Abstract
Background.
Club cell secretory protein (CC16) exerts anti-inflammatory functions in lung disease. We sought to determine the relation of serum CC16 deficits and genetic variants that control serum CC16 to lung function among children with cystic fibrosis (CF).
Methods.
We used longitudinal data from CF children (EPIC Study) with no positive cultures for Pseudomonas aeruginosa prior to enrollment. Circulating levels of CC16 and an inflammatory score (generated from CRP, SAA, calprotectin, G-CSF) were compared between participants with the lowest and highest FEV1 levels in adolescence (LLF and HLF groups, respectively; N=130-per-group). Single nucleotide variants (SNVs) in the SCGB1A1, EHF-APIP loci were tested for association with circulating CC16 and with decline of FEV1 and FEV1/FVC % predicted levels between ages 7–16 using mixed models.
Results.
Compared with the HLF group, the LLF group had lower levels of CC16 (geometric means: 8.2 vs 6.5 ng/ml, respectively; p=0.0002) and higher levels of the normalized inflammatory score (−0.21 vs 0.21, p=0.0007). Participants in the lowest CC16 and highest inflammation tertile had the highest odds for having LLF (p<0.0001 for comparison with participants in the highest CC16 and lowest inflammation tertile). Among seven SNVs associated with circulating CC16, the top SNV rs3741240 was associated with decline of FEV1/FVC and, marginally, FEV1 (p=0.003 and 0.025, respectively; N=611 participants, 20,801 lung function observations).
Conclusions.
Serum CC16 deficits are strongly associated with severity of CF lung disease and their effects are additive with systemic inflammation. The rs3741240A allele is associated with low circulating CC16 and, possibly, accelerated lung function decline in CF.
Keywords: Club cell secretory protein, cystic fibrosis, lung function
Introduction
Cystic fibrosis (CF) is an autosomal recessive condition due to pathogenic variants in cystic fibrosis transmembrane conductance regulator (CFTR). CF is associated with dysfunction of multiple organs (e.g., lung, pancreas, intestine, liver), but the primary cause of morbidity and mortality is progressive obstructive lung disease due in large part to chronic endobronchial inflammation. However, there is wide variation in lung disease severity among individuals with the same CFTR genotype and the genes, proteins and pathways mediating the inflammatory response are ideal potential candidate modifiers.
Club cell secretory protein (CC16, also known as CC10 and CCSP) is a homodimeric pneumoprotein encoded by SCGB1A1. CC16 is produced mainly by club cells and other epithelial cells in distal airways, but it can be readily measured in circulation(1–3). The biological functions of CC16 and the molecular pathways that mediate its functions have not been established. However, experimental studies support a critical role for CC16 in preventing structural lung alterations and remodeling(4) and in reducing airway inflammation in response to oxidative stress and infections(5). Accordingly, CC16 deficits in blood and airways have been associated with the prevalence, severity, and progression of obstructive lung diseases such as asthma and chronic obstructive pulmonary disease (COPD)(6–14).
The few studies exploring the role of CC16 in CF lung disease have found significant airway CC16 deficits in CF patients(15) that correlate with airway inflammation(16) and disease severity(17). However, none of these studies evaluated circulating CC16 (a biomarker that can be measured in readily available blood samples), or whether the association of low levels of CC16 with lung disease progression is augmented in the presence of higher levels of inflammation. Moreover, a genome-wide association study (GWAS)(18) identified multiple protein quantitative trait loci (pQTLs) for serum CC16 levels in two genomic regions on chromosome 11, one near SCGB1A1 and one 25 Mb away near EHF-APIP, the latter of which is also near some of the top GWAS hits for lung function in CF(19, 20).
We hypothesized that CF lung disease severity is associated with low serum CC16 and with genetic variants associated with serum CC16 levels.
Methods
Study Population
The Early Pseudomonas Infection Control Observational Study(21) (hereafter EPIC) was a multicenter, longitudinal observational cohort study designed to investigate risk factors for Pseudomonas aeruginosa (Pa) acquisition and to characterize the clinical course of disease in young children with CF. Children age ≤ 12 years with an established diagnosis of CF were eligible if they met one of two criteria: they had either no prior respiratory cultures positive for Pa (Criterion 1) or at least a 2-year history of Pa negative cultures following a history of a positive culture for Pa (Criterion 2). Between 2004 and 2006, 1704 children with CF were enrolled at 59 U.S. CF Centers(22) (mean age at enrollment [SD]: 5.7 [3.6] years), the majority of whom were recruited under Criterion 1 (65%)(21). Demographic and questionnaire data (including maternal smoking, maternal education, and household income) were collected prospectively via the US Cystic Fibrosis Foundation Patient Registry and spirometric tests were completed at each clinical encounter (typically quarterly) up to 2016, for an average of 10.6 years of follow-up per subject (SD 3.3 years). Blood was collected and serum banked annually, independent of clinical status. However, for this study we did not use any serum samples collected during exacerbations, which were defined by the use of intravenous antibiotic treatment in the hospital or at home, as captured from the CF Registry(23). Informed consent was obtained from all parents/guardians and IRB approval at each participating site.
