Abstract
During endospore formation in Bacillus subtilis, over two dozen polypeptides are localized to the developing spore and coordinately assembled into a thick multilayered structure called the spore coat. Assembly of the coat is initiated by the expression of morphogenetic proteins SpoIVA, CotE, and SpoVID. These morphogenetic proteins appear to guide the assembly of other proteins into the spore coat. For example, SpoVID forms a complex with the SafA protein, which is incorporated into the coat during the early stages of development. At least two forms of SafA are found in the mature spore coat: a full-length form and a shorter form (SafA-C30) that begins with a methionine encoded by codon 164 of safA. In this study, we present evidence that the expression of SafA-C30 arises from translation initiation at codon 164. We found only a single transcript driving expression of SafA. A stop codon engineered just upstream of a predicted ribosome-binding site near codon M164 abolished formation of full-length SafA, but not SafA-C30. The same effect was observed with an alanine substitution at codon 1 of SafA. Accumulation of SafA-C30 was blocked by substitution of an alanine codon at codon 164, but not by a substitution at a nearby methionine at codon 161. We found that overproduction of SafA-C30 interfered with the activation of late mother cell-specific transcription and caused a strong sporulation block.
In response to nutrient depletion, Bacillus subtilis can differentiate to produce a dormant cell type known as the endospore (reviewed in references 7, 17, 21, and 23). The endospore can withstand physical and chemical insults such as dehydration, exposure to organic solvents, lysozyme, and extreme temperatures and pressures. These properties of Bacillus subtilis spores can be attributed to the physical and chemical makeup of the structures that encase the spore (6, 10, 21). There are two conspicuous structures surrounding the spore, as viewed by thin-section electron microscopy: a thick central region of modified peptidoglycan (the cortex) and an exterior, multilayer protein structure (the spore coat). The coat is composed of over two dozen proteins, organized into three layers: an amorphous undercoat, a thin lamellar inner coat, and a thick striated outer coat (as reviewed in references 6 and 10).
Assembly of the spore coat is one of several complex morphological changes that occur during endospore formation. Endospore formation begins with an asymmetric cell division that produces a small cell and a large cell (forespore and mother cell, respectively). After the asymmetric division, the membranes of the mother cell engulf the forespore, producing a small cell within a larger cell (7, 17, 21, 23). The two cells have different developmental fates. The forespore develops into the endospore, while the mother cell provides an environment that nurtures development and provides structural components of the endospore. Assembly of the coat is initiated by the expression in the mother cell of a group of morphogenetic proteins, SpoIVA, CotE, and SpoVID, which control the assembly of several of the coat structural components. As a result, their absence has profound impacts on coat structure and function. The production of the coat morphogenetic proteins occurs in the initial stages of the assembly process and is driven by the early mother cell-specific regulator ςE. Some coat structural components are also produced under the control of ςE, but most of the proteins found in the mature spore coat are produced during later stages of endospore development, when ςK becomes active, replacing ςE in the mother cell (6, 10). Although the synthesis of the different coat polypeptides is temporally regulated, their order of assembly and localization within the coat layers may rely mainly on specific protein-protein interactions, as well as a variety of posttranslational modifications, including proteolytic processing, secretion, cross-linking, and glycosylation (6, 10).
The SafA gene is predicted to encode a 45-kDa protein. This protein is found in the spore coat; however, a 30-kDa protein composed of the C-terminal region of SafA is also found in spore coats (20). Takamatsu et al. determined the N-terminal amino acid sequence of this 30-kDa form of SafA and found that the N-terminal residue is a methionine encoded by codon 164 of the safA gene (26, 27). They hypothesized that the 30-kDa form of SafA is produced from proteolytic cleavage of the full-length SafA protein (26, 27). However, in this work, we provide evidence that the 30-kDa form of SafA is produced primarily by initiation of translation at codon 164 of full-length safA mRNA transcript. This is the first report implicating alternative initiation of translation as the mechanism for the generation of a structural component of the bacterial endospore coat. The internal initiation of translation of the safA message must be subjected to precise control, because the slight overproduction of the 30-kDa form of SafA interferes with the transcriptional activity of the late mother cell regulator ςK and severely compromises sporulation.
MATERIALS AND METHODS
Bacterial strains, media, and general techniques.
The B. subtilis strains used in this study are listed in Table 1. The Escherichia coli strain DH5α (Bethesda Research Laboratories) was used for transformation and amplification of all plasmid constructs. Luria-Bertani medium was routinely used for growth and maintenance of E. coli and B. subtilis strains, with appropriate antibiotic selection when needed (9). Nutrient exhaustion was used to induce sporulation in liquid culture or on plates of Difco sporulation medium (DSM) (19).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or propertiesa | Source or reference |
|---|---|---|
| Strains | ||
| MB24 | Wild-type trpC2 metC3 Spo+ | Laboratory stock |
| AH131 | trpC2 metC3 amyE::Emr | Laboratory stock |
| AOB68 | trpC2 metC3 ΔsafA::Spr | 20 |
| AOB90 | trpC2 metC3 MB24ΩpOZ139 | This work |
| AOB91 | trpC2 metC3 MB24ΩpOZ140 | This work |
| AOB145 | trpC2 metC3 MB24ΩpOZ178 | This work |
| AOB146 | trpC2 metC3 MB24ΩpOZ179 | This work |
| AOB236 | trpC2 metC3 MB24ΩpOZ220 | This work |
| AOB232 | trpC2 metC3 MB24ΩpOZ500 | This work |
