Abstract
Objective
SPON2 is one of the extracellular matrix proteins, which is closely related to the progression of a variety of tumors including non-small cell lung cancer (NSCLC), but its upstream regulation mechanism remains unclear. Our research aims to find the specific regulatory pathway of SPON2 by exploring the potential crosstalk between tumor cells and cancer-associated fibroblasts (CAFs) in tumor microenvironment (TME) of NSCLC.
Methods
We analyzed T1 lung adenocarcinoma samples from TCGA and screened extracellular matrix proteins that indicate poor prognosis. Expression level of SPON2 was verified by qPCR in clinical samples. The exosomes of NSCLC cell supernatant were extracted and identified by nanoparticle tracking analysis (NTA) and transmission electron microscope, western blots. The exosomes and CAFs were co-cultured, and cell migration and Matrigel invasion assay were used to evaluate the effect of CAFs on the migration and invasion of NSCLC cells. The interaction between LncRNA and miRNA was verified by Targetscan prediction, luciferase reporter assay, and RNA binding protein immunoprecipitation (RIP).
Results
We found that the expression of SPON2 was up-regulated in clinical T1a stage NSCLC patients. The expression of lnc HOTAIRM1 (HOTAIRM1) in exosomes secreted by NSCLC tissues increased. After exosomal HOTAIRM1 entered CAFs, HOTAIRM1 can adsorb miR-328-5p to up-regulate the expression of SPON2 in CAFs. Up-regulation of SPON2 in CAFs could promote the migration and invasion of NSCLC cells.
Conclusion
Tumor-derived exosomal HOTAIRM1 can transfer into CAFs and competitively adsorb miR-328-5p, and regulate the SPON2 expression of CAFs cells, ultimately promote the progression of NSCLC. The discovery of this regulatory pathway can provide a new potential therapeutic target for the diagnosis and treatment of NSCLC.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12672-022-00553-7.
Keywords: Lung adenocarcinoma, Tumour microenvironment, CAFs, Exosomes, SPON2
Introduction
Lung cancer is still one of the tumors with high morbidity and mortality [1]. The vigorous implementation of the lung cancer screening program has significantly increased the detection rate of early lung cancer [2]. However, even in the early stage of lung cancer, tumor heterogeneity also leads to the risk of progression and migration, which is the main factor of poor prognosis of lung adenocarcinoma [3]. Therefore, it is of great clinical significance to study the triggering mechanism of metastasis in early lung cancer.
In recent years, the role of TME in the research of tumor cell differentiation, proliferation, invasion and metastasis and drug resistance has attracted more and more attention [4–6]. According to the “seed & soil” theory, the crosstalk between microenvironment stromal cells and tumor cells plays an important regulatory role in the progression of tumor invasion [7, 8]. CAFs is one of the fundamental cell components with the highest proportion in tumor matrix, which has a strong ability of extracellular matrix remodeling, and promotes tumor progression by inducing stromal cell fibrosis and increasing matrix tension [9, 10]. The ability of CAFs to reshape the microenvironment is not a unique phenomenon, and as with other stromal cells, interactions between tumor cells and matrix cells play an important role [11]. Among them, for example, the delivery of small molecules through exosome pathway has attracted increasing attention of researchers.
Exosomes are vesicles with a diameter of 40 to 160 nm (an average of 100 nm) derived from the interior of cells. They contain a large number of substances, such as DNA, RNA, lipids, metabolites, cytoplasmic and cell surface proteins, which play various vital functions in intercellular communication and modulate biological behavior in downstream cells [12, 13]. It has been reported that exosomes derived from NSCLC can activate CAFs cells to secrete extracellular matrix, but the specific mechanism is not completely clear [14]. SPON2 is one of the extracellular matrix proteins. Physiologically, it can bind to bacteria and participate in the phagocytosis of bacteria by macrophages. It is a unique pattern recognition molecule of microbial pathogens in ECM and crucial in the initiation of innate immune response [15].In early studies, overexpression of SPON2 was confirmed to be an independent prognostic biomarker of lung adenocarcinoma. However, the specific mechanism for its overexpression is still unclear [16]. Our preliminary study found that the increased SPON2 expression level in early NSCLC patients was associated with poor prognosis, and an oncogenic long non-coding RNA (LncRNA), HOTAIRM1 in exosomes secreted from NSCLC cells may be associated with regulating SPON2 expression in CAFs. HOTAIRM1 can regulate neuronal differentiation and promote osteogenesis in physiological state, and inhibit T-cell depletion in late sepsis, so as to reduce lung injury and improve the survival rate.HOTAIRM1 has been proved to promote the progression of lung adenocarcinoma [17, 18], and its positive correlation with SPON2 expression attracts our interest, which means that it may also have other cancer promoting pathways.However, the exact mechanism is unclear. The purpose of this study was to explore the role and the exact mechanism of exsomal LncRNA in crosstalk between tumor cells and CAFs in tumor microenvironment.
Materials and methods
Analysis of the correlation between SPON2 and prognosis of lung adenocarcinoma in stage T1a in the TCGA database
Lung adenocarcinoma samples at stage T1a in TCGA were downloaded and extracellular matrix proteins associated with poor prognosis [19] in these samples were analyzed using the R software.The cell subpopulations related to ECM were analyzed by deconvolution calculation.The data of GSE12428 and GSE11969 in GEO database were analyzed by survival analysis database LOGpc [20].
Patient and tissue samples
Clinical specimens were collected from 10 patients underwent operation in Jiangsu Provincial People’s Hospital. The study was approved by the ethics committee of Jiangsu Provincial People’s Hospital (No. 2019-SR-266), and the patient was informed and signed consent forms. The research was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Cell lines and cell culture
A549 and H1299 were purchased from Chinese Academy of Sciences, Shanghai Institute of Biochemistry and Cell Biology (Shanghai, China). A549/Luc cells (A549 cells stably expressing luciferase) were constructed by Synthgene (Nanjing, China). A549 was cultured with DMEM (Dulbecco’s Modified Eagle’s Medium) (Gibco, Rockville, USA) and H1299 was cultured with Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% FBS (fetal bovine serum) (Gibco, Rockville, USA) and 100 µg/mL streptomycin and penicillin (Gibco, Rockville, USA) in a humidified atmosphere at 37 ℃. CAFs were isolated from surgical specimens diagnosed as T1 stage lung adenocarcinoma by pathology. Details of these patients are in the “Information About CAFs from Patients' in the Additional file 1. Tumor tissue was dissected and digested with trypsin and collagenase to obtain single cell suspension for culture, and then maintained in DMEM high glucose culture medium (including 10% FBS, Gibco).GW4869(HY-19,363, MedChemExpress, China) is used to inhibit the release of exosomes.
