Abstract
The quinohemoprotein tetrahydrofurfuryl alcohol dehydrogenase (THFA-DH) from Ralstonia eutropha strain Bo was investigated for its catalytic properties. The apparent kcat/Km and Ki values for several substrates were determined using ferricyanide as an artificial electron acceptor. The highest catalytic efficiency was obtained with n-pentanol exhibiting a kcat/Km value of 788 × 104 M−1 s−1. The enzyme showed substrate inhibition kinetics for most of the alcohols and aldehydes investigated. A stereoselective oxidation of chiral alcohols with a varying enantiomeric preference was observed. Initial rate studies using ethanol and acetaldehyde as substrates revealed that a ping-pong mechanism can be assumed for in vitro catalysis of THFA-DH. The gene encoding THFA-DH from R. eutropha strain Bo (tfaA) has been cloned and sequenced. The derived amino acid sequence showed an identity of up to 67% to the sequence of various quinoprotein and quinohemoprotein dehydrogenases. A comparison of the deduced sequence with the N-terminal amino acid sequence previously determined by Edman degradation analysis suggested the presence of a signal sequence of 27 residues. The primary structure of TfaA indicated that the protein has a tertiary structure quite similar to those of other quinoprotein dehydrogenases.
The xenobiotic cyclic ether tetrahydrofurfuryl alcohol (THFA) is used as a versatile solvent in industry and is released in large amounts during the synthesis of furan resins. We have recently isolated a bacterium able to use THFA (at concentrations of up to 200 mM) as its sole source of carbon and energy which was identified as Ralstonia eutropha and designated strain Bo (56). Enzymatic studies revealed that the degradation of THFA is initiated by an oxidation of the alcohol via the aldehyde to the corresponding carboxylic acid (Fig. 1). In vitro, both steps are catalyzed by an inducible tetrahydrofurfuryl alcohol dehydrogenase (THFA-DH). In contrast to various NAD(P)-dependent alcohol dehydrogenases (ADHs) and aldehyde dehydrogenases described for R. eutropha (23, 31, 42), THFA-DH could be identified as a type I quinohemoprotein ADH containing pyrroloquinoline quinone (PQQ) and a heme c as cofactors (56).
FIG. 1.
Oxidation of THFA to the corresponding carboxylic acid catalyzed by THFA-DH from R. eutropha strain Bo.
NAD(P)-independent ADHs containing PQQ as a prosthetic group are found in many aerobic bacteria (3, 10–13, 20, 21, 25, 40, 48, 53, 54). According to their structure and cofactor content, quinoprotein alcohol dehydrogenases are divided into two main types (24). Class I is represented by enzymes containing PQQ and Ca2+-like methanol dehydrogenase from methylotrophic bacteria (8), whereas class II includes proteins containing an additional heme c designated quinohemoprotein ADHs. Quinohemoprotein ADHs type I are monomeric enzyme of about 72 kDa with an equimolar content of PQQ, Ca2+, and heme c (9, 48, 56). Quinohemoprotein ADHs type II are multimeric proteins of at least three subunits found in bacteria of the genera Acetobacter and Gluconobacter (1, 34). These enzymes contain one PQQ molecule and one heme c in the α-subunit and several heme c binding motifs in the other subunits. It became obvious from the structural data that the type I dehydrogenases resemble the α-subunit of type II enzymes (22, 45).
It is known that quinoprotein ADHs can be used for biotechnological applications, e.g., as biosensors or for the stereoselective oxidation of chiral alcohols (14, 17, 28, 39, 51). The broad substrate spectrum makes them useful for the production of different pure enantiomers (44). In the present paper, we report some of the catalytic features of THFA-DH from R. eutropha strain Bo. Furthermore, the complete amino acid sequence of THFA-DH derived from the isolated gene was determined and analyzed.
MATERIALS AND METHODS
Chemicals.
The chemicals used in the present study were at least of analytical reagent grade and were obtained from Sigma, Merck, or Fluka. Chromatographic materials were obtained from Pharmacia.
Bacterial strains, plasmids, and growth conditions.
Maintenance of strains, growth of mass cultures, and cell harvest of strain Bo (DSM 11098) was done as reported before (56). Escherichia coli XL-1 Blue used for cloning purposes was grown in Luria-Bertani medium or on agar plates containing antibiotics as described by Sambrook et al. (38). The plasmid pUC19 was used for construction of a genomic library of R. eutropha strain Bo. PCR products were ligated into the pGEM-T Easy vector (Promega).
Enzyme assay.
The standard assay for determination of THFA-DH activity was performed as described previously with ferricyanide as an artificial electron acceptor (56) except that the Tris HCl buffer was replaced by 75 mM morpholinepropanesulfonic acid (MOPS)-NaOH, pH 8.2.
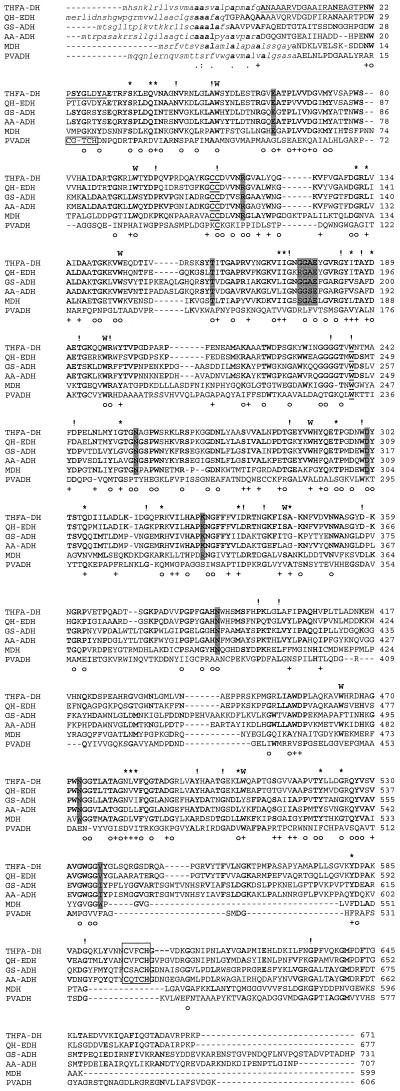
The kinetic constants were determined by fitting the obtained data to the model of Michaelis-Menten with or without substrate inhibition. The data determined for substrates exhibiting inhibition kinetics were analyzed by equation 1.
 |
1 |
Data analysis was done with the SigmaPlot program (Jandel Scientific). Analysis of the reaction mechanism was performed with a modified enzyme assay using 20 mM MOPS-NaOH, pH 8.0, containing 5 mM CaCl2. Semicarbazide (2 mM) was added to the reaction mixture if ethanol was used as a substrate. The observed endogenous ferricyanide reduction was subtracted from the substrate-dependent activity.
