Abstract
DsrA is an 85-nucleotide, untranslated RNA that has multiple regulatory activities at 30°C. These activities include the translational regulation of RpoS and H-NS, global transcriptional regulators in Escherichia coli. Hfq is an E. coli protein necessary for the in vitro and in vivo replication of the RNA phage Qβ. Hfq also plays a role in the degradation of numerous RNA transcripts. Here we show that an hfq mutant strain is defective for DsrA-mediated regulation of both rpoS and hns. The defect in rpoS expression can be partially overcome by overexpression of DsrA. Hfq does not regulate the transcription of DsrA, and DsrA does not alter the accumulation of Hfq. However, in an hfq mutant, chromosome-expressed DsrA was unstable (half-life of 1 min) and truncated at the 3′ end. When expressed from a multicopy plasmid, DsrA was stable in both wild-type and hfq mutant strains, but it had only partial activity in the hfq mutant strain. Purified Hfq binds DsrA in vitro. These results suggest that Hfq acts as a protein cofactor for the regulatory activities of DsrA by either altering the structure of DsrA or forming an active RNA-protein complex.
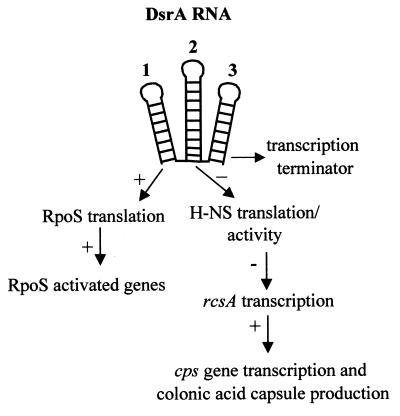
DsrA is a small untranslated RNA that regulates expression of two global transcription factors, H-NS and RpoS (Fig. 1). The DsrA secondary structure is conserved and is predicted to form a three-stem-loop structure (14). The three stem-loops correlate with discrete regulatory activities of DsrA (Fig. 1). The first stem-loop is involved in the anti-antisense regulation of translation of the rpoS mRNA (14). Increased transcription of DsrA at low temperatures leads to increased translation of rpoS and, consequently, increased transcription of some RpoS-dependent genes (27). The second stem-loop is necessary for the regulation of hns translation (13, 14) and possibly its activity (26). The third stem-loop apparently functions as a factor-independent transcription terminator (14, 26). Since many other RNAs require protein cofactors for activity, we looked for proteins that might be necessary for the DsrA-mediated regulation. A strong candidate cofactor was Hfq, an Escherichia coli protein required for replication of the RNA genome of the Qβ phage both in vivo and in vitro (7, 8, 23). Hfq functions by destabilizing an RNA secondary structure on the 3′ end of the positive strand of Qβ (7, 8, 23). In addition, Hfq binds tightly to Qβ RNA, poly(A) RNA (4, 8, 22), and OxyS RNA (37). Hfq also targets several mRNAs for degradation, possibly by increasing polyadenylation (10) or by interfering with ribosome binding (36). Hfq is essential for the efficient translation of RpoS in E. coli (17) and Salmonella enterica serovar Typhimurium (3) and for the instability of the ompA, miaA, mutS, and hfq mRNAs in E. coli (32, 35). Hfq copurifies with H-NS, and overexpression or mutations in Hfq can mask some H-NS− phenotypes (24). In this paper, we show that Hfq is necessary for the regulatory activity of DsrA and binds specifically to DsrA in vitro. In addition, in the absence of Hfq, chromosome-expressed DsrA was unstable, while plasmid-expressed DsrA remained stable.
FIG. 1.
Model of DsrA-mediated regulation. DsrA has three functional domains that correspond to three predicted stem-loops (14, 27). Stem-loop 1 positively regulates RpoS translation and the expression of RpoS-dependent genes. Stem-loop 2 decreases the translation of H-NS and also the ability of H-NS to repress transcription of rcsA and other H-NS-repressed genes (13, 26). RcsA activates transcription of the cps genes involved in colonic acid capsule production (29, 30). Stem-loop 3 functions as a factor-independent transcription terminator (14).
MATERIALS AND METHODS
Bacterial strains and genetic techniques.
All strains used in this paper were derivatives of E. coli K-12 strain SG20250 (1), a derivative of MC4100, unless indicated otherwise. Transductions with P1vir were done as described previously (16). λGN272 is equivalent to λMPM5 (15) and was a gift from D. Gentry. The hfq-1::kan and hfq-2::kan alleles are from strains TX2808 and TX2758, respectively (34). Transformations were done either by electroporation (6) using a GenePulsar II electroporator (Bio-Rad Laboratories, Richmond, Calif.) or by TSS transformation (5).
β-Galactosidase assays.
β-Galactosidase activities of the various lacZ fusions were assayed as described previously (16). Cells were grown with shaking at 30°C in Luria-Bertani (LB) medium supplemented with the appropriate antibiotic. Total β-galactosidase units were plotted against the optical density of the culture at 600 nm (OD600). The slopes of the linear regressions (differential rate of expression) were used as the specific activities of the fusions. Regression fits had an r2 of >0.95. Specific activities varied less than 10% between experiments.
Plasmid construction.
To generate plasmid pSP64-dsrA (pGSO122), a fragment carrying the T7 promoter and the E. coli dsrA gene was amplified by PCR from K-12 genomic DNA using primers (5′-CTT GAA TTC TAA TAC GAC TCA CTA TAG GGA ACA CAT CAG ATT TCC TGG and 5′-TAC AAG CTT AAA TCC CGA CCC TGA GGG GGT). The DNA fragment was digested with EcoRI and HindIII and cloned into the corresponding sites of the pSP64 vector (Promega, Madison, Wis.). To generate plasmid pT3-5S (pGSO123), a fragment carrying the T3 promoter and the E. coli 5S gene was amplified by PCR from K-12 genomic DNA using primers 5′-GCC AAG CTC GAA ATT AAC CCT CAC TAA AGG GTG CCT GGC GGC CGT AGC GCG and 5′-CCC CGG GTT TAA ATG CCT GGC AGT TCC CTA CTC. The DNA fragment was cloned into pCR 2.1 vector according to the manufacturer's protocol (TA Cloning Kit; Invitrogen, Carlsbad, Calif.) (K. M. Wassarman and G. Storz, unpublished data). Plasmid pSP64-oxyS (pGSO100) was described previously (37). The pBADHfq plasmid (pDDS400) was built by directionally cloning a PCR-generated fragment of the hfq gene into the EcoRI and PstI sites of the arabinose-inducible vector pBAD24 (9) using the primers 5′-GGA ATT CAC CAT GGC TAA GGG GCA ATC T (hfq1) and 5′-AAC TGC AGT TAT TCG GTT TCT TCG CTG TCC (hfq2).