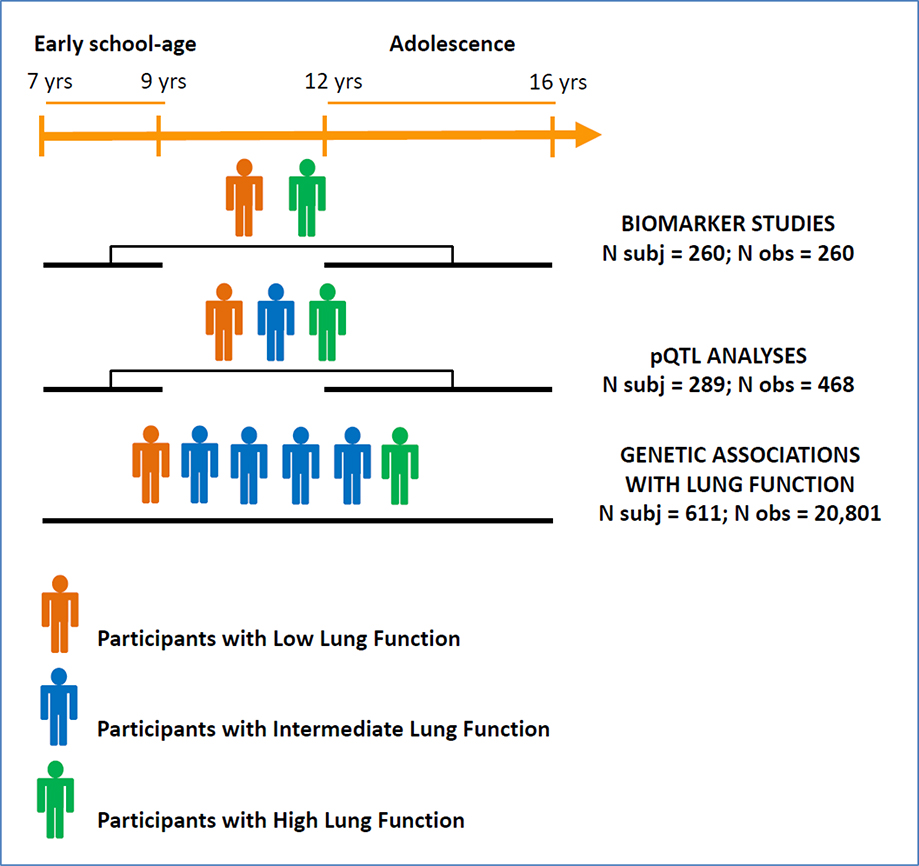
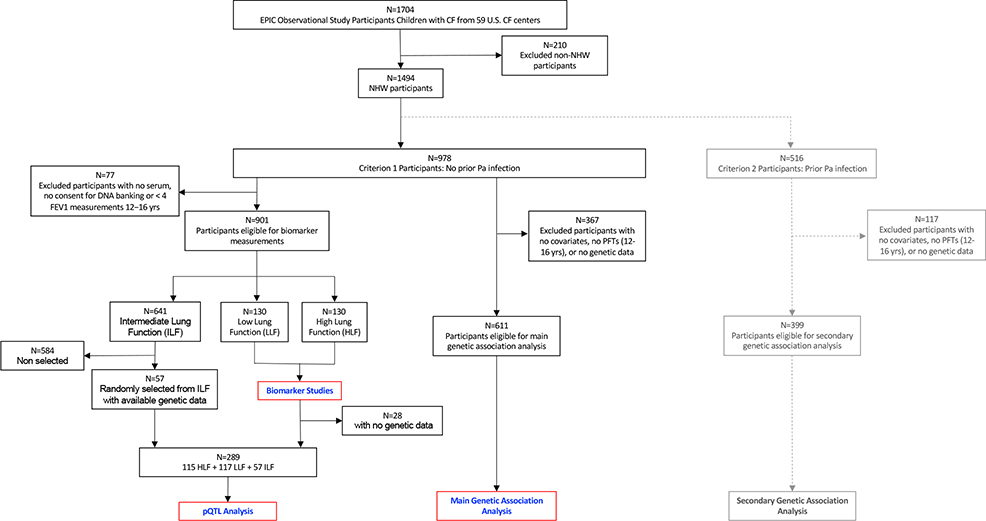
The experimental design of the biomarker and genetic studies is summarized in Figure 1a. Figure 1b summarizes selection of participants for each of the analyses included in the present study. Overall, 88% of EPIC participants were non-Hispanic white (NHW). To reduce phenotypic and genetic modifier heterogeneity, we limited main analyses to NHW participants enrolled under Criterion 1. In secondary analyses, significant genetic associations were also tested among NHW participants enrolled under Criterion 2.
Figure 1a.
Study design
pQTL: protein quantitative trait loci
N obs for Biomarker Studies/pQTL analyses = number of samples analyzed for biomarkers
N obs for Genetic Associations with Lung Function = number of prospective lung function measurements
Figure 1b.
Consort diagram for the different components of the current study.
Biomarker Studies
Case-control study design for biomarker studies.
For biomarker studies, to contain costs of the molecular assays while preserving statistical power we used a nested case-control study design in which cases and controls were selected as participants with the lowest and highest FEV1 % predicted, respectively, between ages 12–16 years (hereafter called “adolescence”). Eligibility criteria included availability of at least four FEV1 measurements for phenotype determination and at least one serum sample between ages 12–16 years. Phenotype determination was completed at the time of the earliest serum sample available between ages 12–16 (see online supplement for additional information on case/control selection). Of 901 participants meeting eligibility criteria, based on power computations we selected the 130 with the lowest (Low Lung Function group, “LLF”) and 130 with the highest (High Lung Function group, “HLF”) % predicted FEV1 based on Global Lung Initiative equations(24), representing 29% of the eligible population.
Of these 260 participants, 131 (50%) had a serum sample available also between age 7–9 years (hereafter “early school-age”). This age interval was chosen to investigate to what extent deficits in CC16 and lung function in adolescence have their origins by early school age.
Molecular assays for serum CC16 and inflammatory biomarkers.
Serum CC16 levels were measured in samples from both early school-age and adolescence at the Asthma and Airway Disease Research Center Immunology Lab at the University of Arizona using a commercially available ELISA kit (BioVendor, Asheville, NC and Modrice, Czech Republic).
Serum levels of four inflammatory biomarkers previously associated with CF lung disease severity(25) were measured in samples from adolescence in the Pediatric Clinical Translational Research Center Core Laboratory at Children’s Hospital Colorado and University of Colorado Anschutz Medical Campus (Aurora, CO), which serves as the Center for Biochemical Markers for the CF Foundation. Proteins were measured using validated commercially available assays: high sensitivity C reactive protein (hsCRP; Siemens Nephelometer assay, Siemens Healthcare, Tarrytown, NY), serum amyloid A (SAA; Milliplex MAP® for Luminex Technology, EMD Millipore, St. Charles, MO), calprotectin (ALPCO Diagnostics, Salem, NH), and granulocyte-colony stimulating factor (G-CSF; Milliplex MAP® for Luminex Technology, EMD Millipore, St. Charles, MO). The lower limits of detection for these assays were: hsCRP, 0.007 mg/L; SAA, 500 ng/mL; calprotectin, 0.4 μg/mL, and G-CSF, 14 pg/mL. All assays were performed in duplicate and mean values were used for analysis.
Statistical analyses for biomarker studies.
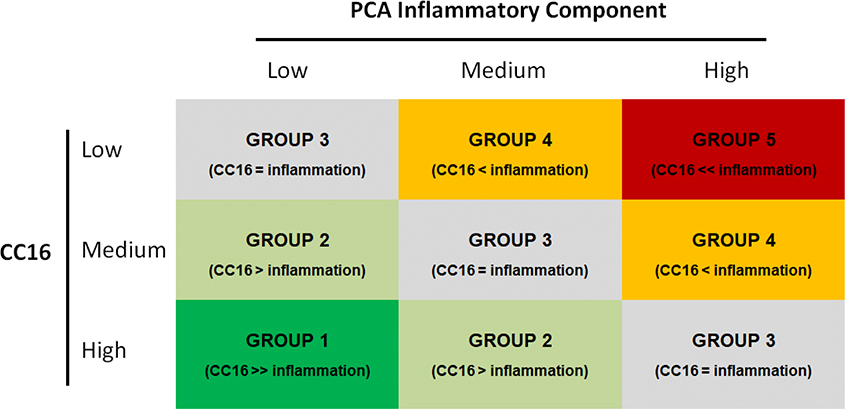
Association of serum CC16 and inflammatory biomarkers with CF lung disease severity was tested in multivariate logistic regressions that used LLF vs HLF at adolescence as the dependent variable and included sex, age, CFTR genotype risk group (minimal vs residual function)(26), maternal smoking during pregnancy, and annual household income as covariates. CC16 was tested both as standardized levels of log-transformed CC16 and as tertiles to evaluate both linear trends and effects of having low CC16 levels (i.e., lowest tertile). Because the four inflammatory biomarkers showed moderate to strong correlations with each other, a principal component analysis was conducted and the first component, which explained 65% of the total variance, was used as a general indicator of systemic inflammation. In order to identify combined effects of low CC16 and high systemic inflammation, we then categorized participants into five mutually exclusive groups based on the combination of tertiles of CC16 and tertiles of the inflammatory component in adolescence. The groups were ordered based on the balance between tertiles of the anti-inflammatory CC16 and of the inflammatory component as shown in Figure 2a, with the hypothesis that risk for low lung function would progressively increase from Group 1 (highest CC16 tertile and lowest inflammatory tertile) to Group 5 (lowest CC16 tertile and highest inflammatory tertile).
