Abstract
Intestinal homeostasis depends on complex interactions between the gut microbiota and host immune system. Emerging evidence indicates that the intestinal microbiota is a key player in autoimmune liver disease (AILD). Autoimmune hepatitis, primary biliary cholangitis, primary sclerosing cholangitis, and IgG4-related sclerosing cholangitis have been linked to gut dysbiosis. Diverse mechanisms contribute to disturbances in intestinal homeostasis in AILD. Bacterial translocation and molecular mimicry can lead to hepatic inflammation and immune activation. Additionally, the gut and liver are continuously exposed to microbial metabolic products, mediating variable effects on liver immune pathologies. Importantly, microbiota-specific or associated immune responses, either hepatic or systemic, are abnormal in AILD. Comprehensive knowledge about host-microbiota interactions, included but not limited to this review, facilitates novel clinical practice from a microbiome-based perspective. However, many challenges and controversies remain in the microbiota field of AILD, and there is an urgent need for future investigations.
Keywords: Gut microbiota, Metabolome, Immunity, Autoimmune liver diseases
Introduction
The human gut microbiota, which refers to a collection of microorganisms residing in the host intestinal tract, has long been recognized to be closely involved in health maintenance and disease development, modulating almost every aspect of physiological processes, including immunity, metabolism, and endocrine and neural activity.[1,2] The microbial community architecture is shaped by various factors, such as diet, lifestyle, geographic location, and health status.[3] On the one hand, as the first line of defense against pathogenic microbes, the intestinal immune system can sustain tolerance to innocuous and commensal flora and affect both the composition and function of gut microbiota. On the other hand, gut-colonizing microorganisms facilitate the establishment of a mature host immune milieu, leading to skewed immune responses under certain conditions.[2] The dynamic crosstalk between microbiota and the immune system has implications for their role in immune-mediated diseases.
Through the portal veins, the liver is the first organ to encounter gut-derived products such as microbial components, metabolites, toxins, and dietary nutrients, making it susceptible to intestinal changes. Anatomically and physiologically, the liver communicates with the gut via the enterohepatic circulation of bile, consisting mainly of bile acids and immunoglobulins, actively interacting with the gut microbial community.[4] Disturbances along this complex and highly regulated gut–liver axis contribute to the pathogenesis of a wide range of liver diseases, including non-alcoholic fatty liver disease, hepatocellular carcinoma, chronic viral hepatitis, and autoimmune liver diseases (AILDs).[5,6]
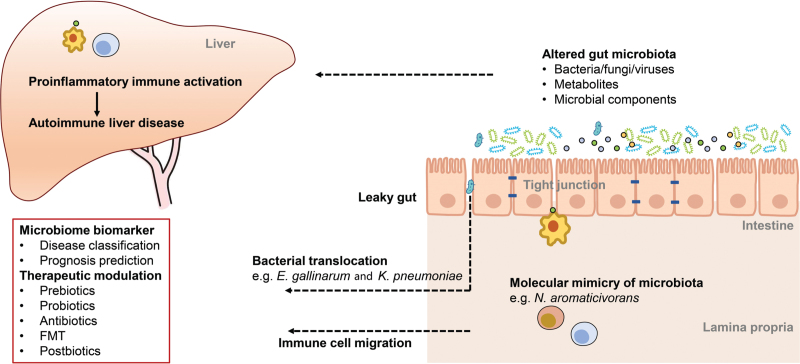
A growing body of preclinical and clinical studies have provided evidence that gut microbiota is closely related to the development of AILDs, including autoimmune hepatitis (AIH), primary biliary cholangitis (PBC), primary sclerosing cholangitis (PSC), and IgG4-related sclerosing cholangitis (IgG4-SC).[2,7–9] For instance, non-obese diabetic (NOD).c3c4 mice developed spontaneous cholangitis in a normal environment, whereas the phenotype diminished in a germ-free environment.[10] Additionally, a mouse model of AIH, established by immunizing leukocyte antigen (HLA)-DR3-transgenic mice with a DNA plasmid coding for human cytochrome P4502D6/formiminotransferase cyclodeaminase fusion protein, demonstrated reduced gut bacterial load and diversity.[11] In this review, we summarize current evidence supporting the importance of gut microbiota in the pathogenesis of AILD and highlight the potential of microbiota-based clinical applications, including diagnostic and prognostic biomarkers and therapeutic interventions [Figure 1].
Figure 1.
The understanding of host–microbiota interactions in AILDs. The compositional and functional alterations in intestinal microbiota lead to increased intestinal barrier permeability. The inflammatory microbes then translocate to the liver, where they can elicit or promote hepatic inflammation, as exemplified by Enterococcus gallinarum in autoimmune hepatitis and Klebsiella pneumoniae in primary sclerosing cholangitis. The cross-reactive antigens in certain microbial organisms contribute to loss of immune tolerance to autoantigens and trigger non-infectious inflammation in genetically predisposed subjects. Additionally, the altered levels of microbiome-related metabolites (eg, succinic acid in IgG4-related sclerosing cholangitis) or microbial components profoundly affect host immune system, establishing a proinflammatory immune milieu. These changed microbiome features are predictive of the disease status and the prognosis of patients with autoimmune liver disease, and therapeutic strategies targeting the human gut microbiome from various aspects have been shown great clinical potential. FMT: Fecal microbiota transplantation.
Altered Gut Microbiota in AILDs
With the advancement of high-throughput sequencing technology, microbial profiles of AILD were distinguished from those of healthy controls (HCs), indicative of the presence of an abnormal gut ecosystem. Reduced bacterial diversity, which evaluates taxa richness and evenness, is a generic feature of AILD gut dysbiosis.[7–9] Indeed, higher microbial diversity suggests competition among colonizing taxa and enables resistance to environmental disturbances, typically reflecting a healthy gut ecosystem. Notably, decreased alpha diversity in patients with AIH and PBC was partially reversed after standard therapy.[8,12] However, longitudinal assessments of microbiota alterations were lacking in PSC and IgG4-SC cohorts.
Sequencing of the members of the gut microbiota in AILD has also prompted the identification of individual bacterial taxa with potential immunomodulatory effects [Table 1]. For example, Veillonella, a gram-negative bacterium involved in lactate fermentation, was enriched in several inflammatory diseases and AILDs, although its underlying mechanisms remain to be studied. Klebsiella pneumoniae, a pore-forming bacterial strain with validated proinflammatory influences, was highly abundant in patients with PSC and inflammatory bowel disease (IBD).[13] Another observation revealed the flourishment of Klebsiella genera in the feces of patients with AIH and PBC, using 16S rRNA amplicon sequencing.[7,8] In contrast to the abundance of pathogenic microorganisms, bacteria depleted in patients can be beneficial. For instance, the abundance of Faecalibacterium prausnitzii, which contributes to gut health maintenance through the production of short-chain fatty acids (SCFAs), was reduced in PBC, PSC, and IgG4-SC.[8,9,14] Particularly, a decrease in F. prausnitzii was observed in the treatment non-responder group of patients with PBC; therefore, F. prausnitzii may have a predictive capacity for long-term prognosis.[15] Although many taxa were consistently altered in several AILDs, some alterations were unique to a particular disease type. For example, Liwinski et al[12] identified a specific decline in Bifidobacterium abundance in AIH, but not in PBC or ulcerative cholangitis. In addition, disease-specific taxonomic changes were detected in PSC and IgG4-SC, including increased abundance of Ruminicoccus gnavus in PSC and reduced abundance of Blautia in IgG4-SC.[9] Nevertheless, individual taxonomic changes are inconsistent across different studies. This can be attributed to variable determinants, including study design, such as stratified disease stage and medication history of enrolled patients, as well as the race and geographic location of the investigated subjects [Table 1].
Table 1.