RNA purification, primer extension, and RNase protection assays.
RNA was extracted from exponentially growing cells with hot phenol using method II of Hinton (11). Primer extension analysis was performed by the extension of the 5′-end 32P-labeled oligonucleotides 5′-GAC CCT GAG GGG GTC GGG ATG (rcsA24) or 5′-AAA CTT GCT TAA GCA AGA AGC ACT TA (dsrA25) following the method of Sambrook (19) with the addition of 1.5 mM MgCl2 and using murine Moloney virus reverse transcriptase (Life Technologies, Rockville, Md.). Oligonucleotides were labeled using [γ-32P]ATP (10 mCi/ml in aqueous solution) (Amersham, Arlington Heights, Ill.) and T4 polynucleotide kinase (Life Technologies) as described previously (19). The RPA II kit (Ambion, Austin, Tex.) was used for RNase protection assays, following the manufacturer's protocol. Antisense RNA probes were made using the MAXIscript T7 in vitro transcription kit (Ambion). The DNA template for the in vitro transcription reaction was a PCR product. The primers used were 5′-TAA TAC GAC TCA CTA TAG GGT CGT TGA ATG CAC AAT AAA A (dsrA21, containing a T7 promoter) and 5′-TAT GGC GAA TAT TTT CTT GTC AGC (dsrA20).
Gel electrophoresis and Western blotting.
Total cellular extracts of E. coli were electrophoretically separated on sodium dodecyl sulfate (SDS)–10% polyacrylamide gels or tris-tricine SDS–16% polyacrylamide gels (20) and electroblotted (31) to a sheet of nitrocellulose or polyvinylidene difluoride membrane. To verify equal protein loading in each lane, the membrane was stained with Ponceau S (Sigma, St. Louis, Mo.) following the manufacturer's protocol. The membrane was probed with rabbit anti-Hfq or anti-RpoS polyclonal antisera. The antibody-antigen complex was visualized with goat anti-rabbit immunoglobin horseradish peroxidase-conjugated antibody (Pierce, Rockford, Ill.) and ECL reagent kit (Pierce) following the manufacturer's protocol. The anti-Hfq rabbit antibody was raised against a chemically synthesized 15-amino-acid peptide (SAQNTSAQQDSEETE) identical to the C′ terminus of the E. coli Hfq protein (Sigma-Genosys, Woodlands, Tex.). The anti-RpoS antibody was a gift from R. Burgess.
Gel mobility shift assays.
The RNAs used for the mobility shift assays were obtained as follows. The pSP64-dsrA and pSP64-oxyS plasmid DNAs were linearized by digestion with HindIII, and the DsrA and OxyS RNAs were generated by in vitro transcription with T7 RNA polymerase (Life Technologies). The pGEM-5S plasmid DNA was linearized by digestion with DraI, and the 5S rRNA was generated by in vitro transcription with T3 RNA polymerase (Life Technologies). The transcripts were radioactively labeled at the 3′ end with [γ-32P]pCp (Amersham Pharmacia Biotech, Piscataway, N.J.) and T4 RNA ligase (Boehringer Mannheim Biochemicals, Indianapolis, Ind.). Full-length transcripts were purified on a 6% polyacrylamide–7 M urea gel and eluted in elution buffer (20 mM Tris-HCl [pH 7.5], 0.5 M sodium acetate, 10 mM EDTA, 1% SDS) at 65°C for 1 h, followed by ethanol precipitation. The RNA concentration was determined by measuring the OD260 of unlabeled transcripts exposed to the same treatment.
The gel mobility shift assays were performed as follows. End-labeled DsrA or 5S transcript (0.2 pmol) and nonspecific competitor yeast RNA (100 ng), without or with the indicated amounts of purified Hfq protein (A. Zhang and G. Storz, personal communication), were incubated in a 10-μl reaction mixture in RNA binding buffer (10 mM Tris-HCl [pH 8.0], 50 mM NaCl, 50 mM KCl, 10 mM MgCl2) for 10 min at 37°C. The samples were then mixed with 2 μl of loading dye (50% glycerol, 0.1% bromophenol blue, 0.1% xylene cyanol), analyzed on a 6% polyacrylamide gel in 0.5× TBE (Tris-borate-EDTA) buffer at 200 V for 1.5 h, and subjected to autoradiography. The competition reactions were performed as described above with purified Hfq protein (3 pmol), end-labeled DsrA transcript (0.2 pmol), and yeast RNA (100 ng) in the absence or presence of the indicated amounts of unlabeled DsrA, OxyS, or 5S transcript.
RESULTS
Hfq is important for DsrA-mediated regulation of RpoS.
Since both DsrA and Hfq regulate the translation of the alternative sigma factor RpoS (3, 17, 27), we explored their possible interactions by testing the ability of DsrA to function in an Hfq− host. Two hfq mutations were transduced into our strains. hfq-1, an hfq null allele, and hfq-2, an insertion in the 3′ end of hfq that produces functional Hfq protein and controls for polar effects on downstream genes (34), were used.
Three lacZ fusions were used as assays for DsrA activity (Table 1). An RpoS::LacZ translational fusion (27), an rcsA90::lacZ transcriptional fusion (an indicator of the anti-H-NS activity [26]), and a cps-2::lacZ transcriptional fusion (an indicator of RcsA expression [28]) were used. Consistent with previous results (2, 3, 17), expression of the RpoS::LacZ translational fusion was decreased 5-fold in a dsrA mutant (11 U for dsrA1 compared to 50 U for wild type) and 19-fold in the hfq-1 mutant (2.5 U for hfq-1 compared to 48 U for the control hfq-2). When DsrA was overproduced, the RpoS::LacZ fusion was expressed at 12-fold-higher levels (610 U) than in cells containing vector alone (50 U). Plasmid-expressed DsrA remained fully functional in a dsrA1 mutant.
TABLE 1.
