Abstract
Background:
As a non-invasive and effective diagnostic method for small intestinal bacterial overgrowth (SIBO), wild-use of breath test (BT) has demonstrated a high comorbidity rate in patients with diarrhea-predominant irritable bowel syndrome (IBS-D) and SIBO. Patients overlapping with SIBO respond better to rifaximin therapy than those with IBS-D only. Gut microbiota plays a critical role in both of these two diseases. We aimed to determine the microbial difference between IBS-D overlapping with/without SIBO, and to study the underlying mechanism of its sensitivity to rifaximin.
Methods:
Patients with IBS-D were categorized as BT-negative (IBSN) and BT-positive (IBSP). Healthy volunteers (BT-negative) were enrolled as healthy control. The patients were clinically evaluated before and after rifaximin treatment (0.4 g bid, 4 weeks). Blood, intestine, and stool samples were collected for cytokine assessment and gut microbial analyses.
Results:
Clinical complaints and microbial abundance were significantly higher in IBSP than in IBSN. In contrast, severe systemic inflammation and more active bacterial invasion function that were associated with enrichment of opportunistic pathogens were seen in IBSN. The symptoms of IBSP patients were relieved in different degrees after therapy, but the symptoms of IBSN rarely changed. We also found that the presence of IBSN-enriched genera (Enterobacter and Enterococcus) are unaffected by rifaximin therapy.
Conclusions:
IBS-D patients overlapping with SIBO showed noticeably different fecal microbial composition and function compared with IBS-D only. The better response to rifaximin in those comorbid patients might associate with their different gut microbiota, which suggests that BT is necessary before IBS-D diagnosis and use of rifaximin.
Registration:
Chinese Clinical Trial Registry, ChiCTR1800017911.
Keywords: Irritable bowel syndrome, Small intestinal bacterial overgrowth, Breath test, Gut microbiota, Rifaximin
Introduction
Irritable bowel syndrome (IBS) is a functional gastrointestinal (GI) disorder with a high prevalence worldwide.[1,2] IBS-like symptoms such as abdominal pain, discomfort, diarrhea, bloating, and flatulence are commonly observed in patients with small intestinal bacterial overgrowth (SIBO) as well.[3] Hydrogen and methane breath test (BT) is an effective and non-invasive diagnostic tool for SIBO.[4] In patients with IBS who meet Rome criteria, BT positive rate is ≥35.5%.[5,6] Studies have confirmed that patients with IBS, especially those with a diarrhea-predominant subtype (IBS-D), are more likely to overlap with SIBO. However, the symptom profiles of IBS or SIBO are non-specific and clinical history alone cannot clearly distinguish the underlying cause.
Rifaximin, a broad-spectrum oral antibiotic with little absorption, is recommended for IBS treatment.[7,8] Rifaximin inhibits bacterial RNA synthesis by binding to the β-subunit of bacterial DNA-dependent RNA polymerase and against a variety of entomopathogens. Although rifaximin is more effective than placebo in patients with non-constipated IBS,[9] the efficacy was modest and symptoms recurred after treatment. In addition, high cost and insufficient efficacy rate limit the clinical use of rifaximin.[10] Patients with IBS overlapping with SIBO presented with higher abdominal bloating scores and severer GI symptoms.[6,11] Eradication of SIBO using absorbable or non-absorbable antibiotics reduces IBS-like symptoms; for example, rifaximin significantly relieves the clinical symptoms of SIBO.[12,13] Therefore, some experienced gastroenterologists[14,15] might recommend BT to exclude SIBO before making an IBS diagnosis or before a rifaximin treatment. However, there is neither any guideline in place that specifies the necessity to do so, nor any study that addresses the pathophysiological mechanism resulting in different curative effects of rifaximin on IBS overlapping with SIBO.
Gut microbiota plays a vital role in IBS. The small intestinal and fecal microbiota of IBS patients has been studied previously.[16–20] The fecal microbial community of IBS exhibits lower α-diversity, with increasing abundance of Firmicutes and decreasing Bacteroidetes at the phylum level, and with increasing Clostridia and decreasing Bacteroidia at the lower taxonomic levels.[16,17] Microbial community changes in SIBO seem to contribute to IBS. Moreover, the changes in small intestinal microbiota may influence fecal flora.[19,20] However, previous studies on small intestinal microbiota of patients with IBS neglect to consider whether the patients have SIBO, and few studies have attempted to characterize the differences in fecal microbials in patients with IBS with or without SIBO.
In this study, we aimed to determine the microbial difference between IBS-D overlapping with/without SIBO, and to study the underlying mechanism of its sensitivity to rifaximin. We elucidated the necessity to eliminate SIBO from IBS diagnosis through BT to provide precise treatment options for better clinical efficacy.
Methods
Study design and subject's recruitment
The trial was performed from April 2015 to February 2019. IBS-D patients whose symptoms fulfilled the diagnostic criteria of Rome III were recruited from the Department of Gastroenterology, Peking University Third Hospital. Healthy volunteers without previous or current GI symptoms and infection were recruited at the same time though advertisements. Both patients and healthy volunteers aged between 18 and 65 years were recruited, as indicated in the flowchart shown in Supplementary Figure 1. After ingestion of 10 g of lactulose in a 20 mL water solution, lactulose, hydrogen, and methane breath test (LHMBT) was performed using the methane-hydrogen breathing analyzer (Quintron Instrument Company, Milwaukee, WI, USA). IBS-D patients were separated into two groups according to the results of LHMBT: patients with BT negative termed as IBSN group, and patients with BT positive termed as IBSP group. Healthy volunteers with BT negative constituted the healthy control (HC) group. All subjects underwent colonoscopy with biopsies in distal ileum and sigmoid. Details are shown in Supplementary Methods. Consecutive patients of IBSN and IBSP were administered with rifaximin (Xifaxan, Alfa Wassermann S.P.A., Italy) 0.4 g twice per day orally for 4 weeks. After 4 weeks, subjects completed LHMBT, IBS-SSS, and fecal sample collected again. During the intervention therapy, patients received a follow-up clinical interview once a week.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki and was approved by the local ethics committee of Peking University Health Science Center (No. IRB00001052-14091). Informed written consent was obtained from all patients prior to their enrollment in this study.
Clinical evaluation
Each subject received routine blood and stool tests to rule out local and systemic organic lesions before treatment. Daily bowel movement frequency and consistency were recorded based on the Bristol Stool Form (BSF) scale. Visceral sensitivity was evaluated by colon rectal distension (CRD) test using a barostat (Distender Series II; G&J Electronics, Ontario, Canada). These procedures have been described in detail in our previous studies.[11,21] GI symptom severity was evaluated by IBS Symptom Severity Scores (IBS-SSS) and Gastrointestinal Symptom Rating Scale (GSRS) before and after treatment. Half-year food frequency questionnaire (FFQ) (Chinese version) was used to estimate the dietary pattern.
Experimental evaluation
Mucosa biopsy tissues from distal ileum and sigmoid were used for intestinal mucosal expression of interlukin-10 (IL10), interlukin-12 (IL12), and tight junction proteins Zona occludens 1 (ZO1) using immunohistochemistry.
Systematic inflammatory tone was assessed by measuring the ratio of IL10/IL12 in both serum and supernatant of peripheral blood mononuclear cells (PBMCs) culturing through Enzyme-Linked Immunosorbent Assay (ELISA) (EBioscience, Human IL10 Platinum ELISA Kit; Human IL12/IL23 p40 Platinum ELISA Kit, Vienna, Austria).[20] Further details can be found in the Supplementary Methods.
Gut microbiota analysis
Details for DNA extraction, PCR amplification, and sequencing and processing of sequencing data are described in the Supplementary Methods.
Alpha diversity indices were calculated in Mothur (version 1.30.1, https://www.mothur.org/wiki/Download_mothur) and compared using Student's t test. The rarefaction curves were plotted in R (version 3.6.0). Partial Least Squares Discriminant Analysis (PLS-DA) was analyzed and plotted using the mixOmics package in R. The ternary plot was performed in GGTERN (http://www.ggtern.com/) software. Difference analyses among groups in each level were performed using Kruskal–Wallis test.
