Abstract
SecG is an auxiliary protein in the Sec-dependent protein export pathway of Escherichia coli. Although the precise function of SecG is unknown, it stimulates translocation activity and has been postulated to enhance the membrane insertion-deinsertion cycle of SecA. Deletion of secG was initially reported to result in a severe export defect and cold sensitivity. Later results demonstrated that both of these phenotypes were strain dependent, and it was proposed that an additional mutation was required for manifestation of the cold-sensitive phenotype. The results presented here demonstrate that the cold-sensitive secG deletion strain also contains a mutation in glpR that causes constitutive expression of the glp regulon. Introduction of both the glpR mutation and the secG deletion into a wild-type strain background produced a cold-sensitive phenotype, confirming the hypothesis that a second mutation (glpR) contributes to the cold-sensitive phenotype of secG deletion strains. It was speculated that the glpR mutation causes an intracellular depletion of glycerol-3-phosphate due to constitutive synthesis of GlpD and subsequent channeling of glycerol-3-phosphate into metabolic pathways. In support of this hypothesis, it was demonstrated that addition of glycerol-3-phosphate to the growth medium ameliorated the cold sensitivity, as did introduction of a glpD mutation. This depletion of glycerol-3-phosphate is predicted to limit phospholipid biosynthesis, causing an imbalance in the levels of membrane phospholipids. It is hypothesized that this state of phospholipid imbalance imparts a dependence on SecG for proper function or stabilization of the translocation apparatus.
The Sec-dependent translocation of secretory proteins across the inner membrane of Escherichia coli is catalyzed by the action of translocase, a complex of cytoplasmic and inner membrane proteins. The core components of translocase are the integral membrane proteins SecY and SecE and the peripheral membrane protein, SecA. SecY and SecE interact to form a protein-conducting channel that translocates secretory proteins across the membrane, while SecA is an ATPase that binds to the SecYE complex. SecA undergoes cycles of membrane insertion and deinsertion coupled to ATP binding and hydrolysis that are believed to drive the segmental translocation of the secretory protein (13, 15, 18, 32).
A number of genetic and biochemical studies have demonstrated that SecY, SecE, and SecA are necessary and sufficient for preprotein translocation (2, 4, 9, 10, 34). However, translocation with only these three components of translocase does not achieve optimal efficiency; the interaction of additional inner membrane proteins with the SecYE complex enhances translocation activity. SecG is a small protein that copurifies with SecYE and stimulates translocation activity in reconstituted membrane vesicles (8, 35), while SecD, SecF, and YajC form a heterotrimeric complex that also interacts with SecYE (16). Duong and Wickner (16) demonstrated that SecYE can associate with either SecG or SecDFYajC to generate full translocation activity. It appears that both SecG and SecDFYajC modulate the SecA cycle of insertion and deinsertion, but they do so by subtly different mechanisms. SecG stimulates the insertion of SecA after initiation of translocation, while SecDFYajC increases SecA insertion and stabilizes the inserted state. Thus, SecG and SecDFYajC appear to have overlapping roles in fully activating the translocation process (16).
Neither SecG nor SecDFYajC is required for translocation activity, although each stimulates activity in vitro (16, 35). In keeping with these observations, none of these genes are essential for viability of E. coli (19, 33, 36). However, depletion of SecDF has a profound effect on cell growth and protein export. Null mutants of secDF form only minute colonies at 37°C and are unable to form colonies at all at 30°C or lower (36). In contrast, deletion of secG results in a less severe export defect and a cold-sensitive phenotype that is manifest only at very low temperatures (20°C) (33). Importantly, both the cold sensitivity and the severity of the ΔsecG export defect have been shown to be strain dependent. Indeed, only the initial deletion strains (including KN370) containing the secG deletion are cold sensitive; no other strain could be constructed that produced this phenotype (6, 19). We previously proposed that the cold sensitivity of KN370 was due to the combination of ΔsecG::kan with another unidentified mutation (19). In this report, that hypothesis is verified and the mutation in KN370 that acts in concert with ΔsecG::kan to produce a cold-sensitive phenotype is identified.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
All bacterial strains used are E. coli K-12 derivatives and are listed in Table 1. Bacteria were grown in Luria-Bertani (LB) or M63 minimal media, with kanamycin (50 mg/liter), ampicillin (125 mg/liter), or tetracycline (25 mg/liter) added when appropriate (40). The dl-α-glycerophosphate (Sigma) was a generous gift from John Cronan (University of Illinois at Urbana-Champaign). Plasmid pBAD18 has been described elsewhere (23). Plasmids pAF69 and pAF70 were constructed by PCR amplification of glpE and glpR, respectively, from the chromosome of MG1655, using primers with restriction sites near the termini (glpE-1, GCAATGCCCGGGTACCGTAAAGAAAGAGAGACGCATG; glpE-2, CCTAGCCTGCAGAAGCTTGACAGTATAAAGCGTTACGC; glpR-1, GCAATGCCCGGGTACCATTCCAGGGATTTATAAATG; and glpR-2, CCTAGCCTGCAGAAGCTTGCACAGCTCCAGTTGAATAT; KpnI and HindIII sites are underlined), followed by digestion with KpnI and HindIII and ligation with pBAD18 that had been digested with KpnI and HindIII. Preparation of competent cells, transformation, and genetic manipulations were performed as described previously (37, 40).