An hfq-1 mutation decreased the regulatory activity of DsrA RNA
| Reporter fusiona | Other allelesb | β-Galactosidase sp act (U)c
|
Colony phenotyped | |
|---|---|---|---|---|
| Vector | pDsrA | |||
| RpoS::LacZ | Wild-type | 50 | 610 | Mucoid |
| dsrA1 | 11 | 600 | Nonmucoid | |
| hfq-1 | 2.5 | 20 | Nonmucoid | |
| hfq-2 | 48 | 593 | Mucoid | |
| rcsA90::lacZ | Wild-type | 15 | 103 | Mucoid |
| hfq-1 | 10 | 12 | Nonmucoid | |
| hfq-2 | 14 | 90 | Mucoid | |
| cps-2::lacZ | Wild-type | 12 | 90 | Mucoid |
| hfq-1 | 10 | 15 | Nonmucoid | |
All strains are single-copy λ(imm21) lysogens at att λ. Strains were grown in LB medium at 30°C. The RpoS::LacZ fusion is a translational fusion. The cps-2 and rcsA90::lacZ fusions are transcriptional fusions.
Mutations were transduced into the fusion strains by P1-mediated transduction.
Total β-galactosidase units were plotted against the OD600 of the culture. The slope of the curve between OD values of 0.2 and 1.0 (approximately log phase) was used as the specific activity of each fusion. Slopes had an r2 of >0.95. Specific activities varied less than 10% between experiments.
Colony phenotypes for strains with pDsrA were determined after growth overnight on LB plates at 30°C. vector (pACYC184) or pDsrA (dsrA cloned into pACYC184) (26) were used.
In the absence of Hfq, overexpression of DsrA increased RpoS::LacZ expression 8-fold (2.5 U compared to 20 U), but this level of expression was 30-fold down from what is seen in the wild-type strain (50 and 610 Units). This suggested that DsrA stimulates RpoS translation in the absence of Hfq but at a reduced level. Since the dsrA mutant reduced activity to 11 U while the hfq-1 mutation alone reduced activity to 2.5 U, not all Hfq-dependent stimulation of RpoS::LacZ was DsrA dependent. This could be due either to the pleiotropic nature of the hfq mutation (34) or the existence of another small RNA that affected RpoS translation (N. Majdalani, J. Murrow, S. Chen, K. Stjohn, and S. Gottesman, Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999, abstr. H-109, p. 350, 1999).
Previous work demonstrated that the expression of the RpoS::LacZ translational fusion correlated with the amount of RpoS protein when DsrA was expressed from the chromosome (14, 27). To ensure that pDsrA was affecting translation of the wild-type rpoS gene, RpoS protein levels were assayed by SDS-polyacrylamide gel electrophoresis and Western blotting. RpoS protein levels are increased during exponential growth at 30°C in LB broth when DsrA is expressed from a plasmid (Fig. 2). Expression of DsrA from a plasmid in the hfq-1 mutant had only a minor effect on the amount of RpoS (Fig. 2).
FIG. 2.
Western blot analysis of RpoS levels in hfq-1 mutant and DsrA-overexpressing strains. Cells were grown at 30°C in LB broth to OD600 of 0.5. Total cellular extracts of the strains were electrophoretically separated on a SDS–10% polyacrylamide gel, and electroblotted to a polyvinylidene difluoride membrane. The membrane was probed with rabbit anti-RpoS polyclonal antisera and visualized as described in Materials and Methods. Lanes 1 and 2 contain wild-type cells with either vector (pACYC184) or pDsrA. Lanes 3 and 4 contain isogenic hfq-1 mutants with either vector (pACYC184) or pDsrA. Since long exposures were used to visualize RpoS in the hfq mutant strains, lanes 1 and 2 are beyond the linear range of the Western blot. The position of RpoS is indicated.
Hfq is important for DsrA-mediated regulation of H-NS.
Plasmid-expressed DsrA increased expression of genes normally repressed by H-NS, including rcsA, proU, and papA (26). DsrA has no effect on these H-NS-regulated genes in an hns mutant strain (26). Activation of these genes by DsrA therefore is indirect and is mediated through down regulation of H-NS repression (Fig. 1) (26).
To monitor the anti-H-NS regulatory activity of DsrA, we assayed the rscA90::lacZ and cps-2::lacZ transcriptional fusions (Table 1). RcsA is an unstable positive regulator of colonic acid capsule expression (30). In an hns mutant host (or when DsrA is expressed from a plasmid in an hns+ host), rcsA transcription was increased, leading to activation of the colonic acid capsule synthesis genes (cps) and a mucoid colony phenotype (26). Activity of the rcsA90::lacZ fusion correlated with expression of RcsA and is therefore a good indicator of rcsA transcription (26). Expression of cps-2::lacZ is a sensitive indicator of the amount of wild-type RcsA (28). Both fusions are on a lambda phage integrated into the chromosome at the lambda att site in otherwise isogenic hosts (26, 28).
Expression of rcsA90::lacZ and cps-2::lacZ was stimulated by DsrA when the RNA was expressed from a multicopy plasmid (Table 1). The hfq-1 mutation interfered with DsrA-dependent stimulation of rcsA90::lacZ (12 U for hfq-1 compared to 90 U for hfq-2) and cps-2::lacZ (15 U for hfq-1 compared to 90 U for wild type). In addition, the colony phenotypes of DsrA-overexpressing strains were no longer mucoid in the hfq-1 mutant strain (Table 1). Thus, Hfq is required for most of the anti-H-NS activity of DsrA. Since some residual DsrA activity was noticed in the hfq-1 mutant strain after 2 days of incubation on MacConkey lactose indicator plates (data not shown), DsrA can function to repress H-NS in the absence of Hfq, albeit at a much reduced level.
Regulated expression of Hfq can complement the hfq-1 mutation but does not increase expression of RpoS::LacZ or rcsA::lacZ in the absence of DsrA.
Overexpression of hfq from a multicopy plasmid complements the hfq-1 mutation and confers other phenotypes (34). To confirm that the hfq-1 mutation was responsible for the loss of DsrA activity, we constructed a plasmid that expressed hfq under control of the araBAD promoter (9). At subsaturating concentrations of arabinose (<1 mM), the araBAD promoter demonstrates different levels of gene expression within individual cells within a population (25). Thus, we determined the optimal concentration of arabinose which complemented the hfq-1 defect in RpoS::LacZ expression. An arabinose concentration of 150 μM restored 80% of the expression of the RpoS::LacZ fusion in an hfq-1 mutant and 100% of the rcsA::lacZ fusion in an hfq-1 mutant containing pDsrA (data not shown). Less complementation was seen with higher or lower concentrations of arabinose. Since this observation agrees with work done by others using a similar araBADHfq construct and assaying RpoS expression (2), 150 μM arabinose was used in the experiments below.