All correlations were analyzed using Spearman's coefficient with pairwise comparisons. The correlations between genera and clinical indicator were visualized using Pheatmap package in R. The correlations between top 50 genera were exported for downstream analysis using Cytoscape (version 3.8.0) software (https://cytoscape.org/) to generate networks, with P < 0.05 and |r| ≥ 0.6 considered significant.
Statistical analysis
Statistical analysis was performed on SPSS 17.0 (SPSS Inc., Chicago, IL, USA) and R using public and inhouse packages. Comparisons of parametric data between more than two groups were performed using one-way analysis of variance (ANOVA), and otherwise, the Mann-Whitney U test was used. Non-parametric data were compared by chi-squared test. All tests were corrected for multiple testing using Benjamini–Hochberg method. P < 0.05, with false delivery rate corrected, was considered statistically significant (unless specified otherwise).
Results
IBSP and IBSN were similar with regard most clinical features except for fat-to-energy ratio
In total, 176 subjects were enrolled, including 49 BT-negative HC (33 males, aged 32.28 ± 11.33 years) and 127 IBS-D patients (92 males, aged 32.61 ± 9.68 years). In patients with IBS-D, 51 were BT-positive (IBSP group; 36 males, aged 30.76 ± 8.94 years) and 76 were BT-negative (IBSN group; 56 males, aged 33.84 ± 10.02 years); the BT-positive rate was 40%. The body mass index (BMI) in IBSP group was significantly lower than that in IBSN group (21.66 ± 3.60 vs. 23.38 ± 3.75 kg/m2, P = 0.01, t = 3.19). For FFQ Supplementary Table 1, more subjects in IBSP were found to be on a high-fat diet compared with IBSN (37% vs. 22%), and their diet contributes to a significant increasing of fat-to-energy ratio when compared with IBSN.
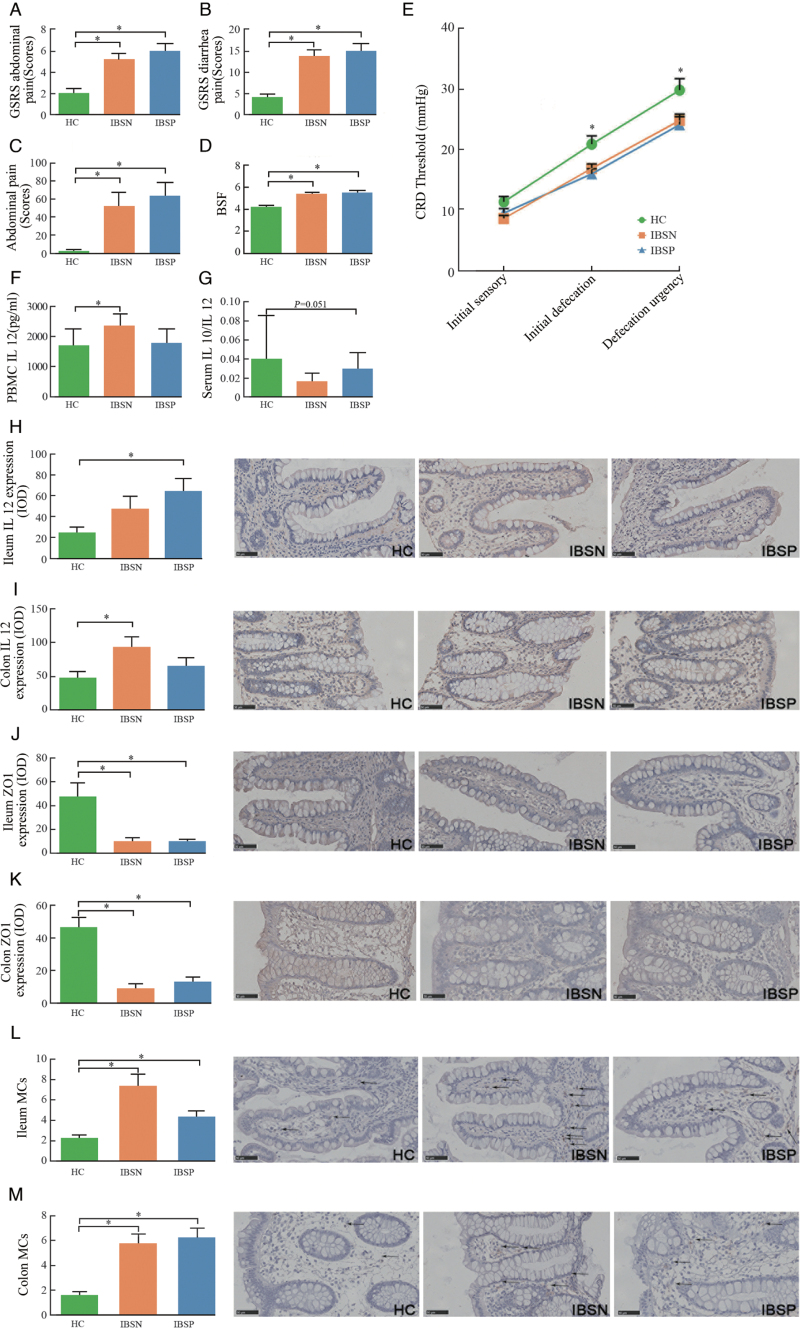
Clinical symptoms were evaluated using IBS symptom severity scale (IBS-SSS) and GSRS for the three groups [Figure 1 and Table 1]. In comparison with HC group, both IBSN and IBSP presented a significant increase in clinical symptom scores, such as abdominal pain and diarrhea [Figure 1A–C]. Watery stools were distinguished in IBSP and IBSN groups according to BSF scores [Figure 1D]. No significant difference was observed between IBSN and IBSP groups according to IBS-SSS and GSRS scores.
Figure 1.
Clinical symptom evaluations and inflammation factors among the three groups. (A,B) Abdominal pain and diarrhea scores evaluated by GSRS; (C) Abdominal pain score evaluated by IBS-SSS; (D) BSF scores; (E) CRD test; ∗IBSN vs. HC, Mann-Whitney U test, P < 0.05; (F) IL12 in PBMCs culturing supernatant; (G) Ratio of IL10/IL12 in serum; (H,I) IL12 expression in ileum and colon; (J,K) ZO1 expression in ileum and colon; (L,M) MCs counts in ileum and colon, shows by arrows. ∗IBSP or IBSN vs. HC, Mann-Whitney U test, P < 0.05. GSRS: Gastrointestinal Symptom Rating Scale; IBS-SSS: IBS Symptom Severity Scores; BSF: Bristol stool form; CRD: Colon rectal distension; PBMCs: Peripheral blood mononuclear cells; IL10: Interlukin-10; IL12: Interlukin-12; ZO-1: Zona occludens 1; MCs: Mast cells; HC: Healthy control; IBS: irritable bowel syndrome; IBSP: breath test positive IBS patients; IBSN: breath test negative IBS patients; Bars: 50 μm.
Table 1.