TABLE 1.
Bacterial strains used in this study
| Strain | Relevant characteristics | Source | Reference |
|---|---|---|---|
| KN370 | FS1576 (C600 recD1009) ΔsecG::kan | Lab stock | 33 |
| MC4100 | F−araD139 Δ(argF-lac)U169 rpsL150 relA1 flhD5301 fruA25 deoC1 thi rbsR22 | Lab stock | 11 |
| Lin205 | fhuA22 ΔphoA8 ompF627 glpA24 fadL701 pit-10 spoT1 relA1 glpD26 glpR8 (Const) glpR7?a | E. coli Genetic Stock Center | 12 |
| MG1655 | F−rph-1 | Lab stock | 22 |
| AF538 | KN370 zgi-203::Tn10 secG+ | Lab stock | 19 |
| AF539 | KN370 zgi-203::Tn10 | Lab stock | 19 |
| AF634 | KN370 yzgL::Tn10 glpR+ | This study | |
| AF635 | MC4100 glpR200 yzgL::Tn10 | This study | |
| AF636 | MC4100 yzgL::Tn10 | This study | |
| AF637 | MC4100 ΔsecG::kan glpR200 yzgL::Tn10 | This study | |
| AF638 | MC4100 ΔsecG::kan Tn10 | This study | |
| AF645 | KN370 zhe-3084::Tn10 | This study | |
| AF646 | KN370 glpD26 glpR8 (Const) glpR7? zhe-3084::Tn10 | This study |
According to the E. coli Genetic Stock Center, glpR7 may or may not be present in this strain.
Growth curves.
Growth curves were performed with a Bioscreen C Microbiology Reader from Labsystems. The Bioscreen C is a computer-controlled shaker-incubator-reader. Customized microplates hold up to 200 individual 400-μl samples. The Bioscreen maintains the samples at the desired temperature, shakes the samples, and measures the optical density at set intervals. Bacterial dilutions were prepared in LB medium prior to inoculation into the microplates, and cell density was determined by viable plate counts from the diluted cultures. For the experiments described here, the temperature was held at 20°C, shaking occurred once every minute for 10 s, and the A600 was measured and recorded every 30 min. The growth curves were continued until all cultures reached stationary phase, usually in 5 days. Data were exported to a Microsoft Excel worksheet for analysis.
Construction of the mini-Tn10 library.
The mini-Tn10 library, constructed in strain MG1655, was a generous gift from Majda Valjavec-Gratian and Thomas Hill (University of North Dakota). P1-mediated transduction was used to introduce the library into strain KN370, with selection for tetracycline-resistant transductants. Following transduction, 200 individual colonies were inoculated into LB medium containing 10 mM sodium citrate and 12 mg of tetracycline/liter. The colonies were inoculated into 20 tubes, with 10 inocula per tube. The mixed cultures were grown overnight at 37°C and then diluted to a starting concentration of approximately 500 CFU/ml. Duplicate samples (400 μl) of each culture were inoculated into microwell plates, and growth rates were analyzed in the Bioscreen C as described above.
Inverse PCR.
Inverse PCR was performed as described previously (31). In short, chromosomal DNA from fast-growing transductants was digested with various restriction enzymes that do not cleave within the mini-Tn10 sequence (BamHI, PstI, KpnI, or XhoI). The digested DNA was ligated in a dilute reaction mixture and then used as a template for PCR amplification. The primers used for PCR were derived from mini-Tn10 sequences (Tn10T-645, GAACCATTTTCAGTGATCCATTGCTGTTGAC and Tn10T-3358, AAAGTGATGATAAAAGGCACCTTTGG) and were oriented such that the resultant PCR product would include the chromosomal DNA. DNA from the samples with the most easily detectable PCR product (from the KpnI digest) were purified and used for DNA sequence analysis.
DNA sequencing.
All PCR DNA to be sequenced was purified by agarose gel electrophoresis followed by Qiaex II gel extraction (Qiagen). DNA sequencing was performed by the Macromolecular Resources Sequi-Net Department at Colorado State University (Fort Collins).
RESULTS
Characterization of the ΔsecG::kan cold-sensitive phenotype.
Previous studies demonstrated that the presence of the ΔsecG::kan allele resulted in a cold-sensitive phenotype only in certain strains, specifically the original deletion strain, KN370, and its derivatives (6, 19, 33). It was also noted that the cold sensitivity manifested by these strains was a leaky phenotype (19). That is, the cold-sensitive strains were able to form single colonies at 20°C, albeit at a slower rate than wild-type strains. After streaking on solid media and incubation at 20°C, the primary streaks grew normally while a marked defect was apparent in streaks further out on the plate. These results suggested that the cold-sensitive strains had a defect in single colony formation but were able to grow well at high cell density (in the initial streak).