| AH2654 | trpC2 metC3 MB24 pMK3 | This work |
| AH2655 | trpC2 metC3 MB24 pOZ194 | This work |
| AH2656 | trpC2 metC3 MB24 pOZ218 | This work |
| AH411 | trpC2 metC3 SPβgerE-lacZ | Laboratory stock |
| AH428 | trpC2 metC3 SPβspoIID-lacZ | Laboratory stock |
| AH685 | trpC2 metC3 SPβsspE-lacZ | Laboratory stock |
| AH2644 | trpC2 metC3 SPβsigK-lacZ | Laboratory stock |
| AH2645 | trpC2 metC2 SPβspoIID-lacZ pMK3 | This work |
| AH2646 | trpC2 metC3 SPβspoIID-lacZ pOZ194 | This work |
| AH2647 | trpC2 metC3 SPβspoIID-lacZ3 pOZ218 | This work |
| AH2648 | trpC2 metC3 SPβsspE-lacZ pMK3 | This work |
| AH2649 | trpC2 metC3 SPβsspE-lacZ pOZ194 | This work |
| AH2650 | trpC2 metC3 SPβsspE-lacZ pOZ218 | This work |
| AH2651 | trpC2 metC3 SPβgerE-lacZ pMK3 | This work |
| AH2652 | trpC2 metC3 SPβgerE-lacZ pOZ194 | This work |
| AH2653 | trpC2 metC3 SPβspoIID-lacZ pOZ218 | This work |
| AH2654 | trpC2 metC3 pMK3 | This work |
| AH2655 | trpC2 metC3 pOZ194 | This work |
| AH2656 | trpC2 metC3 pOZ218 | This work |
| AH2658 | trpC2 metC3 SPβsigK-lacZ pMK3 | This work |
| AH2659 | trpC2 metC3 SPβsigK-lacZ pOZ194 | This work |
| AH2660 | trpC2 metC3 SPβsigK-lacZ pOZ218 | This work |
| safA::gusA transcriptional fusions | ||
| AOB98 | trpC2 metC3 amyE::pOZ150 | This work |
| AOB95 | trpC2 metC3 amyE::pOZ148 | This work |
| AOB105 | trpC2 metC3 amyE::pOZ156 | This work |
| AOB147 | trpC2 metC3 amyE::pOZ176 | This work |
| safA::lacZ translational fusions | ||
| AOB125 | trpC2 metC3 amyE::pOZ168 | This work |
| AOB148 | trpC2 metC3 amyE::pOZ175 | This work |
| AOB135 | trpC2 metC3 amyE::pOZ172 | This work |
| AOB149 | trpC2 metC3 amyE::pOZ174 | This work |
| AOB150 | trpC2 metC3 amyE::pOZ173 | This work |
| AOB200 | trpC2 metC3 amyE::pOZ189 | This work |
| Plasmids | ||
| pCR2.1 TOPO | TA cloning vector | Invitrogen |
| pMK3 | B. subtilis multicopy plasmid | Laboratory stock |
| pAC5 | lacZ translational fusion vector | 24 |
| pOZ115 | safA (with promoter) in pCR2.1 TOPO | This work |
| pMLK83 | gusA transcriptional fusion | 13 |
| pOZ138 | safA155stop in pCR2.1 TOPO | This work |
| pOZ139 | safA-FLAG in pCR2.1 TOPO, Cmr | This work |
| pOZ140 | pOZ138, Cmr | This work |
| pOZ148 | safA −105 to +1234 bp, pMLK83 | This work |
| pOZ150 | safA −554 to +1234 bp, pMLK83 | This work |
| pOZ156 | safA −22 to +1234 bp, pMLK83 | This work |
| pOZ158 | safA M1A in pCR2.1 TOPO | This work |
| pOZ160 | safA M161 in pCR2.1 TOPO | This work |
| pOZ161 | safA M164 in pCR2.1 TOPO | This work |
| pOZ168 | safA::lacZ, in pAC5 | This work |
| pOZ172 | safA M1A::lacZ, in pAC5 | This work |
| pOZ173 | safA M164A::lacZ, in pAC5 | This work |
| pOZ174 | safA M161::lacZ, in pAC5 | This work |
| pOZ175 | safA 155stop::lacZ, in pAC5 | This work |
| pOZ176 | safA +83 to +1234 bp, pMLK83 | This work |
| pOZ178 | pOZ160, Cmr | This work |
| pOZ179 | pOZ161, Cmr | This work |
| pOZ189 | safA M161A,M164A::lacZ, in pAC5 | This work |
| pOZ194 | safA M1A-pMK3 | This work |
| pOZ218 | safA promoter-pMK3 | This work |
| pOZ220 | safA M161A, M164A in pCR2.1 TOPO, Cmr | This work |
| pOZ500 | safA155stop-FLAG in pCR2.1TOPO, Cmr | This work |
Ω denotes that the plasmid was integrated into the B. subtilis chromosome by a Campbell integration event (single crossover in the region of homology).
Spore resistance tests and spore purification.
Spore resistance tests—heat and lysozyme—were performed as previously described (8, 9). Mature spores were harvested 48 h after the onset of sporulation and subjected to a step gradient for purification. Purified spores were used for downstream tests of germinability or for analysis of the coat protein profile on Coomassie-stained sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gels (8–10).
Construction of strains carrying a C-terminal FLAG-tagged version of safA.
Primers saf + 41-d and FLAG-R (which encodes the FLAG tag, DYKDDDDK; see Table 2) were used to amplify the safA coding region, excluding the C-terminal stop codon. The resulting 1,223-bp PCR product, in which the FLAG tag is fused in frame to the 3′ end of safA, was cloned into the pCR2.1 TOPO vector (Invitrogen) to form pOZ130. A chloramphenicol cassette, released from pMS38 (M. Serrano and A. O. Henriques, unpublished data) by digestion with BglII and BamHI, was inserted into the BamHI site of pOZ130 to form pOZ139 (Table 1). Competent cells of MB24 were transformed with pOZ139, selecting for chloramphenicol resistance. Transformants were expected to arise as the result of a single reciprocal crossover (Campbell-type) event at the safA locus. A transformant, whose chromosomal structure in the vicinity of the safA locus was confirmed by PCR analysis, was chosen and named AOB90. Strain AOB90 carries two copies of safA: a functional upstream copy with a C-terminal FLAG tag and a downstream promoterless allele.
TABLE 2.
Primers used for cloning, site-directed mutagenesis, and primer extension in this study
| Primer name | Sequence (5′→3′)a |
|---|---|
| saf + 41-d | TAGGAGGGGAAAACCATGGAAATCCATATCG |
| FLAG-R | TCATCATTTGTCATCGTCATCTTTATAATCCTCATTTTCTTCTTCCGGACGGCCAAACATTTGGTTTACAGAAGGC |
| saf + 1234R | CGTTCCGAAAGATCTCTCATTTTCTTCTTCCGG |
| saf-554d | GATGAATTAGTAGCTGAATCCGGGC |
| saf-105d | CAGGAAGATCTACTCCTTGCCC |
| saf-22d | CTTTTGTTCAGGCAGATCTTGTGAAGAAACATATTG |
| saf + 83d | GGCGATTAGATCTGGAAAATAGC |
| S + 107 | CAGCTATTTTCCAGAGCGAATCGCC |
| S + 187 | CATTCCAGGCATGATTAAGTC |
| safM1A-d | GGAGGGGAAAACGGCGAAAATCCATATCG |
| safM1A-R | CGATATGGATTTTCGCCGTTTTCCCCTCC |
| safM161A-D | CAACAGGAGGCTGCGAGTAATATGGAAAATGC |
| safM161A-R | GCATTTTCCATATTACTCGCAGCCTCCTGTTG |
| safM164A-d | CAACAGGAGGCTATGAGTAATGCGGAAAATGC |
| safM164A-R | GCATTTTCCGCATTACTCATAGCCTCCTGTTG |
| safM161/164-d | CCACAACAGGAGGCTGCGAGTAATGCTGAAAATGCAAATTATCC |
| safM161/164-R | CGGATAATTTGCATTTTCAGCATTACTCGCAGCCTCCTGTTGTGG |
| saf155stop-d | CATATGTATCATATGCAAGACCAATGACCACAACAGGAG |
| saf155stop-R | CCATATTACTCATAGCCTCCTGTTGTGGTCATTGGTCTT |
| saf-121d | GCCAAAAGCCATTCAGGAGGATCCACTCCTTGCCC |
| pAC5R1 | ACGTTGTAAAACGACGGGATCCGCATTTTCCATATTACTA |
| pAC5R2 | ACGTTGTAAAACGACGGGATCCGCATTTTCCGCATTACTCATAGCCTCC |
| pAC5R3 | ACGTTGTAAAACGACGGGATCCGCATTTTCCGCATTACTCGCAGCCTCC |
| safPr-R | GCTATTTTCCAGAGCGAAGCGCCTTTTTGAACG |
Restriction enzyme sites are underlined.