Clinical symptoms and inflammation factor analysis among healthy controls and IBS-D patients.
| Items | HC (n = 49) | IBSN (n = 76) | IBSP (n = 51) | P value |
| IBS-SSS | ||||
| Abdominal pain | 0.00 (0.00, 0.00) | 50.00 (0.00, 81.25)∗ | 50.00 (33.75, 80.00)† | 0.001 |
| Pain frequency | 0 (0.00, 0.00) | 30.00 (30.00, 50.00)∗ | 30.00 (10.00, 40.00)† | 0.001 |
| Abdominal bloating | 0.00 (0.00, 0.00) | 30.00 (0.00, 30.00)∗ | 27.50 (0.00, 26.50)† | 0.001 |
| Dissatisfaction with bowel movement | 20.00 (0.00, 70.00) | 70.00 (30.00, 70.00)∗ | 70.00 (40.00, 80.00)† | 0.001 |
| Disturbance to life | 0.00 (0.00, 20.00) | 70.00 (40.00, 70.00)∗ | 68.00 (40.00, 70.00)† | 0.001 |
| GSRS | ||||
| Abdominal pain | 2 (2, 2) | 5.00 (4, 6)∗ | 6.00 (5.00, 7.75)† | 0.001 |
| Abdominal bloating | 4.00 (3.00, 4.00) | 8.00 (5.00, 10.75)∗ | 9.00 (5.00, 11.00)† | 0.001 |
| Diarrhea | 4.00 (4.00, 5.00) | 13.00 (9.00, 18.00)∗ | 16.00 (9.25, 19.75)† | 0.001 |
| Constipation | 2.00 (2.00, 2.00) | 2.00 (2.00, 3.00) | 3.00 (2.00, 4.00)† | 0.002 |
| Satiety | 2.00 (2.00, 2.00) | 3.00 (2.00, 5.00)∗ | 5.50 (3.25, 8.00)† | 0.001 |
| Inflammation factors | ||||
| Serum IL10 | 1.40 (0.94, 3.17) | 1.34 (0.99, 2.02) | 1.70 (1.12, 2.40) | 0.120 |
| Serum IL12 | 162.80 (113.10, 250.20) | 212.10 (111.10, 335.30) | 151.20 (85.69, 222.20) | 0.130 |
| Serum IL10/IL12 | 0.01 (0.01, 0.03) | 0.01 (0, 0.02)∗ | 0.01 (0.01, 0.02) | 0.050 |
| PBMC IL10 | 46.39 (19.43, 90.47) | 38.96 (15.70, 76.88) | 33.41 (10.00, 108.80) | 0.930 |
| PBMC IL12 | 1380.00 (781.70, 1862.0) | 2282.00 (1163.00, 3096.00)∗ | 1508.00 (769.40, 2516.00) | 0.070 |
| PBMC IL10/IL12 | 0.03 (0.01, 0.05) | 0.02 (0.01, 0.04) | 0.02 (0.01, 0.06) | 0.330 |
Data were described as median (25% percentile, 75% percentile). P value: ANOVA test.
IBSN vs. HC, M-W U test, P < 0.05.
IBSP vs. HC, M-W U test, P < 0.05. ANOVA: Analysis of variance; M-W U test: Mann-Whitney U test; GSRS: Gastrointestinal Symptom Rating Scale; HC: Healthy control; IBS: Irritable bowel syndrome; IBSP: breath test positive IBS patients; IBSN: breath test negative IBS patients; IL10: Interlukin-10; IL12: Interlukin-12; PBMC: Peripheral blood mononuclear cell.
In CRD test, subjects in IBSN and IBSP showed a visceral hypersensitivity compared with HC, which presents as a significant decrease in thresholds for initial defecation, and defecation urgency [Figure 1E]. Additionally, no significant difference was observed in visceral sensitivity between the IBSP and IBSN groups.
IBSN presented a low-grade inflammation
The levels of IL10 and IL12 were used to evaluate the inflammatory condition. When compared with HC group, serum IL12 tended to elevate and IL10 tended to decrease in the IBSN group [Table 1], and thus the ratio of serum IL10/IL12 decreased (P = 0.051; Figure 1G). The IL12 level in PBMC supernatant was significantly higher in IBSN than that in HC (P < 0.05; Figure 1F). Neither the level of IL10 nor that of IL12 presented a significant change between IBSP and HC.
Tissue expression for IL12 differed from IBSP or IBSN to HC group. The level of IL12 increased significantly in IBSP group in ileum, whereas in colon, it showed a significant increase in IBSN group [Figure 1H and 1I].
The level of tight junction protein ZO1, which is associated with gut barrier function, was observed to be significantly reduced in both the ileum and colon in IBSP and IBSN groups when compared with HC group (P < 0.001; Figure 1J and 1K).
Both IBSN and IBSP groups had a higher count of mast cells in the ileum and colon (P < 0.05, separately) when compared with HC groups [Figure 1N and 1M].
No significant difference was found between IBSP and IBSN among those markers.
Different fecal microbiota features in IBSN and IBSP groups
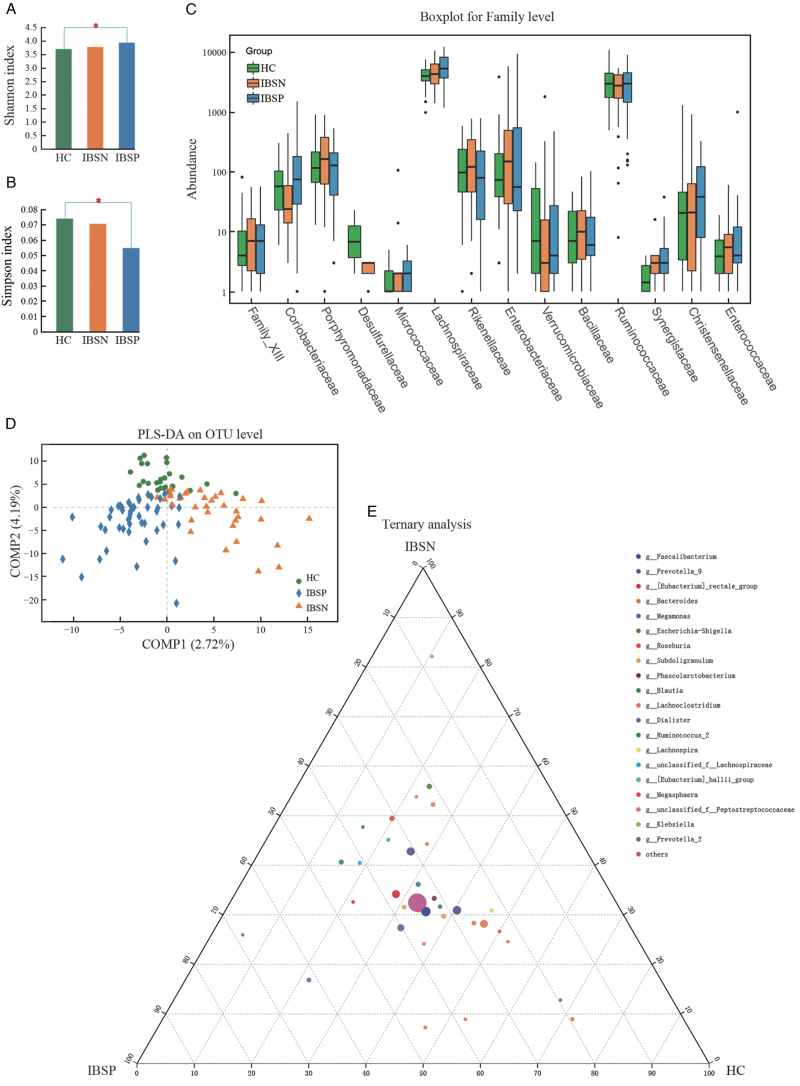
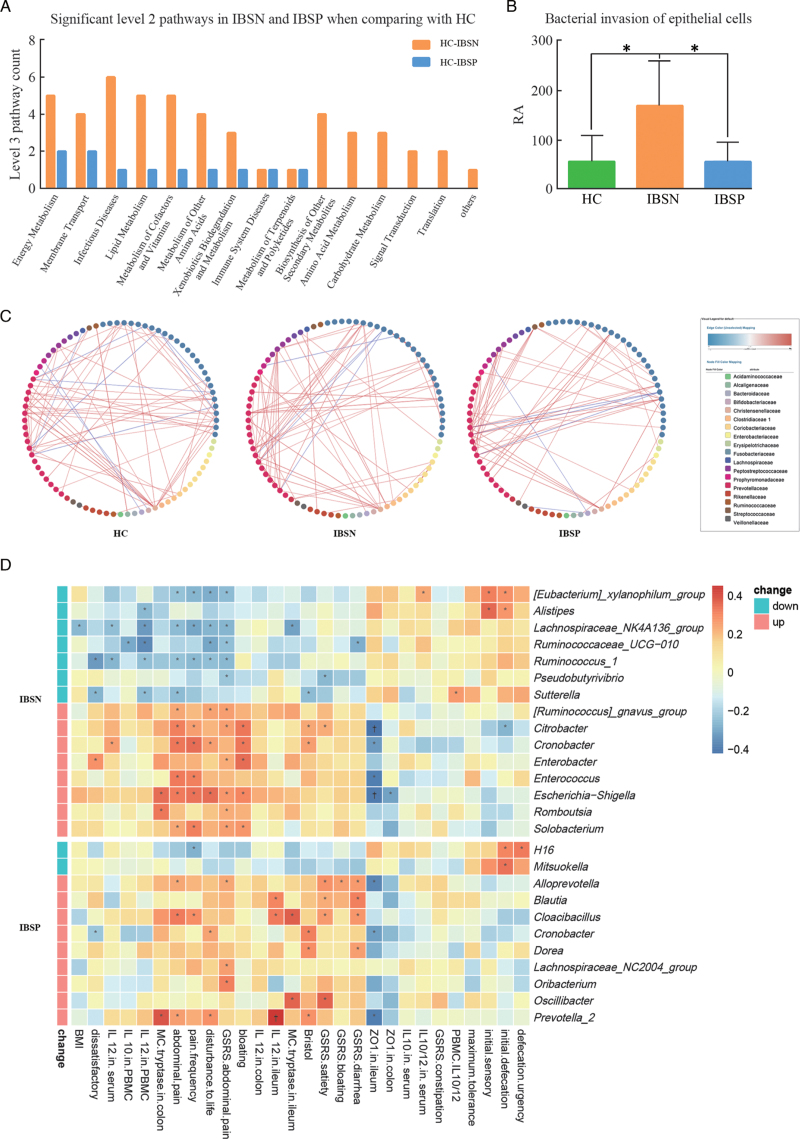
A total of 6,440,193 16S rRNA sequences were obtained from the V3 to V4 regions. The rarefaction curve indicated that a reasonable number of sequence samples were obtained Supplementary Figure 2A and 2B. The community diversity [Figure 2A and 2B] and partial least squares discriminant analyses (PLS-DA; Figure 2D) indicated a compositional distinction of microbiota in IBSN, IBSP, and HC. IBSP had the highest abundance of gut microbes, presenting with the highest Shannon index and the lowest Simpson index among the three groups. There was significant difference among three groups in the beta-diversity evaluated using analysis of similarity (R = 0.0551, P = 0.005). The phylum Proteobacteria enriched significantly in IBSN group compared to HC and IBSP groups