Abstract
The discovery of toxin-antitoxin gene pairs (also called addiction modules) on extrachromosomal elements of Escherichia coli, and particularly the discovery of homologous modules on the bacterial chromosome, suggest that a potential for programmed cell death may be inherent in bacterial cultures. We have reported on the E. coli mazEF system, a regulatable addiction module located on the bacterial chromosome. MazF is a stable toxin and MazE is a labile antitoxin. Here we show that cell death mediated by the E. coli mazEF module can be triggered by several antibiotics (rifampicin, chloramphenicol, and spectinomycin) that are general inhibitors of transcription and/or translation. These antibiotics inhibit the continuous expression of the labile antitoxin MazE, and as a result, the stable toxin MazF causes cell death. Our results have implications for the possible mode(s) of action of this group of antibiotics.
In Escherichia coli cultures, programmed cell death is mediated through a unique genetic system. This system, called an “addiction module,” consists of a pair of genes that specify for two components: a stable toxin and an unstable antitoxin which prevents the lethal action of the toxin. Until recently, such genetic systems for bacterial programmed cell death have been found mainly in E. coli on low-copy-number plasmids, where they are responsible for what is called the postsegregational killing effect. When bacteria lose the plasmid(s) (or other extrachromosomal elements), the cured cells are selectively killed because the unstable antitoxin is degraded faster than is the more stable toxin (6, 9, 14, 27). Thus, the cells are “addicted” to the short-lived product, since its de novo synthesis is essential for cell survival (27). Therefore, these addiction modules have been implicated as having a role in maintaining stability in the host of the extrachromosomal elements on which they are borne (6, 9, 14, 27).
Pairs of genes homologous to some of these extrachromosomal addiction modules have been found on the E. coli chromosome (1, 11, 12, 15–17). Members of our group have reported on the E. coli mazEF system, the first known regulatable prokaryotic chromosomal addiction module (1). The mazEF module consists of two adjacent genes, mazE and mazF, located in the rel operon downstream from the relA gene (17). In the study by members of our group (1), mazEF was found to have the properties required for an addiction module: (i) MazF is toxic and MazE is antitoxic; (ii) MazF is long lived, while MazE is a labile protein degraded in vivo by the ATP-dependent ClpPA serine protease; (iii) MazE and MazF interact; and (iv) MazE and MazF are coexpressed. Moreover, the mazEF system has a unique property: its expression is inhibited by guanosine 3′,5′-bispyrophosphate (ppGpp), which is synthesized under conditions of extreme amino acid starvation by the RelA protein (4). Based on these properties of mazEF and on the requirement for the continuous expression of MazE to prevent cell death, members of our group offered a model for programmed cell death under conditions of nutrient starvation (1). This model was further supported by the results of our previous experiments showing that mazEF-mediated cell death is induced by the artificial overproduction of ppGpp (1, 8), leading to high concentrations that might not be found under normal physiological conditions, even when the cells suffer from extreme nutrient starvation.
Here we ask: in E. coli, can mazEF-mediated cell death be triggered under the specific physiological condition of the inhibition of RNA and/or protein synthesis? To create this condition we briefly treated the bacterial cells with a low concentration of one of several antibiotics known to inhibit transcription and/or translation in E. coli. We found that such antibiotics can indeed trigger mazEF-mediated cell death by reducing the level of MazE.
MATERIALS AND METHODS
Materials and media.
[35S]methionine (>800 Ci/mmol [1 Ci = 37 GBq]) was obtained from Amersham (Little Chalfont, England). The antibiotics rifampin, chloramphenicol, spectinomycin, and ampicillin were obtained from Sigma (St. Louis, Mo.). Polycolonal antibodies against E. coli MazE and TrpR were prepared by injecting purified His-tagged MazE and TrpR proteins into rabbits (13). Bacteria were grown in M9 medium (14) with a mixture of amino acids (20 μg/ml each) or in Luria-Bertani medium (LB) (18).
Bacterial strains.