Summary of key gut microbiota studies in AILDs.
| Studies | Cohort description | Sample | Microbiome assessment technique | Metabolites assessment technique | Main findings |
| Lin et al[25] | China; AIH (24), HC (8) | Fecal sample | Targeted 16S rDNA | NA | AIH (vs. HC): Bifidobacterium↓; Lactobacillus↓ |
| Wei et al[7] | China; Explorative cohort: AIH (91), HC (98); Validation cohort: AIH (28), HC (34) | Fecal sample | 16S rDNA | NA | AIH (vs. HC): Alpha diversity↓; Ruminococcaceae↓; Oscillospira↓; Coprococcus↓; Clostridiales↓; Rikenellaceae↓; Parabacteroides↓; Lactobacillus↑; Streptococcus↑; Klebsiella↑; Veillonella↑ |
| Liwinski et al[12] | Germany; AIH (72), PBC (99), ulcerative colitis (81), HC (95) | Fecal sample | 16S rRNA | NA | AIH (vs. HC): Alpha diversity↓, Faecalibacterium↓, Bifidobacterium↓, Streptococcus↑, Klebsiella↑; AIH (vs. PBC): Facalibacterium↑, Bifidobacterium↓; AIH (vs. UC): Bifidobacterium↓ |
| Elsherbiny et al[84] | Egypt; AIH (15), HC (10) | Fecal sample | 16S rRNA | NA | AIH (vs. HC): Alpha diversity↓, Faecalibacterium↑, Blautia↑, Streptococcus↑, Haemophilus↑, Bacteroides↑, Veillonella↑, Eubacterium↑, Lachnospiraceae↑, Butyricicoccus↑, Prevotella↓, Parabacteroides↓, Dilaster↓ |
| Tang et al[8] | China; Explorative cohort: PBC (60), HC (80); Validating cohort: PBC (19), HC (34); Longitudinal cohort: PBC before and after UDCA (37) | Fecal sample | 16S rRNA | NA | PBC (vs. HC): Alpha diversity↓, Oscillospira↓, Faecalibacterium↓, Sutterella↓, Bacteroides↓, Lactobacillus↑, Streptococcus↑, Klebsiella↑, Veillonella↑, Clostridium↑, Pseudomonas↑, Haemophilus↑, Enterobacteriaceae↑; PBC (before vs. after UDCA): partially restored dysbiosis |
| Chen et al[41] | China, Cross-sectional cohort: PBC (65), HC (109); longitudinal cohort: PBC before and after UDCA (28) | Fecal and serum sample | 16S rRNA | UPLC-MS/MS, targeted bile acids quantification | PBC (vs. HC): unconjugated/conjugated BAs↓, secondary/primary BAs↓, Veillonella↑, Klebsiella↑, Faecalibacterium↓, Oscillospira↓, association between altered bile acids and gut microbes; PBC after UDCA (vs. before UDCA): unconjugated/conjugated BAs↑, Bilophila spp.↑ |
| Furukawa et al[15] | Japan; PBC (76), HC (23) | Fecal sample | 16S rRNA | NA | PBC (vs. HC): Alpha diversity↓, Clostridiales↓, Bifidobacterium↑, Streptococcus↑, Lactobacillus↑, Enterococcus↑; PBC UDCA responders (vs. UDCA non-responder): Faecalibacterium↑ |
| Lammert et al[68] | USA, PBC with non-advanced fibrosis (15), PBC with advanced fibrosis (8) | Fecal sample | 16S rRNA | NMR, targeted SCFAs quantification | PBC with advanced fibrosis (vs. non-advanced): Alpha diversity↓, Weisella↑, SCFAs↑, acetate↑ |
| Sabino et al[66] | Belgium; Explorative cohort: PSC (52), UC (13), Crohn's disease (30), HC (52); Validating cohort: PSC (14), HC (14) | Fecal sample | 16S rRNA | NA | PSC (vs. HC): Alpha diversity↓, Enterococcus↑, Fusobacterium↑, Lactobacillus↑ |
| Kummen et al[14] | Norwegian; PSC-IBD (55), PSC alone (30), UC (36), HC (263) | Fecal sample | 16S rRNA | NA | PSC (vs. HC): Alpha diversity↓, Veillonella↑, Coprococcus↓, Clostridiales. spp↓, Phascolarctobacterium↓, Chrisensenellaceae. spp↓ |
| Quraishi et al[85] | UK; PSC-IBD (11), IBD (10), HC (9) | Pan-colonic biopsies | 16S rRNA | NA | PSC-IBD (vs. HC): Escherichia↑, Lachnospiraceae↑, Megasphera↑, Prevotella↓, Roseburia↓, Bacteroides↓ |
| Vieira-Silva et al[86] | Belgium; PSC (18), CD (29), UC (13), PSC-CD (20), PSC-UC (26), HC (66) | Fecal sample | Quantitative 16S rRNA | NA | PSC-IBD: Bacteroides 2 enterotype↑, Fusobacterium↑ (inflammation associated), Enterococcus↑ (cholangitis associated) |
| Lemoinne et al[18] | France; PSC-IBD (27), PSC alone (22), IBD alone (33), HC (30) | Fecal sample | 16S rRNA and ITS2 | NA | PSC (vs. other groups): Bacterial alpha diversity↓,Veillonella↑, Ruminococcus↓, Blautia↓, Faecalibacterium↓; Fungal biodiversity↑, Exophila↑, Saccharomyces cerevisiae↓ |
| Kummen et al[20] | Norwegian and Germany; PSC (136), IBD alone (93), HC (158) | Fecal and plasma samples | Metagenomics | Targeted LC-MS/MS | PSC (vs. HC): Microbial genes↓, Clostridium species↑, Eubacterium spp.↓, Ruminococuus obeum↓, genes related to vitamin B6 synthesis and BCAAs synthesis↓; Plasma concentrations of vitamin B6 and BCAAs↓ |
| Liu et al[9] | China; IgG4-SC (34), PSC (37), HC (64) | Fecal sample | 16S rRNA | Untargeted LC-MS/MS | IgG4-SC/PSC (vs. HC): Alpha diversity↓, Coprococcus↓, Faecalibacterium↓, Phascolarctobacterium↓, Lactobacillus↑, Fusobacterium↑, Veillonella↑; L-palmitoylcarnitine↑; IgG4-SC (vs. HC): Blautia↓, succinic acid↑; PSC (vs. HC): Eubacterium↓, secondary BAs↓ |
AIH: Autoimmune hepatitis; AILDs: Autoimmune liver diseases; BCAAs: Branched chain amino acids; HCs: Healthy controls; IgG4-SC: IgG4-related sclerosing cholangitis; IBD: Inflammatory bowel disease; PBC: Primary biliary cholangitis; PSC: Primary sclerosing cholangitis; SCFAs: Short-chain fatty acids; UDCA: Ursodeoxycholic acid; UC: Ulcerative colitis; UPLC-MS/MS: Ultra performance liquid chromatography tandem mass spectrometry; BAs: Bile acids; NMR: Nuclear magnetic resonance spectroscopy; ITS2: Internal transcribed spacer 2; LC-MS/MS: Liquid chromatography with tandem mass spectrometry; NA: Not applicable.
Notably, microbial members belonging to fungi, viruses, archaea, and eukaryotes, such as protozoa, also contribute considerably to the overall gut microbiome community. All of these taxa, together with bacteria, form a complex community and tightly interact with each other via diverse mechanisms, including cross-feeding and niche competition. A higher prevalence of anti-Saccharomyces cerevisiae antibodies was found in AILD, particularly in PSC and antimitochondrial antibody (AMA)-negative PBC.[16,17] In agreement with this finding, gut fungal dysbiosis, characterized by altered composition, increased fungal biodiversity, and a disrupted correlation network with bacteria, was reported in PSC.[18]
In addition to taxonomic composition, functional changes in the microbiota have great implications for host health, given the robust modulatory effects of microbial-derived metabolic products on host immunity.[19] First, metagenomic shotgun sequencing provides insights into microbial encoding genes and pathways, which assist in determining dysregulated microbial functional capacities. The functions related to vitamin B6 synthesis and branched-chain fatty acid synthesis were reduced in patients with PSC, and these findings were further corroborated using circulating targeted metabolite analysis.[20] Furthermore, the fecal metabolome represents a functional readout of the gut microbiome, making it a novel tool for examining the links among microbiota composition, microbial metabolites, and host phenotype.[21] For instance, succinic acid, a potential microbial-derived metabolite, was elevated in the feces of patients with IgG4-SC.[9] However, metagenomics or untargeted metabolomics exploring metabolic changes in microbiota remain uninvestigated in AIH and PBC, and future in-depth multi-omics integrative studies are also required to gain deeper insights into the underlying causative mechanisms.
Potential Mechanisms of Microbiota Contributing to AILD
We cannot yet determine whether dysbiosis of the gut microbiome occurs in response to disease status or whether it actively engages in the development and progression of disease pathogenesis. However, emerging evidence from murine models sheds light on how microbiota-derived and microbiota-modified metabolites contribute to AILD.[22,23] As immune-mediated inflammation is an essential aspect of AILD pathology, excessive exposure to gut microbes, associated products, and ensuing hyperactivation of intestinal, hepatic, and possibly systemic immunity involving innate and adaptive components underlies the mechanistic framework of liver-microbiota crosstalk in AILD [Figure 1]. The current understanding of how the microbiota is involved in AILD pathogenesis is discussed below.