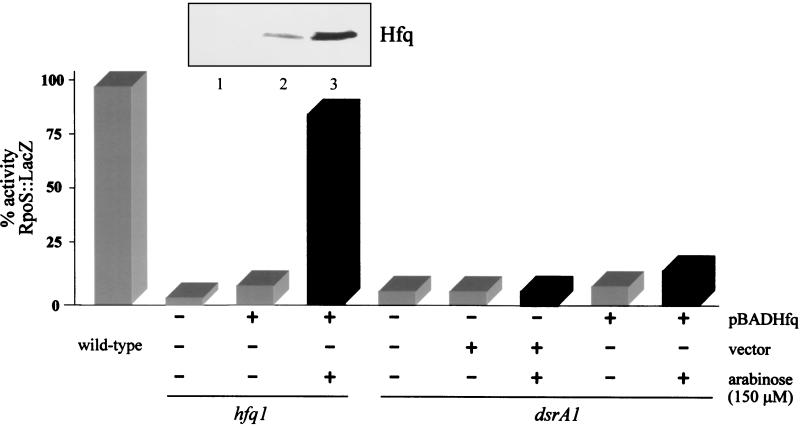
In an hfq-1 mutant, DsrA-mediated regulation of RpoS::LacZ was restored when Hfq was expressed from the plasmid in rich media at 30°C with 150 μM arabinose (Fig. 3). In contrast, increased expression of Hfq in a dsrA1 strain only slightly increased expression of the RpoS::LacZ fusion (Fig. 3). This suggested that both DsrA and Hfq are necessary for the optimal expression of RpoS at low temperatures.
FIG. 3.
DsrA and Hfq are both needed for regulation of RpoS::LacZ at 30°C. Cultures of the various strains grown overnight were diluted 1:100 in LB broth. Cultures were grown at 30°C, and samples were taken at various times during logarithmic growth (0.2 > OD600 < 1.0). Total β-galactosidase units were plotted against the OD600 of the culture. The slopes of the curves between OD values of 0.2 and 1.0 (approximately log phase) were used as the specific activities of the fusions. Slopes had an r2 of >0.95. Specific activities varied less than 10% between experiments. All activities are in relation to that of the wild-type RpoS::LacZ strain during logarithmic growth (100%) (gray bars). Black bars represent the addition of 150 μM arabinose. pBADHfq expresses recombinant Hfq under control of the araBAD promoter. The vector is the araBAD promoter vector pBAD24. pDsrA has DsrA cloned into pACYC184 under control of its own promoter. The presence of hfq-1 and dsrA1 mutations in isogenic strains is indicated. The inset above the bar graph is a Western blot of those strains using anti-Hfq antibody. Lanes 1 to 3 of the gel correspond to the strains and conditions immediately below the inset. Note the induction of Hfq after the addition of arabinose.
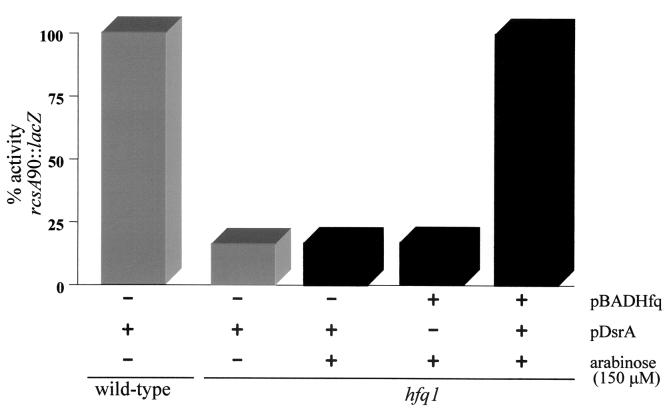
In an hfq-1 mutant expressing wild-type dsrA from the chromosome, induction of the plasmid hfq gene did not affect the expression of an rcsA90::lacZ fusion (Fig. 4) or a cps-2::lacZ fusion (data not shown) and did not make cells mucoid (an indicator of cps gene expression). Induction of the plasmid hfq gene in wild-type cells also did not affect rcsA or capsule production (data not shown). Hfq is therefore apparently not limiting under these conditions. When both pDsrA and pBADHfq are present in an hfq-1 mutant, induction of hfq increased rcsA90::lacZ expression about fivefold (Fig. 4). No increase in rcsA90::lacZ expression was seen following arabinose induction of a strain carrying pDsrA and paraBAD (data not shown). All of Hfq's effects on rcsA and cps expression require pDsrA. In the absence of arabinose, Hfq was detected from the pBADHfq plasmid (Fig. 3) but was not sufficient to complement the tested phenotypes.
FIG. 4.
Hfq is necessary for DsrA-mediated regulation of rcsA::lacZ, but Hfq does not regulate rcsA::lacZ alone. Cultures of the various strains grown overnight were diluted 1:100 in LB broth. Cultures were grown at 30°C, and samples were taken at various times during logarithmic growth (0.2 > OD600 < 1.0). Total β-galactosidase units were plotted against the OD600 of the culture. The slopes of the curves between OD values of 0.2 and 1.0 (approximately log phase) were used as the specific activities of the fusions. Slopes had an r2 of >0.95. Specific activities varied less than 10% between experiments. All activities are in relation to a wild-type rcsA90::lacZ strain containing pDsrA (100%) (gray bars). Black bars represent the addition of 150 μM arabinose. pBADHfq expresses recombinant Hfq under control of the araBAD promoter. pDsrA has DsrA cloned into pACYC184 under control of its own promoter. The presence of the hfq-1 mutation in an isogenic wild-type strain is indicated.
Hfq is necessary for the stability of chromosome-expressed DsrA.