The E. coli strains used in this study were MC4100 relA+ [genotype, araD139 Δ(argF-lac)205 flb-5301 pstF25 rpsL150 deoC1 (wild type) (8)] and its derivatives MC4100 relA+ mazEF::kan (ΔmazEF) (8) and MC4100 relA+ clpP::cat (ΔclpP). The latter was constructed here by P1 transduction from the strain MC4100 clpP::cat (1).
Activation by antibiotics of mazEF-mediated killing.
Cells were grown in M9 (with a mixture of 20 μg of each amino acid/ml) or LB medium (18) with shaking (160 rpm) at 37°C. At mid-logarithmic phase (optical density at 600 nm [OD600], 0.4 to 0.6), to each sample we added one of the antibiotics at the following final concentrations: 5 to 25 μg of rifampin/ml in M9 and 15 to 25 μg of rifampin/ml in LB, 15 μg of chloramphenicol/ml, 200 μg of spectinomycin/ml or 100 μg of ampicillin/ml both in M9 and in LB. The results of the experiments shown in Fig. 1A to D were observed under conditions in which we used the minimal concentrations of antibiotics at which they were effective. The cells were incubated at 37°C for 10 min, washed in LB, diluted, plated on LB plates, and incubated at 37°C for 18 h. Note that both the LB liquid medium used for washing and diluting and the LB plates were prewarmed to 37°C. Cell survival was calculated by comparing the colony-forming ability of cells treated by antibiotics to that of untreated cells.
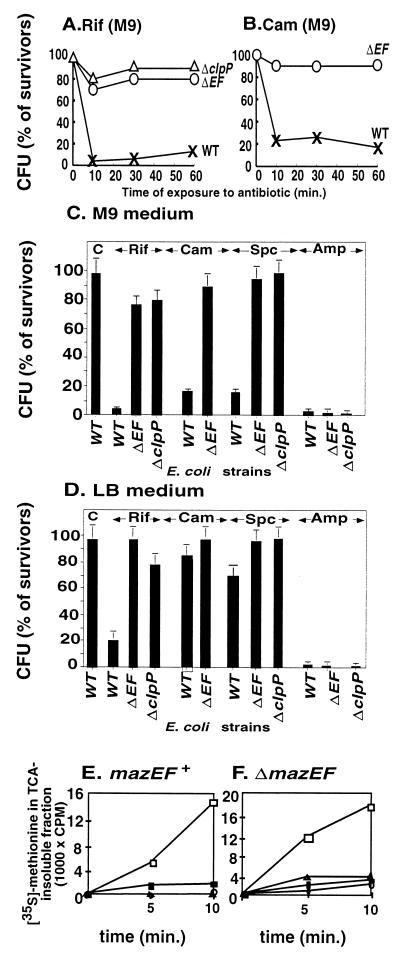
FIG. 1.
Antibiotics that inhibit transcription and/or translation in E. coli induce mazEF-dependent cell death. Viability plotted against the time of exposure to rifampin (A) and chloramphenicol (B) in M9 medium of E. coli MC4100 relA+ (WT) (x) and its ΔmazEF (ΔEF) (O) and ΔclpP (Δ) derivatives is shown. (C) The viability of E. coli MC4100 relA+ (WT) and its ΔmazEF (ΔEF) and ΔclpP derivatives in M9 medium either untreated (C, control) or treated for 10 min with rifampin (Rif), chloramphenicol (Cam), spectinomycin (Spc), or ampicillin (Amp). (D) As for panel C but in LB medium. (E) The effects in M9 medium of the antibiotics (untreated cells, □; rifampin, ■; chloram phenicol, ●; spectinomycin, ▴) on protein synthesis in E. coli MC4100 relA+. (F) As for panel E but in the derivative strain MC4100 relA+ ΔmazEF. The effects of the antibiotics on mazEF-mediated killing and on protein synthesis were measured as described in Materials and Methods.
Assay for the effect of antibiotics on protein synthesis.