Bacterial translocation
The integrity of the gut epithelium is required to establish equilibrium between host immunity and the microbiome. Impairment of the gut barrier and the subsequent translocation of microbial organisms have long been correlated with various acute and chronic liver diseases. In support of this hypothesis, increased intestinal permeability and translocation of the cytolytic Enterococcus faecalis to the liver were detected after ethanol administration in mice.[24]
In AILD, studies have provided more evidence to reveal the fundamental role of aberrant translocation of microbiota-derived products in disease development. Reduced expression of tight junction proteins and higher serum levels of bacterial lipopolysaccharides have been observed in AIH.[25] A recent study elegantly proposed a direct link between breaches of the epithelial barrier and the consequent liver and systemic autoimmune pathologies.[26]Enterococcus gallinarum DNA was found in the AIH liver, and the levels of serum IgG antibodies against E. gallinarum RNA were elevated in most of the recruited patients with AIH. Importantly, the detection of higher cellular autoantigens expressed on primary human hepatocytes activated by E. gallinarum is likely to establish a causal effect of microbiota aberrancies in AIH.[26] Nakamoto et al[13] reported that a strain of K. pneumoniae isolated from the feces of patients with PSC-IBD disrupted intestinal epithelial integrity, triggered bacterial translocation, and promoted hepatic Th17 immune responses in response to 3,5-diethoxycarbonyl-1,4-dihydrocollidine feeding, ultimately leading to aggravation of cholangitis. Consistently, a higher presence of enteric bacteria was detected in bile duct samples of patients with PSC, particularly those with more severe stenosis.[27] Deficiency of toll-like receptor 2 (TLR-2), a vital receptor facilitating the expression of tight junction proteins, exacerbates gut leakiness and leads to subsequent bacterial translocation to the liver in dnTGFβRII mice, a well-characterized murine model of PBC. Importantly, the cholangiopathies could in part be mitigated after microbiota clearance with antibiotic treatment.[28]
In addition to the disruption of the intestinal physical barrier, antibody deficiency is also a primary contributor to the abnormal localization of intestinal flora. Among all antibody isotypes, IgA antibodies represent a key aspect of immune defense at the mucosal barrier.[1] As a case in point, many studies argued that secretory IgA antibodies (sIgA) within the intestinal lumen controlled the colonization and motility of pathogenic bacteria while mediating host immune tolerance against certain commensal taxa.[29] Patients with PBC exhibited a marked reduction in duodenal sIgA levels relative to non-disease controls, which may alter mucosal immunity and serve as a portal for bacterial access to the hepatobiliary system.[30]
Overall, these findings suggest that the gut microbiota of patients with AILD exhibits abnormal contact with the host immune system and is capable of triggering immunopathology within the liver via various approaches.
Molecular mimicry
Previous epidemiological studies of large cohorts have established a link between prior bacterial infection and subsequent development of AILD, highlighting the possibility of an infectious etiology.[31] Molecular mimicry was proposed early as a possible mechanism for elucidating microbial effects on immune-mediated inflammation. Supporting this hypothesis, serum antibodies in patients with PBC have been reported to react with various microbial components, such as beta-galactosidase of Lactobacillus delbrueckii[32] and lipoylated E2 subunits of the pyruvate dehydrogenase complex (PDC-E2) epitope from Novosphingobium aromaticivorans.[33] Immune cells can be cross-activated by microbial antigens and thus become autoreactive. The autoantigen of PBC, which is PDC-E2, has been identified. Accordingly, researchers have revealed that Escherichia coli infection in genetically predisposed PBC mice (NOD.B6 Idd10/Idd18 mice harboring loci essential for bile duct injury) disrupted immune tolerance against PDC-E2 and led to biliary pathology.[34] Similarly, infection of mice with N. aromaticivorans induced signature AMA in serum and infiltration of natural killer T (NKT) cells in the liver that initiated immune responses against small bile ducts.[35]
Molecular mimicry has also been observed in other diseases. Atypical perinuclear anti-neutrophil cytoplasmic antibodies (p-ANCA), present in the sera of patients with AIH and PSC, recognize both β-tubulin isotype 5 (TBB-5) and bacterial cell division protein FtsZ.[36,37] Intriguingly, exposure to gut microbiota is an important prerequisite for this cross-reactivity, reinforced by the observation that IL-10–/– mice with normal gut microbiota developed p-ANCA with dual reactivities, but not wild-type mice in a germ-free context.[36] Collectively, these findings indicate that abnormal immune responses against enteric bacteria-derived antigens with structural resemblance to host antigens play a pivotal role in AILD pathogenesis.
Microbiota-derived metabolites
In addition to cross-reactivation with foreign antigens, host immune responses are also controlled by various microbial metabolic products, such as bile acids, indole derivatives, SCFAs, and sex hormone derivatives.[38] Among them, bile acids are produced in the liver, modified by gut microbiota, and reabsorbed into the blood via enterohepatic circulation and significantly influence local and systemic immunity.[39] Well-known receptors mediating bile acid signaling are the nuclear receptor farnesoid X receptor (FXR) and transmembrane receptor G protein-coupled BA receptor (TGR5), both of which are closely involved in host immunity and metabolism.[40] Bile acids with distinct molecular structures differentially activate these bile acid receptors and various downstream signaling pathways.[39] The altered profiles of bile acids have been delineated in AILD, particularly in PBC and PSC. In general, reduced secondary bile acid levels and elevated primary bile acid levels are features of cholestatic AILD.[9,41–43] In accordance with these observations, the anti-inflammatory effects of secondary bile acids were elucidated, as exemplified by the finding that deficiency of secondary bile acids following gut dysbiosis, mostly lithocholic acid (LCA) and deoxycholic acid (DCA), aggravated intestinal inflammation.[44] Additionally, the conjugation pattern of bile acids also differs in AILD, although the potential mechanism merits further investigation. As a case in point, the conjugated/unconjugated ratio increased in PBC.[41] Overall, intestinal dysbiosis is partially responsible for the above changes, mediated by variations in encoded bile acid metabolism enzymes, particularly 7α-dehydroxylase and bile salt hydrolase.[39] Further validation of the effect of bacterial bile acid metabolism on AILD has been conducted. Microbiota depletion via antibiotic treatment exacerbated the cholestatic phenotype in multidrug resistance gene 2 knockout (Mdr2–/–) mice via reduced FXR signaling activated by secondary bile acids, such as LCA and DCA.[43] Importantly, the crosstalk between bile acids and gut microbiome is a two-way interaction. Bile acids can also regulate the composition and function of gut microbiota through intrinsic detergent antimicrobial properties, as well as indirect effects on immunity modulated by bile acid receptor signaling.[2] Reduced levels of bile acids, particularly secondary bile acids, are associated with bacterial overgrowth, which has also been observed in PBC and other advanced liver diseases.[45,46]
Moreover, the microbiota is implicated in the observed sexual bias in autoimmunity, which is particularly intriguing given the female predominance in AIH and PBC and their abnormal gut microbiota.[47] The gut microbiota deconjugates host estrogen through secretion of β-glucuronidase and produces estrogenic metabolites such as equol and enterodiol from dietary phytoestrogens.[48] Such changes in relevant metabolic pathways are consequently manifested by changes in sex hormone profiles. Fecal microbiota transplantation (FMT) from adult male mice to immature females led to microbiota community alteration and robust autoimmunity protection in a NOD-mouse model of type 1 diabetes; particularly, these protective effects were reliant on androgen receptor activation.[49] Interestingly, increased androgen levels in NOD mice were observed with the enrichment of particular bacterial consortia, which were capable of eliciting higher androgen production; thereby, a positive feedback loop may exist between gut microbiota and host sex hormones.[47] For example, testosterone treatment completely suppressed liver inflammation in female mice, whereas male mice became susceptible to liver injury after castration.[50] These findings provide intriguing insights into AILD, where sex hormones are closely related to disease pathogenesis.