Cell transfection
miR-328-5p inhibitor, mimic and SPON2 and HOTAIRM1 expression plasmid were synthesized from Nanjing Relgene biological company(Inhibitor is an RNA that can reduce miRNA expression through base complementation, and mimic is an RNA with the same sequence as miRNA that can improve expression); HOTAIRM1 overexpression lentivirus was synthesized from Shanghai Hanbio biological company, and HOTAIRM1 siRNA was purchased from Guangzhou RiboBio biological company. Lipofectamine RNA IMAX was used to transfect siRNA, miR-328-5p inhibitor and mimic, and lip3000 and P300 (Invitrogen, USA, L3000-015) were used to transfect plasmids overexpressing SPON2.In tumor cells, we used lentivirus transduction for HOTAIRM1 overexpression, while for miR-328-5p knockdown and overexpression, both tumor cells and CAFs used mimics and inhibitors. Since CAFs cannot be stably subcultured, siRNA and plasmid are used for knockdown and silencing, whether HOTAIRM1 or SPON2. After screening the best viral titer and puromycin concentration overexpressing HOTAIRM1, the cell transduction process was carried out according to the specification. Puromycin (ST551, Beyotime, China) is used to screen and maintain stable lentivirus strains.
5 Western blot analysis
Western blot analysis was performed as described in our previous study [11]. The protein concentration was quantified by the BCA protein quantification Kit(P0012, Beyotime, China), refer to the instructions of the kit for experimental operation. The primary antibodies in this study were purchased from Abcam: rabbit anti-CD9(ab236630, 1:500), TSG101(ab30871, 1:500), SPON2(ab171955, 1:1,000), ago2(ab186733, 1:500) and glyceraldehyde-3-phosphate dehydrogenase(GAPDH)(ab8245, 1:5,000). The horseradish peroxidase labeled goat anti-rabbit IgG antibody (ab205718, 1:10,000) and goat anti-mouse (ab6789, 1:10,000) were available as the secondary antibodies. Image J software was used to quantify each protein band.
Extraction and identification of exosomes
The lung adenocarcinoma cells were cultured in DMEM or RPMI 1640 supplemented with 10% FBS without exosomes for 24 h, and the culture supernatant was collected. Exosomes were isolated and purified from conditioned media of lung adenocarcinoma cells according to the standard isolation method of exosome with ultracentrifugation. Bicinchoninic Acid Assay (BCA) method was carried out to measure the total protein concentration of isolated exosomes, which was completed by a professional kit (P0012, Beyotime, China). Transmission electron microscopy (Hitachi ht7700, Tokyo, Japan) was used to verify the morphology of the isolated exosomes. Nanoparticle tracking analysis(NTA) was completed by Shanghai XP Biomed Ltd. and exosome characteristic proteins CD9 and TSG101 were identified by Westerns Blot. Dio (green) and DIL (red) are used to label exosome membrane and CAFs cell membrane to verify that exosomes are ingested by CAFs. Exosomes were ingested by CAFs and photographed by confocal microscope.
RNA extraction and quantitative real-time PCR analysis
The extraction and reverse transcription of total RNA were performed according to the previous report [11]. The expression levels of SPON2, HOTAIRM1 and miR-328-5p were analyzed by quantitative real-time PCR with the glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene or U6 as a standard control. GAPDH was served as reference for the total RNA of PCR cells, U6 for the miRNA, and λ polyA (Code No. 3789, takara, Japan) for the exosome LncRNA. Primers of SPON2, HOTAIRM1, miR-328-5p and U6 were as Table 1. These primers were synthesized and purified by RiboBio (Guangzhou, China).
Table 1.
The sequences of oligonucleotides and primers
| Primer | Forward primer | Reverse primer |
|---|---|---|
| HOTAIRM1 | AGGGGGTTGAAATGTGGGTGA | CTTGAAAGTGGAGAAATAAAGTGCC |
| SPON2 | TCCTTTAACACGCGAGGCTT | TCTACCCCGTTCCTCATCGT |
| miR-328-5p | GGGGGCAGGAGGGGC | GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGACCCCTGA |
| U6 | TCGGCAGCACATATACTAA | CGCTTCACGAATTTGCGTGT |
| GAPDH | GACCTCAACTACATGGTT | AACCATGTAGTTGAGG |
Cell migration and invasion assays
The cell invasion and migration assays were performed by 24-well Transwell cell culture chambers with 8-µm sized pores with or without precoated Matrigel (BD Biosciences, San Jose, CA, USA). Same as before [11], A549 cells at a density of 2 × 104 cells/mL, were resuspended with 200µL DMEM medium (serum-free) and seeded into the upper chamber, while the lower chamber was placed with CAFs transfected with different plasmids, cultured with 600 µL DMEM medium (10% FBS, Gibco, Rockville, USA). After 24 h, the cells left in the upper chamber were removed. The cells which invaded or migrated were fixed with Paraformaldehyde, 4% (Solarbio, China) for 30 min, and stained with 0.1% crystal violet solution for 15 min, and then photographed under the inverted microscope.
Immunofluorescence assay and fluorescence in situ hybridization (FISH)
Immunofluorescence assay was performed as described in our previous study [11]. Cy3 labeled HOTAIRM1 and fish Kit (Ruibo biology, Guangzhou, China) were used in accordance with the manufacturer’s instructions. Images were captured using FV10i confocal microscope (Olympus, Japan).
Immunoprecipitation assay
RIP kit was purchased from Magna(Cat. #17-700, USA); The total RNA of CAFs was obtained on the specification. After washing, immunoprecipitated RNA was isolated and qPCR was performed.
Luciferase reporter gene
Luciferase reporter gene detection kit was purchased from Promega (the USA). HOTAIRM1 or SPON2 3’UTR fragments covering the binding sites of wild-type (WT) or mutant (MUT) miR-328-5p were prepared to generate pmirglo-HOTAIRM1-wt/mut and pmirglo-SPON2 3’utr-wt. They were co-transfected into CAFs cells with NC (A meaningless RNA as a control for miR-328-5p) or miR-328-5p mimic for 48 h.
Statistical analysis
All statistical analyses in this study were performed using Graphpad prism 8.0 software. Mann-Whitney test was used to compare two groups of clinical patients’ genes expression. Kaplan-Meier analysis was performed for survival analysis. Statistical analysis was performed using the Student’s two-tailed t-test and one-way analysis of variance (ANOVA). Asterisk indicates significant difference (*P < 0.05; **P < 0.01; ***P < 0.001).