We measured the incorporation of [35S]methionine into a trichloroacetic acid (TCA)-insoluble fraction. Cultures were grown in M9 medium without methionine at 37°C to mid-logarithmic phase (OD600, 0.4 to 0.6). To each 1-ml sample of culture a solution of a single antibiotic (25 μg of rifampin/ml, 15 μg of chloramphenicol/ml, and 100 μg of spectinomycin/ml) was added. The culture was labeled with 0.2 μCi/ml in [35S]methionine in a final concentration of 2 μg of unlabeled methionine/ml. At various time intervals, the reactions were stopped by the addition of TCA to a final concentration of 10%, after which the reaction tubes were put in ice. The samples were centrifuged at 14,000 rpm for 5 min in Eppendorf centrifuge 5417C. The pellets were washed twice with 5% TCA and then twice with acetone. The TCA-insoluble counts were determined by using a scintillation counter (BETAmatic I/II; KONTRON).
Determination of the cellular levels of E. coli MazE and TrpR.
The cultures were grown in LB or M9 media with shaking at 37°C. When the cultures reached an OD600 of 0.25 (time zero), one of the following antibiotics at the specified concentration was added to each culture: 200 μg of rifampin/ml, 50 μg of chloramphenicol/ml, or 200 μg of spectinomycin/ml. Over a period of 90 min, equal volumes (100 μl) of samples that were grown in M9 or LB were withdrawn and then immediately centrifuged at 3,000 rpm at room temperature for 10 min in Eppendorf centrifuge 5417C. The collected cells were resuspended in 0.5 ml of TE buffer (20 mM Tris, 1 mM EDTA [pH 8.0]), lysed by sonication for 30 s, and centrifuged at 14,000 rpm at 4°C for 30 min in Eppendorf centrifuge 5417C. The supernatants were loaded on 16.5% Tricine–SDS polyacrylamide gels. Electrophoresis was carried out at 150 V overnight. Proteins were transferred onto a nitrocellulose membrane at 100 V for 1.5 h. Western analysis was carried out using MazE or TrpR polyclonal antibodies as primary antibodies which were prepared in rabbits by injecting His-tagged purified MazE and TrpR proteins (13). The secondary antibody was horseradish peroxidase goat anti-rabbit immunoglobulin G. MazE and TrpR were detected through the enhanced chemiluminescence reaction after an exposure to a sensitive film.
RESULTS
Antibiotics that inhibit transcription and/or translation in E. coli trigger mazEF-mediated death.
As a transcriptional inhibitor we chose the antibiotic rifampin, known to inhibit the initiation of RNA synthesis through its interaction with the β-subunit of RNA polymerase (7, 25). As translational inhibitors we chose the antibiotics chloramphenicol and spectinomycin, which are known to affect the machinery of translation elongation: chloramphenicol acts on the 50S ribosomal subunit to inhibit the petidyl transferase reaction, and spectinomycin affects tRNA translocation (7, 24). We compared the viability of wild-type E. coli MC4100 relA+ to the viability of its ΔmazEF and ΔclpP derivatives after exposing each of these strains to antibiotics in M9 medium at 37°C over 60 min. Even after only a short exposure (10 min) to rifampin (Fig. 1A and C), chloramphenicol (Fig. 1B and C), or spectinomycin (data not shown and Fig. 1C), it was clear that cell death was both mazEF mediated and clpP dependent. In each case the antibiotics caused most (85 to 95%) of the wild-type E. coli cells to die. In contrast, under identical conditions, we observed almost no killing of the ΔmazEF and ΔclpP derivatives. In addition, we observed similar results when we tested the effect of these antibiotics on the cell viability of E. coli B (BL21) and its ΔmazEF derivatives (data not shown). Four important points should be noted. (i) Cells rendered nonviable by the antibiotics do not form colonies (even after several weeks); however, no lysis was observed in the test tubes and by a microscope (data not shown). (ii) About 10% of the subpopulation of cells survive antibiotic treatment, and that cannot be attributed to antibiotic resistance and/or to resistance to mazEF-mediated cell death. When we re-treated the surviving subpopulation with the same antibiotic we found the same results: 90% of the culture suffered mazEF-mediated death, and 10% survived (data not shown). (iii) Although in the presence of the antibiotics under study, ΔmazEF mutants remain viable (Fig. 1A to C), they still remain sensitive to these drugs: protein synthesis (Fig. 1E to F) and cell growth rates (data not shown) are inhibited equally in the wild-type and their ΔmazEF derivative strains. (iv) Obviously, rifampin did not trigger mazEF-dependent killing in rifampin-resistant mutants, nor did chloramphenicol or spectinomycin in chloramphenicol-resistant or spectinomycin-resistant mutants respectively (data not shown). Therefore, as we shall explain below, we suggest that the mazEF-mediated death may be a consequence of the inhibition of transcription and/or translation that can be triggered by these antibiotics. This idea is supported by the fact that wild-type and ΔmazEF cells were affected similarly by the cell wall synthesis inhibitor ampicillin (7) (Fig. 1C and D).