The enantioselectivity of THFA-DH during the conversion of racemic substrates was estimated as previously described (44) (in equation 2, A and B are the different enantiomers).
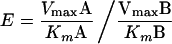 |
2 |
Purification of THFA-DH.
Crude extracts were prepared as previously described (56) by suspending cell paste of R. eutropha strain Bo grown on THFA (1 g/ml) in 50 mM Tris HCl, pH 8.0, containing 5 mM CaCl2, 0.5 mM dithiothreitol, and 1 mM EDTA. THFA-DH was purified from cell extracts of R. eutropha strain Bo as described previously (56).
Electrophoresis.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis was done according to the method of Laemmli (27) with 12.5% polyacrylamide gels. Electrophoresis was carried out under a constant current of 25 mA per gel and maximum voltage.
Standard DNA techniques.
DNA was treated by standard methods (38). Genomic DNA was isolated from cells of R. eutropha strain Bo grown in mineral salts medium with 0.2% fructose (wt/vol) as described by Marmur (33). Plasmid DNA was isolated from E. coli with the Qiagen plasmid purification kits. The molecular masses of DNA fragments were estimated by using restricted λ DNA or pGEM vector as molecular mass markers (Promega). Restriction enzymes purchased from MBI Fermentas or Roche were used according to the manufacturer's instructions. DNA fragments were isolated from agarose gels using Qiaex gel extraction kits (Qiagen). The standard PCR was performed by using Taq DNA polymerase (Roche) in an appropriate buffer, an annealing temperature optimized for the primer combination used, and 30 cycles. Hybridization experiments were carried out using the DIG DNA labelling and detection kit (Roche) according to the manufacturer's instructions.
DNA sequences were determined by primer walking and the dideoxynucleotide chain-termination method with an ABI 377 Sequencer, version 4.0 (Applied Biosystems), a Cycle Sequencing kit (Pharmacia), and universal, reverse, or sequence-specific primers. All primers were obtained from a commercial supplier (Gibco or Metabion).
Cloning of the gene encoding THFA-DH.
Degenerated primers were designed from the N-terminal amino acid sequence of THFA-DH (primer 1, 5′AAYGARGCIGGIACICCIAAYTGG3′) and from highly conserved sequence regions of other quinohemoprotein ADHs comprising the region between nucleotides 1087 and 1109 (primer 2, 5′AARAAICCITTYTTIGGIGCRTG3′) and the region between nucleotides 1903 and 1919 (primer 3, 5′CCRTGRCARAARACRCA3′) (quinohemoprotein ethanol dehydrogenase [QH-EDH]; Comamonas testosteroni numbering). The obtained specific PCR products were cloned in the pGEM-T Easy vector and sequenced. A genomic library of R. eutropha strain Bo was constructed by partial digestion of genomic DNA with Sau3A. DNA fragments of 2.5 to 6 kb were isolated from agarose gels and cloned in the BamHI-restricted pUC19 vector (MBI Fermentas). After transformation in E. coli XL-1 Blue, about 6,300 clones were obtained, and their isolated plasmids were stored as a mixture at −20°C.
Computer-aided analysis of gene sequences.
DNA sequences were analyzed using computer programs DNASIS (version 5.00) and Clone Manager (version 4.0) (Scientific & Educational Software). The sequences were aligned with the program Fasta3 of the European Molecular Biology Laboratory outstation, Hinxton (http://www2.ebi.ac.uk/fasta3/), or BlastX at the homepage of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/blast/). The multiple alignment of various sequences available at EMBL and GenBank databases was done with the program ClustalW at the European Molecular Biology Laboratory service homepage (http://www2.ebi.ac.uk/clustalw/). The theoretical molecular mass and isoelectric point were estimated with program tools located at http://www-biol.univ-mrs.fr/cgi-bin/abim/.
Nucleotide sequence accession number.
The nucleotide sequence of tfaA is available from GenBank under accession number AF277373.
RESULTS
Catalytic properties of THFA-DH. (i) Kinetic parameters.
We previously reported that THFA-DH from R. eutropha strain Bo exhibited a broad substrate spectrum (56). To obtain further information about the catalytic properties of THFA-DH, we performed initial rate measurements in more detail. In addition to the substrates previously reported to be converted by THFA-DH, the following compounds were also oxidized to a significant extent (values in parentheses are Vmax values in units per milligram [THFA (43.6)]: n-heptanol (53.8), 2-propanol (9.5), tetrahydropyran-2-alcohol (32), propionaldehyde (73.8), 1,3-propandiol (61.2), 1,2-pentandiol (44.6), 1,5-pentandiol (73.4), and 1,7-heptandiol (73.7). Although THFA-DH was isolated from R. eutropha strain Bo enriched and grown on THFA as a substrate, the purified enzyme showed the highest catalytic efficiency under in vitro conditions with n-pentanol, whereas THFA was converted with a comparable low rate (Table 1). Ethanol was the only primary alcohol investigated which was a poor substrate of THFA-DH. No dye-linked THFA-DH activity was detectable in crude extracts of R. eutropha strain Bo grown on ethanol, 2-propanol, and 1,5-pentandiol. However, the purified enzyme showed detectable activities with these substrates in the in vitro enzyme assay (see above).