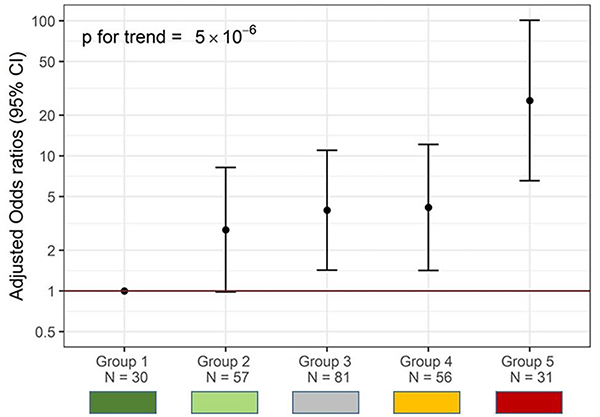
Figure 2a.
Categorization of participants into 5 groups based on tertiles of CC16 and tertiles of the PCA component of inflammatory biomarkers in adolescence.
The 5 groups are defined as:
- Group 1 (Dark Green, N=30): participants in the highest CC16 tertile and lowest inflammatory tertile
- Group 2 (Light Green, N=57): other participants with CC16 tertile higher than inflammatory tertile
- Group 3 (Gray, N=81): participants with CC16 tertile equal to inflammatory tertile
- Group 4 (Orange, N=56): participants with CC16 tertile lower than inflammatory tertile (excluding participants in Group 5)
- Group 5 (Red, N=31): participants in the lowest CC16 tertile and highest inflammatory tertile
Genetic Studies
Targeted single molecule molecular inversion probe sequencing.
Targeted single molecule molecular inversion probe (smMIP) sequencing was completed for two regions of chromosome 11 previously shown to harbor pQTLs of serum CC16 level(18): 11:62103403-62199817 near SCGB1A1 and 11:34699611-35005768 near EHF-APIP. Analyses were limited to 56 single nucleotide variants (SNVs) that had minor allele frequency (MAF) ≥ 5%, had <15% missing values, were previously validated in dbSNP, and were in Hardy-Weinberg equilibrium. Genetic studies included pQTL and genetic association analyses.
Statistical analyses for identification of pQTLs.
For pQTL analyses, we used data from 289 participants: 232 of the 260 participants included in the biomarker studies who were also genotyped plus an additional 57 randomly selected participants with Intermediate lung function in order to have a representation of individuals who did not meet the extreme criteria for either the LLF or HLF groups (Figure 1b). All 289 participants had serum CC16 data from adolescence and 179 of them also had serum CC16 levels available from early school-age, for a total of 468 observations (Figure 1a). To maximize statistical power, pQTL analyses were completed using mixed models that included serum CC16 levels from both early school-age and adolescence. To control for the intra-subject serial correlation of repeated CC16 observations, we used random effects models to test each of the SNVs for association with serum CC16. In sensitivity analyses, pQTL results were confirmed using weights(27) generated based on the relative frequencies of individuals in the Low, High, and Intermediate lung function groups to generate estimates of association between SNVs and serum CC16 levels representative of the entire EPIC cohort.
Genetic association analyses.
For genetic association analyses, we used the seven SNVs that, in the above pQTL analyses, reached significance based on a False Discover Rate (FDR) criterion (FDR ≤ 0.05) and survived LD-based pruning (see online supplement for more information). These variants were tested for association with decline of spirometric indices of airway obstruction (FEV1 % predicted and FEV1/FVC % predicted) between ages 7–16 years in random coefficient models that included all 611 NHW EPIC participants recruited under Criterion 1 with complete genetic and covariate data, for a total of 20,801 prospective lung function measurements. We applied FDR criteria also for these analyses on decline of FEV1 and FEV1/FVC. In these analyses, we included a genotype-by-age interaction in random coefficients models to test whether the decline of the specific lung function index of interest was significantly different for participants who were heterozygous and for participants who were homozygous for the effect allele, as compared with participants who were homozygous for the reference allele. Genetic variants that were significant in these models were also tested in the smaller group of 399 NHW participants who were enrolled under Criterion 2 to evaluate their potential effects among cases with more advanced CF lung disease. All models were adjusted for sex, age, CFTR genotype risk group, maternal smoking during pregnancy, and annual household income (additional information provided in the online supplement).
Results
Biomarker Studies
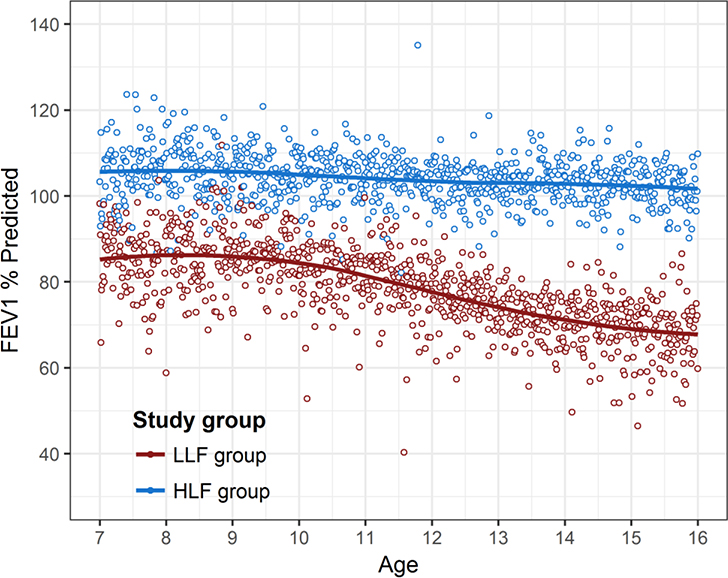
Table E1 summarizes the characteristics of the 260 participants in the LLF and HLF groups who were selected for the biomarker studies. The LLF group demonstrated more maternal smoking, lower annual household income, and lower lung function at both ages with greater deficits in adolescence. Levels of FEV1 % predicted in the LLF and HLF groups were well-separated at age 12 and, although differences in lung function were already partially present earlier in childhood, LLF participants had a significantly steeper decline of FEV1 % predicted between ages 7 to 16 than HLF participants (Figure 3).