The effect on the cellular level of MazE of antibiotics that inhibit transcription and/or translation.
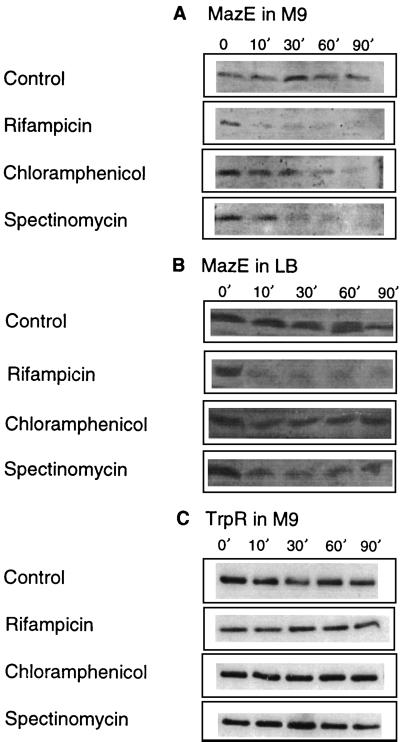
We hypothesized that the antibiotics rifampin, chloramphenicol, and spectinomycin triggered mazEF-mediated killing by inhibiting the continuous expression of the labile antitoxin MazE. As a result, the remaining, stable toxin, MazF, caused cell death. To test this hypothesis, we studied the effect of these antibiotics on the cellular levels of MazE (Fig. 2A). So far, no chromosome-borne component of an addiction module has been detected. Here, we determined the cellular level of MazE by Western analysis using antibodies that we prepared against purified MazE (see Materials and Methods). In our control cultures of untreated cells in M9 medium, we found a constant level of MazE over a period of 90 min. In contrast, upon the addition of rifampin, the level of MazE was already drastically reduced after only 10 min. When we added chloramphenicol or spectinomycin to cells growing in M9 medium, the reduction in the level of MazE started after 10 min, and by 90 min MazE was barely detectable (Fig. 2A). Under our experimental conditions, it was not possible to determine the MazE/MazF ratios, because though we could detect the cellular levels of the chromosomally directed antitoxic protein MazE (with anti-MazE antibodies), we were unable to do so for the toxic protein MazF (with anti-MazF antibodies). Using anti-MazF antibodies, we could detect MazF only when it was either purified or expressed by plasmid-borne mazF. Since in the case of each of the known extrachromosomal addiction modules the antitoxic protein is produced in excess over the toxic protein (for a review, see reference 9), it seems that the level of the toxic MazF protein expressed from the chromosome-borne gene was too low to be detected. Therefore, instead we studied the effect of rifampin, chloramphenicol, and spectinomycin on the cellular levels of another E. coli protein, TrpR (the Trp repressor) (19). We choose TrpR because like MazF (9 kDa), TrpR (12 kDa) is a small E. coli protein found in low intracellular concentrations. Unlike the short-lived MazE, however, TrpR is known to be stable (19) and therefore it resembles the stable MazF (1). As we expected, in cells treated with each of the antibiotics, in contrast to the reduced levels of the labile protein MazE (Fig. 2A), the levels of TrpR remained constant (Fig. 2C). Note that in M9 medium, the cellular level of MazE is significantly lower than it is in LB medium (compare Fig. 2A and B). In LB medium, compared to the control samples, 10 min after the addition of rifampin we detected a significant reduction in the level of MazE, which remained at very low levels during the whole course of the experiment (10 to 90 min) (Fig. 2B). Similarly, 10 min after we added chloramphenicol or spectinomycin, we also found reduced levels of MazE. However, these reduced levels were still significantly higher than those in cells treated with rifampin, and they remained constant over the whole period of the experiment (Fig. 2B).
