SUMMARY
The transcriptional regulator Runx2 (runt-related transcription factor 2) has essential but distinct roles in osteoblasts and chondrocytes in skeletal development. However, Runx2-mediated regulatory mechanisms underlying the distinctive programming of osteoblasts and chondrocytes are not well understood. Here, we perform an integrative analysis to investigate Runx2-DNA binding and chromatin accessibility ex vivo using neonatal osteoblasts and chondrocytes. We find that Runx2 engages with cell-type-distinct chromatin-accessible regions, potentially interacting with different combinations of transcriptional regulators, forming cell-type-specific hotspots, and potentiating chromatin accessibility. Genetic analysis and direct cellular reprogramming studies suggest that Runx2 is essential for establishment of chromatin accessibility in osteoblasts. Functional enhancer studies identify an Sp7 distal enhancer driven by Runx2-dependent binding and osteoblast-specific chromatin accessibility, contributing to normal osteoblast differentiation. Our findings provide a framework for understanding the regulatory landscape encompassing Runx2-mediated and cell-type-distinct enhancer networks that underlie the specification of osteoblasts.
Graphical Abstract
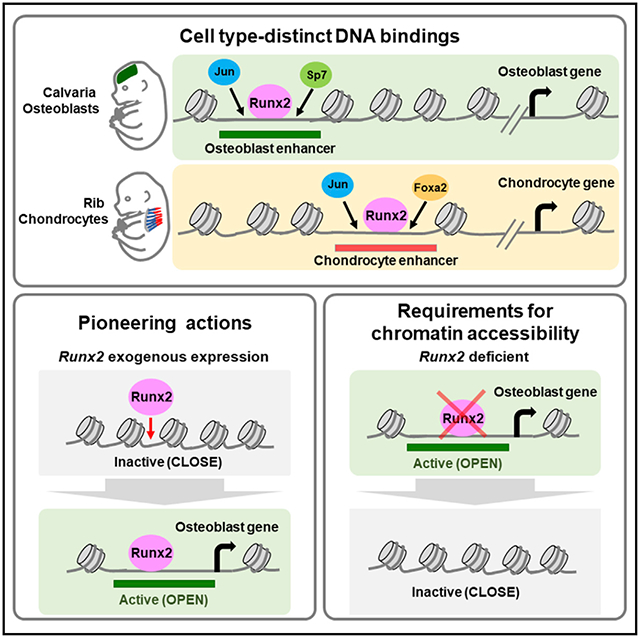
In brief
Hojo et al. investigate the gene-regulatory landscape underlying specification of skeletal cell types in neonatal mice. Runx2, an osteoblast determinant, engages with cell-type-distinct chromatin-accessible regions and is essential for establishment of chromatin accessibility in osteoblasts. The study provides insights into enhancer networks in skeletal development.
INTRODUCTION
The mammalian skeleton is formed from distinct lineages of progenitor cells (Olsen, 2019). Specification of skeletal cell types is critically dependent on master transcriptional regulators. SRY-box-containing gene 9 (Sox9) is initially required for establishment of skeletal progenitors and subsequent differentiation to chondrocytes (Akiyama et al., 2002). Sp7/Osx is essential for specification of osteoblast precursors (Nakashima et al., 2002). Runt-related transcription factor 2 (Runx2) is required for osteoblast specification and chondrocyte hypertrophy (Ducy et al., 1997; Komori et al., 1997; Mundlos et al., 1997; Otto et al., 1997; Yoshida et al., 2004).
In the osteoblast program, skeletal progenitors are committed to Runx2-positive osteoblast precursors that transition to Runx2 and Sp7 double-positive osteoblast precursors before adopting a mature osteoblast phenotype (Olsen, 2019). In Runx2-deficient mutant mice, osteoblast differentiation is arrested prior to activation of Sp7 (Nakashima et al., 2002). However, Runx2-positive osteoblast precursors are present in Sp7-deficient mice but fail to progress to mature osteoblasts (Nakashima et al., 2002). These data suggest that Runx2 initiates osteogenesis upstream of Sp7 early in the regulatory hierarchy of osteoblast development.