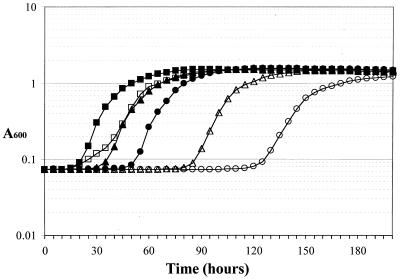
To characterize the ΔsecG::kan-related growth defect more carefully, growth rates in liquid media were determined for the isogenic strains AF538 (secG+) and AF539 (ΔsecG::kan), both derivatives of KN370. Surprisingly, in a standard growth curve experiment in which overnight cultures were diluted to a starting A600 of 0.1, there was no discernable difference in the growth rates of the two strains, even at 20°C (data not shown). Because the growth pattern on plates suggested that the cold-sensitive phenotype was exacerbated when cells were more dilute, the effect of increasing dilution on the growth rate in liquid media was examined (Fig. 1). Overnight cultures were diluted such that the starting concentration of bacteria ranged from 3 to 4 CFU/ml to 37,500 CFU/ml in 10-fold increments. At the highest bacterial concentration (37,500 CFU/ml), there was a slight difference in the growth curve (Fig. 1), with AF539 (ΔsecG::kan) growing more slowly in early exponential growth phase and then at the same rate as AF538 (secG+) in late exponential phase. As the cultures were diluted further, a delay before the onset of exponential growth became more pronounced (Fig. 1). In all cases, however, the maximal growth rate of AF539 was indistinguishable from that of AF538. Therefore, the cold sensitivity is a concentration-dependent defect in the ability of these ΔsecG::kan cells to enter exponential growth at the maximal rate.
FIG. 1.
Growth curves of AF538 and AF539. Strains were diluted from overnight cultures to starting concentrations of 37,500 CFU/ml (squares), 375 CFU/ml (triangles), or 3 to 4 CFU/ml (circles) and grown at 20°C in LB. AF538 is represented by closed symbols and AF539 is shown by open symbols. Although absorbance was measured every 30 min, for clarity only readings for each 5-h increment are shown.
To ensure that the eventual wild-type growth of AF539 was not due to acquisition of a suppressor mutation, AF539 was allowed to grow to saturation at 20°C from the most dilute sample, and then serial dilutions were performed and the growth rate was measured again. The pattern of dilution-dependent extension of lag phase was completely reproducible, indicating that this growth pattern is an inherent characteristic of the strain (data not shown). To determine whether the defect was due to an inability to emerge from lag phase, or if it was a concentration-dependent growth defect regardless of growth phase, bacteria in exponential growth at 20°C were diluted and the growth rates were measured. Again, the more dilute the cells, the longer the apparent lag phase before exponential growth began (data not shown). These results indicated that AF539 exhibits a cold-sensitive growth defect that manifests as an inability to grow at maximal rate when the cell culture is dilute.
The growth defect of KN370 can be rescued by a single gene.
The observed growth defect of AF539 was clearly dependent on the deletion of secG, as its isogenic partner, AF538, did not exhibit this cold sensitivity. To confirm our previous hypothesis that a second mutation contributes to this phenotype (19) and to identify the second mutation, the presumed mutation was replaced with a wild-type allele, thereby restoring normal growth characteristics to the strain. To this end, a mini-Tn10 library was constructed from a wild-type strain (MG1655) and transduced into KN370 (AF539 could not be used because it is tetracycline resistant). If the growth defect is due to a single other mutation in combination with ΔsecG::kan, then some transductants should receive a wild-type copy of that gene and thereby regain normal growth characteristics.
Preliminary investigation revealed that mixing nine colonies of AF539 (slow grower) and one colony of AF538 (fast grower) would yield a mixed culture that grew with a phenotype intermediate to the two, but that was easily distinguishable from AF539; that is, one fast grower in a mix of 10 colonies will overtake the culture quickly enough that its phenotype will predominate. Accordingly, transductants were pooled into 20 cultures with 10 colonies each (200 total individual transductants) and incubated at 37°C. After overnight growth, the pooled cultures were diluted and growth rates at 20°C were measured to identify pools that contained fast growers. Six such cultures were identified. Although each culture represented a mix of transductants, the fast grower should have predominated. Therefore, each culture was plated for single colonies, and one colony was isolated for further analysis.
There are two ways a colony could become a fast grower in this experiment. One is the replacement of the unidentified mutant gene with a wild-type copy, and the other is replacement of ΔsecG::kan with wild-type secG. To eliminate the latter isolates, colonies from each of the fast-growing pools were isolated and examined for kanamycin resistance. Two of the six had become kanamycin sensitive, demonstrating that ΔsecG::kan had been replaced by a wild-type copy of secG. However, the four remaining colonies were kanamycin resistant, indicating that the ΔsecG::kan allele was still present. These four isolates were examined in pure culture for their growth characteristics. Three cultures repeated the fast-growth phenotype, while one did not. Therefore, in these three isolates, wild-type growth characteristics were restored while the strain retained the secG deletion.
To ensure that the enhanced growth of these isolates was due to replacement of a single mutant gene in KN370, the mini-Tn10 from each isolate was transduced back into KN370 and tetracycline resistance was selected. Fifty colonies from each transduction were patched on solid media at 20°C and examined for cold sensitivity. The majority (>90%) of colonies from each group of transductants appeared to be cold resistant. As this test is not as sensitive as the growth curves, a colony of each was grown in LB media, diluted, and analyzed for growth. All three were found to be fast growers. These results confirmed our hypothesis that a single mutant gene in combination with ΔsecG::kan was responsible for the cold-sensitive phenotype of KN370 and its derivatives.
Cold sensitivity can be rescued by glpR+.