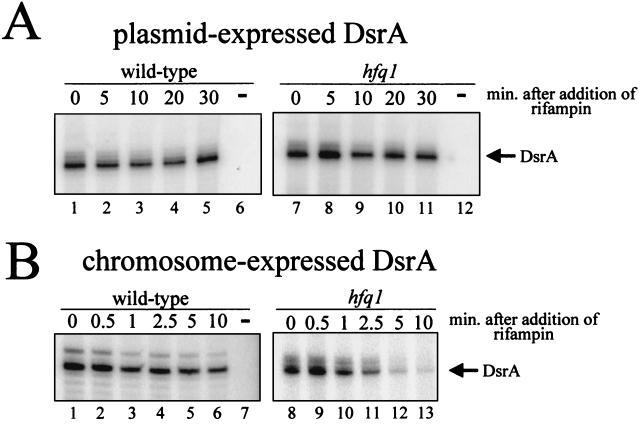
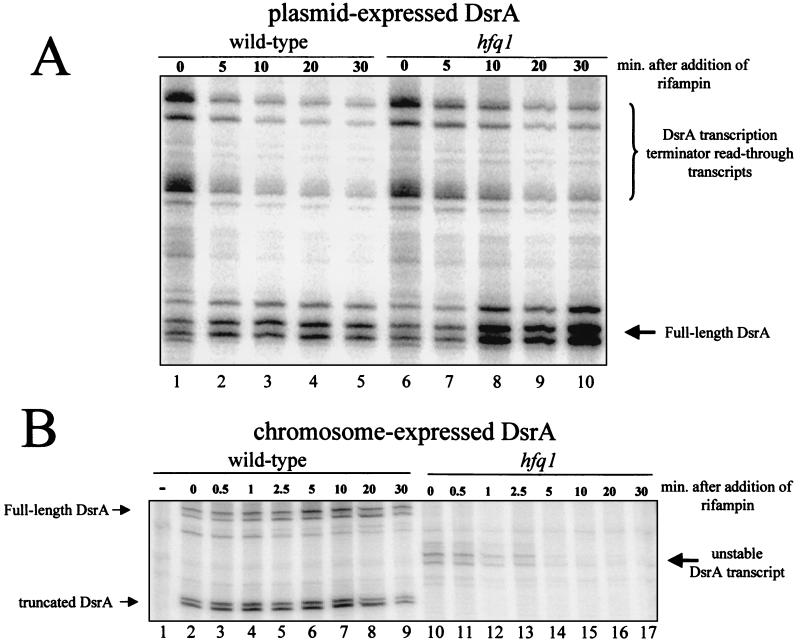
Hfq affects the accumulation of ompA, miaA, hfq, and other mRNAs (32, 33, 35). We therefore determined the amount and stability of chromosome-expressed DsrA RNA by both a RNase protection assay and a primer extension assay (see Fig. 6). In wild-type cells, chromosome-expressed DsrA was less stable than plasmid-expressed DsrA with a half-life between 6 and 30 min (Fig. 5B, 6B, and 7). The lower value was calculated from the primer extension assay (Fig. 6B). The higher value was calculated from the RNase protection assay (Fig. 5B). The differences in the half-lives calculated by the two assays were consistent between experiments using the same RNA samples. Accurate calculation of the stability of chromosome-expressed DsrA was difficult, since the amount of DsrA increased after the addition of rifampin in the RNase protection experiments, peaking after ∼10 min (Fig. 5B). This increase was not seen in the primer extension assays (Fig. 6B).
FIG. 6.
Primer extension assay of DsrA stability in an hfq-1 strain. Cells were grown at 30°C in LB broth to an OD600 of 0.5 and treated with rifampin to block new transcription. Samples were taken at the indicated times, and RNA was extracted. DsrA was detected using a primer extension assay and DsrA-specific primers. Lanes marked with a minus sign contain RNA isolated from a dsrA deletion mutant. (A) DsrA expressed from a plasmid and the chromosome. (B) DsrA expressed only from the chromosome. Note that the chromosome- and plasmid-expressed dsrA genes have the same transcription start site. Only the chromosome-expressed DsrA has a decreased half-life in the Hfq− host.
FIG. 5.
RNase protection assay of DsrA stability in an hfq-1 strain. Cells were grown at 30°C in LB broth to an OD600 of 0.5 and treated with rifampin to block new transcription. Samples were taken at the indicated times, and RNA was extracted. DsrA was detected using a RNase protection assay and DsrA-specific probe. The minus sign in panel B indicates that the RNA was isolated from a dsrA deletion mutant. (A) DsrA expressed from the chromosome and the plasmid pDDS164. (B) DsrA expressed only from the chromosome. The positions of full-length, processed, read-through, and unstable DsrA transcripts are indicated. Processed DsrA transcripts were also seen when DsrA was expressed from the plasmid but are not shown. Note that in lane 1, no DsrA is detected in RNA isolated from a DsrA deletion strain.
FIG. 7.
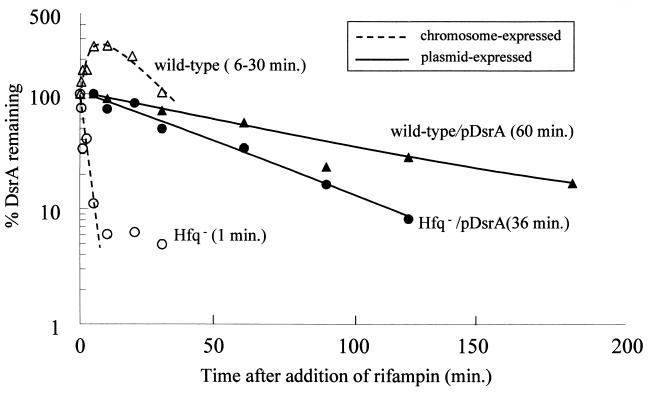
DsrA half-life in wild-type and Hfq− strains. DsrA amounts from a RNase protection assay were quantitated on a Storm 840 phosphorimager. Dashed lines are chromosome-expressed DsrA. Solid lines are plasmid-expressed DsrA. The numbers in parentheses are half-lives calculated from the curve fit equations. Hfq− is an isogenic strain containing the hfq-1::kan mutation.
In the hfq-1 strain, chromosome-expressed DsrA was dramatically less stable than in the wild type, with a half-life of <1 min in both the RNase and primer extension assays (Fig. 6B, 7B, and 8). In addition, the DsrA transcript was significantly shorter in the hfq-1 strain than in the wild type (Fig. 5B). This difference in size was not detected in primer extension experiments (data not shown), suggesting a truncation in the 3′ end of the DsrA transcript.
FIG. 8.