TABLE 1.
Kinetic parameters for several substrates of THFA-DH from R. eutropha strain Boa
| Substrate | Parameter
|
||
|---|---|---|---|
| Km (mM) | kcat/Km × 104 (M−1 s−1) | Ki (mM) | |
| Primary alcohols | |||
| Ethanolb | 3.1 | 3 | — |
| n-Propanol | 0.036 | 248 | ND |
| n-Butanol | 0.015 | 594 | 14.7 |
| n-Pentanol | 0.012 | 788 | 3.2 |
| n-Hexanol | 0.014 | 495 | 1.4 |
| n-Heptanol | 0.016 | 413 | 1.9 |
| Aldehydes | |||
| Formaldehydeb | 1.93 | 2.3 | — |
| Acetaldehydeb | 0.075 | 120 | — |
| Propionaldehyde | 0.057 | 159 | 8.2 |
| Butyraldehyde | 0.033 | 244 | 0.95 |
| Pentanal | 0.061 | 121 | 0.33 |
| Secondary alcohols | |||
| 2-Propanol | 1.9 | 0.6 | ND |
| 2-Butanolb | 1.12 | 3.1 | — |
| 2-Pentanol | 0.12 | 99 | 8.6 |
| 2-Hexanol | 0.08 | 167 | 1.6 |
| 2-Heptanol | 0.058 | 204 | 1.0 |
| 2-Octanol | 0.076 | 129 | 1.3 |
| O-heterocyclic compounds | |||
| THF-2-alcohol | 0.039 | 138 | 2.2 |
| THF-3-alcohol | 0.056 | 98 | ND |
| Furfurylalcohol | 0.025 | 185 | 1.9 |
| Furfuraldehyde | 0.147 | 16 | 11.0 |
| Tetrahydropyran-2-alcohol | 0.011 | 386 | 1.5 |
| Diols | |||
| 1,2-Butandiolb | 1.03 | 5 | — |
| 1,3-Butandiolb | 0.49 | 15 | — |
| 1,4-Butandiolb | 0.25 | 30 | — |
| PEG 6000b | 0.55 | 7 | — |
Activities were determined with homogeneous THFA-DH (18.8 nM) in 75 mM MOPS-NaOH, pH 8.2, and 5 mM CaCl2 under standard assay conditions with appropriate substrate concentrations. The apparent kinetic constants were determined by nonlinear curve fittings based on Michaelis-Menten kinetics. ND, not determined; —, no substrate inhibition detected under the conditions used.
The curves were fitted based on the model of Michaelis and Menten. All other curves were fitted based on the model of substrate inhibition as described in Materials and Methods.
Initial rate measurements of in vitro substrate conversion of THFA-DH showed that in most cases the obtained data can be explained by a strong substrate inhibition, although some substrates were not inhibitory (Table 1 and 2) and these curves fitted the kinetic model without substrate inhibition.
TABLE 2.
Enantioselective oxidation of chiral alcohols by homogeneous THFA-DH from R. eutropha strain Boa
| Substrate | Kmb (mM) | Vmax (U/mg) | Kcat/Km × 104 (M−1 s−1) | Ki (mM) | E |
|---|---|---|---|---|---|
| (±)-2-Butanol | 1.12 | 28.2 | 3.1 | — | |
| R-(−)-2-Butanol | 1.14 | 17.5 | 1.9 | — | |
| S-(+)-2-Butanol | 0.59 | 32.0 | 6.6 | — | 3.5 |
| (±)-2-Heptanol | 0.058 | 96.8 | 204.2 | 1.03 | |
| R-(−)-2-Heptanol | 1.16 | 22.1 | 2.3 | 0.82 | |
| S-(+)-2-Heptanol | 0.046 | 131.6 | 350.0 | 0.93 | 150.2 |
| (±)-2-Octanol | 0.076 | 80.4 | 129.4 | 1.34 | |
| R-(−)-2-Octanol | 0.269 | 18.2 | 8.3 | 6.62 | |
| S-(+)-2-Octanol | 0.051 | 101.3 | 243.0 | 0.98 | 29.4 |
| (±)-Solketal | ND | ||||
| R-(−)-Solketal | 0.11 | 3.1 | 3.4 | — | |
| S-(+)-Solketal | 0.74 | 5.6 | 0.9 | — | 0.27 |
The kinetic parameters were determined using ferricyanide as an artificial electron acceptor and under the conditions described in Table 1, footnote a. The enantiomeric ratios are expressed in E values as described in Materials and Methods. —, no substrate inhibition detected under the conditions used.
The curves were fitted to the model of Michaelis and Menten with or without substrate inhibition. ND, not determined.
(ii) Kinetic mechanism.
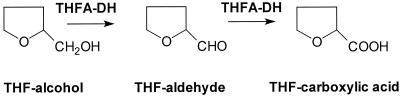
Due to the observed lack of substrate inhibition, the kinetic mechanism of THFA-DH was investigated with ethanol or acetaldehyde as a substrate and ferricyanide as an electron acceptor. Analysis was performed by varying the concentration of ethanol or acetaldehyde while maintaining the concentration of ferricyanide at different values in the range of its apparent Km value and vice versa. The double-reciprocal plots obtained from the data of initial rate measurements are depicted in Fig. 2. The fact that the plot lines are parallel suggested that a ping-pong kinetic mechanism should be assumed for the THFA-DH catalyzed reaction. Because n-pentanol as the best substrate of THFA-DH had an inhibitory effect, analysis of the kinetic mechanism was more difficult. However, at low substrate concentrations, the lines of double-reciprocal plots were parallel (data not shown), thus indicating that this substrate is also converted by a ping-pong mechanism.
FIG. 2.