Preparation of B. subtilis whole-cell extracts and immunoblotting.
For immunoblotting, samples (10 ml for the French press, 1 ml for lysozyme lysis) of DSM cultures of various strains were collected at intervals after the onset of sporulation. Whole-cell lysates were prepared by French press as described by Ozin et al. (20) or by gentle lysozyme lysis as described by Kodama et al. (14). Proteins were resolved on SDS-PAGE (12 or 7.5% polyacrylamide) gels as indicated in the figure legends. Immunoblotting was performed as in reference 20 with the following antibody concentrations: anti-SafA, 1:15,000 (20); anti-β-galactosidase, 1:50,000 (Promega); anti-FLAG, 1:30,000 (Sigma-Aldrich).
Construction of strains carrying transcriptional fusions of safA to the gusA gene.
The safA gene, including 554 bp upstream from its start point of transcription, was amplified by PCR generated from primers saf-554d and saf + 1234R (Table 2). This 1,788-bp DNA fragment was cloned into the TA cloning site of pCR2.1 TOPO (Invitrogen) to produce pOZ115 (Table 1). The 5′ region, from −554, −105, −22, or +83 nucleotides from the +1 start site of transcription, up to codon 387 of safA, was amplified from pOZ115 (Table 1) by using the reverse primer saf + 1234R paired with each of the direct primers saf-554d, saf-105d, saf-22, and saf + 83 (Table 2). The oligonucleotides were engineered with either a BamHI site or a BglII site so that the PCR products could be cloned into the BamHI site of pMLK83 (13). The resulting amyE integrational plasmids are pOZ150, pOZ148, pOZ156, and pOZ176, respectively. The orientation of the safA insert was confirmed by restriction digest and PCR analyses.
Competent cells of AH131 (Table 1) were transformed with linearized plasmids pOZ150, pOZ148, pOZ156, and pOZ176, with selection for kanamycin resistance (Kanr), and the resulting transformants were screened for erythromycin sensitivity (Ems). The Kanr Ems phenotype indicated a marker replacement recombinational event, and appropriate transformants were identified and named AOB98, AOB95, AOB105, and AOB147, respectively. PCR analysis confirmed that they were the result of a double crossover (allele replacement) event involving the Em cassette at the amyE locus (AH131) and the indicated plasmids (Table 1).
Construction of plasmids containing mutations in the SafA coding region.
Plasmid pOZ115 (Table 1) served as a template for site-directed mutagenesis. Three independent alanine substitutions (M1A, M164A, and M161A) and a nonsense mutation (TGA) at codon 155 were made following the manufacturer's protocol for the Quick Change system (Stratagene) with the following primer sets: safM1A-d and safM1A-R, safM164A-d and safM164A-R, safM161A-d and safM161A-R, and saf155stop-d and saf155stop-R (Table 2). The mutations in the resulting plasmids (pOZ158, pOZ161, pOZ160, and pOZ138, respectively) were sequenced (Table 1).
A chloramphenicol resistance gene was extracted from pMS38 (Serrano and Henriques, unpublished) by digestion with BglII and BamHI and cloned into the BamHI sites of pOZ160, pOZ161, and pOZ138 to form pOZ178, pOZ179, and pOZ140, respectively (Table 1). We used the Quick Change site-directed mutagenesis system to make a double alanine substitution at codons 161 and 164 by using primers safM161/164-d and safM161/164-R (Table 2) and with pOZ178 (Table 1) as a template. The double mutation in the resulting plasmid, pOZ220, was confirmed by sequencing.
Construction of strains containing alanine substitutions in safA at codons 164 and 161.
Competent cells of a safA deletion mutant, AOB68 (Table 1), were transformed independently with plasmids pOZ178, pOZ179, pOZ140, and pOZ220 (Table 1). Transformants were isolated with selection for chloramphenicol resistance (Cmr). The resulting clones, AOB145, AOB146, AOB91, and AOB236 (Table 1), were shown by PCR analysis to arise from a Campbell integration event (single crossover) in the regions of homology upstream from the safA gene. The presence of the correct mutations was confirmed by sequencing of the recombinant chromosomes.
Mapping the 5′ terminus of safA mRNA.
B. subtilis cells were grown in DSM, and 25 ml was harvested at 2.5 and 4.5 h after the onset of sporulation. RNA for primer extension analysis was prepared by using the RNeasy Maxi kit (Qiagen) according to the manufacturer's directions. Primer extension was performed essentially as previously described (11) with primers S + 107 and S + 187 (Table 2) 5′ end labeled with γ-32P (Promega). The same 5′-end-labeled primers were used to generate a sequence ladder by the dideoxy chain termination method with pOZ115 (Table 1) as a DNA template and the f-mol Cycle Sequencing kit (Promega) (22). The products of the primer extension were subjected to electrophoresis in a 6% (wt/vol) polyacrylamide slab gel containing 8 M urea and were detected on a PhosphorImager (Molecular Dynamics).
Construction of strains carrying safA::lacZ translational fusions ectopically inserted at the amyE locus.
Primer set saf-121d and saf-pAC5R1 (Table 2) was used to PCR amplify the safA promoter region up to codon 167 independently from pOZ115, pOZ138, pOZ158, and pOZ160 (Table 1). Primers saf118d and saf-pAC5R2 (contains safA M164A substitution) or saf-pAC5R3 (contains safA M161A and M164A substitutions) (Table 2) were used to PCR amplify the same 663-bp region from pOZ115. The six resulting PCR products, flanked by engineered BamHI sites, were cloned into the BamHI site of pAC5 (Table 1) to create safA::LacZ translational fusion plasmids pOZ168 (safA::LacZ), pOZ175 (safA 155stop::LacZ), pOZ172 (safA M1A::LacZ), pOZ174 (safA M161A::LacZ), pOZ173 (safA M164A::LacZ), and pOZ189 (safA::M161A, M164A::LacZ), respectively. In all cases, the orientation of the insert was confirmed to be in the same direction as lacZ by PCR and restriction digest analysis.