Immune Crosstalk Between Gut and Liver
The gut and liver immune systems are constantly exposed to variable proinflammatory insults from the intestinal content, poised for rapid responses mainly involving innate immune responses. The innate immune system is generally tolerant to microbial presence under steady-state conditions, coordinated by several redundant innate immune cell subsets and intracellular pathways with polyreactive specificities. In the context of AILD, innate immune tolerance is gradually lost, and the host innate immune system reacts to the deleterious antigenic load. PAMP receptors, including TLRs and NOD-like receptors (NLRs), are broadly expressed in liver parenchymal and stromal cells such as Kupffer cells. Their activation and propagation mediate a proinflammatory program in AILD pathogenesis.[51]
The role of the gut microbiota in unconventional T cell modulation is indispensable. Mucosal-associated invariant T (MAIT) cells, a novel subset of innate-like T cells restricted by MHC class I (MR1), are markedly activated and expand in response to commensal-derived riboflavin derivatives.[52] Growing evidence also reveals that MAIT cells contribute to antimicrobial responses and are considered the “firewall” against pathogens lining the enterohepatic epithelial barrier.[53] Mechanistically, liver MAIT cells can upregulate the expression of CD40L and IFN-γ in response to E. coli-exposed liver macrophages, B cells, and biliary epithelial cells.[54] Consistently, the frequency and anti-bacterial program of MAIT cells were downregulated in the liver and blood of patients with PBC[54,55] and the peripheral blood of patients with PSC[56] compared with those in non-diseased controls. Notably, these defects were persistent in PBC even in the biochemical remission phase,[55] indicating the persistence of impaired defense against pathogens and related toxic products. In contrast, MAIT cells can adopt a proinflammatory fate in AIH. Increased MAIT cells were recruited from the periphery to the liver in AIH and exhibited enhanced granzyme B activity, a possible contributing factor to hepatocyte injury.[57] Interestingly, abnormal MAIT cell distribution in AIH remained unchanged after the standard immunosuppressive regime.[57] The failure of standard treatment to reverse abnormal immune milieu in AIH and PBC underscores the importance and necessity of identifying other novel microbiome-based therapeutic targets. Furthermore, the different changes in MAIT cell responses among PBC, PSC, and AIH showed a contextual influence on the functional outcome of specific immune cell subsets.
Unconventional T cells, such as γδ T cells, also participate in liver pathogenesis via immunomodulatory programs, particularly in response to bacterial stimulation. For example, accumulation of γδ T cells in the liver of Mdr2–/– mice produces high levels of IL-17 to perpetuate biliary duct lesions after exposure to translocated Lactobacillus gasseri from the gut.[58] Notably, unconventional T cells generally function synergistically as a network rather than as an individual cell subset. For instance, γδ T cells and MAIT cells secrete type 17 cytokines and are closely involved in immune surveillance and barrier integrity maintenance.[52,58]
The disturbed balance between the regulatory and effector aspects of adaptive immunity is well delineated in AILD pathogenesis, and intestinal homeostasis is required for balance maintenance. For example, pathogenic Th17 cell responses implicated in AIH pathogenesis[59] are to a great extent induced and educated by the intestinal microbiota. A recent investigation conducted by Renand et al[57] provided a detailed description of the circulating and hepatic immune profiles in adult patients with AIH. First, CD8+CD161+ cells were increased in AIH blood and liver. Intriguingly, CD8+CD161+ cells were previously reported to be enriched within the gut and shown to have potent antiviral memory.[60]
The migration of immune cells primed in the gut to the liver is also postulated as a potential mechanism mediating gut microbiota–liver immune crosstalk in AILD pathogenesis. Lymphocyte recruitment to specific tissues from the periphery is regulated by interactions between a range of chemokines and adhesive molecules expressed in tissues and their corresponding receptors on specific immune cell lineages. In particular, integrin α4β7 and C-C motif chemokine receptor 9 (CCR9) are highly expressed on effector memory T cells with intestine-homing capability, and accordingly, their corresponding ligands’ mucosal address in cell-adhesion molecule-1 (MAdCAM1) and C-C chemokine ligand 25 (CCL25) are also highly expressed in the gut endothelium.[61] AILD liver was marked by increased expression of chemokines or adhesive molecules with heightened attraction for gut-primed cells in intestinal homeostasis disruption. For example, PSC livers exhibited aberrant expression of gut-specific molecules MAdCAM1 and CCL25, and α4β7+ CCR9+ T cells accounted for 20% of liver infiltrating lymphocytes.[62]
In addition to changes in chemokine molecule expression, immune cell migration can also be antigen dependent. A higher proportion of memory T cells in the liver and colon demonstrated shared clonotypes in patients with PSC and IBD compared with those in normal liver and colon, illustrating their reactivity to similar antigens.[63] Using novel transgenic mice that express ovalbumin on enterocytes and biliary epithelial cells, Seidel et al[64] demonstrated that, following the development of antigen-dependent colitis or dextran sodium sulfate colitis, the gut-primed antigen-specific CD8+ T cells were transferred to and infiltrated into the liver and subsequently elicited immune-mediated cholangitis, affirming the notion of gut-priming immune cells in liver autoimmunity.
Antigen-presenting cells also contribute to immune cell migration between the gut and the liver and mainly act via two mechanisms. First, intestinal inflammation is sufficient to induce the activation of liver dendritic cells (DCs), cooperating with the transferred antigen-specific T cells, and results in inflammatory immune responses against the common antigen.[64] Second, activated DCs primed in the gut can also migrate to the liver, where they can directly lead to non-specific immune attacks against parenchymal cells.[65] As a case in point, the interdependence of microbiota, intestinal immunity (DCs), and hepatic immune responses (NKT cells) were well defined in the concanavalin A (ConA)-induced hepatitis model.[65] First, pathogenic bacteria exacerbated liver injury in the ConA treatment group. Second, the number of intestinal DCs increased following ConA treatment. Considering the similar DC activation patterns in the Peyer's patches and liver, the observation indicated that intestinal DCs possessed homing abilities to adjacent liver organs. Finally, after stimulation with pathogenic bacteria, DCs promote NKT cell effector functions by targeting hepatocytes.
Clinical Applications
Clinical role of microbiome biomarkers
Diagnostic signature
Several seminal studies on the microbiota of patients with AILD have uncovered its potential for clinical applications in disease status classification. The diagnosis of AILD, particularly AIH, is based on invasive liver biopsy, which bears procedural risks and has relatively low acceptance by patients. Instead, mathematical models based on microbiome profiles exhibit high accuracy in disease discrimination. First, microbiome-derived signatures can effectively distinguish patients with AILD from HCs. Here, we reviewed several cohort studies with relatively large sample sizes. A cross-sectional study of biopsy-proven AIH reported the use of four genera, including Veillonella, Lactobacillus, Oscillospira, and Clostridiales, to discriminate patients from HCs with an area under the curve (AUC) of 0.78 in the explorative cohort, and yielded an AUC of 0.81 in the validation cohort.[7] In PBC, a microbiome-derived signature of 12 genera displayed an AUC of 0.86 in the test cohort and was subsequently validated with an AUC of 0.84 in validation data.[8] Similarly, a diagnostic model of PSC encompassing Enterococcus, Lactobacillus, and Fusobacterium yielded a precise classification of 95% among the tested subjects and was independently validated with an AUC of 0.72.[66] Another PSC cohort reported better performance using disease-associated genera, which showed an AUC of 0.78 in explorative analysis.[14] Furthermore, microbial metabolic activities are generally more stable than taxonomic composition; therefore, microbiota-derived metabolites also serve as surrogate biomarkers for AILD and outperform taxonomic signatures under certain circumstances. In IgG4-SC and HC differentiation, the fecal metabolites associated with disease status were better biomarkers than the differential microbial genera.[9] In brief, microbiota provides novel tools for disease differentiation in AILD, as exemplified by how fecal metabolites discriminate IgG4-SC and PSC.[9] Given the influence of geographic location, race, and lifestyle on the gut microbiota, it may be imprudent to extrapolate the model to a different population. Therefore, a meta-analysis integrating cohort data from different geographic regions may be more effective in deriving reproducible models. Random forest models built on differential microbiome data discriminated PSC and HC in both German and Norwegian cohorts, with an average AUC of 0.88,[67] thus establishing a reproducible microbiota model independent of geographic location.
Implications for evaluation of disease severity and prognosis
Multiple cross-sectional studies have documented the association between the AILD disease phenotype and perturbation of gut microbiota. The enrichment of pathogenic taxa and depletion of commensal taxa reflect disease severity. For example, Veillonella was enriched in AIH with advanced hepatic inflammation and in PSC with a higher Mayo risk score.[7,14] A decline in Bifidobacterium abundance is associated with increased disease activity in AIH.[12]Blautia was depleted in IgG4-SC with higher transaminase levels and in PSC with an increased degree of cholestasis.[9,18]
Microbiome-derived metabolites are also indicative of disease severity. For instance, fecal acetate and butyrate levels were higher in advanced fibrosis patients with PBC.[68] Succinic acid was positively associated with transaminase in patients with IgG4-SC. Moreover, reduced levels of secondary bile acids, specifically DCA and LCA, are associated with heightened cholestasis in PSC.