Result
1 SPON2 is up-regulated in T1 stage lung adenocarcinoma
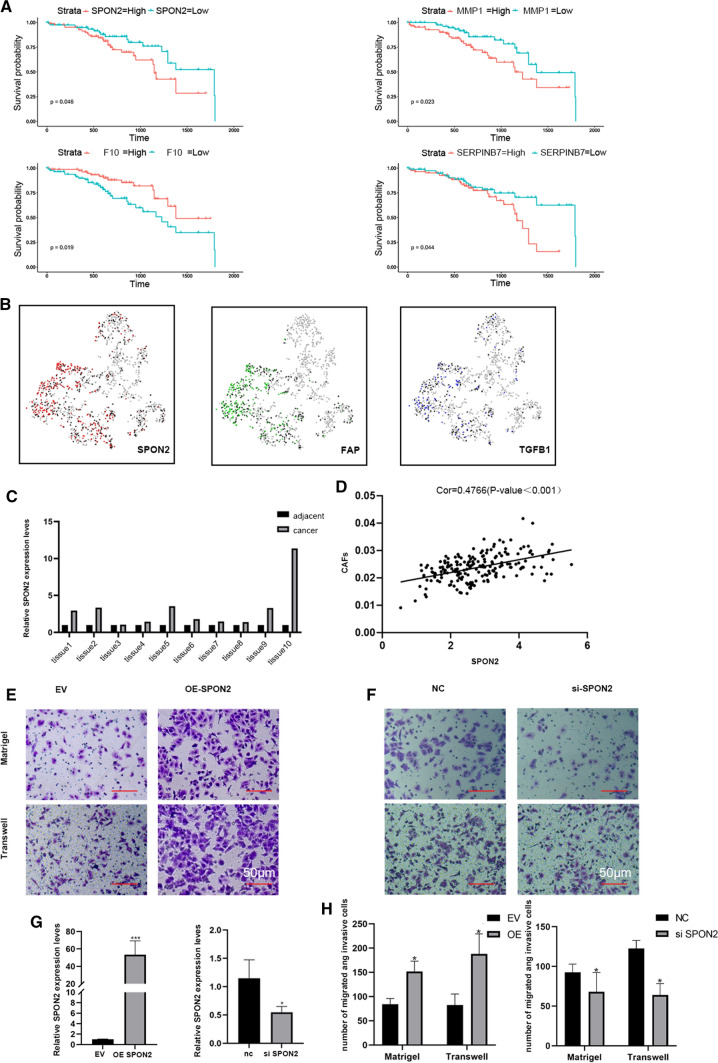
Analysis of 191 cases of T1 stage lung adenocarcinoma samples from the TCGA database revealed that the high expression of four tumor related extracellular matrix proteins was associated with poor clinical prognosis (Fig. 1A). The analysis of LOGpc further confirmed that the high expression of SPON2 was associated with the poor prognosis of early lung adenocarcinoma (Additional file 3: Fig. S1). Using R software to analyze the data of SPON2 and Lambrechts et al., we found that the calculation results suggest that SPON2 coincides with TGFB1 and FAP, which means that SPON2 is related to them [21]. (Fig. 1B). Then, qPCR was performed to verify the total RNAs extracted from ten pairs of clinical samples resected during surgery, and the results showed that the expression of SPON2 in tumor tissues was significantly higher than that of the adjacent normal tissues (Fig. 1C). Interestingly, the deconvolution calculation results of TCGA database revealed that SPON2 expression is closely related to infiltration of CAFs (Fig. 1D).
Fig. 1.
High expression of SPON2 in CAFs affects the prognosis of lung adenocarcinoma. A TCGA database analyzed the survival curve of up-regulated ECM in NSCLC. B Single cell sequencing data were used to analyze the correlation between SPON2, TGFB1 and FAP. C The expression of SPON2 in tumor and adjacent normal tissues was detected by qPCR. D The correlation between SPON2 and CAFs was analyzed by deconvolution method. (E–H) The migration and invasion abilities were measured by transwell assay in A549 cells (200× magnification). Scale bar = 100 μm. G Efficiency of SPON2 knockdown and overexpression
High expression of SPON2 in CAFs promoted the migration and invasion of NSCLC
Two NSCLC cell lines, A549 and H1299, were spread in Transwell and Matrigel chambers respectively, and co-cultured with SPON2-overexpressing CAFs for 24 and 48 h. Figure 1G shows the efficiency of SPON2 gene knockout and overexpression.The results showed that overexpression of SPON2 in CAFs significantly promoted migration and invasion of NSCLC compared with the EV group (Fig. 1E). Inhibition of SPON2 in CAFs reduced the aggressiveness of co-cultured tumor cells compared with the NC group (Fig. 1F). Figure 1G shows the transfection efficiency of overexpression and knockdown of SPON2.The migration and invasion function of H1299 is shown in Additional file 3: Fig. S2A–C.