FIG. 2.
Antibiotics that inhibit transcription and/or translation in E. coli affect the level of MazE. (A) The level of E. coli MazE in M9 medium. (B) The level of E. coli MazE in LB medium. (C) The level of E. coli TrpR protein in M9 medium. The experiments were carried out with E. coli strain MC4100 relA+ for 90 min in the presence of rifampicin, chloramphenicol, or spectinomycin or without any antibiotics (control) as described in Materials and Methods.
The effect on mazEF-mediated cell death in LB medium of antibiotics that inhibit transcription and/or translation.
As in M9 medium, we also tested the effects of adding the antibiotic rifampin, spectinomycin, or chloramphenicol to cultures in LB medium (Fig. 1D). In LB, the difference in the effect on MazE levels between cells treated by rifampin and cells treated by chloramphenicol or spectinomycin (Fig. 2B) is reflected in the differing effect(s) of these drugs on cell viability (Fig. 1D). mazEF-mediated killing triggered by rifampin left only 5% survivors in M9 medium (Fig. 1C) and 20% survivors in LB medium (Fig. 1D). On the other hand, in LB medium chloramphenicol did not trigger mazEF-dependent death (Fig. 1D). This was true in LB even when we increased the concentration of chloramphenicol more than 10 times, to 200 μg/ml (data not shown). In the case of spectinomycin in LB, we observed only 30% mazEF-mediated killing (Fig. 1D). It appears that in LB medium, even after the addition of either chloramphenicol or spectinomycin, the levels of MazE remained high enough to prevent cell death by MazF (Fig. 2D).
DISCUSSION
The main finding of this work is that E. coli mazEF-mediated cell death can be triggered by several antibiotics, such as rifampin, chloramphenicol, or spectinomycin (Fig. 1A to C) that are general inhibitors of transcription and/or translation (7, 24, 25). We have also shown that these antibiotics reduce the cellular level of the antitoxic labile protein MazE (Fig. 2A) and seem thereby to permit the lethal action of the toxic protein MazF. The effect of the antibiotics both on cell death and on the reduction in the cellular level of MazE is particularly apparent in M9 medium (Fig. 1C and 2A). In our experiments, we determined two parameters. One is cell death which was determined by cell viability after 18 h (Fig. 1A to C) and which is the result of multiple processes. The other is the level of the MazE protein, which we determined at several time points during the first 90 min of our experiments (Fig. 2A) and which represents only one step in the process of cell death. We observed a correlation between the initial reduction in the levels of MazE and the overall loss in cell viability. This correlation is valid not only in the case of treatment by rifampin, where MazE was not detectable after 10 min, but even in the cases of treatment by chloramphenicol and spectinomycin, where the reduction in MazE was seen only after 60 to 90 min (Fig. 2A). In addition, our results clearly show that even in M9 medium the transcription inhibitor rifampin triggered MazE degradation and cell death more effectively than did chloramphenicol or spectinomycin. As we mentioned earlier, ppGpp, the signal molecule for nutrient starvation (4), leads to the inhibition of mazEF transcription (1). Both ppGpp and rifampin bind to the β-subunit of RNA polymerase (5, 22). Therefore, rifampin may have been more effective than chloramphenicol or spectinomycin. In addition, we observed that E. coli strain MC4100 (carrying the mutation relA1) (3) is less sensitive to the mazEF-mediated cell death triggered by the herein-described antibiotics than is the same E. coli strain carrying the wild-type relA+ gene (data not shown). E. coli MC4100 relA1 produces lower levels of ppGpp than the wild-type relA+ strain (4). It seems, therefore, that ppGpp may be involved in the mazEF-mediated killing even when it is triggered by antibiotics that inhibit transcription and/or translation. This assumption is under our current investigation.