Furthermore, the microbiota represents a source for disease stratification and treatment response prediction. Ursodeoxycholic acid (UDCA) is the standard treatment for PBC; however, some patients fail to respond to UDCA. Therefore, screening candidate non-responders is essential for future precision treatment. Investigations of fecal microbiota in PBC demonstrated that UDCA non-responders exhibited a parallel reduction in Faecalibacterium abundance at baseline, and this decrease can be predictive of long-term prognosis in PBC.[15] Similarly, depletion of Bifidobacterium was observed in patients with AIH without biochemical remission.[12] More recent studies of icteric PBC have revealed the reciprocal interplay between bile acid sequestrant treatment responses and microbiota alterations. In particular, SCFA-producing taxa and serum SCFAs, including valeric acid and caproic acid, were elevated in patients with superior remission.[69] In PSC, decreased microbiota-mediated bile acid modification and its subsequent effects on hepatic bile acid synthesis affect disease progression.[43] Specifically, patients with PSC showing a higher ratio of primary/secondary and conjugated/unconjugated bile acids were more likely to progress to end-stage biliary cirrhosis. Serum C4 level, suggestive of hepatic bile acid synthesis and concordant with microbiota-mediated bile acid profile changes, was another predictor of patient survival.[43]
Advances in high-throughput sequencing technology over the past few decades and reduced costs have dramatically promoted the clinical implementation of microbiota knowledge. However, there are several limitations to the application of microbiota signatures in determining disease status and/or prognosis. Various exogenous and endogenous factors shape the gut microbiome, including genetics, age, sex, disease, and lifestyle, posing a challenge in determining true disease-related microbial information. The human microbiota is temporally dynamic; hence, repeated measurements are required to accurately examine microbiome variations. Furthermore, uniform sequencing platforms across the microbiome research community could yield more reproducible and interpretable microbial signatures. Finally, microbiota data are multi-dimensional; therefore, data analysis and integration are statistically challenging.
Therapeutic manipulation of microbiota
Dietary intervention and prebiotics
Altered food preferences led by a westernized lifestyle can partially explain the increased incidence of autoimmune diseases, as evidenced by a proinflammatory immune phenotype profile in urban residents compared with rural residents.[70] Additionally, diet has a great impact on microbiota composition. Hence, dietary modulation can be an ideal approach for modifying microbiota configuration. As a case in point, higher protein intake was reported to correlate with beneficial Bifidobacterium in AIH, which deserves further study to evaluate therapeutic value.
Other modulating approaches include prebiotics that can support the growth of certain beneficial taxa. Dietary fiber and inulin, both indigestible polysaccharides and fermented by the intestinal microbiota, ameliorated liver lesions in experimental AIH mice.[71,72]
Probiotics
Given that AILD is characterized by a reduced abundance of beneficial taxa, administration of commensal bacteria, also termed probiotics, can be of therapeutic value. Lactobacillus rhamnosus GG, a well-characterized probiotic strain, prevents liver fibrosis in Mdr2–/– mice and bile duct-ligated mice by reversing reduced intestinal FXR signaling and facilitating bile acid excretion through feces and urine.[73]Bifidobacterium animalis subsp. lactis 420 (B420) has been widely acknowledged for its immunomodulatory effects and has been reported to improve intestinal homeostasis in experimental animal models and clinical trials.[74] Correspondingly, B420 supplementation in experimental AIH mice led to improved liver inflammation. More explorative investigations showed that the beneficial effects were partly mediated by microbiota community reconfiguration and subsequent intestinal barrier maintenance, with modulation of receptor-interacting serine/threonine kinase 3 signaling in liver macrophages.[75] Similarly, Akkermansia muciniphila, frequently observed to attenuate immunological pathologies, alleviates liver injuries induced by ConA, concomitant with the reduction of multiple proinflammatory cytokines.[76] However, it is imperative to remember that the beneficial effects observed are primarily based solely on preclinical research. A previous clinical trial that examined the beneficial effects of a probiotic cocktail containing four Lactobacillus species and two Bifidobacterium species on biochemical parameters and symptoms in patients with PSC failed to find relevant evidence.[77] Thus, further clinical studies with larger sample sizes are warranted.
Postbiotics
The mitigation of the effects of altered microbiota-derived metabolites, also referred to as postbiotics, provides a promising source of microbiota-derived therapeutic targets. This can be achieved through various approaches, including the administration of exogenous microbiota-derived metabolites, interference with relevant metabolic pathways in bacteria, and colonization of engineered bacteria with particular functions. A reduced abundance of butyrate-producing taxa, especially Faecalibacterium, was consistently observed in AILD.[7–9] Concordantly, butyrate sodium ameliorated liver injury and improved intestinal permeability in mice with S100/Freund's complete adjuvant-induced AIH, mechanistically via disruption of the TLR-4 signaling pathway and increasing the ratio of regulatory T cells (Tregs)/Th17.[22,72] Additionally, peripheral blood mononuclear cells from patients with AIH showed dysfunctional aryl hydrocarbon receptor (AHR) signaling, specifically aberrant inhibition and heightened non-canonical AHR activation, which drove the imbalance between Treg and Th17 cell subsets.[78] Regarding therapeutic implications, AHR agonists can restore the regulatory ability of Tregs impaired in AIH.[78] In light of their ability to trigger AHR activation, microbiota-derived indole derivatives should be optimized for therapeutic use in future clinical practice.[79]
Antibiotics
Clinical assessments of the efficacy of antibiotics in disease treatment are mainly performed in patients with PSC. With the administration of variable doses of vancomycin and metronidazole in 35 patients with PSC, Tabibian et al[80] scrutinized the reduction in alkaline phosphatase (ALP) level as the primary endpoint in PSC, with Mayo clinical score and total bilirubin (TB) as the secondary endpoints. The results suggested that patients treated with vancomycin reached the primary endpoint and those treated with metronidazole showed reduced Mayo clinical scores and TB levels. Oral vancomycin was also effective in treating pediatric PSC. A trial of 14 children with PSC showed that vancomycin treatment normalized biochemical indices of patients with non-cirrhosis and improved biochemical abnormalities in those with cirrhosis, notably PSC symptoms recurring after vancomycin discontinuation.[81] Interestingly, the autoantibody levels decreased significantly or tested negative after vancomycin treatment,[81] supporting the immunomodulatory effects of vancomycin. Indeed, vancomycin treatment has been reported to interfere with TNF-α signaling and increase peripheral TGF-β and Treg levels in patients with PSC-IBD.[82] More thorough investigations are needed to decipher the mechanisms by which vancomycin alleviates the disease phenotype of PSC.
FMT
FMT has been shown to alter the microbiota community and is effective in the treatment of a myriad of diseases, including Clostridium difficile infection and IBD. An open-label pilot study of patients with PSC and IBD was performed to evaluate the biochemical amelioration and microbiota-modulation effects of FMT. Specifically, the results showed no adverse effects were observed, and 30% of the patients exhibited a ≥50% reduction in ALP level. The increased diversity after FMT and the engraftment of certain taxa were associated with ALP reduction.[83] However, the safety and efficacy of FMT in other AILDs remain elusive and require further exploration.
Concluding Remarks
Accumulating evidence suggests that the gut microbiota is closely linked to the pathogenesis of AILD, and the mechanisms responsible for host–microbiota interactions are increasingly understood. In this review, we highlight several general concepts that can be applied to AILD. Microbiota-dependent immune aberrancies are initiated by intestinal dysbiosis and dysfunctional intestinal barrier. Subsequent mechanisms include bacterial translocation and cross-reactivity between bacterial antigens and autoantigens, as well as microbiota-derived metabolites that promote cascading inflammatory responses. However, the relevant microbial organisms and related types of immune responses are contextually dependent on a combination of several factors, including host genetic predisposition, environmental exposure, and other poorly defined mechanisms. This dependency helps explain the inconsistency observed in the experimental and clinical data. There are still unaddressed problems that need further investigation. The microbiome profiling of AILD mostly provides taxonomic information instead of detailed functional annotations. Given the redundant properties of functional genes across species, taxonomic information is insufficient to infer the relevant mechanism through which microbiota dysbiosis contributes to AILD. Although functional dysbiosis has been previously reported in AILD, the main findings are based on the predicted 16S rRNA data, which lack accuracy and completeness.[7,84] In addition, strain information is lacking in the AILD microbiota studies. Different strains within the same species can exhibit contrasting immunogenic properties.[1] Elucidating the relationship between AILD and the microbiota also requires the integration of other aspects host biology, including genetics and cellular biology.[85] Moreover, stool moisture variation is necessary to be considered in future studies, due to its correlation with faecal microbial load.[86] Furthermore, despite the clinical evidence, the cause–effect relationships between microbiota and AILD are still far from understood. Therefore, more rigorous mechanistic studies are warranted. Patients with preclinical subtypes, such as subclinical PBC, can also be longitudinally tracked to infer a cause-effect relationship.
Altogether, the increasingly comprehensive knowledge of gut microbiota and intestinal homeostasis has transformed our understanding of host–microbiota interactions in AILD. Despite these extraordinary findings, investigations of immune system–microbiota interactions in AILD are still preliminary, and further collective efforts are required for the formidable task of harnessing microbiota for novel and effective therapeutics.