3 Tumor derived exosomal HOTAIRM1 up-regulates the expression of SPON2 in CAFs
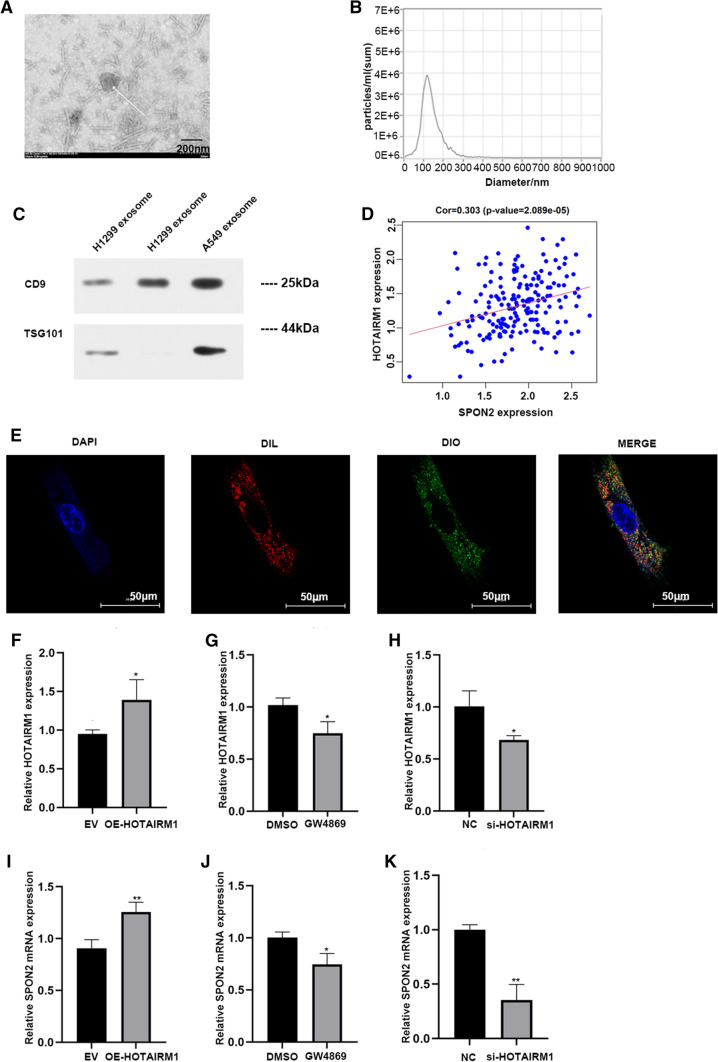
In order to study the interaction between tumor cells and CAFs, exosomes were collected from the supernatant of A549 and H1299 cells, and the isolated exosomes were detected by electron microscopy. A typical exosome is marked with a white arrow in Fig. 2A (scale, 200 nm). Further analysis of the purified particles using nanoparticle tracking analysis (NTA) showed particle sizes in the 50–200 nm range (Fig. 2B). Western blot was used to detect the expression of CD9 and TSG101 in exosomes (Fig. 2C) and the original bands are shown in Additional file 3: Fig. S3. We sequenced the lncRNA chip in the exosomes of A549 and orrelation analysis showed that HOTAIRM1 was positively correlated with SPON2 (Fig. 2D). Exosomes of A549 cells were labeled with DiO and co-cultured with CAFs labeled by DiL for 24 h, and the nucleus of CAFs was dyed blue with DAPI. We confirmed that exosomes from A549 cells were taken up by CAFs by confocal microscopy (Fig. 2E). The exosomes from H1299 cells were taken up by CAFs by confocal microscopy is shown in Additional file 3: Fig. S2D. Subsequently, A549 overexpressing HOTAIEM1 and control group were co-cultured with CAFs, respectively, and qPCR results showed the expression of SPON2 was up-regulated in CAFs in the HOTAIRM1 overexpression group (Fig. 2F, I), which was consistent with the results of previous analysis (Fig. 2D), and the expression of exosomal HOTAIRM1 after overexpression of HOTAIRM1 in A549 cells, is shown in Additional file 3: Fig. S4. In order to confirm that SPON2 in CAFs is regulated by exosomes rather than other pathways, we cultured tumor cells in culture medium containing GW4869 (a reagent that can inhibit exosome release [22]) and co-cultured them with CAFs for 48 h. The results showed that the expression of HOTAIRM1 in CAFs decreased after inhibition of A549 exosome secretion (Fig. 2G, J). Besides, we also knocked down HOTAIRM1 of tumor cells and co-cultured with CAFs. Subsequently, we observed that SPON2 expression in CAFs decreased synchronously (Fig. 2H–K). These synergistic changes supported that SPON2 in CAFs could be regulated by HOTAIRM1 from tumor cell exosomes.
Fig. 2.
Exosomes secreted by A549, H1299 regulate the expression of SPON2 in CAFs. A The structure of exosome was identified by transmission electron microscope (40.0k× magnification). Scale bar = 200 nm. B The size of exosome was detected by nanoparticle tracking analysis. C The protein levels of exosomal markers TSG101, CD9 were analyzed by western blot. D Correlation analysis between HOTAIRM1 and SPON2 (Cor = 0.303, p-value = 2.089e−05). E The fluorescence signal of CAFs cells babeled by DIL co-cultured with A549 exosomes labeled by DIO was detected (400× magnification). Scale bar = 50 μm. F–H After co-cultured with lung cancer cells were knocked-down, overexpressed of HOTAIRM1 or transfected by GW4869, the expression of HOTAIRM1 in CAFs. I–K After co-cultured with lung cancer cells were knocked-down, overexpressed of HOTAIRM1 or transfected by GW4869, the expression of SPON2 in CAFs
HOTAIRM1 regulates the mRNA level of SPON2 by competing with miR-328-5p
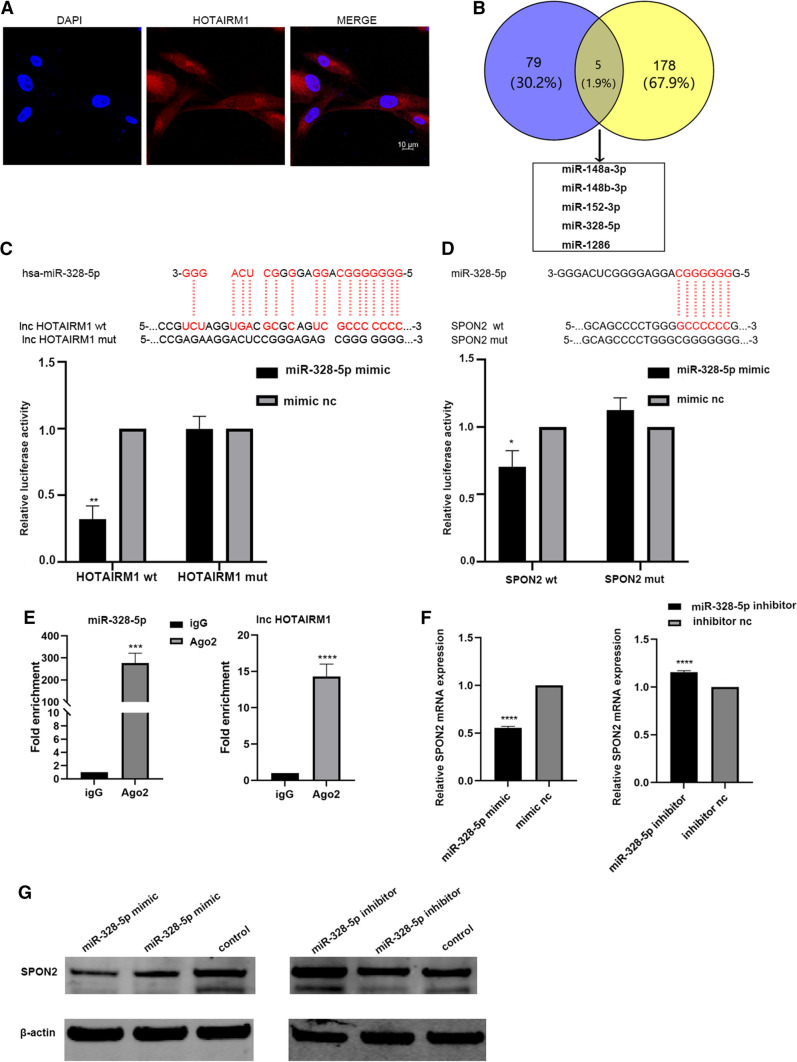