Figure 3.
FEV1 measurements between ages 7–16 years among the 130 participants in the Low Lung Function (LLF) and 130 participants in the High Lung Function (HLF) groups selected for biomarker studies.
The solid lines represent locally weighted scatterplot smoothing (loess) lines. Loess is a non-parametric strategy for fitting smooth curves to data points without assuming a data distribution shape.
When tested in mixed models, LLF participants had a significantly steeper decline of FEV1 % predicted between ages 7–16 years than HLF participants (p<0.0001).
Serum CC16 deficits and severity of CF lung disease.
In adolescence, the LLF group had significantly lower levels of serum CC16 as compared with the HLF group (geometric means: 6.5 vs 8.2 ng/ml, respectively; p=0.0002) (Table 1). Serum CC16 differences between participants in the LLF vs HLF groups remained significant after adjustment for sex, age, CFTR genotype risk group, maternal smoking during pregnancy, and annual household income (adjusted OR [95% CI] for being in the LLF group associated with 1-SD increase in CC16: 0.53, 0.39–0.73, p=0.0001). Consistent with these results, as compared with participants in the highest CC16 tertile, after adjustment for covariates participants in the lowest CC16 tertile had faster declines of FEV1 and FEV1/FVC % predicted levels between ages 7–16 years (p=0.025 and 0.037, respectively) and greater than 3-fold increased odds of being in the LLF group (adjOR: 3.3, 1.7–6.4, p=0.0004). The adjOR of being in the LLF group associated with the medium CC16 tertile was 1.7 (0.9–3.2, p=0.099), with a significant linear trend across CC16 tertiles (p=0.0004). In line with these results, using a linear regression model with the same covariates as above, we also observed a significant inverse association between serum CC16 levels in adolescence and frequency of exacerbations (i.e., number of exacerbations recorded between ages 7 to 16 divided by the number of years of follow-up). Among the 140 participants who had at least one exacerbation, after adjustment for sex, age, CFTR genotype risk group, maternal smoking during pregnancy, and annual household income, individuals in the lowest CC16 tertile had on average 0.27 more exacerbations per year (0.02–0.52; p=0.033) than individuals in the highest tertile. In contrast, no differences were found for participants in the medium CC16 tertile.
Table 1.
Serum levels of CC16 and inflammatory biomarkers in adolescence among participants from the LLF and HLF groups.
| Low Lung Function LLF (N=130) | High Lung Function HLF (N=130) | P | unadjOR # (95% CI) | P | adjOR # * (95% CI) | P | ||
|---|---|---|---|---|---|---|---|---|
| Serum CC16, geom. mean (95% CI) in ng/ml | 6.5 (6.0, 7.2) | 8.2 (7.6, 8.9) | 0.0002 | 0.59 (0.45, 0.79) | 0.0003 | 0.53 (0.39, 0.73) | 0.0001 | |
| Serum CC16 tertiles: | 0.004 | |||||||
| Low CC16, N (%) | 55 (42.3%) | 32 (24.6%) | 2.76 (1.49, 5.11) | 0.001 | 3.30 (1.71, 6.37) | 0.0004 | ||
| Medium CC16, N (%) | 42 (32.3%) | 45 (34.6%) | 1.50 (0.82, 2.74) | 0.190 | 1.71 (0.90, 3.25) | 0.099 | ||
| High CC16, N (%) | 33 (25.4%) | 53 (40.8%) | Reference | -- | Reference | -- | ||
| Inflammatory Biomarkers | ||||||||
| Serum hsCRP, geom. mean (95% CI) in mg/L | 0.66 (0.49, 0.89) | 0.32 (0.25, 0.42) | 0.0003 | 1.60 (1.23, 2.08) | 0.0005 | 1.59 (1.21, 2.11) | 0.001 | |
| Serum SAA, geom. mean (95% CI) in μg/ml | 8.0 (5.7, 11.3)^ | 3.9 (3.1, 5.0) | 0.0008 | 1.55 (1.19, 2.01) | 0.001 | 1.59 (1.20, 2.10) | 0.001 | |
| Serum calprotectin, geom. mean (95% CI) in μg/ml | 2.7 (2.3, 3.2) | 1.8 (1.6, 2.1) | 0.0006 | 1.57 (1.20, 2.05) | 0.0009 | 1.67 (1.24, 2.23) | 0.0006 | |
| Serum G-CSF, geom. mean (95% CI) in pg/ml | 21.4 (17.9, 25.6)^^ | 15.1 (13.4, 17.0)^ | 0.001 | 1.51 (1.16, 1.96) | 0.002 | 1.67 (1.25, 2.23) | 0.0005 | |
| Normalized PCA inflammatory component, mean (95% CI) | 0.21 (−0.02, 0.44)^^ | −0.21 (−0.29, −0.13)^ | 0.0007 | 2.21 (1.33, 3.65) | 0.002 | 2.51 (1.45, 4.36) | 0.001 |
N=129
N=126
Results from logistic regression models with LLF vs HLF as the dependent variable. For biomarkers analyzed on a continuous scale, odds ratios are expressed for 1-SD increase in biomarker levels
Odds ratios adjusted for sex, age at blood collection, CFTR genotype risk group, maternal smoking during pregnancy, and annual household income.
Serum CC16 levels correlated between early school-age and adolescence (Spearman correlation coefficient: 0.67, p<0.0001), indicating tracking over time. Consistent with results from adolescence, CC16 levels were also lower in early school-age for participants in the LLF group than for those in the HLF group (geom. means: 6.8 vs 8.3 ng/ml, respectively; p=0.035). Similarly, being in the lowest CC16 tertile in early school-age was associated with a greater than 3-fold increased odds of having LLF in adolescence (adjOR: 3.6, 1.4–9.6, p=0.009). These results suggest that CC16 deficits in CF adolescents associated with low lung function may be partly established by school age.
Combined effects of CC16 deficits and systemic inflammation on severity of CF lung disease.
In adolescence, serum concentrations of the inflammatory biomarkers CRP, SAA, calprotectin, and G-CSF were all significantly higher in participants in the LLF compared to the HLF group (Table 1). These biomarkers showed moderate to strong correlations with each other with Spearman correlation coefficients between 0.42 and 0.78 (all p < 0.0001), while only SAA correlated significantly (although weakly) with CC16 (Figure E1). When a principal component analysis was conducted among the four inflammatory biomarkers, participants in the LLF group were found to have levels of the inflammatory principal component that were on average 0.42 SDs higher than participants in the HLF group (p=0.0007, Table 1). When we analyzed the combined effects of CC16 deficits and systemic inflammation as illustrated in Figure 2a, we observed that individuals in Group 5 (low CC16, high inflammation) had higher odds for having LLF than any of the other groups, with a strikingly increasing trend from Group 1 (high CC16, low inflammation) up to Group 5 (p for linear trend < 0.0001) (Figure 2b). In ROC analyses, the inclusion of this 5-group variable significantly increased the performance of a base model in classifying participants as LLF versus HLF, improving the area under the curve (AUC) from 0.649 to 0.740 (p=0.001) (Figure E2).