Competent cells of AH131 (Table 1) were transformed independently with ScaI-linearized pOZ168, pOZ175, pOZ172, pOZ174, pOZ173, or pOZ189 with selection for chloramphenicol resistance (Cmr). Appropriate Cmr Ems transformants were identified and named AOB125, AOB148, AOB135, AOB149, AOB150, and AOB200, respectively (Table 1). PCR analysis confirmed that they were the result of an allele replacement event at the amyE locus of the AH131 recipient strain (Table 1). The presence of the correct mutations in the safA::lacZ constructs was confirmed by cycle sequencing of the relevant regions of the recombinant chromosomes.
Construction of a strain overproducing the C-terminal form of SafA or the SafA promoter.
We used primers saf-554d (BamHI) and saf + 1234R (BglII) to PCR amplify a 1,788-bp fragment, containing safA M1A plus the promoter region from a plasmid template, pOZ158 (Table 1). The resulting PCR products, flanked by engineered BamHI-BglII sites, were cloned into the BamHI site of plasmid pMK3 (24), which replicates in B. subtilis, thereby creating pOZ194 (Table 1). We also cloned the promoter region of safA plus 30 bp of the coding region into pMK3 by PCR amplifying a 584-bp fragment and cloning it first into pCR 2.1 TOPO, to create pOZ217. The insert was then excised from pOZ217 by digestion with EcoRI, gel purified, and ligated into the EcoRI site of pMK3 to create pOZ218 (Table 1).
Plasmids pOZ194 (safA M1A-pMK3), pOZ218 (safA promoter region in pMK3), and the pMK3 vector were introduced into the B. subtilis wild-type strain MB24 by transformation followed by selection for neomycin resistance (Nmr), creating the congenic strains AH2655, AH2656, and AH2654, respectively (Table 1). The following fusions to the lacZ gene were used to monitor sporulation-specific gene expression: SPβspoIID-lacZ (9); SPβsspE-lacZ, found in a screen similar to that described in reference 9; SPβsigK-lacZ (16); and SPβgerE-lacZ (5). They were introduced into MB24 by either transformation or specialized transduction. MB24 lysogens of SPβspoIID-lacZ (AH428), SPβsspE-lacZ (AH685), SPβsigK-lacZ (AH2644), and SPβgerE-lacZ (AH411) were also transformed with pOZ194, pOZ218, and pMK3, to produce strains AH2645 to AH2653 and AH2658 to AH2660 (Table 1).
Enzyme assays.
The activities of β-galactosidase and β-glucuronidase were determined with the substrates o-nitrophenyl-β-d-galactopyranoside (ONPG) and p-nitrophenyl-β-d-glucopyranoside (PNPG), respectively, as previously described (9). In both cases, cells were permeabilized by treatment with toluene. Enzyme-specific activity is expressed in nanomoles of substrate (ONPG or PNPG) hydrolyzed per milligram (dry weight) per minute.
RESULTS
A single promoter for safA.
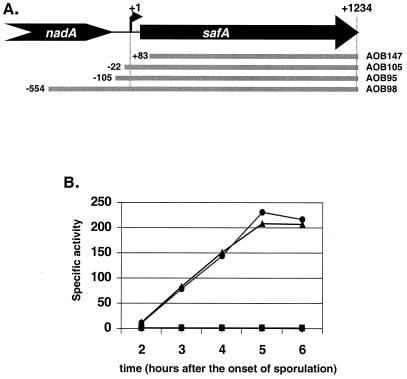
Takamatsu et al. (27) used Northern blots and primer extension analysis to identify a ςE-dependent transcript of the safA gene. We refer to the promoter driving expression of this transcript as safA P1. In order to seek other promoters of safA expression, we fused a series of DNA fragments from the safA region to a promoterless gusA reporter gene and looked for the minimal region required for gusA expression. All of the DNA fragments from the safA gene contained the C terminus of the safA gene and extended upstream to various points up to 554 bp upstream from the start point of the safA P1. These fusions were moved into the amyE locus of a wild-type strain, and the β-glucuronidase activity of the resulting strains was measured during sporulation (Fig. 1). The safA DNA fragment that extended 105 bp upstream from the start point of transcription directed gusA expression, whereas the fragment that extended only 22 bp upstream failed to promote gusA expression. Therefore, any promoter located upstream or within safA requires sequences located at least 23 bp upstream from the start point of safA P1.
FIG. 1.
Defining the promoter region(s) in safA. (A) Illustration of regions of safA (solid bars) that were cloned into a gusA transcriptional fusion plasmid construct, pMLK83 (Table 1), to make the indicated B. subtilis strains, AOB147, AOB105, AOB95, and AOB98. Numbers indicate the distance from the P1 +1 start site of transcription. (B) Measurement of transcriptional activity of safA-gusA. Circles, AOB98; triangles, AOB95; squares, AOB105; diamonds, AOB147.
In another attempt to seek evidence for a second promoter of safA transcription, we used primers that annealed 107 and 187 bp downstream from the safA P1 start site in primer extension analyses. We detected the transcript that initiated at P1, but detected no other transcripts in these analyses (data not shown). In these primer extension experiments, we could not have detected a transcript that was initiated more than 145 bp downstream from the start site, P1. However, since our gusA reporter fusions showed that sequences located at least 23 bp upstream from the start site, P1, are required for all promoter activity in the safA gene, we concluded that it is unlikely that a second transcript is initiated more than 145 bp downstream from the P1 start. Therefore, we conclude that the alternative forms of the SafA protein are produced from a transcript derived from a single promoter.
Accumulation of the 30-kDa short form of SafA does not require expression of the full-length form.