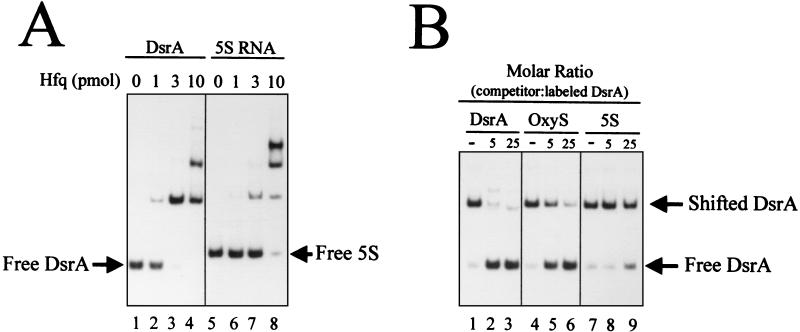
Gel mobility shift analysis of DsrA binding to Hfq. (A) 3′-end γ-32P-labeled transcript (0.2 pmol) of DsrA or 5S RNA and nonspecific competitor yeast RNA (100 ng) were incubated without (lanes 1 and 5) or with the indicated amounts of Hfq protein (lanes 2 to 4 and 6 to 8). (B) 3′-end γ-32P-labeled DsrA (0.2 pmol) and yeast RNA (100 ng) were incubated with purified Hfq protein (3 pmol). Unlabeled transcript of DsrA (lanes 2 and 3), OxyS (lanes 5 and 6), or 5S (lanes 8 and 9) were added as competitors.
Hfq does not affect the transcription or stability of plasmid-expressed DsrA.
The amount and stability of the plasmid-expressed DsrA were also determined. The pDsrA plasmid has dsrA under control of its own promoter with the same transcription start site as chromosome-expressed DsrA (Fig. 6) (26). Two sets of DsrA transcripts were detected with the RNase protection assay in wild-type cells (Fig. 5B). Based on the results with molecular weight markers, the larger transcripts corresponded to the three-stem-loop DsrA RNA. The second, faster-migrating set of transcripts was also detected with primer extension assays (data not shown). The size was consistent with the absence of the first stem-loop of DsrA (responsible for RpoS regulation [14]) and the 5′ half of the second stem-loop (responsible for H-NS regulation [13]). Although this shorter transcript is stable, it is not known whether it has any regulatory activities.
When grown in rich media at 30°C, the amount of plasmid-expressed DsrA was equal in the hfq-1 and wild-type strains in both the RNase protection assay (Fig. 5A, lanes 1 and 6) and primer extension assay (Fig. 6A, lanes 1 and 7). In addition, the expression of a dsrA::lacZ fusion is unchanged in a wild-type strain compared to an isogenic hfq-1 mutant (specific activities of 51 ± 4 and 46 ± 5 U, respectively). Thus, Hfq does not regulate dsrA transcription.
The multiple bands seen in both assays are consistent with multiple transcription start sites (26).
The half-life of plasmid-expressed DsrA, assessed by measuring DsrA levels at various times after inhibiting transcription with rifampin, was >30 min for both hfq+ and hfq-1 strains (Fig. 5A, 6A, and 7). This agrees with previous results showing that DsrA is stable (14). A 60% decrease in the stability of plasmid-expressed DsrA in an hfq mutant was observed during very long (up to 3 h) experiments (Fig. 7). This is unlikely to be physiologically significant, because both half-lives are still well above the 20-min doubling time of exponentially growing E. coli grown in rich media.
The RNase protection assay detected DsrA transcription terminator read-through products from the plasmid-expressed DsrA (Fig. 5A). In both the hfq+ and hfq-1 strains, these transcripts were unstable with a half-life of ∼2 min (Fig. 5A).
Hfq binds DsrA in vitro.
The loss of stability of chromosomally expressed DsrA in an hfq-1 mutant coupled with our genetic data suggested a direct interaction between DsrA and Hfq. To examine Hfq binding to DsrA, 0.2 pmol of end-labeled DsrA RNA was incubated with purified Hfq protein and examined by a gel mobility shift assay (Fig. 8). Incubation with 3 pmol of Hfq led to complete retardation of DsrA in the presence of 100 ng of yeast tRNA (Fig. 8A). Hfq binding to the control 5S rRNA was less efficient, requiring 10 pmol of Hfq for complete retardation (Fig. 8A). Multiple complexes were observed for both DsrA and 5S RNAs, possibly due to different multimeric forms of the Hfq protein binding to the RNAs.
OxyS RNA binds to Hfq in vitro and in vivo (37). To test the strength and specificity of the DsrA-Hfq complex, the labeled DsrA RNA and Hfq protein were incubated in the presence of excess unlabeled DsrA, OxyS, or 5S RNA (Fig. 9B). The DsrA binding to Hfq was competed by fivefold molar excess unlabeled DsrA. Between 5- and 25-fold more OxyS RNA was necessary to get a comparable competition. This suggests that Hfq binds more tightly to DsrA than OxyS. 5S rRNA was a poor competitor, requiring more than 25-fold molar excess to partially compete with the DsrA-Hfq interaction (Fig. 9B). These results are indicative of a specific interaction between DsrA and Hfq that is approximately equal in strength to the OxyS-Hfq interaction.
FIG. 9.
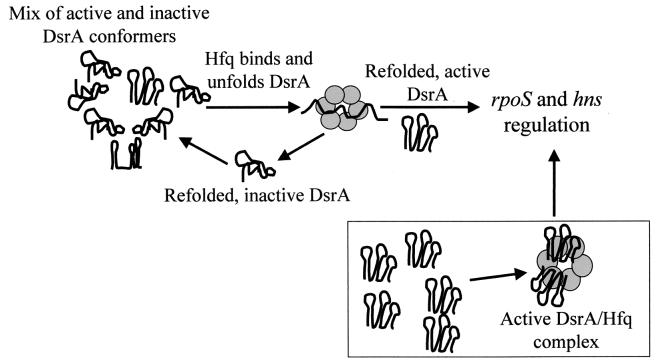
Model of Hfq's role in DsrA-mediated regulation. When initially synthesized, DsrA folds into active and inactive forms. Hfq binds to DsrA molecules, unfolds them, and allows them to refold into active forms and inactive forms. Active DsrA forms regulatory complexes with its targets and leaves the cycle. Alternatively, Hfq and DsrA could form an RNA-protein complex that is active for the regulation of rpoS and hns (lower box). These models are not mutually exclusive.