In order to explore the specific regulating mechanism of HOTAIRM1 to SPON2, the localization of HOTAIRM1 in CAFs was firstly confirmed by fluorescence in situ hybridization (FISH) experiment. HOTAIRM1 was distributed in the cytoplasm and nucleus, and mainly in the cytoplasm (Fig. 3A). In recent years, many studies reported that LncRNA in cytoplasm plays a biological role mainly through competitive combination with miRNA. We analyzed the possible pathway of HOTAIRM1 by online software, and also found that HOTAIRM1 could work through the mechanism of competing endogenous RNAs (ceRNA). The miRNAs predicted to bind to HOTAIRM1 and SPON2 were crossed by online software [23], and finally five miRNAs were selected, which can bind SPON2 and HOTAIRM1 at the same time in the prediction results (Fig. 3B), software prediction results in “Predicted miRNA Binding to SPON2 and HOTAIRM1” (Additional file 2). The wild type and mutant double luciferase reporter gene plasmids were constructed at the 3 ‘UTR region binding sites of miR-328-5p, HOTAIRM1 and SPON2, respectively (Fig. 4C, D). Further verification by luciferase reporter gene experiment showed that only miR-328-5p could bind to SPON2. Compared with the control group, the luciferase activity in the co-transfection group of wild-type luciferase plasmid and miR-328-5p simulants decreased significantly, which proved that miR-328-5p could bind to the 3’UTR region of HOTAIRM1 and SPON2 (Fig. 3C, D). The ago2 RIP experiment results showed that HOTAIRM1 could sponge-adsorb miR-328-5p (Fig. 3E). Subsequently, miR-328-5p simulants were transfected into CAFs. The qPCR results showed that the expression of SPON2 decreased significantly, which proved that miR-328-5p could inhibit the expression of SPON2 (Fig. 3F, G) (The original WB band is shown in Additional file 3: Figs. S5, S6).
Fig. 3.
HOTAIRM1 exerts its biological function by competitive adsorption of miR-328-5p. A Fluorescence in situ hybridization(FISH) detects HOTAIRM1 position. B Online software predicts miRNAs and their intersections associated with HOTAIRM1 and SPON2. C, D Luciferase reporter gene detected the binding sites of miR-328-5p with HOTAIRM1 and SPON2. E RIP experiment verified that HOTAIRM1 and miR-328-5p were involved in the formation of RISC complex. F, G miR-328-5p inhibits SPON2 expression was detected to inhibit the expression of SPON2.
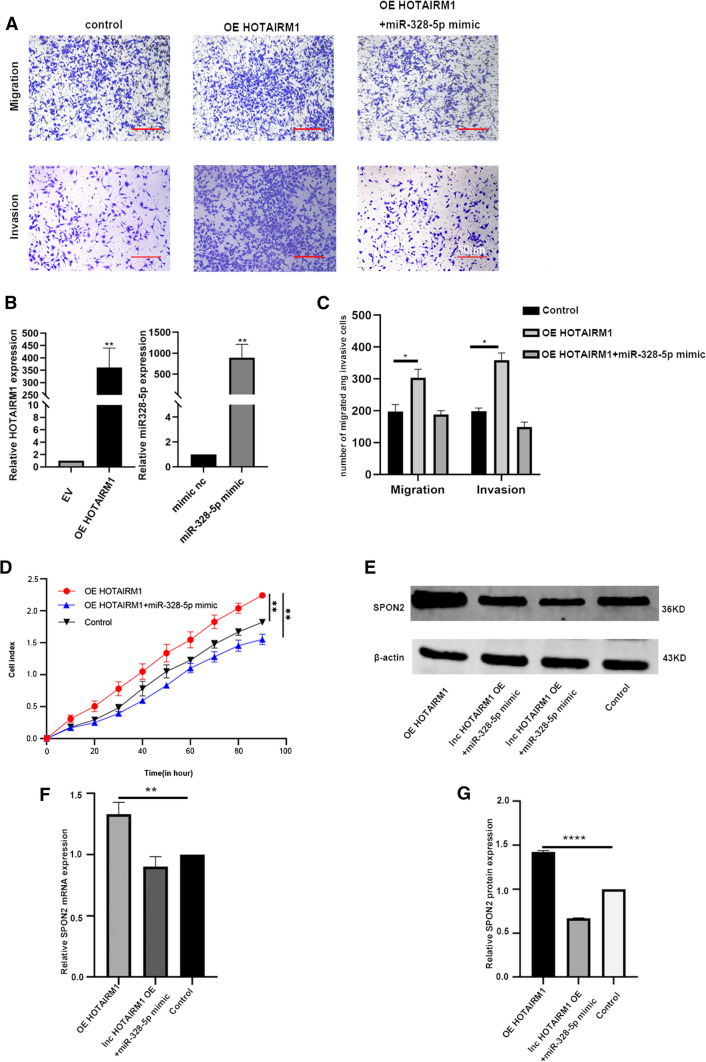
Fig. 4.
miR-328-5p promotes the migration and invasion of A549 cells by regulating the expression of SPON2 in CAFs. A The migration and invasion abilities were measured by Transwell and Matrigel assay in A549 cells (200× magnification). Scale bar = 50 μm. B PCR verifies the efficiency of overexpression of HOTAIRM1 and miR-328-5p simulants. C RTCA curve shows the invasion of A549. D, F The protein expression levels of SPON2 were analyzed by western blot and qPCR in A549 cells. E The mRNA expression levels of SPON2 were analyzed by western blot and qPCR in A549 cells. Error bars represent standard deviations and asterisks show significant differences from corresponding control according to Student’s t-test (*P < 0.05, **P < 0.01, ***P < 0.001)
The depletion of miR-328-5p eliminated the effect of HOTAIRM1 on the up-regulation of SPON2 expression
.