Figure 2b.
ORs for being in the Low Lung Function (LLF) group in adolescence across the five groups generated by the combination of CC16 and inflammatory tertiles. Group 1 (High CC16 tertile, Low inflammatory tertile) is the reference. Group 5 includes participants in the highest inflammatory and lowest CC16 tertiles.
ORs adjusted for sex, age at serum collection, CFTR genotype risk group, maternal smoking during pregnancy, and annual household income.
P values for the comparison of Group 5 with Group 1 (p<0.0001), Group 2 (p<0.0001), Group 3 (p=0.001), and Group 4 (p=0.002)
Genetic Studies
Effects of genetic variants on serum CC16 levels (pQTL Analyses).
We identified a set of 18 SNVs that were significantly associated with serum CC16 levels after FDR adjustment (Table E2). The associations of CC16 levels with all these SNVs, with the exception of rs17157266, were confirmed in secondary weighted analyses (see Methods), indicating that results were robust to the case-control study design. LD-based pruning reduced the number of independently segregating SNVs to 7 (Table 2). The top pQTL, rs3741240, explained up to 19% of the variability in CC16 levels, with the A allele being associated with lower serum CC16 levels (Figure E3).
Table 2.
The seven genetic variants significantly associated with circulating CC16 levels§ among 289 EPIC participants after FDR adjustment and LD-based pruning.
| Position | rs ID | Alt. Allele | Ref. Allele | MAF^ | Gene | Coef. (95% CI)* | P | FDR | R-squared |
|---|---|---|---|---|---|---|---|---|---|
| 62186542 | rs3741240 | A | G | 0.335 | SCGB1A1 | −0.634 (−0.778, −0.490) | 6.2 × 10−18 | 4.3 × 10−16 | 0.190 |
| 62197427 | rs2077224 | A | C | 0.357 | AHNAK | 0.315 (0.158, 0.471) | 8.3 × 10−5 | 0.002 | 0.048 |
| 62190700 | rs2302364 | C | T | 0.172 | SCGB1A1 | −0.411 (−0.616, −0.205) | 8.9 × 10−5 | 0.002 | 0.046 |
| 34937697 | rs2016814 | C | T | 0.203 | PDHX, APIP | −0.281 (−0.458, −0.104) | 0.002 | 0.022 | 0.028 |
| 34909926 | rs1571133 | G | T | 0.323 | APIP | −0.234 (−0.394, −0.074) | 0.004 | 0.031 | 0.023 |
| 34842506 | rs568529 | A | C | 0.295 | -- | −0.233 (−0.405, −0.060) | 0.008 | 0.038 | 0.019 |
| 34780278 | rs906902 | A | G | 0.393 | -- | −0.207 (−0.363, −0.051) | 0.009 | 0.039 | 0.016 |
Standardized levels of log-transformed CC16 were used in analyses
MAF: minor allele frequency
The coefficient indicates the change in SDs of CC16 levels associated with one-allele change of the specific genetic variant from additive models.
Associations between genetic variants and lung function decline (Genetic Association Analyses).
Genetic and lung function data were available for 611 EPIC participants recruited under Criterion 1. None of the 7 SNVs identified in pQTL analyses was, after FDR adjustment, significantly associated with changes in FEV1 % predicted between ages 7–16 years. However, rs3741240 was associated with significantly greater FEV1/FVC decline between these ages (Table 3). The FEV1/FVC ratio among carriers of the AA genotype declined on average 0.4% predicted per year faster than among carriers of the GG genotype (95% CI: 0.70%, −0.14%; p = 0.003; p[FDR] = 0.045). This effect was consistent with the observation that FEV1 % predicted levels of carriers of the AA genotype declined on average 0.6% per year faster than those of carriers of the GG genotype, although this association was only marginally significant and did not pass the FDR threshold. Trajectories of FEV1 and FEV1/FVC levels between ages 7–16 years across the rs3741240 genotypes are summarized in Figure E4.
Table 3.
Estimated effects of the 7 pQTL SNVs on changes in FEV1 and FEV1/FVC % predicted between ages 7–16 years. Models include 611 NHW EPIC participants recruited under Criterion 1 with 20,801 lung function measurements.
| rs ID | Estimated effects on FEV1 % predicted change between ages 7–16 yrs * | Estimated effects on FEV1/FVC % predicted change between ages 7–16 yrs * | |||
|---|---|---|---|---|---|
| Coefficient (95% CI) | p | Coefficient (95% CI) | p | ||
| 62186542 | rs3741240 | ||||
| GG | Ref | Ref | |||
| A/G | −0.08 (−0.40, 0.24) | 0.611 | −0.10 (−0.28, 0.08) | 0.285 | |
| AA | −0.57 (−1.07, −0.07) | 0.025 | −0.42 (−0.70, −0.14) | 0.003 | |
| 62197427 | rs2077224 | ||||
| CC | Ref | Ref | |||
| A/C | −0.06 (−0.38, 0.27) | 0.735 | 0.12 (−0.06, 0.30) | 0.200 | |
| AA | 0.05 (−0.46, 0.55) | 0.853 | 0.05 (−0.23, 0.33) | 0.735 | |
| 62190700 | rs2302364 | ||||
| TT | Ref | Ref | |||
| C/T | −0.31 (−0.64, 0.02) | 0.067 | −0.17 (−0.36, 0.01) | 0.067 | |
| CC | −0.60 (−1.69, 0.50) | 0.285 | −0.65 (−1.25, −0.04) | 0.037 | |
| 34937697 | rs2016814 | ||||
| TT | Ref | Ref | |||
| C/T | 0.37 (−0.41, 1.15) | 0.354 | −0.18 (−0.61, 0.26) | 0.419 | |
| CC | 0.39 (−0.37, 1.15) | 0.312 | −0.28 (−0.70, 0.14) | 0.187 | |
| 34909926 | rs1571133 | ||||
| TT | Ref | Ref | |||
| G/T | 0.72 (0.22, 1.23) | 0.005 | 0.03 (−0.26, 0.31) | 0.846 | |
| GG | 0.67 (0.17, 1.17) | 0.009 | −0.12 (−0.40, 0.16) | 0.410 | |
| 34842506 | rs568529 | ||||
| CC | Ref | Ref | |||
| A/C | −0.03 (−0.60, 0.54) | 0.924 | −0.14 (−0.46, 0.18) | 0.386 | |
| AA | 0.13 (−0.43, 0.69) | 0.650 | −0.04 (−0.36, 0.27) | 0.787 | |
| 34780278 | rs906902 | ||||
| GG | Ref | Ref | |||
| A/G | 0.17 (−0.17, 0.50) | 0.330 | 0.19 (0.004, 0.38) | 0.046 | |
| AA | 0.28 (−0.18, 0.74) | 0.231 | 0.02 (−0.23, 0.28) | 0.859 | |
Models adjusted for sex, age at lung function measurement, CFTR genotype risk group, maternal smoking during pregnancy, and annual household income.