Analysis of the reaction mechanism of THFA-DH from R. eutropha strain Bo in the in vitro enzyme assay with ethanol or acetaldehyde as a substrate and ferricyanide as an electron acceptor. The initial rate measurements are depicted as Lineweaver-Burk plots. The activity was determined with 20 mM MOPS-NaOH, pH 8.0, containing 5 mM CaCl2 and 18.8 nM THFA-DH. In case ethanol was used as a substrate, 2 mM semicarbazide was added to the reaction mixture. (A) Initial rates at varying ethanol concentrations, depending on the ferricyanide concentration used: 0.05 (⧫), 0.1 (□), 0.25 (●), and 0.5 (○) mM. (B) Initial rates at varying ferricyanide concentrations, depending on the ethanol concentration used: 0.5 (⧫), 1.0 (□), 2.0 (●), 3.0 (○), and 5.0 (▴) mM. (C) Initial rates at varying acetaldehyde concentrations, depending on the ferricyanide concentration used: 0.02 (⧫), 0.04 (□), 0.06 (●), 0.1 (○), and 0.3 (▴) mM. (D) Initial rates at varying ferricyanide concentrations depending on the acetaldehyde concentration used: 0.02 (⧫), 0.04 (□), 0.06 (●), 0.1 (○), and 0.3 (▴) mM.
(iii) Enantioselectivity.
It was previously reported that the quinohemoprotein ADHs isolated from C. testosteroni, Acetobacter pasteurianus, and Acetobacter aceti exhibited an enantiomeric preference if racemic mixtures of chiral alcohols were used as substrates (15, 17, 29, 30, 43, 44). Thus, we examined if THFA-DH from R. eutropha strain Bo also showed a preference for one enantiomer of different chiral alcohols. Initial rate measurements performed with secondary aliphatic alcohols indicated that the enzyme had a high preference for the oxidation of the S-(+)-enantiomers (Table 2). Although the catalytic efficiency for conversion of the cyclic ether solketal was low, THFA-DH unequivocally preferred the R-(−)-enantiomer (Table 2).
Cloning and sequencing of the gene encoding THFA-DH.
To obtain more information on the primary structure of THFA-DH from R. eutropha strain Bo, the encoding gene was cloned and sequenced. Two DNA fragments of 0.95 and 1.8 kb were obtained by PCR and identified by sequencing to encode parts of THFA-DH comprising the N-terminal sequence previously determined by Edman degradation (56). These fragments were used as a probe to screen the genomic library of R. eutropha strain Bo. Three positive clones were obtained carrying plasmids pRE21-17, pRE28-15, and pRE18-87, which contained 6.1-, 6.3-, and 6.0-kb fragments of genomic DNA from strain Bo inserted into the pUC19 vector.
Sequencing of the inserts of pRE21-17, pRE28-15, and pRE18-87 led to the identification of a large open reading frame designated as tfaA. This open reading frame started with the translation start codon ATG (nucleotides 104 to 106) and ended with a TGA termination codon (nucleotides 2198 to 2200). A putative ribosome binding site (AGGAG) is located six nucleotides upstream of the translation start codon (data not shown). The protein encoded by tfaA is composed of 698 amino acids. The N-terminal amino acid sequence of THFA-DH previously determined by Edman degradation (56) corresponded exactly to the deduced sequence from Ala-28 to Ala-57 (Fig. 3). These data indicated that THFA-DH contains a leader peptide which is cleaved off during maturation of the protein (Fig. 3). Analysis of the N-terminal amino acid sequence deduced from tfaA by the computer program PSORT (36) or SignalP (version 1.1) suggested a signal peptide for translocation of the protein into the periplasm with a possible cleavage site between Gly-27 and Ala-28 (Fig. 3). The presence of a leader peptide was also supported by sequence alignments with other quinohemoprotein ADHs, all known to contain a signal sequence (Fig. 3). A theoretical molecular mass of 72,689 Da was calculated for TfaA from the deduced amino acid sequence (without leader peptide, PQQ, and heme cofactor). These data confirmed our previous molecular mass determination of about 73 kDa (56). Furthermore, the calculated isoelectric point of 9.22 corresponded to 9.1, previously determined by isoelectric focusing (56). The G+C content of the tfaA gene was 67.5 mol%, close to the 65 mol% determined for the genomic DNA of R. eutropha strain Bo (U. Lechner, personal communication). These results together with the homology studies described below strongly suggested that we have cloned the DNA fragment encoding THFA-DH from R. eutropha strain Bo. Southern blot analysis of genomic DNA from strain Bo showed that the tfaA gene was present as a single copy (data not shown).
FIG. 3.
Multiple sequence alignment of TfaA from R. eutropha strain Bo (THFA-DH) with various quinoprotein ADHs: QH-EDH from C. testosteroni (Q46444); GS-ADH, quinoprotein ADH from Gluconobacter suboxydans (O05542); AA-ADH, quinoprotein ADH from A. aceti (P18278); MDH, methanol dehydrogenase from Methylobacterium extorquens (P16027); PVADH from Pseudomonas sp. P77931. Signal sequences are depicted in lowercase letters. Amino acids which were conserved in all investigated sequences are indicated on top of the alignment by !, similar residues are marked by ∗, and residues which are conserved in most sequences are indicated by boldface letters. Amino acids identical in all sequences, excluding PVADH, are indicated at the bottom of the alignment by ○, and similar residues are shown by +. The heme c binding sites are boxed, and amino acids supposed to be involved in PQQ binding are indicated by shaded boxes. The disulfide ring-forming cysteine residues and the opposite tryptophan residue are underlined. The tryptophan residues corresponding to tryptophan-docking motifs are indicated by a boldface W above the sequences. The N-terminal sequence previously determined by Edman degradation is underlined.
Analysis of the sequence located upstream and downstream of the tfaA gene (each, 0.8 kb) did not reveal an open reading frame which might encode a protein involved in THFA degradation or PQQ biosynthesis (data not shown).
Protein sequence comparison.