To identify the gene that contributes to the cold sensitivity of KN370, inverse PCR was used to locate the site of insertion of two of the three mini-Tn10s. Both mapped to approximately 76.5 min on the E. coli chromosome, one in glpG and the other in yzgL (5). Cotransduction frequencies indicated that the gene that rescues the growth defect was approximately 95 to 98% linked to each of the mini-Tn10s. Cloning of chromosomal DNA fragments demonstrated that a 6-kb KpnI fragment, which contains only five intact genes, glpR, glpG, glpE, glpD, and yzgL, was able to complement the cold sensitivity of KN370. As the mini-Tn10s were located in two of these genes, only three candidate genes remained, glpD, glpE, and glpR. The glpE and glpR wild-type genes were amplified from MG1655 by PCR and subcloned into pBAD18. Recombinant plasmids were transformed into KN370, and growth was examined at 20°C in the presence and absence of arabinose. The plasmid containing glpR rescued growth of KN370 in the presence of arabinose. This result suggested that a mutation in glpR contributed to the growth defect in KN370 and, further, that the glpR mutation was recessive because the growth defect was complemented by plasmid-borne wild-type glpR.
KN370 has a constitutive mutation in glpR.
The glpR gene encodes the repressor for the glp regulon, which is involved in metabolism of glycerol phosphate (38). KN370 is a derivative of C600 (3), which is known to have a mutation in glpR, glpR200 (17). To confirm that KN370 did in fact contain the glpR200 mutation, glpR was PCR amplified from KN370 as well as from fast-growing derivatives, and DNA sequence analysis was performed. KN370 glpR had a mutation in amino acid 55 from Gly to Ala, while the fast-growing derivatives did not contain this mutation. Because KN370 was complemented by the wild-type copy of glpR, it seemed likely that this mutation resulted in an inactive GlpR repressor and constitutive synthesis of the glp regulon. Indeed, the original characterization of glpR200 in C600 demonstrated that it does result in constitutive expression of the regulon (17). Further, this same amino acid alteration has been identified independently as glpR2 and characterized as a constitutive mutation (Donald Pettigrew, Texas A&M University, personal communication).
The combination of ΔsecG::kan and glpR200 results in a cold-sensitive phenotype.
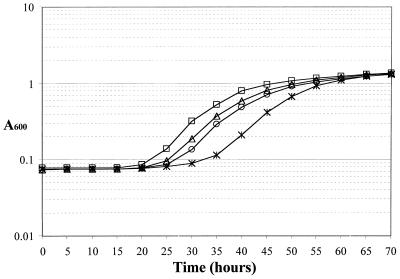
To verify that the glpR200 mutation in combination with ΔsecG::kan results in cold sensitivity, both mutations were introduced into MC4100 by P1-mediated transduction. As shown previously, ΔsecG::kan alone does not produce a cold-sensitive phenotype in MC4100 (Fig. 2). Furthermore, glpR200 alone does not result in cold sensitivity (Fig. 2). However, the two mutations in combination did cause a cold-sensitive growth defect in the MC4100 background (Fig. 2). The lag in growth was not as striking as it was in KN370, but it was reproducible and was detectable by streaking as well as by growth curve analysis. The growth defect was more apparent when the length of time required to reach mid-log phase (A600 = 0.3) was calculated. The parent and singly mutant strains reached this point in 30 to 35 h, while the doubly mutant strain required 42.5 h to reach the same absorbance. This result explains the puzzling prior observations that ΔsecG::kan resulted in cold sensitivity in some strains but not in others. Only when glpR200 and ΔsecG::kan were both present in the same strain was a growth defect observed.
FIG. 2.
Growth curves for MC4100 containing ΔsecG::kan and glpR200. Strains were diluted from overnight cultures to a starting concentration of approximately 400 CFU/ml and grown in LB medium at 20°C. Symbols: AF636 (secG+ glpR+), open triangles; AF638 (ΔsecG::kan), open squares; AF635 (glpR200), open circles; AF637 (ΔsecG::kan glpR200), asterisks. Absorbance readings for each 5-h increment are shown.
Growth of KN370 is improved by increasing the intracellular concentration of glycerol-3-phosphate.
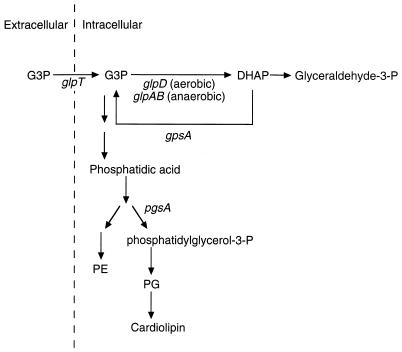
Why does the combination of glpR200 and ΔsecG::kan result in cold sensitivity? One possible explanation is that constitutive expression of the glp regulon results in depletion of the intracellular pool of glycerol-3-phosphate, due to overexpression of glpD (the aerobic glycerol-3-phosphate dehydrogenase) and subsequent channeling of glycerol-3-phosphate into metabolic pathways (29). It is possible that this depletion limits the glycerol-3-phosphate available for phospholipid biosynthesis, leading to perturbations in the phospholipid levels (Fig. 3). Under these circumstances, the function of SecG may become critical. If this hypothesis is correct, the growth defect should be alleviated either by addition of glycerol-3-phosphate to the media or by introduction of a null mutation in glpD.
FIG. 3.