Associations significant after FDR adjustment are reported in bold font. The FDR significance level is 0.0033.
Genetic and lung function data were available from an additional group of 399 EPIC participants recruited under Criterion 2 (i.e., who had isolation of Pa from respiratory cultures prior to enrollment). As expected, these participants differed from the main population of participants recruited under Criterion 1 in terms of both a higher proportion of high risk CFTR genotypes and more severe lung function impairment (Table E3). In this group of participants, rs3741240 had no effects on decline of percent predicted levels of FEV1 and FEV1/FVC (Table E4).
Discussion
We found serum CC16 deficits to be strongly associated with lung disease severity, assessed as decreased lung function, in children with CF. The impact of low serum CC16 was additive with that of systemic inflammation, as children who had both low levels of circulating CC16 and high levels of inflammatory biomarkers had the highest risk of lung function deficits. Consistent with previous GWAS studies(18), we found that SNVs on chromosome 11 nearest the genes SCGB1A1 and EHF-APIP were significantly associated with serum CC16 levels. The SNV rs3741240, which explained the largest amount of variability in serum CC16 levels, was associated with lung function worsening between ages 7–16 years, although this association was restricted to participants with no prior cultures positive for Pa.
To date, only a few studies have addressed airway CC16 in CF(15–17). However, no study has linked circulating CC16 with lung function in CF in the context of longitudinal data. We found marked deficits of serum CC16 among CF patients with low lung function. Indeed, inclusion in the lowest CC16 tertile increased the odds for having low lung function by more than three-fold. Similar to observations in individuals without CF(28), we also found significant tracking of serum CC16 levels between early school-age and adolescence (at mean ages of 8 and 14 years, respectively) and that CC16 deficits at both age intervals were consistently associated with low lung function in adolescence. These observations suggest that a trajectory of low CC16 levels may be already established early in life in these patients.
Airway and systemic inflammation are hallmarks of CF(29, 30) and, in line with previous research(25), we found the acute phase reactants CRP and SAA and the markers of neutrophilic inflammation calprotectin and G-CSF to be strongly associated with lung function deficits. Anti-inflammatory properties, inhibition of NF-κB activation, and reduction of lung infiltration by activated leukocytes are some of the postulated mechanisms by which CC16 may exert its protective effects in the lung(31–34). This is consistent with our finding that CF patients with a highly skewed inflammatory imbalance – i.e., who were in the lowest tertile for the anti-inflammatory protein CC16 and in the highest tertile for inflammatory biomarkers – had the greatest risk for low lung function.
In line with a previous GWAS study in individuals with COPD(18), we report the existence of pQTLs for circulating CC16 levels harbored around the encoding gene SCGB1A1 and the EHF-APIP genomic region in patients with CF. The variant rs3741240, located in the 5’ untranslated region of SCGB1A1, explained up to 19% of variability in circulating levels of CC16, with carriers of the AA genotype having mean CC16 levels in adolescence (Figure E3) that were nearly 50% lower than those we observed in healthy adolescents from our population-based Tucson Children’s Respiratory Study(28). Moreover, we found that the rs3741240 A allele was also associated with accelerated decline of FEV1/FVC (and marginally with accelerated decline of FEV1) % predicted levels between ages 7–16 years. Consistent with these results, this SNV was previously reported to be associated with decline of lung function among participants in the Lovelace Smokers cohort(35, 36). However, in our study rs3741240 had no association with lung function among participants who had prior isolation of Pa from respiratory cultures (i.e., enrolled under Criterion 2), a group of patients with a different distribution of CFTR risk genotypes and more severe lung disease. These results may indicate that genetic variation linked to circulating CC16 influences lung function in individuals with milder forms of CF lung disease perhaps before airway epithelial damage of club cells is established, as it may be the case for patients with severe disease and/or a positive history of prior Pseudomonal infections. Larger studies will be needed to elucidate conclusively these hypotheses as we did not have information on serum CC16 among Criterion 2 participants in our study.
CC16 may protect against severe CF lung disease through several mechanisms. Animal models indicate that CC16 deficient mice are susceptible to structural lung abnormalities(4), lung infiltration by activated leukocytes(34), and enhanced inflammatory responses to noxious exposures including respiratory infections(37). In line with these potential mechanisms, although exacerbations were not the primary outcome of interest for our study, we also observed that participants with low circulating CC16 had an increased frequency of exacerbations, which are in turn a known risk factor for accelerated decline of lung function in CF patients(38, 39). Further, we found combined effects of CC16 deficits and systemic inflammation on lung function deficits in our CF patients. Combining information from these biomarkers significantly improved the discriminatory ability of our model for concomitant lung function deficits in ROC analysis, although it should be noted that the predictive performance of these biomarkers is likely overestimated in a case-control study based on extreme phenotypes (i.e. groups with worst vs best lung function) like ours.
With the development of highly effective CFTR modulator treatment (HEMT), it is conceivable that the association of CC16 deficits with lung disease in CF would not be as strong due to reductions in airway inflammation. However, as survival improves and prevention of irreversible airflow limitation becomes a major goal of CF care, insights into potential disease modification through lung protective strategies will remain important, especially for the 7–10% of CF patients not currently eligible for HEMT therapy(40). Should the association between CC16 deficits and severe lung disease in CF be causal, the use of this protein could be evaluated in novel therapeutic interventions via either exogenous (e.g., administration of recombinant CC16(41)) or endogenous augmentation strategies. Among the latter, we have recently reported that retinoids increase CC16 secretion in human bronchial epithelial cells(42), an observation with potentially important implications in CF because circulating vitamin A is frequently low and inversely related to systemic inflammation in patients with CF(43).
Our study has some limitations. We evaluated concurrent associations between serum CC16 and severity of lung disease and did not test whether low CC16 can identify CF patients at risk for subsequent rapid progression of lung disease. However, the discovery of robust biomarkers of concomitant disease is a necessary first step leading to future longitudinal studies evaluating their possible use in risk prediction. We only included NHW participants in our analyses and cannot conclude whether our results are generalizable to other racial or ethnic groups. This limitation reduces the potential impact of our findings on other under-studied CF populations. Similarly, because we did not have an independent replication cohort and could not test how robust and reproducible genetic results are in other CF populations, our findings of an association between rs3741240 and lung function decline should be interpreted with caution and will require future validation. We focused our genetic analyses on the two loci (SCGB1A1 and EHF-APIP) that were known to harbor CC16 pQTLs at the time our study was designed. Although additional pQTLs have been recently identified(44), these two loci still capture the genetic variants known to exert the strongest effects on serum CC16 levels. Finally, although experimental(4) and Mendelian randomization(44) studies support a direct, protective role of CC16 in the lung, observational data like ours cannot provide conclusive evidence regarding whether the association of protein levels with disease is causal in nature. We cannot exclude, for example, that airway epithelial damage in CF patients is the main etiology of reduced production of CC16 and that deficits in CC16 levels are a consequence, rather than a cause, of lung function deficits in CF.