Footnotes
How to cite this article: Liu Q, He W, Tang R, Ma X. Intestinal homeostasis in autoimmune liver diseases. Chin Med J 2022;135:1642–1652. doi: 10.1097/CM9.0000000000002291
References
- 1.Ansaldo E, Farley TK, Belkaid Y. Control of immunity by the microbiota. Annu Rev Immunol 2021; 39:449–479. doi: 10.1146/annurev-immunol-093019-112348. [DOI] [PubMed] [Google Scholar]
- 2.Jones RM, Neish AS. Gut microbiota in intestinal and liver disease. Annu Rev Pathol 2021; 16:251–275. doi: 10.1146/annurev-pathol-030320-095722. [DOI] [PubMed] [Google Scholar]
- 3.Vujkovic-Cvijin I, Sklar J, Jiang L, Natarajan L, Knight R, Belkaid Y. Host variables confound gut microbiota studies of human disease. Nature 2020; 587:448–454. doi: 10.1038/s41586-020-2881-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chopyk DM, Grakoui A. Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology 2020; 159:849–863. doi: 10.1053/j.gastro.2020.04.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albillos A, de Gottardi A, Rescigno M. The gut-liver axis in liver disease: pathophysiological basis for therapy. J Hepatol 2020; 72:558–577. doi: 10.1016/j.jhep.2019.10.003. [DOI] [PubMed] [Google Scholar]
- 6.Dean G, Hanauer S, Levitsky J. The role of the intestine in the pathogenesis of primary sclerosing cholangitis: evidence and therapeutic implications. Hepatology 2020; 72:1127–1138. doi: 10.1002/hep.31311. [DOI] [PubMed] [Google Scholar]
- 7.Wei Y, Li Y, Yan L, Sun C, Miao Q, Wang Q, et al. Alterations of gut microbiome in autoimmune hepatitis. Gut 2020; 69:569–577. doi: 10.1136/gutjnl-2018-317836. [DOI] [PubMed] [Google Scholar]
- 8.Tang R, Wei Y, Li Y, Chen W, Chen H, Wang Q, et al. Gut microbial profile is altered in primary biliary cholangitis and partially restored after UDCA therapy. Gut 2018; 67:534–541. doi: 10.1136/gutjnl-2016-313332. [DOI] [PubMed] [Google Scholar]
- 9.Liu Q, Li B, Li Y, Wei Y, Huang B, Liang J, et al. Altered faecal microbiome and metabolome in IgG4-related sclerosing cholangitis and primary sclerosing cholangitis. Gut 2022; 71:899–909. doi: 10.1136/gutjnl-2020-323565. [DOI] [PubMed] [Google Scholar]
- 10.Schrumpf E, Kummen M, Valestrand L, Greiner TU, Holm K, Arulampalam V, et al. The gut microbiota contributes to a mouse model of spontaneous bile duct inflammation. J Hepatol 2017; 66:382–389. doi: 10.1016/j.jhep.2016.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuksel M, Wang Y, Tai N, Peng J, Guo J, Beland K, et al. A novel “humanized mouse” model for autoimmune hepatitis and the association of gut microbiota with liver inflammation. Hepatology 2015; 62:1536–1550. doi: 10.1002/hep.27998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liwinski T, Casar C, Ruehlemann MC, Bang C, Sebode M, Hohenester S, et al. A disease-specific decline of the relative abundance of Bifidobacterium in patients with autoimmune hepatitis. Aliment Pharmacol Ther 2020; 51:1417–1428. doi: 10.1111/apt.15754. [DOI] [PubMed] [Google Scholar]
- 13.Nakamoto N, Sasaki N, Aoki R, Miyamoto K, Suda W, Teratani T, et al. Gut pathobionts underlie intestinal barrier dysfunction and liver T helper 17 cell immune response in primary sclerosing cholangitis. Nat Microbiol 2019; 4:492–503. doi: 10.1038/s41564-018-0333-1. [DOI] [PubMed] [Google Scholar]
- 14.Kummen M, Holm K, Anmarkrud JA, Nygård S, Vesterhus M, Høivik ML, et al. The gut microbial profile in patients with primary sclerosing cholangitis is distinct from patients with ulcerative colitis without biliary disease and healthy controls. Gut 2017; 66:611–619. doi: 10.1136/gutjnl-2015-310500. [DOI] [PubMed] [Google Scholar]
- 15.Furukawa M, Moriya K, Nakayama J, Inoue T, Momoda R, Kawaratani H, et al. Gut dysbiosis associated with clinical prognosis of patients with primary biliary cholangitis. Hepatol Res 2020; 50:840–852. doi: 10.1111/hepr.13509. [DOI] [PubMed] [Google Scholar]
- 16.Muratori P, Muratori L, Guidi M, Maccariello S, Pappas G, Ferrari R, et al. Anti-Saccharomyces cerevisiae antibodies (ASCA) and autoimmune liver diseases. Clin Exp Immunol 2003; 132:473–476. doi: 10.1046/j.1365-2249.2003.02166.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu C, Deng C, Zhang S, Song G, Li L, Li X, et al. Clinical significance and prevalence of anti-Saccharomyces cerevisiae antibody in Chinese patients with primary biliary cirrhosis. Clin Exp Med 2013; 13:245–250. doi: 10.1007/s10238-012-0207-4. [DOI] [PubMed] [Google Scholar]
- 18.Lemoinne S, Kemgang A, Ben Belkacem K, Straube M, Jegou S, Corpechot C, et al. Fungi participate in the dysbiosis of gut microbiota in patients with primary sclerosing cholangitis. Gut 2020; 69:92–102. doi: 10.1136/gutjnl-2018-317791. [DOI] [PubMed] [Google Scholar]
- 19.Agus A, Clément K, Sokol H. Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut 2021; 70:1174–1182. doi: 10.1136/gutjnl-2020-323071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kummen M, Thingholm LB, Rühlemann MC, Holm K, Hansen SH, Moitinho-Silva L, et al. Altered gut microbial metabolism of essential nutrients in primary sclerosing cholangitis. Gastroenterology 2021; 160:1784–1798. doi: 10.1053/j.gastro.2020.12.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zierer J, Jackson MA, Kastenmüller G, Mangino M, Long T, Telenti A, et al. The fecal metabolome as a functional readout of the gut microbiome. Nat Genet 2018; 50:790–795. doi: 10.1038/s41588-018-0135-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu JL, Zou JY, Hu, Chen DZ, Chen L, Lu FB, et al. Sodium butyrate ameliorates S100/FCA-induced autoimmune hepatitis through regulation of intestinal tight junction and toll-like receptor 4 signaling pathway. Immunol Lett 2017; 190:169–176. doi: 10.1016/j.imlet.2017.08.005. [DOI] [PubMed] [Google Scholar]
- 23.Xue R, Zhang H, Pan J, Du Z, Zhou W, Zhang Z, et al. Peripheral dopamine controlled by gut microbes inhibits invariant natural killer T cell-mediated hepatitis. Front Immunol 2018; 9:2398.doi: 10.3389/fimmu.2018.02398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan Y, Llorente C, Lang S, Brandl K, Chu H, Jiang L, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 2019; 575:505–511. doi: 10.1038/s41586-019-1742-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin R, Zhou L, Zhang J, Wang B. Abnormal intestinal permeability and microbiota in patients with autoimmune hepatitis. Int J Clin Exp Pathol 2015; 8:5153–5160. [PMC free article] [PubMed] [Google Scholar]
- 26.Manfredo Vieira S, Hiltensperger M, Kumar V, Zegarra-Ruiz D, Dehner C, Khan N, et al. Translocation of a gut pathobiont drives autoimmunity in mice and humans. Science 2018; 359:1156–1161. doi: 10.1126/science.aar7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pohl J, Ring A, Stremmel W, Stiehl A. The role of dominant stenoses in bacterial infections of bile ducts in primary sclerosing cholangitis. Eur J Gastroenterol Hepatol 2006; 18:69–74. doi: 10.1097/00042737-200601000-00012. [DOI] [PubMed] [Google Scholar]
- 28.Ma HD, Zhao ZB, Ma WT, Liu QZ, Gao CY, Li L, et al. Gut microbiota translocation promotes autoimmune cholangitis. J Autoimmun 2018; 95:47–57. doi: 10.1016/j.jaut.2018.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Inamine T, Schnabl B. Immunoglobulin A and liver diseases. J Gastroenterol 2018; 53:691–700. doi: 10.1007/s00535-017-1400-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Floreani A, Baragiotta A, Pizzuti D, Martines D, Cecchetto A, Chiarelli S. Mucosal, IgA defect in primary biliary cirrhosis. Am J Gastroenterol 2002; 97:508–510. doi: 10.1111/j.1572-0241.2002.05521.x. [DOI] [PubMed] [Google Scholar]