DISCUSSION
The untranslated RNA DsrA regulates at least two global regulatory networks by affecting mRNA translational efficiency of the global transcription factors RpoS and H-NS (13, 14). In this paper, we show that DsrA-mediated regulation of RpoS and H-NS was absent or severely reduced in an hfq-1 mutant (Table 1). Hfq is not required for DsrA synthesis, and overexpression of Hfq does not complement a dsrA1 mutation. In addition, plasmid-expressed DsrA accumulated in an hfq-1 mutant but was only weakly active for regulation. A direct interaction between DsrA and Hfq was detected in vitro using a gel mobility shift assay. This binding was specific and was approximately equal to the binding of Hfq to another regulatory RNA, OxyS, which was shown to bind Hfq in vivo (37). Since both OxyS and DsrA could compete with each other for binding to Hfq in vitro, it is likely that DsrA also binds to Hfq in vivo. This observation is consistent with a model that OxyS binding to Hfq may compete with the binding of other RNAs (37). According to this model, overexpression of OxyS could decrease RpoS translation in an Hfq-dependent manner by competing with DsrA.
How do Hfq and DsrA affect gene expression?
Our data are consistent with two models that explain the necessity for both DsrA and Hfq to obtain maximal translation of RpoS at low temperatures (Fig. 9). First, DsrA and Hfq could form a complex that binds to the RpoS mRNA leader, destabilizing the putative 5′ translation inhibitory stem-loop structure (2). In this case, the function of Hfq might be to bind to DsrA and unfold the first stem-loop, which is necessary for the regulation of RpoS translation (14). The role of Hfq might be analogous to Rop, in which two phenylalanines intercalate into the stem and facilitate a secondary RNA hybridizing (18). The unfolded DsrA-Hfq complex could then hybridize to the RpoS mRNA leader, preventing the formation of the translational inhibitory RNA secondary structure (2). It is interesting to note that among the Hfq proteins from 11 gram-negative and 1 gram-positive bacteria, there are two absolutely conserved phenylalanines at positions 39 and 42 (data not shown).
A second possibility is that DsrA and Hfq transiently interact, and Hfq functions as an RNA chaperone that is necessary for the proper folding of DsrA into an active conformation. A similar activity was proposed for Hfq interactions with mutS, miaA, hfq, Qβ phage (21, 32) and ompA (35) mRNAs as well as StpA stimulation of RNA self-splicing (38). In the case of Qβ, the binding of Hfq is thought to denature an RNA secondary structure at the 3′ end of the positive strand, allowing the Qβ replicase to synthesize negative strands (21).
We have shown that plasmid-expressed DsrA increased RpoS and Cps expression in an hfq-1 host, albeit only modestly; thus, Hfq is not essential for the regulatory activity of DsrA. With the Hfq chaperone model, a small portion of DsrA might be expected to spontaneously fold into an active conformation in the absence of Hfq. This necessity, but not absolute requirement, for Hfq was also seen with Qβ replication (21). Conversely, an hfq-1 mutant clearly has regulatory effects on RpoS and other genes that are independent of DsrA (Table 1 and reference 34). In addition, since RpoS expression is not regulated by DsrA at 37°C (27) but is regulated by Hfq, it is possible that Hfq can directly affect the formation of the RpoS mRNA 5′ translational inhibitor. At lower temperatures, the secondary structure of the RpoS mRNA 5′ untranslated region is likely to be more stable and Hfq alone might not be able to “melt-out” the structure. DsrA's function might then be to decrease the stability of the RpoS leader by binding to it and forming a stable alternative structure (2, 3). Alternatively, there may be other factors (RNAs) that interact with Hfq at higher temperatures to modulate RpoS translation (Majdalani et al., Abstr. 99th Gen. Meet. Am. Soc. Microbiol. 1999).
Interactions similar to those postulated for RpoS could also explain the DsrA-mediated regulation of H-NS translation and H-NS-regulated genes (13, 26).
Hfq and DsrA stability.
Hfq had a dramatic effect on DsrA stability when DsrA was expressed from the chromosome. In the wild-type strain, chromosome-expressed DsrA had a half-life of 6 to 30 min depending on the assay. In contrast, in the hfq-1 strain, chromosome-expressed DsrA was very unstable with a half-life of ∼1 min in both assays. A degradation product was detected; its presence suggested that DsrA was truncated at the 3′ end in the hfq-1 mutant. This sensitivity to degradation in the absence of Hfq is opposite to what was seen with the ompA, miaA, mutS, and hfq mRNAs which are stabilized in an hfq mutant (32, 35).
As previously shown (14) and confirmed in this paper, plasmid-expressed DsrA was very stable in wild-type cells with a half-life of 60 min. What was surprising was that in the absence of Hfq, plasmid-expressed DsrA remained stable with a half-life of 36 min. This difference might be strain specific, since plasmid-expressed DsrA does not accumulate in some hfq mutant backgrounds (12). One possibility is that factors necessary for the degradation of DsrA are found only within the nucleoid. One prediction of this model is that DsrA would remain unstable in an hfq-1 mutant even when overexpressed from the chromosome by an inducible promoter.
Whether Hfq acts transiently, by altering the structure of DsrA or by forming an active RNA-protein complex within the cell, it will be interesting to discover how Hfq functions to enhance the interaction of DsrA with its targets. The availability of an in vitro model of DsrA-Hfq binding will allow us to study this interaction in more detail.
ACKNOWLEDGMENTS
We thank S. Gottesman, S. Garges, N. Majdalani, G. Storz, and R. Blumenthal for helpful comments and G. Beyer for technical assistance. We also thank M. Winkler for the hfq mutant strains and R. Lease and M. Belfort for unpublished data.
This research was supported in part by grants from NIH (GM56448), The American Cancer Society-Ohio Division, and the Ohio Board of Regents Research Challenge II.