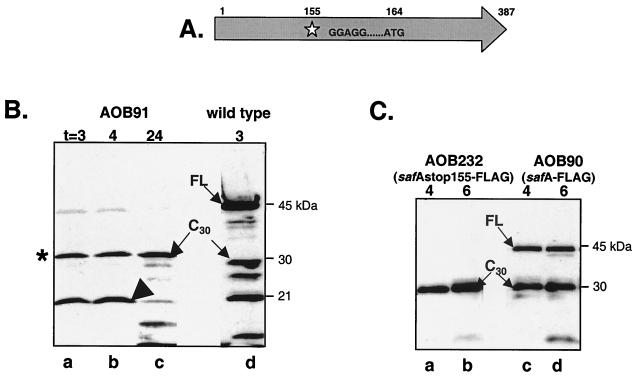
The N-terminal sequence of a 30-kDa form of SafA (SafA-C30) begins with a methionine encoded by codon 164 of safA (26, 27), and the transcriptional analysis suggests that there is only a full-length transcript driving expression of safA (27; this work). Therefore, accumulation of SafA-C30 may arise from proteolytic processing of the full-length protein or from initiation of translation at codon 164 within the full-length transcript. To distinguish between these two possibilities, we constructed a B. subtilis strain, AOB91, carrying a stop codon just upstream of a predicted ribosome-binding site (RBS) preceding M164, to block formation of the full-length form (SafA-FL) (Fig. 2A). We monitored the accumulation of SafA products by immunoblot analysis with anti-SafA antiserum. As we had shown previously (20), no SafA products were detected in an extract from a SafA null mutant (data not shown). However, we found that even in the absence of full-length SafA (SafA-FL), a 30-kDa form of SafA was detectable by immunoblots of extracts from sporulating cells of AOB91 probed with the anti-SafA antibody (Fig. 2B). If this 30-kDa form of SafA (Fig. 2B, lanes a to c) is the same as the 30-kDa form produced in the wild type (SafA-C30; Fig. 2B, lane d), we would conclude that production of the 30-kDa form is not dependent on the production of full-length SafA. We confirmed that the 30-kDa species produced in AOB91 was the same as the SafA-C30 in the wild type by using an anti-FLAG antibody to probe immunoblots of extracts from strains containing C-terminally FLAG epitope-tagged versions of SafA, with and without the stop codon mutation (Fig. 2C). Therefore, production of the 30-kDa form probably results from initiation of translation at codon 164. This model predicts that the AUG codon at 164 is required for SafA-C30 production. This prediction was tested as described below. In addition to the 30-kDa band, a band of about 21 kDa in extracts of AOB91 reacted with anti-SafA antiserum. Presumably, this band results from termination of translation at the TGA nonsense codon introduced into the safA coding sequence, which would produce a polypeptide of about 17.7 kDa (Fig. 2A). Interestingly, a band with about the same apparent mobility is detected in extracts prepared from the wild-type strain, as is a band that migrates between the 30- and 21-kDa species (Fig. 2B, lane d). The band that migrated between the 30- and 21-kDa species was not detected when an anti-FLAG monoclonal antibody was used to monitor the expression of C-terminal FLAG-tagged alleles of SafA (Fig. 2C). Therefore, it may be derived by proteolytic removal of the C-terminal region.
FIG. 2.
Detection of different forms of SafA in a safA-stop codon mutant during sporulation. (A) Diagram of the SafA coding region (amino acid residues 1 to 387) showing the position of the stop codon mutation engineered at codon 155 (star), an internal RBS (GGAGG), and the ATG methionine codon at position 164. (B) Immunoblots of whole-cell extracts (French press method) were prepared at the indicated times (hours) after the onset of sporulation and probed with anti-SafA antibodies. Lanes a to c, AOB91 (safAstop155-FLAG); lane d, wild type (MB24). The asterisk marks the position of the 30-kDa form of SafA (C30), the arrow marks the position of the full-length SafA (FL), and the large arrowhead marks the position of the 21-kDa band described in the text. (C) Whole-cell extracts (lysozyme method) of strains AOB232 (safA with a TGA stop codon at 155 and a C-terminal FLAG tag) (lanes a and b) and AOB90 (safA with a C-terminal FLAG tag) (lanes c and d) were harvested at 4 and 6 h after the onset of sporulation and probed with antibodies against the FLAG epitope.
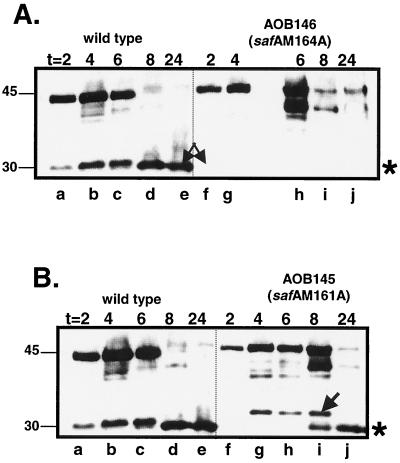
To test whether the methionine at codon 164 is the start site of translation of SafA-C30, we substituted an alanine codon for the methionine codon at 164 (Table 1). We also noted the occurrence of an ATG codon encoding a methionine at codon 161 in frame with M164. Therefore, we also tested whether codon 161 could contribute to internal initiation of translation of the safA transcript by making a single alanine substitution of the methionine at position 161, as well as double alanine substitutions of the methionines at codons 161 and 164. Immunoblots of extracts from the resulting strains demonstrated that SafA-C30 requires the methionine at codon 164 for its accumulation (Fig. 3A, lanes f to j), but not the methionine at codon 161 (Fig. 3B, lanes f to j). We also noted that the alanine substitution at codon M161 resulted in production of a slower-migrating species between 4 and 8 h after the onset of sporulation (Fig. 3B). This species of SafA was detected only in this strain, and its origin is unknown. The double alanine mutations at codons 161 and 164 also prevented accumulation of SafA-C30 product (not shown), and the immunoblots of this strain looked the same as those for the strain with the M164A mutation (Fig. 3A, lanes f to j).
FIG. 3.
Accumulation of SafA in safA M164A and safA M161A mutants. Immunoblots of extracts (lysozyme method) from sporulating cells at the indicated times (hours) after the onset of sporulation, probed with anti-SafA antibodies. (A) Lanes a to e, wild type (MB24); lanes f to j, AOB146 (safA M164A). The arrow marks the position of SafA-C30. This form is missing in lanes f to j. (B) Lanes a to e, wild type (MB24); lanes f to j; AOB145 (safA M161A). The arrow marks the position of a band that is not present in wild-type extracts. In both panels, the asterisk indicates the expected position of SafA-C30. This form accumulates in both the wild type and the AOB145 mutant, but not the AOB146 mutant.
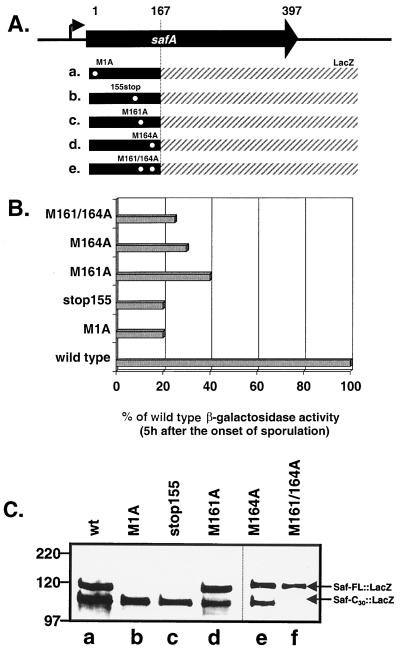
Translational fusions between safA and lacZ were used to examine the effects of mutations on production of SafA translation products. The lacZ coding region was fused just downstream of the putative internal translation initiation site, M164, of safA (Fig. 4A). Mutations were engineered in positions in safA::lacZ that we predicted would block formation of a full-length fusion protein (alanine substituted at position 1, M1A; nonsense codon substituted at codon 155, stop 155; Fig. 4A, a and b) or may interfere with the site of internal initiation of translation (M161A and M164A; Fig. 4A, c to e). At 5 h after the onset of sporulation, the translational activities of all of the fusion protein constructs had reached their peak (not shown); however, the levels of activity of the amino acid-substituted versions of safA::lacZ were 60 to 80% less than that of the wild-type fusion (Fig. 4B). This decrease in activity was observed as a decrease in total fusion protein accumulation in immunoblots probed with a monoclonal antibody against β-galactosidase (Fig. 4C). Western blots showed that the wild-type safA::lacZ construct was translated into a full-length form and a shorter form (Fig. 4C, lane a). The mutations M1A and stop155 blocked the formation of the full-length form of the fusion protein, but not the short form of the protein (Fig. 4C, lanes b and c). In this experiment, both the short and long forms of the SafA::LacZ fusion contain the C-terminal end of SafA (up to codon 167), since lacZ was fused to the C-terminus-encoding end of safA and the proteins were detected with anti-β-galactosidase antibody. Therefore, production of the short form of SafA::LacZ was shown to be independent of translation initiation at codon 1 of SafA. Therefore, production of the shorter form of SafA::LacZ must result from translation initiated downstream from codon 155, where a nonsense codon fails to block SafA-C30 expression.