Pathways for glycerol-3-phosphate and phospholipid metabolism. Abbreviations are as follows: G3P, glycerol-3-phosphate; DHAP, dihydroxyacetone phosphate; PE, phosphatidylethanolamine; PG, phosphatidylglycerol. Genes encoding relevant proteins are indicated. Not all steps of phospholipid biosynthesis are shown. This figure is adapted from previous reviews (14, 29).
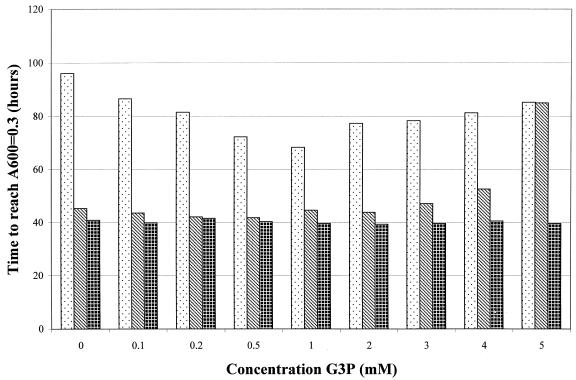
This hypothesis first was tested by growing AF538 (glpR200), AF539 (ΔsecG::kan, glpR200), and AF634 (ΔsecG::kan) at 20°C with increasing concentrations of glycerol-3-phosphate and monitoring growth (Fig. 4). At low levels of glycerol-3-phosphate (<2 mM), growth of the doubly mutant strain was greatly improved, with the length of time required to reach mid-log phase (A600 = 0.3) dropping from 96 to 68 h (Fig. 4). However, increasing the glycerol-3-phosphate concentration above 2 mM resulted in an inhibition of growth. This inhibition occurred in the glpR200 single mutant as well (Fig. 4), while growth of the glpR+ ΔsecG::kan strain was unaltered at any concentration of glycerol-3-phosphate (Fig. 4). The growth inhibition in a glpR mutant strain at higher levels of glycerol-3-phosphate is not surprising, as it is known that excess intracellular glycerol-3-phosphate is detrimental to the bacterial cell (1, 20) and that the glpR200 mutation will result in constitutive expression of GlpT, the transporter for glycerol-3-phosphate, thereby allowing the uptake of excess glycerol-3-phosphate (21, 29). The observation that is important to these studies is that glycerol-3-phosphate did rescue the growth defect of the double mutant, supporting the hypothesis that intracellular pools of glycerol-3-phosphate are limiting in the glpR mutant strain.
FIG. 4.
Growth characteristics in the presence of glycerol-3-phosphate. Strains were grown in LB with the indicated amounts of glycerol-3-phosphate (G3P) at 20°C. Strains were diluted from overnight cultures to a starting concentration of approximately 400 CFU/ml. The bars represent the number of hours required for each culture to reach mid-log phase (A600 = 0.3). AF539 (ΔsecG::kan glpR200), stippled bars; AF538 (glpR200), striped bars; AF634 (ΔsecG::kan), cross-hatched bars. All assays were performed in duplicate, and the standard error never exceeded ±5%.
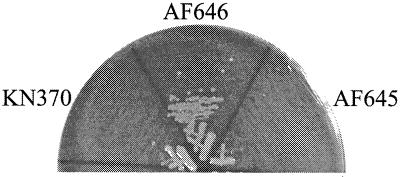
To further test the hypothesis that the growth defect was attributable to constitutive expression of glpD, a glpD mutation was introduced into KN370 and growth at 20°C was assessed. According to this hypothesis, the glpD mutation should prevent the excessive channeling of glycerol-3-phosphate to metabolic pathways, thereby promoting synthesis of phospholipids and alleviating the growth defect. Indeed, strain AF646 containingΔsecG::kan, glpR8 (another glpR constitutive allele), and glpD26 was cold resistant (Fig. 5), while the isogenic glpD+ strain (AF645) remained cold sensitive. This result directly demonstrated that overexpression of glpD was contributory to the growth defect of KN370.
FIG. 5.
Effect of glpD26 on growth. Strains were grown on LB plates at 20°C. AF645 is ΔsecG::kan glpR200, while AF646 is ΔsecG::kan glpR8 glpD26.
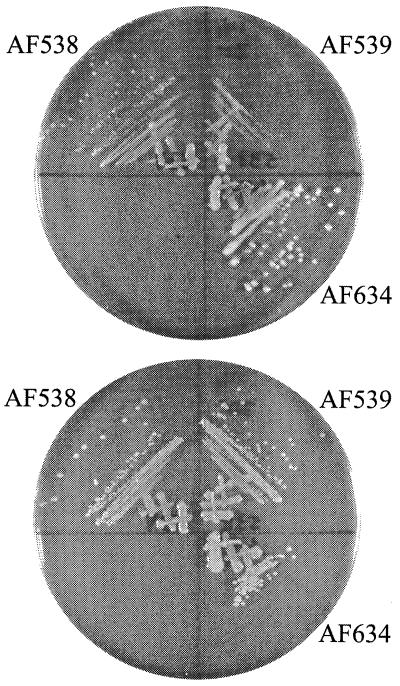
A final demonstration of the relationship between glycerol-3-phosphate metabolism and the growth defect of KN370 was the analysis of the effect of different carbon sources on growth at 20°C. This hypothesis predicts that AF539 should be cold sensitive on glucose minimal medium or other carbon sources unrelated to glycerol-3-phosphate metabolism, while glycerol or glycerol-3-phosphate should permit growth by increasing the intracellular pool of glycerol-3-phosphate. This in fact was the case (Fig. 6). Strain AF539 (glpR200 ΔsecG::kan) was cold sensitive on glucose minimal medium and cold resistant on glycerol and glycerol-3-phosphate media, demonstrating that providing the precursor metabolites for phospholipid biosynthesis alleviated the growth defect.