Supplementary Figure 2C and 2D. The abundance of Enterobacteriaceae, Enterococcaceae, and Lachnospiraceae are increased, and Coriobacteriaceae, Desulfurellaceae and Verrucomicrobiaceae are decreased significantly at the family level in the IBSN group compared with HC group [Figure 2C]. The phylum Tenericutes was significantly enriched in the IBSP group compared with HC and IBSN groups. A higher abundance of Synergistaceae and lower Desulfurellaceae at the family level were found in the IBSP group compared with HC group. Overall, at the genus level, more differences were identified among the three groups [Figure 2E and Supplementary Table 2]. We found significant enrichments of Escherichia-Shigella, Blautia, Klebsiella, Enterococcus, Citrobacter, Enterobacter, and Cronobacter (top five relative abundance [RA]) in IBSN group compared with HC group, but depletion of Lachnospiraceae NK4A136 group, Alistipes, and Ruminococcus_1 in IBSN group compared with HC group. Meanwhile, higher abundance of Blautia, Prevotella_2, Lachnospiraceae NC2004 group, Cronobacter, and Romboutsia, along with lower abundance of Mitsuokella, were found in IBSP group compared with HC group. Further analysis was applied to explore the differences in microbiota between IBSN and IBSP groups. The abundance of Ruminococcaceae_UCG-002, Parabacteroides, Ruminococcus_1, Butyricimonas, Lachnospiraceae_UCG-010, and Odoribacter (top five relative abundance [RA]) was significantly higher in IBSP group, while some pathogens such as Lachnoclostridium, Escherichia-Shigella, Klebsiella, Enterococcus, and Cronobacter (top five RA) were significantly higher in IBSN group. We used Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) to infer the metagenomic function based on the microbial community profiles obtained from the 16S rRNA gene sequences. A comparison among the three groups is shown in Supplementary Table 3. We present a significant level 2 pathway for IBSN and IBSP groups in Figure 4A, where the ordinate represents the number of level 3 pathway belonging to level 2 pathway. In the IBSN group, the predicted Kyoto Encyclopaedia of Genes and Genomes (KEGG) pathways were significantly enriched in infectious diseases, energy metabolism, membrane transport, lipid metabolism, and metabolism of cofactors and vitamins [Figure 3A]. The enriched pathways in IBSP group were the same as those in IBSN group. In level 3 pathways, bacterial invasion of epithelial cells pathway was significantly increased in IBSN group compared with HC (P = 0.002) and IBSP groups (P = 0.001; Figure 3B). Besides, lipopolysaccharide (LPS) biosynthesis proteins and geraniol degradation were significantly decreased in the IBSP group compared to HC group (P = 0.003), and it did not show a remarkable difference between IBSN and HC groups Supplementary Table 3. Microbial inner correlation in IBSP, IBSN, and HC groups is shown in Figure 3C, which suggests the interaction networks among the different fecal microbes. Microbial interaction in HC group was more complex than that in IBSN and IBSP; the most active microbes were Prevotellaceae, Lachnospiraceae, Bifidobacteriaceae, and Bacteroidaceae. In the IBSN and IBSP groups, their microbial inner correlation was less active and limited to one or two families such as Prevotellaceae and Fusobacteriaceae. To determine the association of microbiota and disease, the RA of the genera and all parameters were considered for the correlation analyses [Figure 3D]. Overall, the enriched genera in the IBSN group were positively associated with IBS-SSS and GSRS scores, and negatively correlated with CRD tolerance. On the other hand, those depleted genera in the IBSN group presented controversial correlation. In particular, enriched genera in the IBSN group, especially the Enterobacteriaceae family (Enterobacter, Cronobacter, Citrobacter, Escherichia, Shigella), were significantly positively correlated with IL12 level (in serum or in PBMC supernatant). Sutterella was positively correlated with IL10/IL12 in the PBMC supernatant.
Figure 2.
Different microbiota profiles in IBSN and IBSP. (A) Shannon index analysis, ∗P < 0.05; (B) Simpson index analysis, ∗P < 0.05; (C) Microbial analysis in family level (the family presented are significantly different among the three groups, Kruskal–Wallis test, P < 0.05); (D) PLSDA analysis on OTU level; (E) Ternary analysis in genus level; PLS-DA: Partial least squares discriminant analysis; HC: healthy controls; IBS: irritable bowel syndrome; IBSP: breath test positive IBS patients; IBSN: breath test negative IBS patients.