The amino acid sequence deduced from tfaA was compared to sequences deposited in the EMBL or GenBank database with the Fasta3 or BlastX program, respectively. This database analysis revealed a similarity of the deduced protein (TfaA) to various quinoprotein dehydrogenases. An identity of 43% was obtained to the large subunit of the type II quinohemoprotein ADHs isolated from different Acetobacter species and about 32% identity was determined to the α-subunit of different methanol dehydrogenases (Fig. 3). However, the highest similarity, with 67% identity, was determined to the type I QH-EDH from C. testosteroni (Fig. 3). Interestingly, the identity to polyvinyl alcohol dehydrogenase (PVADH) from Pseudomonas sp. VM15C, the second sequenced type I enzyme, was only 24%.
The C-terminal part of TfaA (about 88 amino acids) should be responsible for binding the heme cofactor. However, this region exhibited an identity of only 13% to the corresponding sequence of the α-subunit of type II quinohemoprotein ADH from acetic acid bacteria (Fig. 3) (22, 46, 47). Furthermore, there was no significant similarity to the β-subunit of type II quinohemoprotein ADHs known to contain heme-binding sites. On the other hand, the C-terminal part of TfaA showed an identity of 30 to 38% to various cytochromes c (references 26 and 40 and data not shown). The multiple sequence alignment summarizes the conserved amino acid residues of the analyzed quinoprotein and quinohemoprotein ADHs (Fig. 3).
The sequence motif Cys-Xaa-Yaa-Cys-His reported to be involved in binding the heme cofactor (35) was discovered in the C-terminal part of TfaA and located between Cys624 and His627 (Fig. 3). Additional residues involved in cofactor binding and β-propeller fold formation were found to be conserved in the sequence deduced for TfaA (Fig. 3) (19). Thus, we could assign the amino acids forming a sandwich structure with the PQQ cofactor. The two cysteine residues of TfaA, Cys109, and Cys110 (Fig. 3), probably form the disulfide ring which was shown to be located at the top of the PQQ cofactor and suggested to be involved in electron transfer during catalysis (4). In case of TfaA, Trp238 (Fig. 3) was identified as the amino acid located at the bottom of the active site interacting by its phenyl ring with the pyridine ring of PQQ (3, 19). The two residues responsible for binding of the Ca2+ ion and the active site base were also conserved in the sequence of TfaA (3, 19).
DISCUSSION
We report here some of the catalytic properties of the THFA-induced type I quinohemoprotein ADH THFA-DH from R. eutropha strain Bo. Furthermore, the complete primary structure of the protein was elucidated and analyzed. Thus, THFA-DH is the third type I quinohemoprotein ADH for which these data are available. Due to its basic properties, PVADH from Pseudomonas sp. strain VM15C should belong to the type I quinohemoprotein dehydrogenases (41). However, the sequence identity to the proteins from C. testosteroni and R. eutropha strain Bo was only about 25%. In contrast to all other quinohemoprotein ADHs described so far, PVADH was suggested to contain a heme-binding motif in the N-terminal region of the protein and should thus not be considered as a typical type I enzyme.
In general, quinohemoprotein dehydrogenases are not very specific with respect to their substrate spectrum. Furthermore, the catalytic efficiency by which different substrates are converted under in vitro conditions shows significant deviations among the enzymes. The more complex type II quinohemoprotein ADHs from acetic acid bacteria have a preference for primary alcohols as substrates, showing the highest level of activity with ethanol (1, 2), but they can also oxidize secondary alcohols, e.g., 2-butanol or solketal (29, 30). In contrast, the substrate spectrum of the type I enzymes is much broader, including primary and secondary alcohols, diols, polyols, and aldehydes. In case of QH-EDH from C. testosteroni, n-butanol or n-pentanol was found to be converted at the highest rate (44). Allyl alcohols and 1,2-propandiol, respectively, were preferred by ADH IIB or IIG of P. putida (48). The PVADH from Pseudomonas sp. VM15C was induced during growth on polyvinyl alcohol and could also oxidize short secondary alcohols. Interestingly, the enzyme was not able to convert primary alcohols (41). Polyethylene glycol (PEG) dehydrogenase isolated from Rhodopseudomonas acidophila converted several kinds of PEG and exhibited the maximum activity with n-propanol (55). The detailed initial rate measurements performed in the present study showed that primary alcohols (except ethanol) were converted by THFA-DH with the highest catalytic efficiency. The substrate inhibition observed for most alcohols and aldehydes tested is in agreement with the data obtained for QH-EDH from C. testosteroni (15).
Investigations of the enantioselectivity of THFA-DH from R. eutropha strain Bo during the oxidation of racemic mixtures of chiral alcohols showed that the enzyme exhibited a stereospecificity. Comparable studies with other quinohemoprotein ADHs were preferentially carried out using industrial relevant alcohols as substrates, e.g., solketal and glycidol (14–17, 29). QH-EDH from C. testosteroni, which showed the highest activity and E value for solketal, had the same preference for the R-(−)-alcohol as that observed for THFA-DH. Likewise, THFA-DH from strain Bo and QH-EDH from C. testosteroni had a preference for the S-(+)-enantiomer of aliphatic chiral alcohols (44). However, the E values determined for different chiral alcohols showed significant deviations.
As far as the catalytic mechanism of quinoprotein dehydrogenases was investigated, the results obtained always indicated that the substrates were converted by a ping-pong reaction mechanism (15, 30, 32, 37). Initial rate measurements performed with THFA-DH from R. eutropha strain Bo with ethanol or acetaldehyde as a substrate also revealed this catalytic mechanism. QH-EDH of C. testosteroni was analyzed in more detail (15). The data obtained indicated that catalysis takes place by a hexa-uni ping-pong mechanism where the aldehyde is released from the enzyme and uncompetitive substrate inhibition occurs in some cases. Due to the high similarity between both enzymes, one might assume that this model is also valid for THFA-DH from R. eutropha strain Bo.
The physiological function of quinoprotein dehydrogenases is to provide the respiratory chain with electrons, and the enzymes are usually located in the periplasm of gram-negative bacteria (19, 40). Due to the observed signal peptide probably responsible for translocation of the protein into the periplasm, THFA-DH seems also to be a periplasmic enzyme (5, 49).