FIG. 6.
Effect of alternate carbon sources on growth of ΔsecG::kan strains. Strains were grown on either glucose minimal (top) or glycerol minimal (bottom) medium at 20°C.
DISCUSSION
Early observations of the phenotypes associated with the ΔsecG::kan mutation were perplexing because the observed cold sensitivity and the severity of the export defect were strain dependent and few strains other than the original secG deletion strain could be constructed that demonstrated these phenotypes (6, 16, 19). To reconcile these data, we proposed that an additional mutation was required for the cold sensitivity of ΔsecG::kan (19), and the results presented here support that hypothesis. The original secG deletion strain, KN370, contains a mutation in glpR; replacement of the mutant allele with wild-type glpR resulted in abrogation of the cold-sensitive phenotype. Further, introduction of both ΔsecG::kan and glpR200 into another strain background, MC4100, also produced a cold-sensitive phenotype. Clearly, the phenotype of the secG deletion mutation varies with the genetic background. Understanding this variation is crucial to understanding the function of SecG.
Several genes were identified previously as high-copy suppressors of the cold sensitivity of KN370. The Bacillus subtilis pgsA and scgR genes were both identified by selection of cold-resistant transformants from a plasmid-borne genomic library (26, 27); the E. coli pgsA and gpsA genes encoded on high-copy-number plasmids also suppressed the cold-sensitive phenotype (27, 39). The identification of these high-copy-number suppressors led to a hypothesis that the effect of secG deletion can be ameliorated by an increase in the level of acidic phospholipids (39, 41).
The evidence presented here suggests an alternative interpretation of those previous results. The current data demonstrate that deletion of secG has a detrimental effect on the cell only in the presence of a glpR constitutive mutation. We propose that the glpR200 mutation leads to constitutive expression of glpD, thereby shunting glycerol-3-phosphate to dihydroxyacetone phosphate which, in turn, limits the amount of glycerol-3-phosphate available for phospholipid biosynthesis (Fig. 3). Consistent with this prediction, the cold-sensitive growth defect of the secG deletion strain was alleviated by addition of glycerol-3-phosphate to the growth media or by introduction of a glpD null mutation. In this scheme, therefore, the previously observed suppression by overexpression of pgsA, gpsA, or scgR is explained not because the amount of acidic phospholipids increases above wild-type levels, but rather because expression of these genes reestablishes normal phospholipid biosynthetic patterns.
The proposed alteration to phospholipid levels is very subtle. It was demonstrated previously that steady-state phospholipid ratios in the secG deletion strain, KN370, are essentially identical to that of MC4100 (39). Our results are consistent with this finding, as we found that KN370 and its derivatives grow at a rate indistinguishable from wild-type strains, once logarithmic growth begins. The growth defect is apparent only in dilute cultures prior to onset of exponential growth. Therefore, we predict that the glpR200 mutation confers a defect in phospholipid metabolism that is not detectable by measurement of steady-state levels. It is possible that there is an alteration in the flux of the pathways, particularly during the transition from stationary phase to log phase. Along these lines, it is interesting to note that phospholipid metabolism is growth-phase regulated, with an increase in cardiolipin and a decrease in phosphatidylglycerol as cells enter stationary phase (14). Additionally, it remains unclear why cell density affects the phenotype of the glpR200 ΔsecG::kan strain, but there is evidence that population density affects the global pattern of cellular metabolites (30), perhaps explaining the density dependence of the cold sensitivity observed here.
Acidic phospholipids (phosphatidylglycerol and cardiolipin) enhance translocation at several steps: they promote the interaction of SecA with the inner membrane (7, 28, 43) and with SecYE (24), they are required for SecA translocation ATPase activity (28, 42), they are involved in the interaction of the signal sequence with the inner membrane (25), and they are important in achieving the proper orientation of membrane proteins (44). SecA insertion and deinsertion is tightly linked to the phospholipid content of the membrane, and it has been shown that an increase in acidic phospholipids stimulates both the insertion and deinsertion steps of the SecA cycle in ΔsecG::kan or secAcsR11 strains (41). Although the precise function of SecG remains unknown, the present studies suggest that SecG may be involved in the function and/or stabilization of the translocation apparatus under conditions that alter the phospholipid content of the membrane.
ACKNOWLEDGMENTS
I am very grateful to Laura Hines for help with early growth curves, John Cronan for helpful discussion and the gift of glycerol-3-phosphate, Tom Hill and Majda Valjavec-Gratian for the gift of the mini-Tn10 library, and Tim Larson, Donald Pettigrew, Stanley Maloy, and E. C. C. Lin for helpful discussion. I am grateful to Tom Hill and Kevin Young for critical reading of the manuscript.
This work was supported by CAREER award MCB-96000851 from the National Science Foundation.