Rescue experiments were carried out to confirm the role of HOTAIRM1/miR-328-5p/SPON2 pathway in promoting the migration and invasion of non-small cell lung cancer. CAFs cotransfected with the overexpressing HOTAIRM1 plasmid, miR-328-5p mimics were co-cultured with A549 for 24 h. After overexpression of HOTAIRM1, the invasion and migration of tumor cells were significantly improved, but after transfection of miR-328-5p simulant, the effect of HOTAIRM1 was weakened (Fig. 4A, C), and The overexpression efficiency of HOTAIRM1 and miR-328-5p was also verified (Fig. 4B). What’s more,the proliferation curve of A549 was also lower than that of the control group (Fig. 4D).The expression levels of SPON2 protein and RNA also increased after overexpression of HOTAIRM1, while miR-328-5p weakened the effect of HOTAIRM1 (Fig. 4E–G). The migration and invasion function of H1299 is shown in Additional file 3: Fig. S2E and the original WB band is shown in Additional file 3: Fig. S7.
Discussion
In recent years, more and more studies have found that extracellular matrix (ECM) plays very important regulatory roles in tumor genesis and development [24, 25]. Abnormalities of extracellular matrix, such as tissue interruption and changes in basic components or morphology of extracellular matrix, can modulate the behavior of stromal cells and promote tumor-related angiogenesis and inflammatory response, which are related to the occurrence and metastasis of cancer [19]. A number of studies have reported that tumor cells can interact with stromal cells to dynamically regulate tumor microenvironment (TME) and eventually induce apoptosis or progression of tumor cells [26–31]. In these crosstalks, changes of proteins expressed by stromal cells are important link [32]. However, due to the extremely complex regulatory process of these proteins in the matrix environment, the molecular mechanism of matrix proteins involved in lung cancer progression is not fully understood.
In this study, an extracellular matrix protein SPON2 was found to be highly expressed in T1 stage lung adenocarcinoma and associated with poor prognosis by analysis of TCGA database. SPON2 (also known as spondin-2) is a highly conserved ECM protein. It is a member of the Mindin-f-Spondin (FS) family of secreted ECM proteins [33] and is widely expressed in spleen and lymph nodes, but not in lung cancer cells [34]. In recent years, SPON2 has been proved to promote cancer progression in many malignant tumors, such as by regulating Notch signaling pathway or activates integrin β1/Pyk2 axis to promote tumor migration and growth and is associated with lymph node metastasis [31, 35–37]. Especially, SHOTARO KURAMITSU et al. found that SPON2 was highly expressed in CAFs, which can increase the proliferation and invasion of gastric cancer cells. More interestingly, it is also associated with peritoneal dissemination in gastric cancer [37]. CAFs is one of the highest cellular components in tumor matrix and has strong ability of remodeling extracellular matrix. It also promotes tumor progression by inducing stromal cell fibrosis and increasing stromal tension [9, 10].
In this study, by deconvolution calculation of T1 stage lung adenocarcinoma in TCGA database, it was found that the expression of SPON2 was closely related to the infiltration of CAFs. Interestingly, it’s contradictory to results of other previous reports that lung cancer cells do not express SPON2 [34], which attracts our interest. What’s more, how CAFs interact with tumor cells and the SPON2 regulation mechanism in CAFs have not been thoroughly studied. Whether tumor cells directly regulate SPON2 expression in CAFs has not been reported. Here we try to explore this phenomenon. It has been recognized that exosome-mediated intercellular communication widely existed between tumor cells and stromal cells [38, 39]. In our recent study, we found exosomes mediated the communication between tumor-associated macrophages (TAMs) and tumor cells and promoted the progression of NSCLC [11]. This also inspired us that tumor cells may regulate SPON2 expression in CAFs through exosome pathway, so as to promote the migration and invasion of tumor cells. Further cell experiments confirmed that exosomes HOTAIRM1 could be absorbed by CAFs, and changes of HOTAIRM1 could lead to corresponding changes of SPON2 expression in CAFs. These results reveal that HOTAIRM1 in exosomes derived from NSCLC cells regulates the expression of SPON2 in CAFs, which provides new evidence for exosome-mediated communication between tumor cells and ECM cells.
HOTAIRM1 is an oncogenic lncRNA, and in recent years, it has been proved to regulate the proliferation and invasiveness of many malignant tumors and affect the prognosis of tumors [40–42].However, some studies have found that HOTAIRM1 played an anti-cancer role in some tumors [43–45] and its specific biological function in lung adenocarcinoma is still unclear. There is an interaction mechanism hypothesis called competitive endogenous RNA (ceRNA) between non coding RNA and RNA (mRNA) with protein coding ability, that is, ncRNA containing miRNA binding sites (such as lncRNA, circRNA, etc.) can release the inhibition of miRNA on target genes by binding with miRNA [46].Given that the common biological mechanism of lncRNA is the ceRNA mechanism, we used bioinformatics methods and molecular experiments for further exploration, and found that after being captured by CAFs, HOTAIRM1 could definitely bind miR-328-5p to regulate SPON2 through the ceRNA mechanism. Although studies have showed that miR-328-5p play a tumor suppressive role in a variety of tumors [47, 48], there are few reports on the function of miR-328-5p in NSCLC. In our study, we found that HOTAIRM1 in lung adenocarcinoma cell exosomes, as upstream of miR-328-5p, can reduce their anti-tumor effects through competitive adsorption of the miRNA. Whether HOTAIRM1 has other biological functions in NSCLC and whether SPON2, as an important cancer-promoting ECM protein, has other regulatory pathways deserve further investigation, and it is now underway in our group.
In our series of experiments, we confirmed that exosomes-mediated communication existed between lung adenocarcinoma cells and stromal CAFs cells. Tumor exosomal HOTAIRM1 can transfer into CAFs and competitively adsorb miR-328-5p through the ceRNA mechanism, and regulate the SPON2 expression of CAFs cells, ultimately promote the progression of NSCLC. This study provides a theoretical basis for migration and invasion of NSCLC from the perspective of tumor microenvironment, and also provides a new idea and target for clinical treatment of NSCLC. Definitive therapy targeting on SPON2 or HOTAIRM1 in tumor microenvironment may improve the therapeutic effect of lung adenocarcinoma.
However, there are some deficiencies in this study. First, the specific pathway of SPON2 activating CAFs and its relationship with FAP and α-SMA was not explored. Second, SPON2 expression was more downregulated after inhibiting lung adenocarcinoma exosome release than we expected, suggesting that other regulatory mechanisms derived from lung adenocarcinoma exosomes may exist.Third, due to limitation of time and funding, the role of SPON2 and corresponding mechanism have only been confirmed in cell experiments, and further verification in animal experiments is lacking. Nevertheless, this was the first and preliminary study to find the role and potential regulatory mechanism of SPON2 in CAFs in progression of lung adenocarcinoma.