In LB medium, only when we used rifampin did we detect both significant mazEF-dependent killing (Fig. 1D) and a reduction in the cellular level of MazE (Fig. 2B). Nevertheless, both of these effects of rifampin were less drastic in LB than they were in M9 medium (Fig. 1C and D and 2A and B). Furthermore, in LB medium, neither chloramphenicol nor spectinomycin had a significant effect on cell viability (Fig. 1D). In addition, the levels of MazE were significantly higher than those in cells treated with rifampin (Fig. 2B) and probably high enough to prevent the killing of MazF. The differences among the actions of the various antibiotics observed in M9 and LB media can be explained by the cellular levels of MazE, which were significantly lower in M9 medium than they were in LB (compare Fig. 2A and B). In addition to other possible explanations, we suggest that in cells growing in the rich LB medium, the high level of low-molecular-weight peptides offers more substrates to compete for degradation by ClpPA, thus somewhat protecting MazE itself from degradation. This possibility is further supported by our results showing similar levels of TrpR in cells grown in either LB or M9 medium (data not shown). Since TrpR is a stable protein (Fig. 2C) and thus not a substrate for ClpPA, it should not be affected by the low-molecular-weight peptides in LB medium.
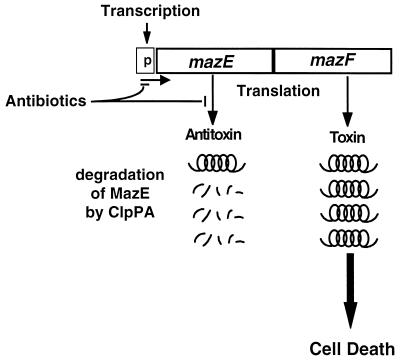
The results that we report here broaden our previous model of programmed cell death mediated by the mazEF system (1). Previously, members of our group based the model only on the artificial overexpression of MazF or ppGpp (1, 8). Here we base our model on conditions that are more similar to physiological conditions in which inhibition of transcription or translation occurs, i.e., conditions of stress. We found that even briefly inhibiting transcription and/or translation by antibiotics was sufficient to induce mazEF-mediated cell death. As illustrated in Fig. 3, the inhibition of protein synthesis will prevent the de novo synthesis of the labile antideath protein MazE, thus triggering mazEF-mediated death. The MazE protein already present in the cell continues to be degraded by the ClpPA protease. As a result, the level of MazE is reduced below the threshold required for antagonizing the toxic protein MazF, leading to cell death. Thus, we suggest that the choice between cell survival and cell death depends on the level of MazE. Inhibiting transcription and/or translation selectively reduced the cellular level of the antitoxic labile protein MazE (Fig. 2A and B) and thus probably permitted the lethal action of the toxic protein MazF (Fig. 3). Our model is further supported by the recent results of members of our group on postsegregational killing mediated by the addiction module phd-doc of plasmid prophage P1 (13a). This module consists of two genes; doc codes for a stable toxin, and phd codes for a labile antitoxin (reviewed in references 9 and 27). In our new study we show the following: (i) the postsegregational killing effect of P1 phd-doc requires the presence of the E. coli mazEF system, and (ii) under conditions of P1 phd-doc postsegregational killing the protein synthesis in E. coli is inhibited. This inhibition is probably caused by the Doc protein, which was recently described as being a translational inhibitor (9; Yarmolinsky, personal communication). Thus, inhibition of protein synthesis either by antibiotics or by the Doc protein triggers the E. coli mazEF-mediated death.
FIG. 3.
A schematic representation of the induction of the E. coli mazEF-mediated cell death by antibiotics that inhibit transcription and/or translation (see the text).