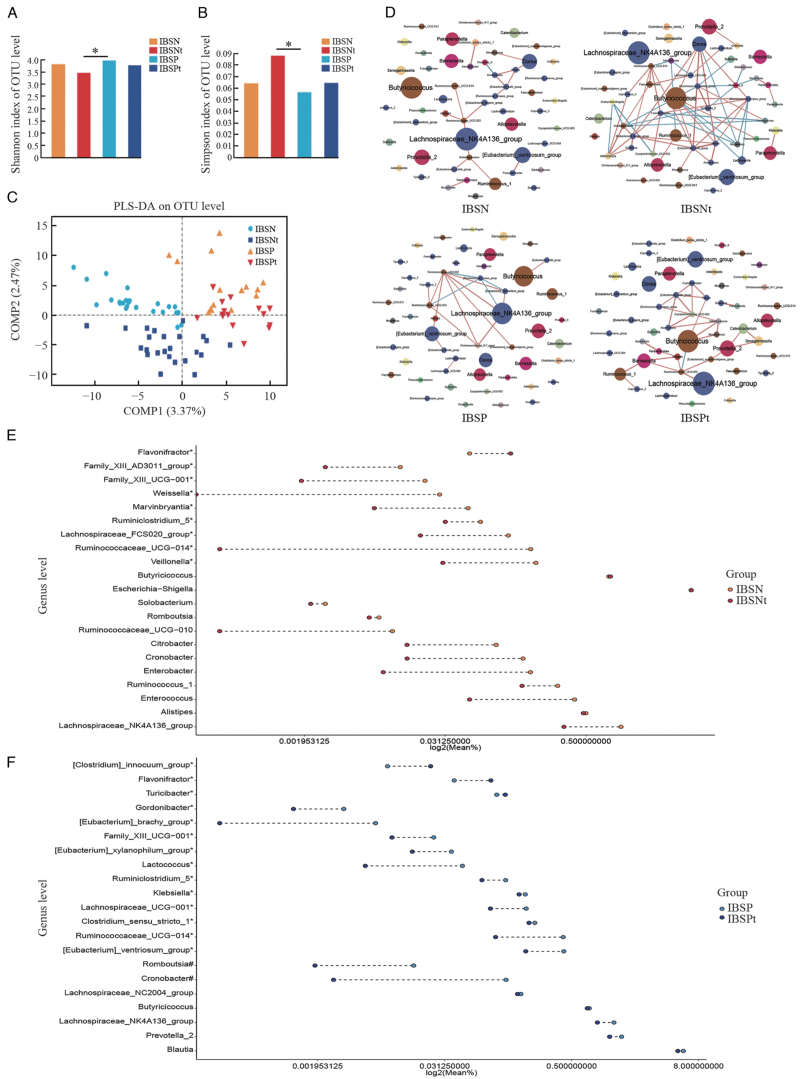
Figure 4.
Microbiota profile changes before and after therapy. (A) Shannon index analysis, ∗P < 0.05; (B) Simpson index analysis, ∗P < 0.05; (C) PLS-DA analysis on OTU level; (D) Microbial interaction network of significant changing genus before and after therapy (Dot color: different phylum level; dot diameter: RA; red line: positive correlation; green line: negative correlation; line color shade: correlation coefficient); (E,F) Significantly changed genus in IBSN and IBSP groups after rifaximin therapy; each dot represents the median RA for genus, ∗significant different genus for IBSN or IBSP in comparison with HC; #significant different genus for IBSP in comparison with HC and significant changes after rifaximin therapy. HC: Healthy control; IBS: irritable bowel syndrome; IBSP: breath test positive IBS patients; IBSPt: IBSP after rifaximin treatment; IBSN: breath test negative IBS patients; IBSNt: IBSN after rifaximin treatment; PLS-DA: Partial least squares discriminant analysis; RA: Relative abundance.
Figure 3.
Microbial function prediction, inner associations, and correlation of microbiota with clinical parameters. (A) Significant level 2 pathways in IBSN and IBSP in comparison with HC. Y-axis: the number or level 3 pathways belonging to level 2; (B) Comparison of the bacterial invasion of epithelial cells pathway in the three groups (∗P < 0.05), (C) Associations among different genus in HC, IBSN, or IBSP; (D) Correlation of significant changing microbiota in IBSN and IBSP with clinical and experimental parameters: (∗P < 0.05; †P < 0.01). HC: Healthy control; IBS: irritable bowel syndrome; IBSP: breath test positive IBS patients; IBSN: breath test negative IBS patients; RA: Relative abundance.
Rifaximin therapy
Clinical symptoms improvement after rifaximin treatment
After rifaximin treatment for 4 weeks, 15 patients in the IBSN group (IBSNt) and 24 patients in IBSP group were recalled and re-donated stool samples. The GI symptoms for treated patients were relieved to varying degrees. The scores for abdominal pain, dissatisfaction with bowel movement, and disturbance to life in IBS-SSS, scores for abdominal pain and diarrhea in GSRS, and BSF scores decreased significantly after treatment in IBSP group (P < 0.05, separately) [Table 2]. On the other hand, in IBSNt patients, only the IBS-SSS abdominal pain scores significantly decreased (P = 0.001); and 0.41% BT-positive patients turned BT-negative after treatment.
Table 2.
Clinical symptoms after rifaximin treatment of the IBS patients.
| Clinical symptoms | IBSN (n = 76) | IBSNt (n = 15) | IBSP (n = 51) | IBSPt (n = 24) | P1 | P2 |
| IBS-SSS | ||||||
| Abdominal pain | 59.11 ± 5.29 | 22.86 ± 6.71 | 62.5 ± 7.02 | 20.91 ± 5.79 | 0.001 | 0 |
| Pain frequency | 40.20 ± 4.64 | 28.57 ± 8.84 | 33.08 ± 4.86 | 26.36 ± 7.41 | 0.326 | 0.505 |
| Bloating | 25.82 ± 2.86 | 15.71 ± 7.82 | 24.50 ± 3.49 | 15.91 ± 5.17 | 0.257 | 0.235 |
| Dissatisfaction with bowel movement | 64.44 ± 10.56 | 39.44 ± 9.73 | 70.00 ± 5.43 | 41.92 ± 6.19 | 0.101 | 0.002 |
| Disturbance to life | 60.13 ± 2.73 | 49.29 ± 8.76 | 59.15 ± 3.22 | 46.36 ± 4.91 | 0.213 | 0.036 |
| Bristol | 5.30 ± 0.13 | 4.79 ± 0.39 | 5.48 ± 0.12 | 4.20 ± 0.13 | 0.111 | 0 |
| GSRS | ||||||
| Abdominal pain | 5.27 ± 0.22 | 4.29 ± 0.78 | 6.06 ± 0.33 | 4.50 ± 0.64 | 0.185 | 0.050 |
| Bloating | 7.81 ± 0.46 | 7.71 ± 1.13 | 8.54 ± 0.55 | 6.70 ± 0.91 | 0.949 | 0.156 |
| Diarrhea | 13.66 ± 0.75 | 11.71 ± 1.85 | 14.88 ± 0.87 | 9.50 ± 0.62 | 0.418 | 0 |
| Constipation | 2.72 ± 0.21 | 2.14 ± 0.14 | 3.46 ± 0.32 | 3.20 ± 0.47 | 0.387 | 0.726 |
| Satiety | 4.06 ± 0.32 | 3.86 ± 0.46 | 6.21 ± 0.51 | 4.70 ± 0.63 | 0.843 | 0.077 |
Data were described as mean ± standard deviation. P1: P value of IBSN vs. IBSNt; P2: P value of IBSP vs. IBSPt. GSRS: Gastrointestinal Symptom Rating Scale; IBS: Irritable bowel syndrome; IBSNt: Patients in IBSN after rifaximin treatment; IBSPt: Patients in IBSP after rifaximin treatment; IBS-SSS: IBS Symptom Severity Scores; IBSP: Breath test positive IBS patients; IBSN: Breath test negative IBS patients.
Different fecal microbial changes in IBSP and IBSN after rifaximin treatment
The alpha-diversity of bacterial communities (Shannon and Simpson indexes) decreased after therapy in both IBSN and IBSP groups [Figure 4A and 4B]. Following therapy, a compositional separation was observed by PLS-DA analysis in both IBSN and IBSP groups [Figure 4C]. There was no significant difference at the phylum level after therapy in both IBSN and IBSP groups by taxonomic analysis, and the same results were found for Firmicutes/Bacteroidetes (F/B) ratio. Analysis for the family level and genus level is shown in Supplementary Figure 3A and 3B. As for genus level, IBSN-enriched genera such as Escherichia-Shigella, Cronobacter, and Enterococcus did not change after therapy [Figure 4E, Supplementary Table 5. IBSP-enriched genera Romboutsia and Cronobacter significantly decreased after rifaximin treatment, and IBSP-depleted genera Alteromonas and Dyella increased after therapy [Figure 4F, Supplementary Table 5. Furthermore, IBSP-depleted Gordonibacter, Butyricimonas, and Parabacteroides increased after treatment.