REFERENCES
- 1.Brill J A, Quinlan-Walshe C, Gottesman S. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1988;170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown L, Elliot T. Mutations that increase expression of the rpoS gene and decrease its dependence on hfq function in Salmonella typhimurium. J Bacteriol. 1997;179:656–662. doi: 10.1128/jb.179.3.656-662.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown L, Elliot T. Efficient translation of the RpoS sigma factor in Salmonella typhimurium requires host factor I, an RNA-binding protein encoded by the hfq gene. J Bacteriol. 1996;178:3763–3770. doi: 10.1128/jb.178.13.3763-3770.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carmichael G G, Weber K, Niveleau A, Wahba A J. The host factor required for RNA phage Qβ RNA replication in vitro. Intracellular location, quantitation, and purification by poly(A)-cellulose chromatography. J Biol Chem. 1975;250:3607–3612. [PubMed] [Google Scholar]
- 5.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franze de Fernandez M T, Eoyang L, August J T. Factor fraction required for the synthesis of bacteriophage Qβ RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 8.Franze de Fernandez M T, Hayward W S, August J T. Bacterial proteins required for replication of phage Qβ ribonucleic acid. Purification and properties of host factor I, a ribonucleic acid-binding protein. J Biol Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- 9.Guzman L M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hajnsdorf E, Regnier P. Host factor Hfq of Escherichia coli stimulates elongation of poly(A) tails by poly(A) polymerase I. Proc Natl Acad Sci USA. 2000;97:1501–1505. doi: 10.1073/pnas.040549897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hinton D M. Transcript analyses of the uvsX-40-41 region of bacteriophage T4. J Biol Chem. 1989;264:14432–14439. [PubMed] [Google Scholar]
- 12.Lease R, Belfort M. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc Natl Acad Sci USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lease R A, Cusick M E, Belfort M. Riboregulation in Escherichia coli: DsrA RNA acts by RNA:RNA interactions at multiple loci. Proc Natl Acad Sci USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McCann M P, Fraley C D, Matin A. The putative ς factor KatF is regulated posttranscriptionally during carbon starvation. J Bacteriol. 1993;175:2143–2149. doi: 10.1128/jb.175.7.2143-2149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller J H. Experiments in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 17.Muffler A, Fischer D, Hengge-Aronis R. The RNA-binding protein HF-I, known as a host factor for Qβ RNA replication, is essential for rpoS translation in Escherichia coli. Genes Dev. 1996;10:1143–1151. doi: 10.1101/gad.10.9.1143. [DOI] [PubMed] [Google Scholar]
- 18.Predki P F, Nayak L M, Gottlieb M B, Regan L. Dissecting RNA-protein interactions: RNA-RNA recognition by Rop. Cell. 1995;80:41–50. doi: 10.1016/0092-8674(95)90449-2. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 21.Schuppli D, Miranda G, Tsui H C, Winkler M E, Sogo J M, Weber H. Altered 3′-terminal RNA structure in phage Qbeta adapted to host factor-less Escherichia coli. Proc Natl Acad Sci USA. 1997;94:10239–10242. doi: 10.1073/pnas.94.19.10239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Senear A W, Steitz J A. Site-specific interaction of Qβ host factor and ribosomal protein S1 with Qβ and R17 bacteriophage RNAs. J Biol Chem. 1976;251:1902–1912. [PubMed] [Google Scholar]
- 23.Shapiro L, Franze de Fernandez M T, August J T. Resolution of two factors required in the Qβ-RNA polymerase reaction. Nature. 1968;220:478–480. doi: 10.1038/220478a0. [DOI] [PubMed] [Google Scholar]
- 24.Shi X, Bennett G N. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J Bacteriol. 1994;176:6769–6775. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siegele D A, Hu J C. Gene expression from plasmids containing the araBAD promoter at subsaturating inducer concentrations represents mixed populations. Proc Natl Acad Sci USA. 1997;94:8168–8172. doi: 10.1073/pnas.94.15.8168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sledjeski D, Gottesman S. A small RNA acts as an antisilencer of the H-NS-silenced rcsA gene of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:2003–2007. doi: 10.1073/pnas.92.6.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sledjeski D D, Gupta A, Gottesman S. The small RNA, DsrA, is essential for the low temperature expression of RpoS during exponential growth in Escherichia coli. EMBO J. 1996;15:3993–4000. [PMC free article] [PubMed] [Google Scholar]
- 28.Stout V. Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J Bacteriol. 1996;178:4273–4280. doi: 10.1128/jb.178.14.4273-4280.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stout V, Gottesman S. Regulation of capsular polysaccharide synthesis in Escherichia coli K12. Mol Microbiol. 1991;5:1599–1606. doi: 10.1111/j.1365-2958.1991.tb01906.x. [DOI] [PubMed] [Google Scholar]
- 30.Stout V, Torres-Cabassa A, Maurizi M R, Gutnick D, Gottesman S. RcsA, an unstable positive regulator of capsular polysaccharide synthesis. J Bacteriol. 1991;173:1738–1747. doi: 10.1128/jb.173.5.1738-1747.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsui H-C T, Feng G, Winkler M E. Negative regulation of mutS and mutH repair gene expression by the Hfq and RpoS global regulators of Escherichia coli K-12. J Bacteriol. 1997;179:7476–7487. doi: 10.1128/jb.179.23.7476-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsui H-C T, Feng G, Winkler M E. Transcription of the mutL repair, miaA tRNA modification, hfq pleiotropic regulator, and hflA region protease genes of Escherichia coli K-12 from clustered Eς32-specific promoters during heat shock. J Bacteriol. 1996;178:5719–5731. doi: 10.1128/jb.178.19.5719-5731.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsui H-C T, Leung H C E, Winkler M. Characterization of broadly pleiotropic phenotypes caused by an hfq insertion mutation in Escherichia coli K-12. Mol Microbiol. 1994;13:35–49. doi: 10.1111/j.1365-2958.1994.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 35.Vytvytska O, Jakobsen J S, Balcunaite G, Andersen J S, Baccarini M, von Gabain A. Host factor I, Hfq, binds to Escherichia coli ompA mRNA in a growth rate-dependent fashion and regulates its stability. Proc Natl Acad Sci USA. 1998;95:14118–14123. doi: 10.1073/pnas.95.24.14118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vytvytska O, Moll I, Kaberdin V R, von Gabain A, Blasi U. Hfq (HF1) stimulates ompA mRNA decay by interfering with ribosome binding. Genes Dev. 2000;14:1109–1118. [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang A, Altuvia S, Tiwari A, Argaman L, Hengge-Aronis R, Storz G. The OxyS regulatory RNA represses rpoS translation and binds the Hfq (HF-1) protein. EMBO J. 1998;17:6061–6068. doi: 10.1093/emboj/17.20.6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang A, Derbyshire V, Salvo-Galloway J L, Belfort M. E. coli protein StpA stimulates self-splicing by promoting RNA assembly in vitro. RNA. 1995;1:783–793. [PMC free article] [PubMed] [Google Scholar]