R. eutropha strain Bo is able to synthesize PQQ; however, the sequence located up- and downstream from the tfaA gene (∼800 bp) did not reveal any similarity to genes encoding enzymes involved in PQQ biosynthesis as was found for Pseudomonas aeruginosa (40). Furthermore, no similarity was obtained to genes which might be involved in the further oxidation of formed aldehydes, as was shown for C. testosteroni and P. aeruginosa (40, 45).
X-ray structures obtained for different methanol dehydrogenases led to the identification of amino acids responsible for cofactor binding and formation of an eight-bladed β-propeller fold maintaining the structure of the α-subunit (18, 19, 50, 52). Homology modelling of different PQQ-dependent dehydrogenases on the structure of methanol dehydrogenase led to the conclusion that the overall topology of these enzymes is similar (6, 7, 24). In the case of QH-EDH from C. testosteroni, the C-terminal part was modelled on cytochromes c for which structural information was available and revealed some structural similarity to these proteins (24). Our analysis of the deduced sequence of TfaA from strain Bo indicated that these topological features are probably conserved in this enzyme. Thus, QH-EDH from C. testosteroni and THFA-DH from R. eutropha strain Bo are closely related and might employ the same catalytic mechanism. The high level of similarity between these enzymes can be a powerful tool to elucidate the structural basis responsible for the observed catalytic deviations.
ACKNOWLEDGMENTS
We thank U. Lechner of our Institute for providing data of the G+C content of R. eutropha strain Bo. This work was partly supported by grants of the Forschungsförderung des Landes Sachsen-Anhalt, the Max Buchner Stiftung, and the Fonds der Chemischen Industrie.
REFERENCES
- 1.Adachi O, Miyagawa E, Shinagawa E, Matsushita K, Ameyama M. Purification and properties of particulate alcohol dehydrogenase from Acetobacter aceti. Agric Biol Chem. 1978;42:2331–2340. [Google Scholar]
- 2.Adachi O, Tayama K, Shinagawa E, Matsushita K, Ameyama M. Purification and characterization of particulate alcohol dehydrogenase from Gluconobacter suboxydans. Agric Biol Chem. 1978;42:2045–2056. [Google Scholar]
- 3.Anthony C, Ghosh M C. The structure and function of the PQQ-containing quinoprotein dehydrogenases. Progr Biophys Mol Biol. 1998;69:1–21. doi: 10.1016/s0079-6107(97)00020-5. [DOI] [PubMed] [Google Scholar]
- 4.Avezoux A, Goodwin M G, Anthony C. The role of the novel disulphide ring in the active site of the quinoprotein methanol dehydrogenase from Methylobacterium extorquens. Biochem J. 1995;307:735–741. doi: 10.1042/bj3070735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyd D, Beckwith J. The role of charged amino acids in the localization of secreted and membrane proteins. Cell. 1990;62:1031–1033. doi: 10.1016/0092-8674(90)90378-r. [DOI] [PubMed] [Google Scholar]
- 6.Cozier G E, Anthony C. Structure of the quinoprotein glucose dehydrogenase of Escherichia coli modelled on that of methanol dehydrogenase from Methylobacterium extorquens. Biochem J. 1995;312:679–685. doi: 10.1042/bj3120679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cozier G E, Giles I G, Anthony C. The structure of the quinoprotein alcohol dehydrogenase of Acetobacter aceti modelled on that of methanol dehydrogenase from Methylobacterium extorquens. Biochem J. 1995;308:375–379. doi: 10.1042/bj3080375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Day D J, Anthony C. Methanol dehydrogenase from Methylobacterium extorquens AM1. Methods Enzymol. 1990;188:210–216. [Google Scholar]
- 9.de Jong G A H, Geerlof A, Stoorvogel J, Jongejan J A, de Vries S, Duine J A. Quinohaemoprotein ethanol dehydrogenase from Comamonas testosteroni: purification, characterization, and reconstitution of the apoenzyme with pyrroloquinoline quinone analogues. Eur J Biochem. 1995;230:899–905. doi: 10.1111/j.1432-1033.1995.tb20634.x. [DOI] [PubMed] [Google Scholar]
- 10.Diehl A, von Wintzingerode F, Görisch H. Quinoprotein ethanol dehydrogenase of Pseudomonas aeruginosa is a homodimer—sequence of the gene and deduced structural properties of the enzyme. Eur J Biochem. 1998;257:409–419. doi: 10.1046/j.1432-1327.1998.2570409.x. [DOI] [PubMed] [Google Scholar]
- 11.Dokter P, Frank J, Duine J A. Purification and characterization of quinoprotein glucose dehydrogenase from Acinetobacter colcoaceticus L.M.D. 79.41. Biochem J. 1986;239:163–167. doi: 10.1042/bj2390163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duine J A, Frank J, Jongejan J A. Enzymology of quinoproteins. Adv Enzymol. 1987;59:169–213. doi: 10.1002/9780470123058.ch4. [DOI] [PubMed] [Google Scholar]
- 13.Fliege R, Tong S, Shibata A, Nickerson K W, Conway T. The Entner-Doudoroff pathway in Escherichia coli is induced for oxidative glucose metabolism via pyrroloquinoline quinone-dependent glucose dehydrogenase. Appl Environ Microbiol. 1992;58:3826–3829. doi: 10.1128/aem.58.12.3826-3829.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geerlof A, Jongejan J A, van Dooren T J G M, Racemakers-Franken P C, van den Tweel W J J, Duine J A. Factors relevant to the production of (R)-(+)-glycidol (2,3-epoxy-1-propanol) from racemic glycidol by enantioselective oxidation with Acetobacter pasteurianus ATCC 12874. Enzyme Microb Technol. 1994;16:1059–1063. doi: 10.1016/0141-0229(94)90143-0. [DOI] [PubMed] [Google Scholar]
