B, … Gillison ML (2014). Genome-wide analysis of HPV integration in human cancers reveals recurrent, focal genomic instability. Genome Research, 24, 185–199. doi: 10.1101/gr.164806.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienkowska-Haba M., Luszczek W., Myers JE., Keiffer TR., DiGiuseppe S., Polk P., … Sapp M. (2018). A new cell culture model to genetically dissect the complete human papillomavirus life cycle. PLoS Pathogens, 14, e1006846. doi: 10.1371/journal.ppat.1006846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner JC,Graham MP, Kumar B, Saunders LM, Kupfer R, Lyons RH, … Carey TE (2010). Genotyping of 73 UM-SCC head and neck squamous cell carcinoma cell lines. Head & Neck, 32, 417–426. doi: 10.1002/hed.21198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd EM (2003). Human papillomavirus and cervical cancer. Clinical Microbiology Reviews, 16, 1–17. doi: 10.1128/CMR.16.1.1-17.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett LC, Lunn G, & Coico R. (2009). Biosafety: Guidelines for working with pathogenic and infectious microorganisms. Current Protocols in Microbiology, 13, 1A.1.1–1A.1.14. doi: 10.1002/9780471729259.mc01a01s13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Martel C, Plummer M, Vignat J, & Franceschi S. (2017). Worldwide burden of cancer attributable to HPV by site, country and HPV type. International Journal of Cancer, 141, 664–670. doi: 10.1002/ijc.30716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham MS, & Kowalczykowski SC (2008). RecBCD enzyme and the repair of double-stranded DNA breaks. Microbiology and Molecular Biology Reviews, 72, 642–671. doi: 10.1128/mmbr.00020-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doorbar J, Egawa N, Griffin H, Kranjec C, & Murakami I. (2015). Human papillomavirus molecular biology and disease association. Reviews in Medical Virology, 25, 2–23. doi: 10.1002/rmv.1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa N, & Doorbar J. (2017). The low-risk papillomaviruses. Virus Research, 231, 119–127. doi: 10.1016/j.virusres.2016.12.017. [DOI] [PubMed] [Google Scholar]
- Gao Z, Yan L, & Li W. (2019). A quantitative method for hepatitis B virus covalently closed circular DNA enables distinguishing direct acting antivirals from cytotoxic agents. Antiviral Research, 168, 197–202. doi: 10.1016/j.antiviral.2019.06.002. [DOI] [PubMed] [Google Scholar]
- Gooi Z, Chan JY, & Fakhry C. (2016). The epidemiology of the human papillomavirus related to oropharyngeal head and neck cancer. Laryngoscope, 126, 894–900. doi: 10.1002/lary.25767. [DOI] [PubMed] [Google Scholar]
- Guidry JT, Myers JE, Bienkowska-Haba M, Songock WK, Ma X, Shi M, … Scott RS (2019). Inhibition of Epstein-Barr virus replication in human papillomavirus-immortalized keratinocytes. Journal of Virology, 93, e01216–18. doi: 10.1128/jvi.01216-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard RA (2003). Human papillomavirus testing methods . Archives of Pathology & Laboratory Medicine, 127, 940–945. doi: . [DOI] [PubMed] [Google Scholar]
- Jeon S, Allen-Hoffmann BL, & Lambert PF (1995). Integration of human papillomavirus type 16 into the human genome correlates with a selective growth advantage of cells. Journal of Virology, 69, 2989–2997. doi: 10.1128/JVI.69.5.2989-2997.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaes R, Woerner SM, Ridder R, Wentzensen N, Duerst M, Schneider A, … von Knebel Doeberitz M. (1999). Detection of high-risk cervical intraepithelial neoplasia and cervical cancer by amplification of transcripts derived from integrated papillomavirus oncogenes. Cancer Research, 59, 6132–6136. [PubMed] [Google Scholar]
- Luft F, Klaes R, Nees M, Durst M, Heilmann V, Melsheimer P, & von Knebel Doeberitz M. (2001). Detection of integrated papillomavirus sequences by ligation-mediated PCR (DIPS-PCR) and molecular characterization in cervical cancer cells. International Journal of Cancer, 92, 9–17. doi: . [DOI] [PubMed] [Google Scholar]
- McBride AA, & Warburton A. (2017). The role of integration in oncogenic progression of HPV-associated cancers. PLoS Pathogens, 13, e1006211. doi: 10.1371/journal.ppat.1006211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers JE, Guidry JT, Scott ML, Zwolinska K, Raikhy G, Prasai K, … Scott RS (2019). Detecting episomal or integrated human papillomavirus 16 DNA using an exonuclease V-qPCR-based assay. Virology, 537, 149–156. doi: 10.1016/j.virol.2019.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olthof NC, Huebbers CU, Kolligs J, Henfling M, Ramaekers FC, Cornet I, … Speel EJ (2015). Viral load, gene expression and mapping of viral integration sites in HPV16-associated HNSCC cell lines. International Journal of Cancer, 136, E207–218. doi: 10.1002/ijc.29112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olthof NC, Speel EJ, Kolligs J, Haesevoets A, Henfling M, Ramaekers FC, … Huebbers CU (2014). Comprehensive analysis of HPV16 integration in OSCC reveals no significant impact of physical status on viral oncogene and virally disrupted human gene expression. PLoS One, 9, e88718. doi: 10.1371/journal.pone.0088718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfenov M, Pedamallu CS, Gehlenborg N, Freeman SS, Danilova L, Bristow CA, … Kucherlapati R. (2014). Characterization of HPV and host genome interactions in primary head and neck cancers. Proceedings of the National Academy of Sciences of the United States of America, 111, 15544–15549. doi: 10.1073/pnas.1416074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pett MR, Herdman MT, Palmer RD, Yeo GS, Shivji MK, Stanley MA, & Coleman N. (2006). Selection of cervical keratinocytes containing integrated HPV16 associates with episome loss and an endogenous antiviral response. Proceedings of the National Academy of Sciences of the United States of America, 103, 3822–3827. doi: 10.1073/pnas.0600078103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern EM (1975). Detection of specific sequences among DNA fragments separated by gel electrophoresis. Journal of Molecular Biology, 98, 503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH, & Pierce AJ (2008). Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Research, 18, 13–18. doi: 10.1101/gr.6858507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, … Munoz N. (1999). Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. Journal of Pathology, 189, 12–19. doi: . [DOI] [PubMed] [Google Scholar]
- Wentzensen N, Vinokurova S, & von Knebel Doeberitz M. (2004). Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Research, 64, 3878–3884. doi: 10.1158/0008-5472.can-04-0009. [DOI] [PubMed] [Google Scholar]
- Yao Y, Nishimura M, Murayama K, Kuranobu N, Tojo S, Beppu M, … Tanaka T. (2019). A simple method for sequencing the whole human mitochondrial genome directly from samples and its application to genetic testing. Scientific Reports, 9, 17411. doi: 10.1038/s41598-019-53449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshinouchi M, Hongo A, Nakamura K, Kodama J, Itoh S, Sakai H, & Kudo T. (1999). Analysis by multiplex PCR of the physical status of human papillomavirus type 16 DNA in cervical cancers. Journal of Clinical Microbiology, 37, 3514–3517. doi: 10.1128/JCM.37.11.3514-3517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]