In order to detect the influence of rifaximin therapy on the gut microbiota, genera were used to construct a microbial interaction network with correlation coefficient >0.6, as shown in Figure 4D. Rifaximin treatment changed the microbial inner correlation from Lachnospiraceae NK4A136 group-dominant to Butyricimonas-dominant in both IBSP and IBSN groups. Moreover, a stronger positive correlation was observed between Gemmiger and Odoribacter, Lactococcus and Enterococcus, and Butyricimonas and Parabacteroides, and a negative correlation was found between Flavonifractor and Dorea.
Discussion
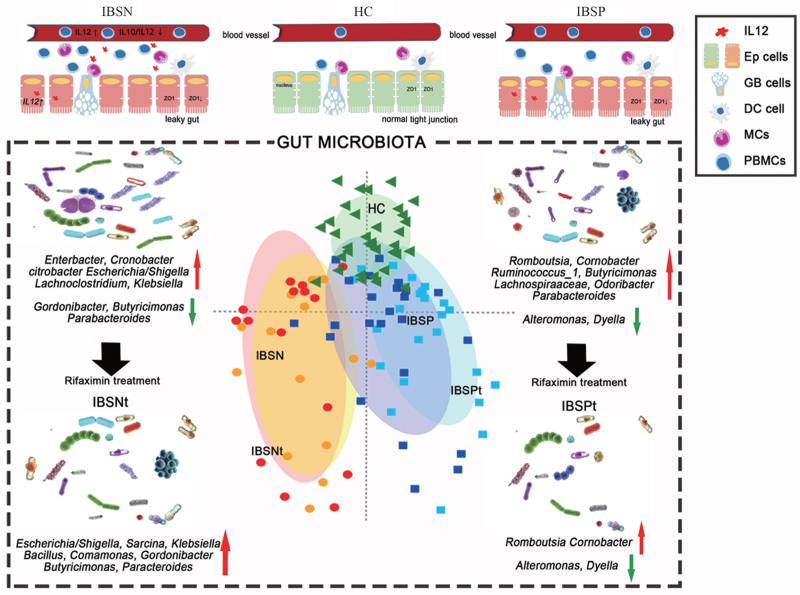
With the use of BT, SIBO has been increasingly detected in patients with IBS. Patients with SIBO and some IBS patients with BT positive respond well to rifaximin, but in some cases those with IBS alone respond modestly.[11,22,23] The underlying reasons are not clear. In this study, we identified that the inflammation features and gut microbiota are different between BT-positive and BT-negative IBS-D (IBSP and IBSN). Fecal microbial variation might contribute to the different therapeutic response to rifaximin in these two groups [Figure 5].
Figure 5.
The probable mechanism of different rifaximin therapeutic response in IBS-D overlapping with and without SIBO. HC: healthy controls; IBS: irritable bowel syndrome; IBSP: breath test positive IBS patients; IBSN: breath test negative IBS patients; SIBO: small intestinal bacterial overgrowth; IBSPt and IBSBt: Patients in IBSP and IBSN group after rifaximin treatment; Ep cells: Epithelium cells; GB cells: Gallbelt cells; DCs: Dendritic cells; MCs: Mast cells; PBMCs: Peripheral blood mononuclear cells.
It is challenging to distinguish whether IBS-D patients are BT positive or negative according to clinical symptoms (IBS-SSS and GSRS), or even using the visceral hypersensitivity status. These two groups were similar with regard most clinical features. However, we identified that systemic and colon inflammation were significantly activated in the BT-negative group presenting with a significant increase for IL12 level in PBMC and colon mucosa. The host gut microbiota contributes to the etiology and symptomology of IBS,[24,25] which are associated with increased epithelial permeability and aberrations in immunity. In our study, the microbiota of BT-negative patients presented a higher proportion of Proteobacteria, a phylum consisting of many pathogenic bacteria. Proteobacteria enriching in the gut can represent an imbalanced and unstable microbial community structure or a state of disease of the host, such as obesity, inflammation, and cancer.[26] Specifically, enriched abundance of Enterobacteriaceae (Enterobacter, Citrobacter, Escherichia-Shigella, Kluyvera), Enterococcaceae, and Lachnospiraceae, most of which are gram-negative, were observed in IBSN group. The Enterobacteriaceae family is well known as enteric pathogens[27,28] and Lachnospiraceae are opportunistic pathogens[29] of the gut. The enrichment in these florae might lead to bacterial invasion of epithelial cells. The enriched genera in BT-negative patients, especially those belonging to the Enterobacteriaceae family (Enterobacter, Cronobacter, Citrobacter, Escherichia-Shigella), were significantly positively correlated with IL12 level (in serum or in PBMC supernatant). Sutterella was positively correlated with IL10/IL12 ratio in the PBMC supernatant. These bacteria are able to activate the immune system.[30] The systemic and local inflammatory processes induced by these bacteria can be activated by different pathways.[31–33] For example, gram-negative bacteria and their bacterial components, especially LPS, induce inflammation responses depending on the release of proinflammatory cytokines from monocytes and macrophages, while other bacteria become aggressive in the crypts, and then are recognized and engulfed by macrophages and dendritic cells or by transcytosis through M cells and active Peyer's patches. These processes contribute a low-degree inflammation status to BT-negative patients. Interestingly, the level of IL12 was increased especially in the colon tissue, which demonstrates that the main local inflammation occurred in the colon rather than small intestine.
Some studies[21,34–36] suggest that SIBO may aggravate the inflammation status in patients with obesity, Crohn's disease, cirrhosis, or even Parkinson's disease. Interestingly, in this study, for IBS-D patients in BT-positive group, their ileum rather than systemic immunity is activated. Our data show that the IL12 level in the ileum was significantly raised in the BT-positive group. For their microbial changes, patients had a higher abundance of Blautia, Prevotella_2, Lachnospiraceae_NC2004 group, Cronobacter, and Romboutsia, and lower abundance of Mitsuokella. This result is consistent with the findings of Wu et al.[6] They found a remarkable increase in Prevotella in patients with IBS-D, especially those who were BT-positive. Moreover, we found that the microbial changes may influence LPS biosynthesis and geraniol degradation as per PICRUSt prediction. The increased abundance of Blautia, Prevotella_2, and Cloacibacillus were positively correlated with IL12 level in the ileum.
In clinical work, it is of great significance to distinguish rifaximin responders from patients with IBS for precision medicine. Two clinical trials[8,37] have shown that rifaximin had a better therapeutic effect on patients with IBS and SIBO in adults and children; however, symptoms of some IBS patients[38] relapse after rifaximin treatment. As an oral GI-targeted antibiotic, rifaximin treatment decreases the total bacterial population in gut. Microbial dysbiosis seems to be the key factor in the efficacy of rifaximin. Our results are consistent with the report that BT-positive patients respond better to rifaximin treatment.[11,22,23] It is shown that in BT-positive group Cronobacter decreased significantly, while Butyricimonas increased after treatment. Cronobacter[39] is associated with outbreaks of life-threatening infections in neonates, which can invade human intestinal cells, replicate in macrophages, and invade the blood–brain barrier. In-vitro studies[40,41] have shown that Cronobacter attachment to and invasion of mammalian intestinal cells, macrophage survival, and serum resistance are comparable with those of Enterobacter cloacae and Citrobacter freundii but are less than those of Salmonella typhimurium. Butyricimonas is a typical butyrate-producing bacterium.[42] Additional sodium butyrate intake through microencapsulation was reported to reduce the frequency of abdominal pain in patients with IBS, which confirms the protective effect of butyrate on IBS symptoms.[43]Butyricimonas might have a specific effect in GI diseases and this effect was reduced immediately after oral probiotic intake. We considered that rifaximin therapy was useful to BT-positive IBS-D patients by reducing the microbial community, suppression of harmful bacteria such as Cronobacter, and regulating the microflora for maintenance of the microecological equilibrium in the gut.