- 15.Geerlof A, Rakels J J L, Straathof A J J, Heijnen J J, Jongejan J A, Duine J A. Description of the kinetic mechanism and the enantioselectivity of quinohaemoprotein ethanol dehydrogenase from Comamonas testosteroni in the oxidation of alcohols and aldehydes. Eur J Biochem. 1994;226:537–546. doi: 10.1111/j.1432-1033.1994.tb20078.x. [DOI] [PubMed] [Google Scholar]
- 16.Geerlof A, Stoorvogel J, Jongejan J A, Leenen E J T M, van Dooren T J G M, van den Tweel W J J, Duine J A. Studies on the production of (S)-(+)-solketal (2,2-dimethyl-1,3-dioxolane-4-methanol) by enantioselective oxidation of racemic solketal with Comamonas testosteroni. Appl Microbiol Biotechnol. 1994;42:8–15. [Google Scholar]
- 17.Geerlof A, van Tol J B A, Jongejan J A, Duine J A. Enantioselective conversions of the racemic C-3-alcohol synthons, glycidol (2,3-epoxy-1-propanol), and solketal (2,2-dimethyl-4-(hydroxymethyl)-1,3-dioxolane) by quinohaemoprotein alcohol dehydrogenases and bacteria containing such enzymes. Biosci Biotechnol Biochem. 1994;58:1028–1036. [Google Scholar]
- 18.Ghosh M, Anthony C, Harlos K, Goodwin M G, Blake C. The refined structure of the quinoprotein methanol dehydrogenase from Methylobacterium extorquens at 1.94 Å. Structure. 1995;3:177–187. doi: 10.1016/s0969-2126(01)00148-4. [DOI] [PubMed] [Google Scholar]
- 19.Goodwin P M, Anthony C. The biochemistry, physiology and genetics of PQQ and PQQ-containing enzymes. Adv Microb Physiol. 1998;40:1–80. doi: 10.1016/s0065-2911(08)60129-0. [DOI] [PubMed] [Google Scholar]
- 20.Görisch H, Rupp M. Quinoprotein ethanol dehydrogenase from Pseudomonas. Antonie Leeuwenhoek. 1989;56:35–45. doi: 10.1007/BF00822582. [DOI] [PubMed] [Google Scholar]
- 21.Groen B, Frank J, Duine J A. Quinoprotein alcohol dehydrogenase from ethanol-grown Pseudomonas aeruginosa. Biochem J. 1984;223:921–924. doi: 10.1042/bj2230921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Inoue T, Sunagawa M, Mori A, Imai C, Fukuda M, Takagi M, Yano K. Cloning and sequencing of the gene encoding the 72-kilodalton dehydrogenase subunit of alcohol dehydrogenase from Acetobacter aceti. J Bacteriol. 1989;171:3115–3122. doi: 10.1128/jb.171.6.3115-3122.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jendrossek D, Steinbüchel A, Schlegel H G. Three different proteins exhibiting NAD-dependent acetaldehyde dehydrogenase activity from Alcaligenes eutrophus. Eur J Biochem. 1987;167:541–548. doi: 10.1111/j.1432-1033.1987.tb13371.x. [DOI] [PubMed] [Google Scholar]
- 24.Jongejan A, Jongejan J A, Duine J A. Homology model of the quinohaemoprotein alcohol dehydrogenase from Comamonas testosteroni. Protein Eng. 1998;11:185–198. doi: 10.1093/protein/11.3.185. [DOI] [PubMed] [Google Scholar]
- 25.Kondo K, Horinouchi S. Characterization of the genes encoding the three-component membrane-bound alcohol dehydrogenase from Gluconobacter suboxydans and their expression in Acetobacter pasteurianus. Appl Environ Microbiol. 1997;63:1131–1138. doi: 10.1128/aem.63.3.1131-1138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kunst F, Ogasawara N, Moszer I, Albertini A M, Alloni G, Azevedo V, Bertero M G, Bessieres P, et al. The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature. 1997;390:249–256. doi: 10.1038/36786. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lisdat F, Ho W O, Wollenberger U, Scheller F W, Richter T, Bilitewski U. Recycling systems based on screen-printed electrodes. Electroanalysis. 1998;10:803–807. [Google Scholar]
- 29.Machado S S, Jongejan A, Geerlof A, Jongejan J A, Duine J A. Entropic and enthalpic contributions to the enantioselectivity of quinohaemoprotein alcohol dehydrogenases from Acetobacter pasteurianus and Comamonas testosteroni in the oxidation of primary and secondary alcohols. Biocatal Biotransform. 1999;17:179–207. [Google Scholar]
- 30.Machado S S, Wandel U, Jongejan J A, Straathof A J J, Duine J A. Characterization of the enantioselective properties of the quinohemoprotein alcohol dehydrogenase of Acetobacter pasteurianus LMG 1635. 1. Different enantiomeric ratios of whole cells and purified enzyme in the kinetic resolution of racemic glycidol. Biosci Biotechnol Biochem. 1999;63:10–20. doi: 10.1271/bbb.63.10. [DOI] [PubMed] [Google Scholar]
- 31.Madyastha K M, Gururaja T L. An NAD(P)(+)-dependent secondary-alcohol dehydrogenase of Alcaligenes eutrophus—purification, characterization, and its application for the production of chiral alcohols. Biotechnnol Appl Biochem. 1996;23:245–253. [Google Scholar]
- 32.Marcinkeviciene J, Johansson G. Kinetic studies of the active sites functioning in the quinohemoprotein fructose dehydrogenase. FEBS Lett. 1993;318:23–26. doi: 10.1016/0014-5793(93)81319-u. [DOI] [PubMed] [Google Scholar]
- 33.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 34.Matsushita K, Yakushi T, Takaki Y, Toyama H, Adachi O. Generation mechanism and purification of an inactive form convertible in vivo to the active form of quinoprotein alcohol dehydrogenase in Gluconobacter suboxydans. J Bacteriol. 1995;177:6552–6559. doi: 10.1128/jb.177.22.6552-6559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer T E, Kamen M D. New perspectives on c-type cytochromes. Adv Protein Chem. 1982;35:105–212. doi: 10.1016/s0065-3233(08)60469-6. [DOI] [PubMed] [Google Scholar]