Conclusion
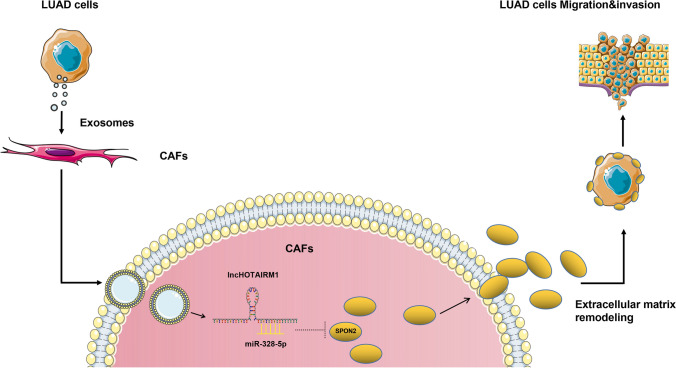
We studied the expression characteristics and biological function of SPON2 in lung adenocarcinoma, and confirmed that SPON2 is an extracellular matrix protein secreted by CAFs with high specific expression and promoting the invasion and metastasis of lung adenocarcinoma, and its expression level is significantly positively correlated with the degree of migration and invasion; SPON2 can enhance the promotion of CAFs on the invasion and progression of lung adenocarcinoma cells. Secondly, this study found that SPON2 can be derived from the exosome HOTAIRM1 of lung adenocarcinoma cells, which can affect the expression by competitive adsorption of miR-328-5p, so as to promote the invasion and metastasis of lung adenocarcinoma. Figure 5 is the schematic diagram of our mechanism.
Fig. 5.
Schematic representation of a deduced regulatory network in which HOTAIRM1 regulates SPON2 through competitive adsorption of miR-328-5p
Supplementary Information
Additional file 1: Information About CAFs from Patients.
Additional file 2: Predicted miRNA binding to spon2 and HOTAIRM1.
Additional file 3: lSupplementary Figure
Acknowledgements
This work was supported in part by key medical research project of Jiangsu Provincial Health Commission (K2019002), Ethicon Excellence in Surgery Grant (HZB-20190528-13),Postgraduate Research & Practice Innovation Program of Jiangsu Province (KYCX20_1435) and Jiangsu Province Natural Science Foundation (BK20201492).
Author contributions
(I) Conception and design: RY, JW; (II) Administrative support: LC; (III) Provision of study materials or patients: JW, RY, ZC, CB; (IV) Collection and assembly of data: ZC, CB, JH, XL; (V) Data analysis and interpretation: JW, YX, XX, RY; (VI) Manuscript writing: All authors; (VII) All authors read and approved the final manuscript.
Availability of data and materials
All data in our study were available from the TCGA database (http://cancergenome.nih.gov/).
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Rong Yin, Email: rong_yin@njmu.edu.cn.
Jun Wang, Email: drwangjun@njmu.edu.cn.
References
- 1.Bray F, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Duffy SW, Field JK. Mortality reduction with low-dose CT screening for lung cancer. 2020; 572–573. [DOI] [PubMed]
- 3.de Sousa VML, Carvalho L. Heterogeneity in lung cancer. Pathobiol J ImmunoPathol Mol Cell Biol. 2018;85(1–2):96–107. doi: 10.1159/000487440. [DOI] [PubMed] [Google Scholar]
- 4.Bu L, et al. Biological heterogeneity and versatility of cancer-associated fibroblasts in the tumor microenvironment. Oncogene. 2019;38(25):4887–901. doi: 10.1038/s41388-019-0765-y. [DOI] [PubMed] [Google Scholar]
- 5.Chen X, Song E. Turning foes to friends: targeting cancer-associated fibroblasts. Nat Rev Drug Discov. 2019;18(2):99–115. doi: 10.1038/s41573-018-0004-1. [DOI] [PubMed] [Google Scholar]
- 6.Hui L, Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Lett. 2015;368(1):7–13. doi: 10.1016/j.canlet.2015.07.039. [DOI] [PubMed] [Google Scholar]
- 7.Su S, et al. A positive feedback loop between mesenchymal-like cancer cells and macrophages is essential to breast cancer metastasis. Cancer Cell. 2014;25(5):605–20. doi: 10.1016/j.ccr.2014.03.021. [DOI] [PubMed] [Google Scholar]
- 8.Hoshino A, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015;527(7578):329–35. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhowmick NA, Neilson EG, Moses HL. Stromal fibroblasts in cancer initiation and progression. Nature. 2004;432(7015):332–7. doi: 10.1038/nature03096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 11.Li X, et al. Tumor-associated macrophages secret exosomal miR-155 and miR-196a-5p to promote metastasis of non-small-cell lung cancer. Translational Lung Cancer Research. 2021;10(3):1338–54. doi: 10.21037/tlcr-20-1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. New York: Science; 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Doyle LM, Wang MZ. Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells. 2019; 8(7). [DOI] [PMC free article] [PubMed]
- 14.Zhang J, et al. Secretome of activated fibroblasts induced by exosomes for the discovery of biomarkers in non-small cell lung cancer. Small. 2021;17(4):e2004750. doi: 10.1002/smll.202004750. [DOI] [PubMed] [Google Scholar]
- 15.Feinstein Y, et al. F-spondin and mindin: two structurally and functionally related genes expressed in the hippocampus that promote outgrowth of embryonic hippocampal neurons. Development. 1999;126(16):3637–48. doi: 10.1242/dev.126.16.3637. [DOI] [PubMed] [Google Scholar]
- 16.Yuan X, et al. Spondin2 is a new prognostic biomarker for lung adenocarcinoma. Oncotarget. 2017;8(35):59324–32. doi: 10.18632/oncotarget.19577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen T, et al. LncRNA HOTAIRM1 inhibits the proliferation and invasion of lung adenocarcinoma cells via the miR-498/WWOX Axis. Cancer Manage Res. 2020;12:4379–90. doi: 10.2147/CMAR.S244573. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Xiong F, et al. Clinicopathologic features and the prognostic implications of long noncoding RNA HOTAIRM1 in non-small cell lung cancer. Genetic Test Mol Biomarkers. 2020;24(1):47–53. doi: 10.1089/gtmb.2019.0102. [DOI] [PubMed] [Google Scholar]
- 19.Lim SB, et al. An extracellular matrix-related prognostic and predictive indicator for early-stage non-small cell lung cancer. Nat Commun. 2017;8(1):1734. doi: 10.1038/s41467-017-01430-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambrechts D, et al. Phenotype molding of stromal cells in the lung tumor microenvironment. Nat Med. 2018;24(8):1277–89. doi: 10.1038/s41591-018-0096-5. [DOI] [PubMed] [Google Scholar]