REFERENCES
- 1.Ackerman R S, Cozzarelli N R, Epstein W. Accumulation of toxic concentrations of methylglyoxal by wild-type Escherichia coli K-12. J Bacteriol. 1974;119:357–362. doi: 10.1128/jb.119.2.357-362.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Akimaru J, Matsuyama S, Tokuda H, Mizushima S. Reconstitution of a protein translocation system containing purified SecY, SecE, and SecA from Escherichia coli. Proc Natl Acad Sci USA. 1991;88:6545–6549. doi: 10.1073/pnas.88.15.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Appleyard R K. Segregation of new lysogenic types during growth of a doubly lysogenic strain derived from Escherichia coli K12. Genetics. 1954;39:440–452. doi: 10.1093/genetics/39.4.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieker K L, Phillips G J, Silhavy T J. The sec and prl genes of Escherichia coli. J Bioenerg Biomembr. 1990;22:291–310. doi: 10.1007/BF00763169. [DOI] [PubMed] [Google Scholar]
- 5.Blattner F R, Plunkett G R, Bloch C A, Perna N T, Burland V, Riley M, Collado Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mau B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 6.Bost S, Belin D. A new genetic selection identifies essential residues in SecG, a component of the Escherichia coli protein export machinery. EMBO J. 1995;14:4412–4421. doi: 10.1002/j.1460-2075.1995.tb00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Breukink E, Demel R A, de Korte-Kool G, de Kruijff B. SecA insertion into phospholipids is stimulated by negatively charged lipids and inhibited by ATP: a monolayer study. Biochemistry. 1992;31:1119–1124. doi: 10.1021/bi00119a021. [DOI] [PubMed] [Google Scholar]
- 8.Brundage L, Fimmel C J, Mizushima S, Wickner W. SecY, SecE, and band 1 form the membrane-embedded domain of Escherichia coli preprotein translocase. J Biol Chem. 1992;267:4166–4170. [PubMed] [Google Scholar]
- 9.Brundage L, Hendrick J P, Schiebel E, Driessen A J M, Wickner W. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell. 1990;62:649–657. doi: 10.1016/0092-8674(90)90111-q. [DOI] [PubMed] [Google Scholar]
- 10.Cabelli R J, Chen L, Tai P C, Oliver D B. SecA protein is required for secretory protein translocation into E. coli membrane vesicles. Cell. 1988;55:683–692. doi: 10.1016/0092-8674(88)90227-9. [DOI] [PubMed] [Google Scholar]
- 11.Casadaban M J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and mu. J Mol Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- 12.Cozzarelli N R, Freedberg W B, Lin E C. Genetic control of L-alpha-glycerophosphate system in Escherichia coli. J Mol Biol. 1968;31:371–387. doi: 10.1016/0022-2836(68)90415-4. [DOI] [PubMed] [Google Scholar]
- 13.Danese P N, Silhavy T J. Targeting and assembly of periplasmic and outer-membrane proteins in Escherichia coli. Annu Rev Genet. 1998;32:59–94. doi: 10.1146/annurev.genet.32.1.59. [DOI] [PubMed] [Google Scholar]
- 14.Dowhan W. Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem. 1997;66:199–232. doi: 10.1146/annurev.biochem.66.1.199. [DOI] [PubMed] [Google Scholar]
- 15.Duong F, Eichler J, Price A, Leonard M R, Wickner W. Biogenesis of the gram-negative bacterial envelope. Cell. 1997;91:567–573. doi: 10.1016/s0092-8674(00)80444-4. [DOI] [PubMed] [Google Scholar]
- 16.Duong F, Wickner W. Distinct catalytic roles of the SecYE, SecG and SecDFyajC subunits of preprotein translocase holoenzyme. EMBO J. 1997;16:2756–2768. doi: 10.1093/emboj/16.10.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elvin C M, Hardy C M, Rosenberg H. Pi exchange mediated by the GlpT-dependent sn-glycerol-3-phosphate transport system in Escherichia coli. J Bacteriol. 1985;161:1054–1058. doi: 10.1128/jb.161.3.1054-1058.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fekkes P, Driessen A J M. Protein targeting to the bacterial cytoplasmic membrane. Microbiol Mol Biol Rev. 1999;63:161–173. doi: 10.1128/mmbr.63.1.161-173.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flower A M, Hines L L, Pfennig P L. SecG is an auxiliary component of the protein export apparatus of Escherichia coli. Mol Gen Genet. 2000;263:131–136. doi: 10.1007/s004380050039. [DOI] [PubMed] [Google Scholar]
- 20.Freedberg W B, Kistler W S, Lin E C C. Lethal synthesis of methylglyoxal by Escherichia coli during unregulated glycerol metabolism. J Bacteriol. 1971;108:137–144. doi: 10.1128/jb.108.1.137-144.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Freedberg W B, Lin E C C. Three kinds of controls affecting the expression of the glp regulon in Escherichia coli. J Bacteriol. 1973;115:816–823. doi: 10.1128/jb.115.3.816-823.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guyer M S, Reed R R, Steitz J A, Low K B. Identification of a sex-factor-affinity site in E. coli as gamma delta. Cold Spring Harbor Symp Quant Biol. 1981;45:135–140. doi: 10.1101/sqb.1981.045.01.022. [DOI] [PubMed] [Google Scholar]