After 4 weeks of rifaximin therapy, enriched genera such as Enterobacter and Enterococcus in BT-negative IBS-D patients did not decrease significantly, and other IBSN-enriched microbes such as Flavonifractor even increased after therapy. As mentioned above, these bacteria are enteric pathogens that lead to intestinal and systemic inflammation and VH. This might be the reason for negligible recovery of symptoms in BT-negative group. We strictly excluded those participants who had a history of GI infection over the past 3 years. However, there is a lack of studies that explore the cause of increase of bacteria belonging to the antimicrobial spectrum of rifaximin after therapy. We hypothesize that antibiotic resistance may have developed in those bacteria, that rifaximin is more efficacious against antagonistic bacteria, or that longer therapeutic time is needed.
There are some limitations that need to be addressed in future studies. First, more patients must be studied to verify the microbial results after therapy. Second, the inflammation status was represented by IL10 and IL12 in this study, but more inflammatory factors or inflammatory cells must be evaluated. Finally, long-term rifaximin therapy (>12 weeks) or diet management must be included in the evaluation, so that the changes in the microbiota can be tracked over time.
In summary, IBS-D patients who were BT-positive presented a different gut microbial composition compared with BT-negative patients. The former also responded better to rifaximin therapy. Thus, it is necessary to use BT to exclude SIBO before IBS diagnosis for precision medicine. Some patients with IBS-D may have responded poorly to rifaximin owing to the absence of obvious changes of Enterobacteriaceae after therapy. However, the detailed mechanism requires to be further analyzed.
Funding
The studies were supported by the National Natural Science Foundation of China (No. 81670491) and the Michigan Medicine-PKUHSC Joint Institute for Translational and Clinical Research (No. BMU20140478).
Conflicts of interest
None.
Supplementary Material
Footnotes
How to cite this article: Liu Z, Zhu S, He M, Li M, Wei H, Zhang L, Sun Q, Jia Q, Hu N, Fang Y, Song L, Zhou C, Tao H, Kao JY, Zhu H, Owyang C, Duan L. Patients with breath test positive are necessary to be identified from irritable bowel syndrome: a clinical trial based on microbiomics and rifaximin sensitivity. Chin Med J 2022;135:1716–1727. doi: 10.1097/CM9.0000000000002294
Zuojing Liu, Shiwei Zhu, and Meibo He contributed equally to this study.
Supplemental digital content is available for this article.
References
- 1.Lovell RM, Ford AC. Global prevalence of and risk factors for irritable bowel syndrome: a meta-analysis. Clin Gastroenterol Hepatol 2012; 10:712–721. doi: 10.1016/j.cgh.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 2.Zhang L, Duan L, Liu Y, Leng Y, Zhang H, Liu Z, et al. A meta-analysis of the prevalence and risk factors of irritable bowel syndrome in Chinese community (in Chinese). Chin J Intern Med 2014; 53:969–975. doi: 10.3760/cma.j.issn.0578-1426.2014.12.011. [PubMed] [Google Scholar]
- 3.Shah A, Talley NJ, Jones M, Kendall BJ, Koloski N, Walker MM, et al. Small intestinal bacterial overgrowth in irritable bowel syndrome: a systematic review and meta-analysis of case-control studies. Am J Gastroenterol 2020; 115:190–201. doi: 10.14309/ajg.0000000000000504. [DOI] [PubMed] [Google Scholar]
- 4.Pimentel M, Saad RJ, Long MD, Rao SSC. ACG clinical guideline: small intestinal bacterial overgrowth. Am J Gastroenterol 2020; 115:165–178. doi: 10.14309/ajg.0000000000000501. [DOI] [PubMed] [Google Scholar]
- 5.Rezaie A, Heimanson Z, McCallum R, Pimentel M. Lactulose breath testing as a predictor of response to rifaximin in patients with irritable bowel syndrome with diarrhea. Am J Gastroenterol 2019; 114:1886–1893. doi: 10.14309/ajg.0000000000000444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu KQ, Sun WJ, Li N, Chen YQ, Wei YL, Chen DF. Small intestinal bacterial overgrowth is associated with diarrhea-predominant irritable bowel syndrome by increasing mainly prevotella abundance. Scand J Gastroenterol 2019; 54:1419–1425. doi: 10.1080/00365521.2019.1694067. [DOI] [PubMed] [Google Scholar]
- 7.Tuteja AK, Talley NJ, Stoddard GJ, Verne GN. Double-blind placebo-controlled study of rifaximin and lactulose hydrogen breath test in gulf war veterans with irritable bowel syndrome. Dig Dis Sci 2019; 64:838–845. doi: 10.1007/s10620-018-5344-5. [DOI] [PubMed] [Google Scholar]
- 8.Lembo A, Pimentel M, Rao SS, Schoenfeld P, Cash B, Weinstock LB, et al. Repeat treatment with rifaximin is safe and effective in patients with diarrhea-predominant irritable bowel syndrome. Gastroenterology 2016; 151:1113–1121. doi: 10.1053/j.gastro.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 9.Ford AC, Harris LA, Lacy BE, Quigley E, Moayyedi P. Systematic review with meta-analysis: the efficacy of prebiotics, probiotics, synbiotics and antibiotics in irritable bowel syndrome. Aliment Pharmacol Ther 2018; 48:1044–1060. doi: 10.1111/apt.15001. [DOI] [PubMed] [Google Scholar]
- 10.Shah ED, Saini SD, Chey WD. Value-based pricing for rifaximin increases access of patients with irritable bowel syndrome with diarrhea to therapy. Clin Gastroenterol Hepatol 2019; 17:2687–2695. doi: 10.1016/j.cgh.2019.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu ZJ, Wei H, Duan LP, Zhu SW, Zhang L, Wang K. Clinical features of irritable bowel syndrome with small intestinal bacterial overgrowth and a preliminary study of effectiveness of rifaximin (in Chinese). Natl Med J China 2016; 96:1896–1902. doi: 10.3760/cma.j.issn.0376-2491.2016.24.005. [DOI] [PubMed] [Google Scholar]
- 12.Barkin JA, Keihanian T, Barkin JS, Antequera CM, Moshiree B. Preferential usage of rifaximin for the treatment of hydrogen-positive smallintestinal bacterial overgrowth. Rev Gastroenterol Peru 2019; 39:111–115. doi: 10.14309/00000434-201610001-01074. [PubMed] [Google Scholar]
- 13.Yang J, Lee HR, Low K, Chatterjee S, Pimentel M. Rifaximin versus other antibiotics in the primary treatment and retreatment of bacterial overgrowth in IBS. Dig Dis Sci 2008; 53:169–174. doi: 10.1007/s10620-007-9839-8. [DOI] [PubMed] [Google Scholar]
- 14.Allegretti JR, Kassam Z, Chan WW. Small intestinal bacterial overgrowth: should screening be included in the pre-fecal microbiota transplantation evaluation? Dig Dis Sci 2018; 63:193–197. doi: 10.1007/s10620-017-4864-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Black CJ, Ford AC. Predicting response to rifaximin in irritable bowel syndrome with diarrhea: is the answer blowing in the wind? Gastroenterology 2020; 158:1508–1510. doi: 10.1053/j.gastro.2020.02.018. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Zhang L, Wang X, Wang Z, Zhang J, Jiang R, et al. Similar fecal microbiota signatures in patients with diarrhea-predominant irritable bowel syndrome and patients with depression. Clin Gastroenterol Hepatol 2016; 14:1602–1611. doi: 10.1016/j.cgh.2016.05.033. [DOI] [PubMed] [Google Scholar]