- 21.Catalano M, O’Driscoll L. Inhibiting extracellular vesicles formation and release: a review of EV inhibitors. J Extracell vesicles. 2020;9(1):1703244. doi: 10.1080/20013078.2019.1703244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paraskevopoulou MD, et al. DIANA-LncBase v2: indexing microRNA targets on non-coding transcripts. Nucleic Acids Res. 2016;44(D1):D231-8. doi: 10.1093/nar/gkv1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Winkler J, et al. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun. 2020;11(1):5120. doi: 10.1038/s41467-020-18794-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Erdogan B, Webb DJ. Cancer-associated fibroblasts modulate growth factor signaling and extracellular matrix remodeling to regulate tumor metastasis. Biochem Soc Trans. 2017;45(1):229–36. doi: 10.1042/BST20160387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shree T, et al. Macrophages and cathepsin proteases blunt chemotherapeutic response in breast cancer. Genes Dev. 2011;25(23):2465–79. doi: 10.1101/gad.180331.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pyonteck SM, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–72. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khazaie K, et al. The significant role of mast cells in cancer. Cancer Metastasis Rev. 2011;30(1):45–60. doi: 10.1007/s10555-011-9286-z. [DOI] [PubMed] [Google Scholar]
- 28.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat Rev Cancer. 2009;9(4):239–52. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309–22. doi: 10.1016/j.ccr.2012.02.022. [DOI] [PubMed] [Google Scholar]
- 30.Huang C, et al. Tumor cell-derived SPON2 promotes M2-polarized tumor-associated macrophage infiltration and cancer progression by activating PYK2 in CRC. J Exp Clin Cancer Res CR. 2021;40(1):304. doi: 10.1186/s13046-021-02108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Breznik B, Motaln H, Lah T, Turnšek Proteases and cytokines as mediators of interactions between cancer and stromal cells in tumours. Biol Chem. 2017;398(7):709–19. doi: 10.1515/hsz-2016-0283. [DOI] [PubMed] [Google Scholar]
- 32.He Y, et al. The extracellular matrix protein mindin is a pattern-recognition molecule for microbial pathogens. Nat Immunol. 2004;5(1):88–97. doi: 10.1038/ni1021. [DOI] [PubMed] [Google Scholar]
- 33.Manda R, et al. Identification of genes (SPON2 and C20orf2) differentially expressed between cancerous and noncancerous lung cells by mRNA differential display. Genomics. 1999;61(1):5–14. doi: 10.1006/geno.1999.5939. [DOI] [PubMed] [Google Scholar]
- 34.Žlajpah M, et al. Differential expression of extracellular matrix-related genes DCN, EPHA4, FN1, SPARC, SPON2 and SPP1 in colorectal carcinogenesis. Oncol Rep. 2019;42(4):1539–48. doi: 10.3892/or.2019.7274. [DOI] [PubMed] [Google Scholar]
- 35.Kang H, et al. SPON2 is upregulated through notch signaling pathway and promotes tumor progression in gastric cancer. Cancers. 2020; 12(6). [DOI] [PMC free article] [PubMed]
- 36.Kuramitsu S, et al. Cancer-associated fibroblast-derived Spondin-2 promotes motility of gastric cancer cells. Cancer Genomics Proteomics. 2021;18(4):521–9. doi: 10.21873/cgp.20277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian X, et al. Tumor-derived exosomes, myeloid-derived suppressor cells, and tumor microenvironment. J Hematol Oncol. 2019;12(1):84. doi: 10.1186/s13045-019-0772-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whiteside TL. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin Immunol. 2018;35:69–79. doi: 10.1016/j.smim.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Q, et al. Over-expressed lncRNA HOTAIRM1 promotes tumor growth and invasion through up-regulating HOXA1 and sequestering G9a/EZH2/Dnmts away from the HOXA1 gene in glioblastoma multiforme. J Exp Clin Cancer Res CR. 2018;37(1):265. doi: 10.1186/s13046-018-0941-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li X, et al. LncRNA HOTAIRM1/HOXA1 axis promotes cell proliferation, migration and invasion in endometrial cancer. Onco Targets Ther. 2019;12:10997–1015. doi: 10.2147/OTT.S222334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li D, et al. The HOTAIRM1/miR-107/TDG axis regulates papillary thyroid cancer cell proliferation and invasion. Cell Death Dis. 2020;11(4):227. doi: 10.1038/s41419-020-2416-1. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Chao H, et al. HOTAIRM1 suppresses cell proliferation and invasion in ovarian cancer through facilitating ARHGAP24 expression by sponging miR-106a-5p. Life Sci. 2020;243:117296. doi: 10.1016/j.lfs.2020.117296. [DOI] [PubMed] [Google Scholar]
- 43.Hamilton MJ, et al. HOTAIRM1 lncRNA is downregulated in clear cell renal cell carcinoma and inhibits the hypoxia pathway. Cancer Lett. 2020;472:50–8. doi: 10.1016/j.canlet.2019.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen T, et al. LncRNA HOTAIRM1 inhibits the proliferation and invasion of lung adenocarcinoma cells via the miR-498/WWOX Axis. Cancer Manage Res. 2020;12:4379–90. doi: 10.2147/CMAR.S244573. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 45.Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–52. doi: 10.1038/nature12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tang J, et al. LncRNA LOC146880 promotes esophageal squamous cell carcinoma progression via miR-328-5p/FSCN1/MAPK axis. Aging. 2021;13(10):14198–218. doi: 10.18632/aging.203037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Luo T, et al. miR-328-5p inhibits MDA-MB-231 breast cancer cell proliferation by targeting RAGE. Oncol Rep. 2018;39(6):2906–14. doi: 10.3892/or.2018.6353. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, et al. LncRNA ITGB2-AS1 promotes the progression of clear cell renal cell carcinoma by modulating miR-328-5p/HMGA1 axis. Hum Cell. 2021;34(5):1545–57. doi: 10.1007/s13577-021-00563-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Information About CAFs from Patients.
Additional file 2: Predicted miRNA binding to spon2 and HOTAIRM1.
Additional file 3: lSupplementary Figure
Data Availability Statement
All data in our study were available from the TCGA database (http://cancergenome.nih.gov/).