Among the strengths of our study are the availability of a large, multicenter, prospective cohort of CF patients across the US with genetic data and annual serum collection, the selection of participants with no previous Pa infection to reduce clinical heterogeneity, the extensive lung function data collected over a decade of follow-up, and the remarkable magnitude and consistency of biomarker associations.
The results of our study warrant further longitudinal evaluation of the role of CC16 deficits in CF lung disease and their potential clinical implications. If CC16 is causally linked to lung function deficits, exogenous or endogenous (e.g., via vitamin A) augmentation strategies could potentially serve as novel therapies for CF lung disease.
Supplementary Material
Highlights:
Deficits in circulating levels of club cell secretory protein (CC16) are strongly associated with severity of CF lung disease
Patients with low levels of CC16 and high level of systemic inflammation have the highest odds for having lung function deficits
The SCGB1A1 and EHF-APIP loci on chromosome 11 harbor multiple single nucleotide variants (SNVs) robustly associated with circulating levels of CC16
The SNV rs3741240, which explains the largest amount of variability in circulating CC16, may be associated with lung function decline in CF
Support:
This work was supported by award CFFT377201 from the Cystic Fibrosis Foundation Therapeutics and award AI135108 from the National Institute of Allergy and Infectious Diseases, US National Institutes of Health. The Pediatric CTRC Core Laboratory at Children’s Hospital Colorado and University of Colorado Anschutz Medical Campus (Aurora, CO) is supported by NIH/NCATS Colorado CTSA Grant UL1 TR002535.
The funding sources had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; and in the decision to submit the article for publication.
Footnotes
Conflict of Interest statement
MJE, EEB, KJB, MH, RLG, MR, SDS, MJB, WJM, and SG report receiving grant support through their institution for the present work. SDS reports receiving honoraria from DKBMed. WJM reports receiving consulting fees from the Cystic Fibrosis Foundation, receiving speaker honoraria from the American College of Chest Physicians, and participating on the CFF DSMB.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Broeckaert F, Bernard A. Clara cell secretory protein (CC16): characteristics and perspectives as lung peripheral biomarker. Clin Exp Allergy. 2000;30(4):469–75. [DOI] [PubMed] [Google Scholar]
- 2.Mukherjee AB, Zhang Z, Chilton BS. Uteroglobin: a steroid-inducible immunomodulatory protein that founded the Secretoglobin superfamily. Endocr Rev. 2007;28(7):707–25. [DOI] [PubMed] [Google Scholar]
- 3.Lakind JS, Holgate ST, Ownby DR, Mansur AH, Helms PJ, Pyatt D, et al. A critical review of the use of Clara cell secretory protein (CC16) as a biomarker of acute or chronic pulmonary effects. Biomarkers. 2007;12(5):445–67. [DOI] [PubMed] [Google Scholar]
- 4.Zhai J, Insel M, Addison KJ, Stern DA, Pederson W, Dy A, et al. Club Cell Secretory Protein Deficiency Leads to Altered Lung Function. Am J Respir Crit Care Med. 2019;199(3):302–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamez AS, Gras D, Petit A, Knabe L, Molinari N, Vachier I, et al. Supplementing defect in Club Cell Secretory Protein attenuates airway inflammation in COPD. Chest. 2014. [DOI] [PubMed] [Google Scholar]
- 6.Guerra S, Halonen M, Vasquez MM, Spangenberg A, Stern DA, Morgan WJ, et al. Relation between circulating CC16 concentrations, lung function, and development of chronic obstructive pulmonary disease across the lifespan: a prospective study. Lancet Respir Med. 2015;3(8):613–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vestbo J, Edwards LD, Scanlon PD, Yates JC, Agusti A, Bakke P, et al. Changes in forced expiratory volume in 1 second over time in COPD. N Engl J Med. 2011;365(13):1184–92. [DOI] [PubMed] [Google Scholar]
- 8.Park HY, Churg A, Wright JL, Li Y, Tam S, Man SF, et al. Club cell protein 16 and disease progression in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2013;188(12):1413–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lomas DA, Silverman EK, Edwards LD, Miller BE, Coxson HO, Tal-Singer R. Evaluation of serum CC-16 as a biomarker for COPD in the ECLIPSE cohort. Thorax. 2008;63(12):1058–63. [DOI] [PubMed] [Google Scholar]
- 10.Bernard A, Marchandise FX, Depelchin S, Lauwerys R, Sibille Y. Clara cell protein in serum and bronchoalveolar lavage. Eur Respir J. 1992;5(10):1231–8. [PubMed] [Google Scholar]
- 11.Pilette C, Godding V, Kiss R, Delos M, Verbeken E, Decaestecker C, et al. Reduced epithelial expression of secretory component in small airways correlates with airflow obstruction in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2001;163(1):185–94. [DOI] [PubMed] [Google Scholar]
- 12.Braido F, Riccio AM, Guerra L, Gamalero C, Zolezzi A, Tarantini F, et al. Clara cell 16 protein in COPD sputum: a marker of small airways damage? Respir Med. 2007;101(10):2119–24. [DOI] [PubMed] [Google Scholar]
- 13.Ye Q, Fujita M, Ouchi H, Inoshima I, Maeyama T, Kuwano K, et al. Serum CC-10 in inflammatory lung diseases. Respiration. 2004;71(5):505–10. [DOI] [PubMed] [Google Scholar]
- 14.Tsoumakidou M, Bouloukaki I, Thimaki K, Tzanakis N, Siafakas NM. Innate immunity proteins in chronic obstructive pulmonary disease and idiopathic pulmonary fibrosis. Exp Lung Res. 2010;36(6):373–80. [DOI] [PubMed] [Google Scholar]
- 15.Gray RD, MacGregor G, Noble D, Imrie M, Dewar M, Boyd AC, et al. Sputum proteomics in inflammatory and suppurative respiratory diseases. Am J Respir Crit Care Med. 2008;178(5):444–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Starosta V, Ratjen F, Rietschel E, Paul K, Griese M. Anti-inflammatory cytokines in cystic fibrosis lung disease. Eur Respir J. 2006;28(3):581–7. [DOI] [PubMed] [Google Scholar]