- 17.Duan R, Zhu S, Wang B, Duan L. Alterations of gut microbiota in patients with irritable bowel syndrome based on 16S rRNA-targeted sequencing: a systematic review. Clin Transl Gastroenterol 2019; 10:e12.doi: 10.14309/ctg.0000000000000012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chung CS, Chang PF, Liao CH, Lee TH, Chen Y, Lee YC, et al. Differences of microbiota in small bowel and faeces between irritable bowel syndrome patients and healthy subjects. Scand J Gastroenterol 2016; 51:410–419. doi: 10.3109/00365521.2015.1116107. [DOI] [PubMed] [Google Scholar]
- 19.Dlugosz A, Winckler B, Lundin E, Zakikhany K, Sandström G, Ye W, et al. No difference in small bowel microbiota between patients with irritable bowel syndrome and healthy controls. Sci Rep 2015; 5:8508.doi: 10.1038/srep08508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee KJ, Tack J. Altered intestinal microbiota in irritable bowel syndrome. Neurogastroenterol Motil 2010; 22:493–498. doi: 10.1111/j.1365-2982.2010.01482.x. [DOI] [PubMed] [Google Scholar]
- 21.Roland BC, Lee D, Miller LS, Vegesna A, Yolken R, Severance E, et al. Obesity increases the risk of small intestinal bacterial overgrowth (SIBO). Neurogastroenterol Motil 2018; 30.doi: 10.1111/nmo.13199. Epub 2017 Sep 21. [DOI] [PubMed] [Google Scholar]
- 22.Rangan V, Ballou S, Shin A, Camilleri M. Beth Israel Deaconess Medical Center GI Motility Working Group, Lembo A. Use of treatments for irritable bowel syndrome and patient satisfaction based on the IBS in America survey. Gastroenterology 2020; 158: 786.e-788.e. doi: 10.1053/j.gastro.2019.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pimentel M. Review of rifaximin as treatment for SIBO and IBS. Expert Opin Investig Drugs 2009; 18:349–358. doi: 10.1517/13543780902780175. [DOI] [PubMed] [Google Scholar]
- 24.Holvoet T, Joossens M, Vázquez-Castellanos JF, Christiaens E, Heyerick L, Boelens J, et al. Fecal microbiota transplantation reduces symptoms in some patients with irritable bowel syndrome with predominant abdominal bloating: short- and long-term results from a placebo-controlled randomized trial. Gastroenterology 2021; 160:145.e–157.e. doi: 10.1053/j.gastro.2020.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Jia Q, Zhang L, Zhang J, Pei F, Zhu S, Sun Q, et al. Fecal microbiota of diarrhea-predominant irritable bowel syndrome patients causes hepatic inflammation of germ-free rats and berberine reverses it partially. Biomed Res Int 2019; 2019:4530203.doi: 10.1155/2019/4530203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salem AE, Singh R, Ayoub YK, Khairy AM, Mullin GE. The gut microbiome and irritable bowel syndrome: state of art review. Arab J Gastroenterol 2018; 19:136–141. doi: 10.1016/j.ajg.2018.02.008. [DOI] [PubMed] [Google Scholar]
- 27.Serhan CN, Hong S, Gronert K, Colgan SP, Devchand PR, Mirick G, et al. Resolvins: A family of bioactive products of omega-3 fatty acid transformation circuits initiated by aspirin treatment that counter proinflammation signals. J Exp Med 2002; 196:1025–1037. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu B, Werlin EC, Chen M, Mottola G, Chatterjee A, Lance KD, et al. Perivascular delivery of resolvin D1 inhibits neointimal hyperplasia in a rabbit vein graft model. J Vasc Surg 2018; 68:188S–200S. doi: 10.1016/j.jvs.2018.05.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tam VC. Lipidomic profiling of bioactive lipids by mass spectrometry during microbial infections. Semin Immunol 2013; 25:240–248. doi: 10.1016/j.smim.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Broz P, Ohlson MB, Monack DM. Innate immune response to Salmonella typhimurium, a model enteric pathogen. Gut Microbes 2014; 3:62–70. doi: 10.4161/gmic.19141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guadamuro L, Delgado S, Redruello B, Flórez AB, Suárez A, Martínez-Camblor P, et al. Equol status and changes in fecal microbiota in menopausal women receiving long-term treatment for menopause symptoms with a soy-isoflavone concentrate. Front Microbiol 2015; 6:777.doi: 10.3389/fmicb.2015.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alexander M, Turnbaugh PJ. Deconstructing mechanisms of diet-microbiome-immune interactions. Immunity 2020; 53:264–276. doi: 10.1016/j.immuni.2020.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liwinski T, Zheng D, Elinav E. The microbiome and cytosolic innate immune receptors. Immunol Rev 2020; 297:207–224. doi: 10.1111/imr.12901. [DOI] [PubMed] [Google Scholar]
- 34.Pande C, Kumar A, Sarin SK. Small-intestinal bacterial overgrowth in cirrhosis is related to the severity of liver disease. Aliment Pharmacol Ther 2009; 29:1273–1281. doi: 10.1111/j.1365-2036.2009.03994.x. [DOI] [PubMed] [Google Scholar]
- 35.Yang C, Guo X, Wang J, Fan H, Huo X, Dong L, et al. Relationship between small intestinal bacterial overgrowth and peripheral blood ET, TLR2 and TLR4 in ulcerative colitis. J Coll Physicians Surg Pak 2020; 30:245–249. doi: 10.29271/jcpsp.2020.03.245. [DOI] [PubMed] [Google Scholar]
- 36.Losurdo G, Salvatore DF, Indellicati G, Lillo C, Ierardi E, Di Leo A. The influence of small intestinal bacterial overgrowth in digestive and extra-intestinal disorders. Int J Mol Sci 2020; 21:3531.doi: 10.3390/ijms21103531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fodor AA, Pimentel M, Chey WD, Lembo A, Golden PL, Israel RJ, et al. Rifaximin is associated with modest, transient decreases in multiple taxa in the gut microbiota of patients with diarrhoea-predominant irritable bowel syndrome. Gut Microbes 2018; 10:1–28. doi: 10.1038/sj.bjp.0706882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao J, Gillilland M, Owyang C. Rifaximin, gut microbes and mucosal inflammation: unraveling a complex relationship. Gut Microbes 2014; 5:571–575. doi: 10.4161/gmic.32130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forsythe SJ. Updates on the Cronobacter genus. Annu Rev Food Sci Technol 2018; 9:23–44. doi: 10.1146/annurev-food-030117-012246. [DOI] [PubMed] [Google Scholar]
- 40.Serafini F, Strati F, Ruas-Madiedo P, Turroni F, Foroni E, Duranti S, et al. Evaluation of adhesion properties and antibacterial activities of the infant gut commensal Bifidobacterium bifidum PRL2010. Anaerobe 2013; 21:9–17. doi: 10.1016/j.anaerobe.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 41.Emami CN, Mittal R, Wang L, Ford HR, Prasadarao NV. Recruitment of dendritic cells is responsible for intestinal epithelial damage in the pathogenesis of necrotizing enterocolitis by Cronobacter sakazakii. J Immunol 2011; 186:7067–7079. doi: 10.4049/jimmunol.1100108. [DOI] [PubMed] [Google Scholar]
- 42.Woting A, Clavel T, Loh G, Blaut M. Bacterial transformation of dietary lignans in gnotobiotic rats. FEMS Microbiol Ecol 2010; 72:507–514. doi: 10.1111/j.1574-6941.2010.00863.x. [DOI] [PubMed] [Google Scholar]
- 43.Jiminez JA, Uwiera TC, Abbott DW, Uwiera R, Inglis GD. Butyrate supplementation at high concentrations alters enteric bacterial communities and reduces intestinal inflammation in mice infected with Citrobacter rodentium. mSphere 2017; 2:e243–e217. doi: 10.1128/mSphere.00243-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.