FIG. 4.
Activity and accumulation of safA::lacZ wild-type and mutant translation fusions. (A) Diagram of the safA coding region and the safA::lacZ fusion construct junction. The positions of the alanine substitutions and the stop codon mutation are symbolized by white dots and labeled M1A (a), 155stop (stop codon at residue 155) (b), M161A (c), M164A (d), and M161A and M164A (e), respectively. (B) Comparison of the translational activity of wild-type safA::LacZ and various mutants in the safA portion of safA::LacZ. The error in measurement of β-galactosidase activity is ±5%. (C) Immunoblot of extracts (lysozyme method) taken at 5 h after the onset of sporulation from strains containing the wild-type or mutant versions of safA::LacZ. The blot was probed with monoclonal antibodies against β-galactosidase. Lane a, wild-type safA::LacZ (strain AOB125); lane b, wild-type safA M1A::LacZ (AOB135); lane c, safA-stop155::LacZ (AOB148); lane d, safA M161A::LacZ (AOB149); lane e, safA M164A::LacZ (AOB150); lane f, safA M161A, M164A::LacZ (AOB200).
The substitution of alanine codons at both 161 and 164 completely eliminated the production of the short form of SafA::LacZ, whereas the long form accumulated (Fig. 4C, lane f). Surprisingly, single substitutions of alanines at either 161 or 164 had little or no effect on accumulation of the short form (Fig. 4C, lanes d and e, respectively). We interpret these results to indicate that production of the short form can result from initiation of translation at codon 161 or 164. The large size of the fusion proteins probably prevented us from detecting a difference in size of fusion proteins initiated at codons 161 and 164. The idea that translation can be initiated at position 161 or 164 appears to contradict our previous results, in which we substituted an alanine codon at position 164 in an otherwise wild-type allele of safA and were unable to detect accumulation of the SafA-C30 with anti-SafA antisera. However, the SafA product initiated at codon 161 may be unstable and only accumulates when fused to β-galactosidase.
Overexpression of SafA-C30 causes a stage IV sporulation block.
We examined the effects of mutations in safA on spore coat function and structure by performing heat and lysozyme tests, SDS-PAGE analysis of proteins extracted from the spore coat, germination assays, and analysis of spore coat ultrastructure by electron microscopy. We determined that expression of just SafA-C30 (AOB91) is not sufficient for lysozyme resistance and has the same phenotype as a safA deletion mutant (not shown). Thus, expression of the full-length protein is required for assembly of the spore coat. In contrast, single alanine substitutions at codons 161 and 164 (AOB145 and AOB146) and double alanine substitutions at codons 161 and 164 (AOB236) had no detectable effect on spore coat structure or function (not shown).
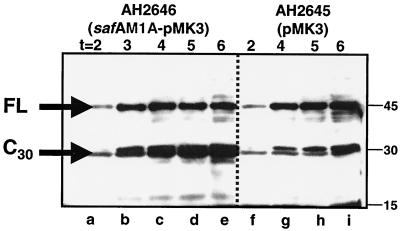
Inactivation of genes encoding coat components does not always result in an observable defect; however, overexpression may reveal detectable phenotypes, as reviewed in references 6 and 10. For instance, a CotT deletion mutant bears no detectable phenotype, but the overexpression of cotT causes thickening of the inner coat layers and severely impairs germination (1, 4). Since we did not detect any obvious coat defects by blocking the accumulation of SafA-C30 (see the section above; strains AOB145, AOB146, and AOB236), we overexpressed SafA-C30 by cloning safA, with an M1A alanine substitution, into pMK3, a plasmid that can replicate in B. subtilis (24), to form pOZ194 (Table 1). We moved pOZ194 and a vector-only control into a wild-type background (strains AH2655 and AH2654, respectively; Table 1), and confirmed the overexpression of SafA-C30 on a Western blot probing against SafA (Fig. 5). We estimated that the SafA-C30 form accumulated to levels two to five times higher than those in a wild-type strain. Derivatives of the wild-type strain carrying pMK3 (AH2654) or a related plasmid carrying the safA promoter region (AH2656) formed about 108 heat-resistant spores/ml and thus sporulated with the same efficiency as the parental MB24 strain (Table 3). However, strain AH2655 (which carries pOZ194) formed 103 heat-resistant spores/ml (Table 3). Thus, the overproduction of SafA-C30 causes a severe block in sporulation. This effect is probably not due to titration of important transcription factors by the safA promoter, since multiple copies of the safA promoter did not interfere with spore formation.
FIG. 5.
SafA-C30 is overproduced during sporulation on a multicopy plasmid. Immunoblots of extracts (lysozyme method) from sporulating cells at the indicated times (hours) after the onset of sporulation, probed with anti-SafA antibodies. Lanes a to e, wild-type strain containing pOZ194, which contains the safA promoter and coding region and an alanine substitution at codon 1 (AH2655); lanes f to i, wild-type strain containing the pMK3 vector-only control (AH2654).
TABLE 3.
Effect of expression of SafA-C30 on a multicopy plasmid on the number of heat-resistant CFU per milliliter 24 h after the onset of sporulation
| Strain | No. of CFU/ml
|
|
|---|---|---|
| Viable cells | Heat resistant | |
| AH2654a | 3.2 × 108 | 1.8 × 108 |
| AH2655b | 3.0 × 107 | 3.3 × 103 |
| AH2656c | 3.6 × 108 | 1.2 × 108 |
Contains plasmid pMK3 (vector only).
Contains plasmid pOZ194 (vector with M1A allele of safA).
Contains plasmid pOZ218 (vector with promoter region from safA).