- 31.Gulamhusein AF, Hirschfield GM. Primary biliary cholangitis: pathogenesis and therapeutic opportunities. Nat Rev Gastroenterol Hepatol 2020; 17:93–110. doi: 10.1038/s41575-019-0226-7. [DOI] [PubMed] [Google Scholar]
- 32.Bogdanos DP, Baum H, Okamoto M, Montalto P, Sharma UC, Rigopoulou EI, et al. Primary biliary cirrhosis is characterized by IgG3 antibodies cross-reactive with the major mitochondrial autoepitope and its Lactobacillus mimic. Hepatology 2005; 42:458–465. doi: 10.1002/hep.20788. [DOI] [PubMed] [Google Scholar]
- 33.Selmi C, Balkwill DL, Invernizzi P, Ansari AA, Coppel RL, Podda M, et al. Patients with primary biliary cirrhosis react against a ubiquitous xenobiotic-metabolizing bacterium. Hepatology 2003; 38:1250–1257. doi: 10.1053/jhep.2003.50446. [DOI] [PubMed] [Google Scholar]
- 34.Wang JJ, Yang GX, Zhang WC, Lu L, Tsuneyama K, Kronenberg M, et al. Escherichia coli infection induces autoimmune cholangitis and anti-mitochondrial antibodies in non-obese diabetic (NOD).B6 (Idd10/Idd18) mice. Clin Exp Immunol 2014; 175:192–201. doi: 10.1111/cei.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mattner J, Savage PB, Leung P, Oertelt SS, Wang V, Trivedi O, et al. Liver autoimmunity triggered by microbial activation of natural killer T cells. Cell Host Microbe 2008; 3:304–315. doi: 10.1016/j.chom.2008.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terjung B, Söhne J, Lechtenberg B, Gottwein J, Muennich M, Herzog V, et al. p-ANCAs in autoimmune liver disorders recognise human beta-tubulin isotype 5 and cross-react with microbial protein FtsZ. Gut 2010; 59:808–816. doi: 10.1136/gut.2008.157818. [DOI] [PubMed] [Google Scholar]
- 37.Terjung B, Spengler U. Atypical p-ANCA in PSC and AIH: a hint toward a “leaky gut”? Clin Rev Allergy Immunol 2009; 36:40–51. doi: 10.1007/s12016-008-8088-8. [DOI] [PubMed] [Google Scholar]
- 38.Ruff WE, Greiling TM, Kriegel MA. Host-microbiota interactions in immune-mediated diseases. Nat Rev Microbiol 2020; 18:521–538. doi: 10.1038/s41579-020-0367-2. [DOI] [PubMed] [Google Scholar]
- 39.Jia W, Xie G, Jia W. Bile acid-microbiota crosstalk in gastrointestinal inflammation and carcinogenesis. Nat Rev Gastroenterol Hepatol 2018; 15:111–128. doi: 10.1038/nrgastro.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fuchs CD, Trauner M. Role of bile acids and their receptors in gastrointestinal and hepatic pathophysiology. Nat Rev Gastroenterol Hepatol 2022; 19:432–450. doi: 10.1038/s41575-021-00566-7. [DOI] [PubMed] [Google Scholar]
- 41.Chen W, Wei Y, Xiong A, Li Y, Guan H, Wang Q, et al. Comprehensive analysis of serum and fecal bile acid profiles and interaction with gut microbiota in primary biliary cholangitis. Clin Rev Allergy Immunol 2020; 58:25–38. doi: 10.1007/s12016-019-08731-2. [DOI] [PubMed] [Google Scholar]
- 42.Tabibian JH, O’Hara SP, Trussoni CE, Tietz PS, Splinter PL, Mounajjed T, et al. Absence of the intestinal microbiota exacerbates hepatobiliary disease in a murine model of primary sclerosing cholangitis. Hepatology 2016; 63:185–196. doi: 10.1002/hep.27927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider KM, Candels LS, Hov JR, Myllys M, Hassan R, Schneider CV, et al. Gut microbiota depletion exacerbates cholestatic liver injury via loss of FXR signalling. Nat Metab 2021; 3:1228–1241. doi: 10.1038/s42255-021-00452-1. [DOI] [PubMed] [Google Scholar]
- 44.Sinha SR, Haileselassie Y, Nguyen LP, Tropini C, Wang M, Becker LS, et al. Dysbiosis-induced secondary bile acid deficiency promotes intestinal inflammation. Cell Host Microbe 2020; 27:659–670. doi: 10.1016/j.chom.2020.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Chen Kiow J, Vincent C, Sidani S, Bouin M. High occurrence of small intestinal bacterial overgrowth in primary biliary cholangitis. Neurogastroenterol Motil 2019; 31:e13691.doi: 10.1111/nmo.13691. [DOI] [PubMed] [Google Scholar]
- 46.Loomba R, Friedman SL, Shulman GI. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021; 184:2537–2564. doi: 10.1016/j.cell.2021.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, et al. Gender bias in autoimmunity is influenced by microbiota. Immunity 2013; 39:400–412. doi: 10.1016/j.immuni.2013.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizzetto L, Fava F, Tuohy KM, Selmi C. Connecting the immune system, systemic chronic inflammation and the gut microbiome: the role of sex. J Autoimmun 2018; 92:12–34. doi: 10.1016/j.jaut.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 49.Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, et al. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013; 339:1084–1088. doi: 10.1126/science.1233521. [DOI] [PubMed] [Google Scholar]
- 50.Schwinge D, Carambia A, Quaas A, Krech T, Wegscheid C, Tiegs G, et al. Testosterone suppresses hepatic inflammation by the downregulation of IL-17, CXCL-9, and CXCL-10 in a mouse model of experimental acute cholangitis. J Immunol 2015; 194:2522–2530. doi: 10.4049/jimmunol.1400076. [DOI] [PubMed] [Google Scholar]
- 51.Arterbery AS, Yao J, Ling A, Avitzur Y, Martinez M, Lobritto S, et al. Inflammasome priming mediated via toll-like receptors 2 and 4, induces Th1-like regulatory T cells in de novo autoimmune hepatitis. Front Immunol 2018; 9:1612.doi: 10.3389/fimmu.2018.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ivanov II, Tuganbaev T, Skelly AN, Honda K. T Cell Responses to the Microbiota. Annu Rev Immunol 2022; 40:559–587. doi: 10.1146/annurev-immunol-101320-011829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Legoux F, Salou M, Lantz O. MAIT cell development and functions: the microbial connection. Immunity 2020; 53:710–723. doi: 10.1016/j.immuni.2020.09.009. [DOI] [PubMed] [Google Scholar]
- 54.Jeffery HC, van Wilgenburg B, Kurioka A, Parekh K, Stirling K, Roberts S, et al. Biliary epithelium and liver B cells exposed to bacteria activate intrahepatic MAIT cells through MR1. J Hepatol 2016; 64:1118–1127. doi: 10.1016/j.jhep.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Setsu T, Yamagiwa S, Tominaga K, Kimura N, Honda H, Kamimura H, et al. Persistent reduction of mucosal-associated invariant T cells in primary biliary cholangitis. J Gastroenterol Hepatol 2018; 33:1286–1294. doi: 10.1111/jgh.14076. [DOI] [PubMed] [Google Scholar]
- 56.von Seth E, Zimmer CL, Reuterwall-Hansson M, Barakat A, Arnelo U, Bergquist A, et al. Primary sclerosing cholangitis leads to dysfunction and loss of MAIT cells. Eur J Immunol 2018; 48:1997–2004. doi: 10.1002/eji.201847608. [DOI] [PubMed] [Google Scholar]
- 57.Renand A, Habes S, Mosnier JF, Aublé H, Judor JP, Vince N, et al. Immune Alterations in patients with type 1 autoimmune hepatitis persist upon standard immunosuppressive treatment. Hepatol Commun 2018; 2:968–981. doi: 10.1002/hep4.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tedesco D, Thapa M, Chin CY, Ge Y, Gong M, Li J, et al. Alterations in intestinal microbiota lead to production of interleukin 17 by intrahepatic T-cell receptor-positive cells and pathogenesis of cholestatic liver disease. Gastroenterology 2018; 154:2178–2193. doi: 10.1053/j.gastro.2018.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao L, Tang Y, You Z, Wang Q, Liang S, Han X, et al. Interleukin-17 contributes to the pathogenesis of autoimmune hepatitis through inducing hepatic interleukin-6 expression. PLoS One 2011; 6:e18909.doi: 10.1371/journal.pone.0018909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fergusson JR, Hühn MH, Swadling L, Walker LJ, Kurioka A, Llibre A, et al. CD161(int) CD8+ T cells: a novel population of highly functional, memory CD8+ T cells enriched within the gut. Mucosal Immunol 2016; 9:401–413. doi: 10.1038/mi.2015.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sun H, Lagarrigue F, Wang H, Fan Z, Lopez-Ramirez MA, Chang JT, et al. Distinct integrin activation pathways for effector and regulatory T cell trafficking and function. J Exp Med 2021; 218:e20201524.doi: 10.1084/jem.20201524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Eksteen B, Grant AJ, Miles A, Curbishley SM, Lalor PF, Hübscher SG, et al. Hepatic endothelial CCL25 mediates the recruitment of CCR9+ gut-homing lymphocytes to the liver in primary sclerosing cholangitis. J Exp Med 2004; 200:1511–1517. doi: 10.1084/jem.20041035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henriksen EK, Jørgensen KK, Kaveh F, Holm K, Hamm D, Olweus J, et al. Gut and liver T-cells of common clonal origin in primary sclerosing cholangitis-inflammatory bowel disease. J Hepatol 2017; 66:116–122. doi: 10.1016/j.jhep.2016.09.002. [DOI] [PubMed] [Google Scholar]