In analogy to the programmed cell death apparatus in eukaryotic cells (20, 26), it seems that the suicide machinery in bacterial cells is always present: it only requires a trigger to activate it. Moreover, at least for E. coli, the straightforward choice is death caused by a stable intracellular toxin (in this case MazF). The choice of life over the “default” death requires a dynamic antagonistic process manifested either by the continued production of the unstable antitoxin (in this case MazE) or by a process that would prevent the degradation of the unstable antitoxin (9). Thus, cell death could be caused by anything that would prevent the continuous expression of the antitoxic protein MazE. Our results, showing that the mazEF system is responsible for approximately 90% killing by rifampin, may illuminate the until now elusive cause of E. coli killing by rifampin (23). Furthermore, though chloramphenicol and spectinomycin are traditionally considered to be bacteriostatic (2, 23), we found that these antibiotics were actually bacteriocidal in M9 medium. It seems likely that until now these antibiotics have not been revealed to be bacteriocidal because they had been tested in rich media like LB (2, 10, 21), where we found that the level of the antitoxic protein MazE remained high (Fig. 2B). Thus, we suggest that the traditional distinction between “bacteriostatic” and “bacteriocidal” should not be taken as absolute and should be reconsidered. Moreover, now we can add “triggering bacterial cell suicide” to the other well-documented modes of action of such antibiotics.
ACKNOWLEDGMENTS
We thank Sudersan Narasimhan for his help. We are deeply grateful to F. R. Warshaw-Dadon (Jerusalem, Israel) for her critical reading of the manuscript.
This research was supported by a grant of the Israel Science Foundation administrated by the Israel Academy of Science and Humanities.
REFERENCES
- 1.Aizenman E, Engelberg-Kulka H, Glaser G. An Escherichia coli chromosomal “addiction module” regulated by ppGpp: a model for programmed cell death. Proc Natl Acad Sci USA. 1996;93:6059–6063. doi: 10.1073/pnas.93.12.6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brock T D. Chloramphenicol. Bacteriol Rev. 1961;25:32–48. doi: 10.1128/br.25.1.32-48.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cashel M, Gentry D R, Hernandez V Z, Vinella D. The stringent response. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B M, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: American Society for Microbiology; 1996. pp. 1458–1496. [Google Scholar]
- 5.Chatterji D, Fujita N, Ishihama A. The mediator for stringent control, ppGpp, binds to the beta-subunit of Escherichia coli RNA polymerase. Genes Cells. 1998;3:279–287. doi: 10.1046/j.1365-2443.1998.00190.x. [DOI] [PubMed] [Google Scholar]
- 6.Couturier M, Bahassi E M, Van Melderen L. Bacterial death by DNA gyrase poisoning. Trends Microbiol. 1998;6:269–275. doi: 10.1016/s0966-842x(98)01311-0. [DOI] [PubMed] [Google Scholar]
- 7.Davies J, Webb V. Antibiotic resistance in bacteria. In: Krause R M, editor. Emerging infections. New York, N.Y: Academic Press; 1998. pp. 239–273. [Google Scholar]
- 8.Engelberg-Kulka H, Reches M, Narasimhan S, Schoulaker-Schwarz R, Klemes Y, Aizenman E, Glaser G. rexB of bacteriophage λ is an anti-cell death gene. Proc Natl Acad Sci USA. 1998;95:15481–15486. doi: 10.1073/pnas.95.26.15481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engelberg-Kulka H, Glaser G. Addiction modules and programmed cell death and antideath in bacterial cultures. Annu Rev Microbiol. 1999;53:43–70. doi: 10.1146/annurev.micro.53.1.43. [DOI] [PubMed] [Google Scholar]
- 10.Fassin W, Hengel R, Klein P. Bakteriostase und Bakterizidie als Alternativen des antibakteriellen Chloramphenicoleffektes. Z Hyg. 1955;141:S363–S375. [PubMed] [Google Scholar]