- 17.Laguna TA, Williams CB, Brandy KR, Welchlin-Bradford C, Moen CE, Reilly CS, et al. Sputum club cell protein concentration is associated with pulmonary exacerbation in cystic fibrosis. J Cyst Fibros. 2015;14(3):334–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim DK, Cho MH, Hersh CP, Lomas DA, Miller BE, Kong X, et al. Genome-wide association analysis of blood biomarkers in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012;186(12):1238–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright FA, Strug LJ, Doshi VK, Commander CW, Blackman SM, Sun L, et al. Genome-wide association and linkage identify modifier loci of lung disease severity in cystic fibrosis at 11p13 and 20q13.2. Nat Genet. 2011;43(6):539–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corvol H, Blackman SM, Boelle PY, Gallins PJ, Pace RG, Stonebraker JR, et al. Genome-wide association meta-analysis identifies five modifier loci of lung disease severity in cystic fibrosis. Nat Commun. 2015;6:8382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenfeld M, Emerson J, McNamara S, Joubran K, Retsch-Bogart G, Graff GR, et al. Baseline characteristics and factors associated with nutritional and pulmonary status at enrollment in the cystic fibrosis EPIC observational cohort. Pediatr Pulmonol. 2010;45(9):934–44. [DOI] [PubMed] [Google Scholar]
- 22.Treggiari MM, Rosenfeld M, Mayer-Hamblett N, Retsch-Bogart G, Gibson RL, Williams J, et al. Early anti-pseudomonal acquisition in young patients with cystic fibrosis: rationale and design of the EPIC clinical trial and observational study’. Contemp Clin Trials. 2009;30(3):256–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juarez-Colunga E, Rosenfeld M, Zemanick ET, Wagner B. Application of multiple event analysis as an alternative approach to studying pulmonary exacerbations as an outcome measure. J Cyst Fibros. 2020;19(1):114–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Quanjer PH, Stanojevic S, Cole TJ, Baur X, Hall GL, Culver BH, et al. Multi-ethnic reference values for spirometry for the 3–95-yr age range: the global lung function 2012 equations. Eur Respir J. 2012;40(6):1324–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sagel SD, Wagner BD, Ziady A, Kelley T, Clancy JP, Narvaez-Rivas M, et al. Utilizing centralized biorepository samples for biomarkers of cystic fibrosis lung disease severity. J Cyst Fibros. 2020;19(4):632–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Green DM, McDougal KE, Blackman SM, Sosnay PR, Henderson LB, Naughton KM, et al. Mutations that permit residual CFTR function delay acquisition of multiple respiratory pathogens in CF patients. Respir Res. 2010;11:140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reilly M, Torrang A, Klint A. Re-use of case-control data for analysis of new outcome variables. Stat Med. 2005;24(24):4009–19. [DOI] [PubMed] [Google Scholar]
- 28.Zhai J, Stern DA, Sherrill DL, Spangenberg AL, Wright AL, Morgan WJ, et al. Trajectories and Early Determinants of Circulating CC16 from Birth to Age 32 Years. Am J Respir Crit Care Med. 2018;198(2):267–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sagel SD, Chmiel JF, Konstan MW. Sputum biomarkers of inflammation in cystic fibrosis lung disease. Proc Am Thorac Soc. 2007;4(4):406–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sagel SD, Sontag MK, Wagener JS, Kapsner RK, Osberg I, Accurso FJ. Induced sputum inflammatory measures correlate with lung function in children with cystic fibrosis. J Pediatr. 2002;141(6):811–7. [DOI] [PubMed] [Google Scholar]
- 31.Laucho-Contreras ME, Polverino F, Gupta K, Taylor KL, Kelly E, Pinto-Plata V, et al. Protective role for club cell secretory protein-16 (CC16) in the development of COPD. Eur Respir J. 2015;45(6):1544–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pang M, Wang H, Bai JZ, Cao D, Jiang Y, Zhang C, et al. Recombinant rat CC16 protein inhibits LPS-induced MMP-9 expression via NF-kappaB pathway in rat tracheal epithelial cells. Exp Biol Med (Maywood). 2015;240(10):1266–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pang M, Liu HY, Li T, Wang D, Hu XY, Zhang XR, et al. Recombinant club cell protein 16 (CC16) ameliorates cigarette smokeinduced lung inflammation in a murine disease model of COPD. Mol Med Rep. 2018;18(2):2198–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Johnson MDL, Younis US, Menghani SV, Addison KJ, Whalen M, Pilon AL, et al. CC16 Binding to alpha4beta1 Integrin Protects against Mycoplasma pneumoniae Infection. Am J Respir Crit Care Med. 2021;203(11):1410–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen H, Leng S, Belinsky SA, Miller BE, Tal-Singer R, Owen CA, et al. Low plasma CC16 levels in smokers are associated with a higher risk for chronic bronchitis. Eur Respir J. 2015;46(5):1501–3. [DOI] [PubMed] [Google Scholar]
- 36.Petersen H, Leng S, Belinsky SA, Miller BE, Tal-Singer R, Owen CA, et al. Low plasma CC16 levels in smokers are associated with a higher risk for chronic bronchitis. Eur Respir J. 2015. [DOI] [PubMed] [Google Scholar]
- 37.Wang SZ, Rosenberger CL, Bao YX, Stark JM, Harrod KS. Clara cell secretory protein modulates lung inflammatory and immune responses to respiratory syncytial virus infection. J Immunol. 2003;171(2):1051–60. [DOI] [PubMed] [Google Scholar]
- 38.Waters V, Stanojevic S, Atenafu EG, Lu A, Yau Y, Tullis E, et al. Effect of pulmonary exacerbations on long-term lung function decline in cystic fibrosis. Eur Respir J. 2012;40(1):61–6. [DOI] [PubMed] [Google Scholar]
- 39.Cogen J, Emerson J, Sanders DB, Ren C, Schechter MS, Gibson RL, et al. Risk factors for lung function decline in a large cohort of young cystic fibrosis patients. Pediatr Pulmonol. 2015;50(8):763–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.https://www.cff.org/Research/Researcher-Resources/Patient-Registry/2019-Patient-Registry-Annual-Data-Report.pdf [
- 41.Levine CR, Gewolb IH, Allen K, Welch RW, Melby JM, Pollack S, et al. The safety, pharmacokinetics, and anti-inflammatory effects of intratracheal recombinant human Clara cell protein in premature infants with respiratory distress syndrome. Pediatric research. 2005;58(1):15–21. [DOI] [PubMed] [Google Scholar]
- 42.Chen Y, Vasquez MM, Zhu L, Lizarraga RE, Krutzsch M, Einspahr J, et al. Effects of Retinoids on Augmentation of Club Cell Secretory Protein. Am J Respir Crit Care Med. 2017;196(7):928–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greer RM, Buntain HM, Lewindon PJ, Wainwright CE, Potter JM, Wong JC, et al. Vitamin A levels in patients with CF are influenced by the inflammatory response. J Cyst Fibros. 2004;3(3):143–9. [DOI] [PubMed] [Google Scholar]
- 44.Milne S, Li X, Hernandez Cordero AI, Yang CX, Cho MH, Beaty TH, et al. Protective effect of club cell secretory protein (CC-16) on COPD risk and progression: a Mendelian randomisation study. Thorax. 2020;75(11):934–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.