- 64.Seidel D, Eickmeier I, Kühl AA, Hamann A, Loddenkemper C, Schott E. CD8 T cells primed in the gut-associated lymphoid tissue induce immune-mediated cholangitis in mice. Hepatology 2014; 59:601–611. doi: 10.1002/hep.26702. [DOI] [PubMed] [Google Scholar]
- 65.Chen J, Wei Y, He J, Cui G, Zhu Y, Lu C, et al. Natural killer T cells play a necessary role in modulating of immune-mediated liver injury by gut microbiota. Sci Rep 2014; 4:7259.doi: 10.1038/srep07259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sabino J, Vieira-Silva S, Machiels K, Joossens M, Falony G, Ballet V, et al. Primary sclerosing cholangitis is characterised by intestinal dysbiosis independent from IBD. Gut 2016; 65:1681–1689. doi: 10.1136/gutjnl-2015-311004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rühlemann M, Liwinski T, Heinsen FA, Bang C, Zenouzi R, Kummen M, et al. Consistent alterations in faecal microbiomes of patients with primary sclerosing cholangitis independent of associated colitis. Aliment Pharmacol Ther 2019; 50:580–589. doi: 10.1111/apt.15375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lammert C, Shin A, Xu H, Hemmerich C, O’Connell TM, Chalasani N. Short-chain fatty acid and fecal microbiota profiles are linked to fibrosis in primary biliary cholangitis. FEMS Microbiol Lett 2021; 368:fnab038.doi: 10.1093/femsle/fnab038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li B, Zhang J, Chen Y, Wang Q, Yan L, Wang R, et al. Alterations in microbiota and their metabolites are associated with beneficial effects of bile acid sequestrant on icteric primary biliary Cholangitis. Gut Microbes 2021; 13:1946366.doi: 10.1080/19490976.2021.1946366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Temba GS, Kullaya V, Pecht T, Mmbaga BT, Aschenbrenner AC, Ulas T, et al. Urban living in healthy Tanzanians is associated with an inflammatory status driven by dietary and metabolic changes. Nat Immunol 2021; 22:287–300. doi: 10.1038/s41590-021-00867-8. [DOI] [PubMed] [Google Scholar]
- 71.Yamaguchi A, Teratani T, Chu PS, Suzuki T, Taniki N, Mikami Y, et al. Hepatic adenosine triphosphate reduction through the short-chain fatty acids-peroxisome proliferator-activated receptor (-uncoupling protein 2 axis alleviates immune-mediated acute hepatitis in inulin-supplemented mice. Hepatol Commun 2021; 5:1555–1570. doi: 10.1002/hep4.1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hu ED, Chen DZ, Wu JL, Lu FB, Chen L, Zheng MH, et al. High fiber dietary and sodium butyrate attenuate experimental autoimmune hepatitis through regulation of immune regulatory cells and intestinal barrier. Cell Immunol 2018; 328:24–32. doi: 10.1016/j.cellimm.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 73.Liu Y, Chen K, Li F, Gu Z, Liu Q, He L, et al. Probiotic Lactobacillus rhamnosus GG prevents liver fibrosis through inhibiting hepatic bile acid synthesis and enhancing bile acid excretion in mice. Hepatology 2020; 71:2050–2066. doi: 10.1002/hep.30975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hibberd AA, Yde CC, Ziegler ML, Honoré AH, Saarinen MT, Lahtinen S, et al. Probiotic or synbiotic alters the gut microbiota and metabolism in a randomised controlled trial of weight management in overweight adults. Benef Microbes 2019; 10:121–135. doi: 10.3920/bm2018.0028. [DOI] [PubMed] [Google Scholar]
- 75.Zhang H, Liu M, Liu X, Zhong W, Li Y, Ran Y, et al. Bifidobacterium animalis ssp. lactis 420 mitigates autoimmune hepatitis through regulating intestinal barrier and liver immune cells. Front Immunol 2020; 11:569104.doi: 10.3389/fimmu.2020.569104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wu W, Lv L, Shi D, Ye J, Fang D, Guo F, et al. Protective effect of akkermansia muciniphila against immune-mediated liver injury in a mouse model. Front Microbiol 2017; 8:1804.doi: 10.3389/fmicb.2017.01804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vleggaar FP, Monkelbaan JF, van Erpecum KJ. Probiotics in primary sclerosing cholangitis: a randomized placebo-controlled crossover pilot study. Eur J Gastroenterol Hepatol 2008; 20:688–692. doi: 10.1097/MEG.0b013e3282f5197e. [DOI] [PubMed] [Google Scholar]
- 78.Vuerich M, Harshe R, Frank LA, Mukherjee S, Gromova B, Csizmadia E, et al. Altered aryl-hydrocarbon-receptor signalling affects regulatory and effector cell immunity in autoimmune hepatitis. J Hepatol 2021; 74:48–57. doi: 10.1016/j.jhep.2020.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Agus A, Planchais J, Sokol H. Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 2018; 23:716–724. doi: 10.1016/j.chom.2018.05.003. [DOI] [PubMed] [Google Scholar]
- 80.Tabibian JH, Weeding E, Jorgensen RA, Petz JL, Keach JC, Talwalkar JA, et al. Randomised clinical trial: vancomycin or metronidazole in patients with primary sclerosing cholangitis - A pilot study. Aliment Pharmacol Ther 2013; 37:604–612. doi: 10.1111/apt.12232. [DOI] [PubMed] [Google Scholar]
- 81.Davies YK, Cox KM, Abdullah BA, Safta A, Terry AB, Cox KL. Long-term treatment of primary sclerosing cholangitis in children with oral vancomycin: an immunomodulating antibiotic. J Pediatr Gastroenterol Nutr 2008; 47:61–67. doi: 10.1097/MPG.0b013e31816fee95. [DOI] [PubMed] [Google Scholar]
- 82.Abarbanel DN, Seki SM, Davies Y, Marlen N, Benavides JA, Cox K, et al. Immunomodulatory effect of vancomycin on Treg in pediatric inflammatory bowel disease and primary sclerosing cholangitis. J Clin Immunol 2013; 33:397–406. doi: 10.1007/s10875-012-9801-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allegretti JR, Kassam Z, Carrellas M, Mullish BH, Marchesi JR, Pechlivanis A, et al. Fecal microbiota transplantation in patients with primary sclerosing cholangitis: a pilot clinical trial. Am J Gastroenterol 2019; 114:1071–1079. doi: 10.14309/ajg.0000000000000115. [DOI] [PubMed] [Google Scholar]
- 84.Elsherbiny NM, Rammadan M, Hassan EA, Ali ME, El-Rehim ASA, Abbas WA, et al. Autoimmune hepatitis: shifts in gut microbiota and metabolic pathways among egyptian patients. Microorganisms 2020; 8:1011.doi: 10.3390/microorganisms8071011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Quraishi MN, Acharjee A, Beggs AD, Horniblow R, Tselepis C, Gkoutos G, et al. A Pilot integrative analysis of colonic gene expression, gut microbiota, and immune infiltration in primary sclerosing cholangitis-inflammatory bowel disease: association of disease with bile acid pathways. J Crohns Colitis 2020; 14:935–947. doi: 10.1093/ecco-jcc/jjaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Vieira-Silva S, Sabino J, Valles-Colomer M, Falony G, Kathagen G, Caenepeel C, et al. Quantitative microbiome profiling disentangles inflammation- and bile duct obstruction-associated microbiota alterations across PSC/IBD diagnoses. Nat Microbiol 2019; 4:1826–1831. doi: 10.1038/s41564-019-0483-9. [DOI] [PubMed] [Google Scholar]