- 23.Guzman L-M, Belin D, Carson M J, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hendrick J P, Wickner W. SecA protein needs both acidic phospholipids and SecY/E protein for functional high-affinity binding to the Escherichia coli plasma membrane. J Biol Chem. 1991;266:24596–24600. [PubMed] [Google Scholar]
- 25.Keller R C, Killian J A, de Kruijff B. Anionic phospholipids are essential for alpha-helix formation of the signal peptide of prePhoE upon interaction with phospholipid vesicles. Biochemistry. 1992;31:1672–1677. doi: 10.1021/bi00121a014. [DOI] [PubMed] [Google Scholar]
- 26.Kontinen V P, Helander I M, Tokuda H. The secG deletion mutation of Escherichia coli is suppressed by expression of a novel regulatory gene of Bacillus subtilis. FEBS Lett. 1996;389:281–284. doi: 10.1016/0014-5793(96)00602-3. [DOI] [PubMed] [Google Scholar]
- 27.Kontinen V P, Tokuda H. Overexpression of phosphatidylglycerophosphate synthase restores protein translocation in a secG deletion mutant of Escherichia coli at low temperature. FEBS Lett. 1995;364:157–160. doi: 10.1016/0014-5793(95)00378-m. [DOI] [PubMed] [Google Scholar]
- 28.Lill R, Dowhan W, Wickner W. The ATPase activity of SecA is regulated by acidic phospholipids, SecY, and the leader and mature portions of precursor proteins. Cell. 1990;60:271–280. doi: 10.1016/0092-8674(90)90742-w. [DOI] [PubMed] [Google Scholar]
- 29.Lin E C C. Dissimilatory pathways for sugars, polyols, and carboxylates. In: Neidhardt F C, et al., editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 307–342. [Google Scholar]
- 30.Liu X, Ng C, Ferenci T. Global adaptations resulting from high population densities in Escherichia coli cultures. J Bacteriol. 2000;182:4158–4164. doi: 10.1128/jb.182.15.4158-4164.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyristis M, Bryant A E, Sloan J, Awad M M, Nisbet I T, Stevens D L, Rood J I. Identification and molecular analysis of a locus that regulates extracellular toxin production in Clostridium perfringens. Mol Microbiol. 1994;12:761–777. doi: 10.1111/j.1365-2958.1994.tb01063.x. [DOI] [PubMed] [Google Scholar]
- 32.Manting E H, Driessen A J M. Escherichia coli translocase: the unravelling of a molecular machine. Mol Microbiol. 2000;37:226–238. doi: 10.1046/j.1365-2958.2000.01980.x. [DOI] [PubMed] [Google Scholar]
- 33.Nishiyama K, Hanada M, Tokuda H. Disruption of the gene encoding p12 (SecG) reveals the direct involvement and important function of SecG in the protein translocation of Escherichia coli at low temperature. EMBO J. 1994;13:3272–3277. doi: 10.1002/j.1460-2075.1994.tb06628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nishiyama K, Kabuyama Y, Akimaru J, Matsuyama S, Tokuda H, Mizushima S. SecY is an indispensable component of the protein secretory machinery of Escherichia coli. Biochim Biophys Acta. 1991;1065:89–97. doi: 10.1016/0005-2736(91)90015-z. [DOI] [PubMed] [Google Scholar]
- 35.Nishiyama K, Mizushima S, Tokuda H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 1993;12:3409–3415. doi: 10.1002/j.1460-2075.1993.tb06015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pogliano J A, Beckwith J. SecD and SecF facilitate protein export in Escherichia coli. EMBO J. 1994;13:554–561. doi: 10.1002/j.1460-2075.1994.tb06293.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Schweizer H, Boos W, Larson T J. Repressor for the sn-glycerol-3-phosphate regulon of Escherichia coli K-12: cloning of the glpR gene and identification of its product. J Bacteriol. 1985;161:563–566. doi: 10.1128/jb.161.2.563-566.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shimizu H, Nishiyama K, Tokuda H. Expression of gpsA encoding biosynthetic sn-glycerol-3-phosphate dehydrogenase suppresses both the LB− phenotype of a secB null mutant and the cold-sensitive phenotype of a secG null mutant. Mol Microbiol. 1997;26:1013–1021. doi: 10.1046/j.1365-2958.1997.6392003.x. [DOI] [PubMed] [Google Scholar]
- 40.Silhavy T J, Berman M L, Enquist L W. Experiments with gene fusions. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 41.Suzuki H, Nishiyama K, Tokuda H. Increases in acidic phospholipid contents specifically restore protein translocation in a cold-sensitive secA or secG null mutant. J Biol Chem. 1999;274:31020–31024. doi: 10.1074/jbc.274.43.31020. [DOI] [PubMed] [Google Scholar]
- 42.Tokuda H, Kim Y J, Mizushima S. In vitro protein translocation into inverted membrane vesicles prepared from Vibrio alginolyticus is stimulated by the electrochemical potential of Na+ in the presence of Escherichia coli SecA. FEBS Lett. 1990;264:10–12. doi: 10.1016/0014-5793(90)80751-4. [DOI] [PubMed] [Google Scholar]
- 43.Ulbrandt N D, London E, Oliver D B. Deep penetration of a portion of Escherichia coli SecA protein into model membranes is promoted by anionic phospholipids and by partial unfolding. J Biol Chem. 1992;267:15184–15192. [PubMed] [Google Scholar]
- 44.van Klompenburg W, Nilsson I M, von Heijne G, de Kruijff B. Anionic phospholipids are determinants of membrane protein topology. EMBO J. 1997;16:4261–4266. doi: 10.1093/emboj/16.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]