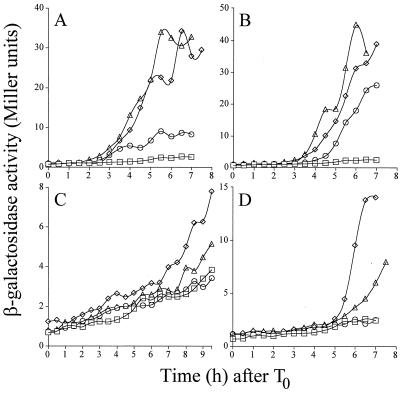
To characterize the developmental block of strain AH2655, we examined the effect of overproducing SafA-C30 on different classes of sporulation-specific gene expression. To do that, we first constructed a collection of congenic strains bearing fusions of different sporulation promoters to the lacZ gene and plasmid pMK3, pOZ194, or pOZ218. Then, we monitored β-galactosidase production by the different reporter strains throughout sporulation in DSM. We found that the presence of pOZ194 reduced expression of the ςE-dependent spoIID-lacZ fusion (Fig. 6A), but had little effect on the expression of the ςG-dependent sspE-lacZ fusion (Fig. 6B). In contrast, expression of the ςK-dependent gerE-lacZ fusion was completely abolished (Fig. 6D), as was expression of sigK-lacZ (Fig. 6C). The control plasmids pOZ218 and pMK3, which were shown not to inhibit spore formation, also did not greatly reduce the expression of the reporter gene fusions tested (Fig. 6).
FIG. 6.
The effect of overproducing SafA-C30 on sporulation-specific gene expression. β-Galactosidase production was monitored throughout sporulation in DSM cultures of a collection of congenic strains bearing fusions of ςE-, ςG-, and ςK-dependent promoters to the lacZ gene and containing plasmid pMK3, pOZ194, or pOZ218 (ςE-dependent spoIID-lacZ fusion in panel A, ςG-dependent sspE-lacZ fusion in panel B, ςK-dependent sigK-lacZ fusion in panel C, and ςK-dependent gerE-lacZ fusion in panel D). Symbols represent strains containing vector-only control, pMK3 (diamonds); safA M1A-pMK3, pOZ194 (circles); or safA promoter region-pMK3, pOZ218 (triangles). Strains without a lacZ fusion are represented by squares.
ςE is required for the completion of the engulfment sequence, at the end of which ςG is activated in the forespore (7, 12, 15, 17). Therefore, we deduced that even though the activity of ςE appears reduced when measured with the spoIID-lacZ fusion (Fig. 6A), it must be sufficient for the cells to complete engulfment and activate ςG (12). The transcriptional activity of ςG is required for the activation of ςK in the mother cell, which then autoregulates its own production (7, 15, 17, 21, 23). However, the expression pattern of the sspE-lacZ fusion indicated that normal levels of ςG activity have been attained. Therefore, the lack of ςK-dependent transcription of gerE-lacZ cannot be explained by an indirect effect on ςG. The activity of ςG is required for synthesis of the primordial cell wall of the spore, but in the absence of ςK activity, the synthesis of the protective cortex and coat layers never takes place (7, 21, 23). We infer from these results that pOZ194 blocks sporulation no sooner than stage III (engulfment) and no later than stage IV (synthesis of the germ cell wall) (21). Moreover, electron microscopy studies of sporulating cells taken at 4 h after the onset of sporulation showed that approximately the same proportion of cells in the pMK3-containing cells and the pOZ194 (SafA-C30- overproducing)-containing strain had completed engulfment (not shown). These observations support our conclusion that pOZ194 blocks sporulation after engulfment. We suggest that the overexpression of SafA-C30 arrests development because it interferes with ςK synthesis or its activity. There is no evidence that SafA-C30 acts in a direct way to reduce mother cell-specific transcription; therefore, the effect of SafA-C30 on transcription may be indirect.
DISCUSSION
SafA accumulates as multiple forms during sporulation. The SafA-C30 form is encoded by the C terminus of the safA gene (20, 27; this work). In this study, we have shown that blocking expression of the full-length SafA, by placing a stop codon just upstream of M164, did not block the accumulation of a 30-kDa SafA species (Fig. 2B). Moreover, an alanine substitution of M164 specifically eliminated the accumulation of SafA-C30 (Fig. 3A). These and the other data presented support a mechanism in which initiation of translation at codon 164 produces SafA-C30. The safA gene appears to be transcribed from a single promoter; therefore, translation is initiated at both codons 1 and 164 of a single primary transcript. It is not known whether translation is initiated from both positions on every transcript or whether posttranscriptional modifications determine which initiation site will be used on each transcript. The kinetics of synthesis of both translation products is also not well established. In Fig. 3, for example, it appears that SafA-FL accumulates before SafA-C30. However, this result may be an artifact, because the SafA antiserum may react more strongly to the N-terminal region of the protein. The FLAG-tagged versions of SafA-FL and SafA-C30 appeared to accumulate simultaneously when probed with anti-FLAG antibody (data not shown). Therefore, SafA-FL and SafA-C30 may be synthesized simultaneously. Moreover, the amount of SafA-FL falls in the later stages of sporulation. If SafA-FL is degraded during sporulation more rapidly than SafA-C30, the relative rates of synthesis of these two proteins cannot easily be determined from the apparent rates of accumulation. Recently, Takamatsu et al. (25) presented evidence that SafA is proteolytically processed by YabG. They showed that the 45-kDa form of SafA and a 31-kDa form of SafA accumulate early during sporulation, but disappear as new forms with sizes of 42 and 30 kDa accumulate, and this appearance of the 42- and 30-kDa forms was dependent on YabG. Our evidence supports the model that SafA-C30 is made primarily from translation initiation at codon 164. However, other SafA products may arise from proteolytic events. For example, the 45-kDa form (SafA-FL) may be processed to the 42-kDa form. We do not know whether or which of the 30- or 31-kDa forms of SafA described by Takamatsu et al. (25) corresponds to the SafA-C30 described by us. However, since our data show that SafA-C30 is made primarily from translation initiation at codon 164, we speculate that SafA-C30 is the form that they call the 31-kDa form. Furthermore, the 31-kDa form may be processed at its carboxy terminus by the protease to produce the form they call the 30-kDa form. In some of our experiments, the addition of an epitope tag or fusion of β-galactosidase to the carboxy-terminal end of SafA may have inhibited proteolytic processing.
SafA is the first example of a coat protein that is expressed as two forms as a result of internal initiation of translation. There are examples in other bacterial systems where alternatively translated gene products may be involved in subtle functional adaptations and for regulation of protein activity. For example, CheA, a histidine protein kinase of the chemotaxis signal transduction system, is synthesized as two forms by in-frame initiation sites within the cheA gene (18). The full-length form (CheAL) is essential for signal transduction, but the function of the short form (CheAs) is unclear. CheAs is coexpressed with CheAL in a variety of enteric clinical isolates, and it is proposed to be important for chemotactic responses in specialized environmental niches. In contrast, the holins of lambda phages use internal initiation of translation (dual start motifs) for expression of a long form and a short form, differing by only a few codons. The long form interacts with the short form, an interaction that regulates the activity of the short form by holding it in an inactivated state (2, 3, 28)
The long and short forms of SafA appear to have different roles. The full-length form of SafA is necessary for formation of an intact coat, since the stop codon mutant (AOB91; Fig. 2A) had the same phenotype as a safA deletion mutant, AOB68 (20). The function of the 30-kDa short form is less clear. Blocking accumulation of SafA-C30 with single or double alanine substitutions at methionines 161 and 164 did not have a detectable effect on spore coat function or morphology. However, overexpression of SafA-C30 on a multicopy plasmid drastically blocked sporulation at stage IV. Further work is required to discover the role of SafA-C30.