- 11.Gerdes K, Gultyaev A P, Franch T, Pederson K, Milkkelsen N D. Antisense RNA-regulated programmed cell death. Annu Rev Genet. 1997;19:49–61. doi: 10.1146/annurev.genet.31.1.1. [DOI] [PubMed] [Google Scholar]
- 12.Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol Microbiol. 1998;29:1065–1076. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- 13.Harlow E, Lane D. antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1998. Immunization; pp. 53–137. [Google Scholar]
- 13a.Hazan R, Sat B, Reches M, Engelberg-Kulka H. Postsegregational killing mediated by the P1 phage “addiction module” phd-doc requires the Escherichia coli programmed cell death system mazEF. J Bacteriol. 2001;183:2046–2050. doi: 10.1128/JB.183.6.2046-2050.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen R B, Gerdes K. Programmed cell death in bacteria: proteic plasmid stabilization systems. Mol Microbiol. 1995;17:205–210. doi: 10.1111/j.1365-2958.1995.mmi_17020205.x. [DOI] [PubMed] [Google Scholar]
- 15.Masuda Y, Miyakawa K, Nishimura Y, Ohtsubo E. chpA and chpB, Escherichia coli chromosomal homologs of the pem locus responsible for stable maintenance of plasmid R100. J Bacteriol. 1993;175:6850–6856. doi: 10.1128/jb.175.21.6850-6856.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masuda Y, Ohtsubo E. Mapping and disruption of the chpB locus in Escherichia coli. J Bacteriol. 1994;176:5861–5863. doi: 10.1128/jb.176.18.5861-5863.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Metzger S, Dror I B, Aizenman E, Schreiber G, Toone M, Friesen J D, Cashel M, Glaser G. The nucleotide sequence and characterization of the relA gene of Escherichia coli. J Biol Chem. 1988;263:15699–15704. [PubMed] [Google Scholar]
- 18.Miller J H. Experiments in molecular genetics. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1972. pp. 205–210. [Google Scholar]
- 19.Pittard A J. Biosynthesis of the aromatic amino acids. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B M, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C.: American Society for Microbiology; 1996. pp. 458–484. [Google Scholar]
- 20.Raff M. Cell suicide for beginners. Nature. 1998;306:119–122. doi: 10.1038/24055. [DOI] [PubMed] [Google Scholar]
- 21.Rahal J J, Jr, Simberkoff M S. Bactericidal and bacteriostatic action of chloramphenicol against meningeal pathogens. Antimicrob Agents Chemother. 1979;16:13–18. doi: 10.1128/aac.16.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reddy P S, Raghavan A, Chatterji D. Evidence for a ppGpp-binding site on Escherichia coli RNA polymerase: proximity relationship with the rifampicin-binding domain. Mol Microbiol. 1995;15:255–265. doi: 10.1111/j.1365-2958.1995.tb02240.x. [DOI] [PubMed] [Google Scholar]
- 23.Schlessinger D, Eisenstein B. Biological basis for antibacterial action. In: Schaechter M, Engleberg N C, Eisenstein B, Medoff G, editors. Microbial Disease. 3rd ed. New York, N.Y: Williams & Wilkins; 1998. pp. 52–61. [Google Scholar]
- 24.Spahn C M T, Prescott C D. Throwing a spanner in the works: antibiotics and the translation apparatus. J Mol Biol. 1996;74:423–439. doi: 10.1007/BF00217518. [DOI] [PubMed] [Google Scholar]
- 25.Wehrli W, Staehelin M. Actions of the rifampicins. Bacteriol Rev. 1971;35:290–309. doi: 10.1128/br.35.3.290-309.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weil M, Jacobson M D, Coles H S, Davies T J, Gardner R L, Raff K D, Raff M C. Constitutive expression of the machinery for programmed cell death. J Cell Biol. 1996;133:1053–1059. doi: 10.1083/jcb.133.5.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yarmolinsky M B. Programmed cell death in bacterial population. Science. 1995;267:836–837. doi: 10.1126/science.7846528. [DOI] [PubMed] [Google Scholar]