- 36.Nakai K, Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci. 1999;24:34–35. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- 37.Olsthoorn A J J, Duine J A. On the mechanism and specificity of soluble, quinoprotein glucose dehydrogenase in the oxidation of aldose sugars. Biochemistry. 1998;37:13854–13861. doi: 10.1021/bi9808868. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Schmidt B. Oxygen-independent oxidases—a new class of enzymes for application in diagnostics. Clin Chim Acta. 1997;266:33–37. doi: 10.1016/s0009-8981(97)00164-2. [DOI] [PubMed] [Google Scholar]
- 40.Schobert M, Görisch H. Cytochrome c550 is an essential component of the quinoprotein ethanol oxidation system in Pseudomonas aeruginosa: cloning and sequencing of the genes encoding cytochrome c550 and an adjacent acetaldehyde dehydrogenase. Microbiology. 1999;145:471–481. doi: 10.1099/13500872-145-2-471. [DOI] [PubMed] [Google Scholar]
- 41.Shimao M, Tamogami T, Nishi K, Harayama S. Cloning and characterization of the gene encoding pyrroloquinoline quinone-dependent poly(vinyl alcohol) dehydrogenase of Pseudomonas sp. strain VM15C. Biosci Biotechnol Biochem. 1996;60:1056–1062. doi: 10.1271/bbb.60.1056. [DOI] [PubMed] [Google Scholar]
- 42.Steinbüchel A, Schlegel H G. A multifunctional fermentative alcohol dehydrogenase from the strict aerobe Alcaligenes eutrophus: purification and properties. Eur J Biochem. 1984;141:555–564. doi: 10.1111/j.1432-1033.1984.tb08229.x. [DOI] [PubMed] [Google Scholar]
- 43.Stigter E C A, Dejong G A H, Jongejan J A, Duine J A, Vanderlugt J P, Somers W A C. Electron transfer between a quinohemoprotein alcohol dehydrogenase and an electrode via a redox polymer network. Enzyme Microb Technol. 1996;18:489–494. [Google Scholar]
- 44.Stigter E C A, van den Lugt J P, Somers W A C. Enantioselective oxidation of secondary alcohols by quinohaemoprotein alcohol dehydrogenase from Comamonas testosteroni. J Mol Catal B Enzym. 1997;2:291–297. [Google Scholar]
- 45.Stoorvogel J, Kraayveld D E, Vansluis C A, Jongejan J A, Devries S, Duine J A. Characterization of the gene encoding quinohaemoprotein ethanol dehydrogenase of Comamonas testosteroni. Eur J Biochem. 1996;235:690–698. doi: 10.1111/j.1432-1033.1996.00690.x. [DOI] [PubMed] [Google Scholar]
- 46.Takemura H, Kondo K, Horinouchi S, Beppu T. Induction by ethanol of alcohol dehydrogenase activity in Acetobacter pasteurianus. J Bacteriol. 1993;175:6857–6866. doi: 10.1128/jb.175.21.6857-6866.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tamaki T, Fukaya M, Takemura H, Tayama K, Okumura H, Kawamura Y, Nishiyama M, Horinouchi S, Beppu T. Cloning and sequencing of the gene cluster encoding two subunits of membrane-bound alcohol dehydrogenase from Acetobacter polyoxogenes. Biochim Biophys Acta. 1991;1088:292–300. doi: 10.1016/0167-4781(91)90066-u. [DOI] [PubMed] [Google Scholar]
- 48.Toyama H, Fujii A, Matsushita K, Shinagawa E, Ameyama M, Adachi O. Three distinct quinoprotein alcohol dehydrogenases are expressed when Pseudomonas putida is grown on different alcohols. J Bacteriol. 1995;177:2442–2450. doi: 10.1128/jb.177.9.2442-2450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.von Heijne G. Patterns of amino acids near signal-sequence cleavage sites. Eur J Biochem. 1983;133:17–21. doi: 10.1111/j.1432-1033.1983.tb07424.x. [DOI] [PubMed] [Google Scholar]
- 50.White S, Boyd G, Mathews F S, Xia Z X, Dai W W, Zhang Y F, Davidson V L. The active site structure of the calcium-containing quinoprotein methanol dehydrogenase. Biochemistry. 1993;32:12955–12958. doi: 10.1021/bi00211a002. [DOI] [PubMed] [Google Scholar]
- 51.Wollenberger U, Neumann B. Quinoprotein glucose dehydrogenase modified carbon paste electrode for the detection of phenolic compounds. Electroanalysis. 1997;9:366–371. [Google Scholar]
- 52.Xia Z X, Dai W W, Xiong J P, Hao Z P, Davidson V L, White S, Mathews F S. The three-dimensional structures of methanol dehydrogenase from two methylotrophic bacteria at 2.6 Å resolution. J Biol Chem. 1992;267:22289–22297. [PubMed] [Google Scholar]
- 53.Yamada M, Sumi K, Matsushita K, Adachi O, Yamada Y. Topological analysis of quinoprotein glucose dehydrogenase in Escherichia coli and its ubiquinone-binding site. J Biol Chem. 1993;268:12812–12817. [PubMed] [Google Scholar]
- 54.Yamanaka H, Kawai F. Purification and characterization of constitutive polyethylene glycol (PEG) dehydrogenase of a PEG 4000-utilizing Flavobacterium sp. no. 203. J Ferment Bioeng. 1989;67:324–330. [Google Scholar]
- 55.Yasuda M, Cherepanov A, Duine J A. Polyethylene glycol dehydrogenase activity of Rhodopseudomonas acidophila derives from a type I quinohaemoprotein alcohol dehydrogenase. FEMS Microbiol Lett. 1996;138:23–28. [Google Scholar]
- 56.Zarnt G, Schräder T, Andreesen J R. Degradation of tetrahydrofurfuryl alcohol by Ralstonia eutropha is initiated by an inducible pyrroloquinoline quinone-dependent alcohol dehydrogenase. Appl Environ Microbiol. 1997;63:4891–4898. doi: 10.1128/aem.63.12.4891-4898.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]