ACKNOWLEDGMENTS
We gratefully acknowledge Jan Pohl at the Emory University Microchemical Facility, Hong Yi at the Emory University Neurology Electron Microscopy Core facility, and Craig Samford for expert technical assistance, as well as Manuel Santos for helpful discussions.
Teresa Costa was the recipient of a Ph.D. fellowship (BD71167/2000) from the “Fundação para a Ciência e a Tecnologia” (FCT). This work was supported by PHS grant GM54395 from the National Institutes of Health to C. P. Moran, Jr.
REFERENCES
- 1.Aronson A I, Song H Y, Bourne N. Gene structure and precursor processing of a novel Bacillus subtilis spore coat protein. Mol Microbiol. 1989;3:437–444. doi: 10.1111/j.1365-2958.1989.tb00189.x. [DOI] [PubMed] [Google Scholar]
- 2.Barenboim M, Chang C Y, dib Hajj F, Young R. Characterization of the dual start motif of a class II holin gene. Mol Microbiol. 1999;32:715–727. doi: 10.1046/j.1365-2958.1999.01385.x. [DOI] [PubMed] [Google Scholar]
- 3.Blasi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 4.Bourne N, FitzJames P C, Aronson A I. Structural and germination defects of Bacillus subtilis spores with altered contents of a spore coat protein. J Bacteriol. 1991;173:6618–6625. doi: 10.1128/jb.173.20.6618-6625.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cutting S, Panzer S, Losick R. Regulatory studies on the promoter for a gene governing synthesis and assembly of the spore coat in Bacillus subtilis. J Mol Biol. 1989;207:393–404. doi: 10.1016/0022-2836(89)90262-3. [DOI] [PubMed] [Google Scholar]
- 6.Driks A. Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Henriques A O, Beall B W, Moran C P., Jr CotM of Bacillus subtilis, a member of the α-crystallin family of stress proteins, is induced during development and participates in spore outer coat formation. J Bacteriol. 1997;179:1887–1897. doi: 10.1128/jb.179.6.1887-1897.1997. . (Erratum, 179:4455.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henriques A O, Beall B W, Roland K, Moran C P., Jr Characterization of cotJ, a ςE-controlled operon affecting the polypeptide composition of the coat of Bacillus subtilis spores. J Bacteriol. 1995;177:3394–3406. doi: 10.1128/jb.177.12.3394-3406.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henriques A O, Moran C P., Jr Structure and assembly of the bacterial endospore coat. Methods. 2000;20:95–110. doi: 10.1006/meth.1999.0909. [DOI] [PubMed] [Google Scholar]
- 11.Inoue T, Cech T R. Secondary structure of the circular form of the Tetrahymena rRNA intervening sequence: a technique for RNA structure analysis using chemical probes and reverse transcriptase. Proc Natl Acad Sci USA. 1985;82:648–652. doi: 10.1073/pnas.82.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karmazyn-Campelli C, Bonamy C, Savelli B, Stragier P. Tandem genes encoding sigma-factors for consecutive steps of development in Bacillus subtilis. Genes Dev. 1989;3:150–157. doi: 10.1101/gad.3.2.150. [DOI] [PubMed] [Google Scholar]
- 13.Karow L M, Piggot P J. Construction of gusA transcriptional fusion vectors for Bacillus subtilis and their utilization for studies of spore formation. Gene. 1995;163:69–74. doi: 10.1016/0378-1119(95)00402-r. [DOI] [PubMed] [Google Scholar]
- 14.Kodama T, Takamatsu H, Asai K, Kobayashi K, Ogasawara N, Watabe K. The Bacillus subtilis yaaH gene is transcribed by SigE RNA polymerase during sporulation, and its product is involved in germination of spores. J Bacteriol. 1999;181:4584–4591. doi: 10.1128/jb.181.15.4584-4591.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroos L, Zhang B, Ichikawa H, Yu Y T. Control of sigma factor activity during Bacillus subtilis sporulation. Mol Microbiol. 1999;31:1285–1294. doi: 10.1046/j.1365-2958.1999.01214.x. [DOI] [PubMed] [Google Scholar]
- 16.Kunkel B, Sandman K, Panzer S, Youngman P, Losick R. The promoter for a sporulation gene in the spoIVC locus of Bacillus subtilis and its use in studies of temporal and spatial control of gene expression. J Bacteriol. 1988;170:3513–3522. doi: 10.1128/jb.170.8.3513-3522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 18.McNamara B P, Wolfe A J. Coexpression of the long and short forms of CheA, the chemotaxis histidine kinase, by members of the family Enterobacteriaceae. J Bacteriol. 1997;179:1813–1818. doi: 10.1128/jb.179.5.1813-1818.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson W L, Setlow P. Sporulation, germination, and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biology methods for Bacillus. Chichester, United Kingdom: John Wiley and Sons; 1990. pp. 391–450. [Google Scholar]
- 20.Ozin A J, Henriques A O, Yi H, Moran C P., Jr Morphogenetic proteins SpoVID and SafA form a complex during assembly of the Bacillus subtilis spore coat. J Bacteriol. 2000;182:1828–1833. doi: 10.1128/jb.182.7.1828-1833.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piggot P J, Coote J G. Genetic aspects of bacterial endospore formation. Bacteriol Rev. 1976;40:908–962. doi: 10.1128/br.40.4.908-962.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- 24.Sullivan A M, Yasbin R E, Young F E. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
- 25.Takamatsu H, Imamura A, Kodama T, Asai K, Ogasawara N, Watabe K. The yabG gene of Bacillus subtilis encodes a sporulation specific protease which is involved in the processing of several spore coat proteins. FEMS Microbiol Lett. 2000;192:33–38. doi: 10.1111/j.1574-6968.2000.tb09355.x. [DOI] [PubMed] [Google Scholar]
- 26.Takamatsu H, Kodama T, Imamura A, Asai K, Kobayashi K, Nakayama T, Ogasawara N, Watabe K. The Bacillus subtilis yabG gene is transcribed by SigK RNA polymerase during sporulation, and yabG mutant spores have altered coat protein composition. J Bacteriol. 2000;182:1883–1888. doi: 10.1128/jb.182.7.1883-1888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Takamatsu H, Kodama T, Nakayama T, Watabe K. Characterization of the yrbA gene of Bacillus subtilis, involved in resistance and germination of spores. J Bacteriol. 1999;181:4986–4994. doi: 10.1128/jb.181.16.4986-4994.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Young R, Blasi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]