In the chondrocyte program, Runx2 is weakly expressed in proliferating columnar chondrocytes but markedly up-regulated as chondrocytes exit the cell cycle, forming prehypertrophic and then hypertrophic chondrocytes. Ectopic expression of Runx2 in columnar chondrocytes accelerates chondrocyte hypertrophy (Stricker et al., 2002; Takeda et al., 2001; Ueta et al., 2001), and removal of Runx2 activity results in loss of normal hypertrophic cartilage mineralization (Yoshida et al., 2004). Restoration of Runx2 in chondrocytes in Runx2-null mutants rescues chondrocyte hypertrophy and mineralization but not the adjacent periosteal bone program, suggesting that Runx2 acts cell autonomously and independently in regulating cartilage and bone development (Takeda et al., 2001).
These genetic studies have provided broad insights into the distinct cell-type-specific roles of Runx2. Increasing evidence also suggests the existence of lineage-specific transcriptional regulators that may act cooperatively with Runx2 (Karsenty, 2008). However, the gene-regulatory landscape by which Runx2 executes two distinct skeletal programs has not been resolved. Here we examined the interactions of Runx2 in developing cartilage and bones ex vivo relative to lineage-specific programs of chromatin accessibility. Together with functional studies of identified enhancer elements, our findings provide insights into the regulatory mechanisms governing mammalian skeletal development.
RESULTS
Identification of cell-type-distinct open chromatin signatures in skeletal development
To identify cell-type-distinct signatures in mammalian skeletal cells, we mapped Runx2 expression within specific skeletal cell types using transgenic reporter strains: Col2a1-ECFP (Chokalingam et al., 2009), Col10a1-mCherry (Maye et al., 2011), and Sp7-EGFP (Rodda and McMahon, 2006). Histological analysis confirmed reporter activities and Runx2 expression in the ribs and calvariae at post-natal day 1 (P1). Runx2 was weakly expressed in Col2a1-ECFP-positive columnar chondrocytes, and its expression was elevated in Col10a1-mCherry-positive hypertrophic chondrocytes. Runx2 expression was also high in Sp7-EGFP-positive osteoblast precursors and osteoblasts (Figures S1A and S1B).
We investigated chromatin accessibility and transcriptional profiles in skeletal cell types by assaying for transposase-accessible chromatin using high-throughput sequencing (ATAC-seq) and RNA sequencing (RNA-seq), respectively, in fluorescence-activated cell sorting (FACS)-purified Col2a1-ECFP-positive rib chondrocytes (C2-Cho), Col10a1-mCherry-positive rib hypertrophic chondrocytes (C10-Cho), and Sp7-EGFP-positive calvarial osteoblasts (S7-Ob) in newborn mice at P1 (Tables S1 and S2). Principal-component analysis (PCA) of the RNA-seq and ATAC-seq data revealed distinct transcriptomes and accessible chromatin signatures among these cell types (Figure 1A). Col10a1 and Col1a1 encode key collagen matrix proteins secreted by hypertrophic chondrocytes and osteoblasts, respectively. C10-Cho chromatin profiling and S7-Ob chromatin profiling detected cell-type-specific chromatin accessibility in regions specifically flanking Col10a1 and Col1a1, respectively (Figure 1B). In contrast, chromatin accessibility flanking Col2a1, which encodes collagen secreted predominantly by non-hypertrophic chondrocytes, was observed in C2-Cho and C10-Cho chondrocytes; however, the position of accessible regions differed between the two (Figure 1B, top), suggesting regulatory differences between early-stage columnar chondrocytes and late-stage chondrocytes, which initiate hypertrophic chondrocyte development. As expected, the cell-type-distinct expression of each gene reflected the cell-type-distinct patterns of chromatin accessibility (Figure 1B). These results highlight the differences in the chromatin landscape between chondrocytes and osteoblasts as well as the dynamic differences in chromatin accessibility associated with chondrocyte differentiation.
Figure 1. Identification of cell-type-distinct chromatin accessibility in neonatal chondrocytes and osteoblasts.
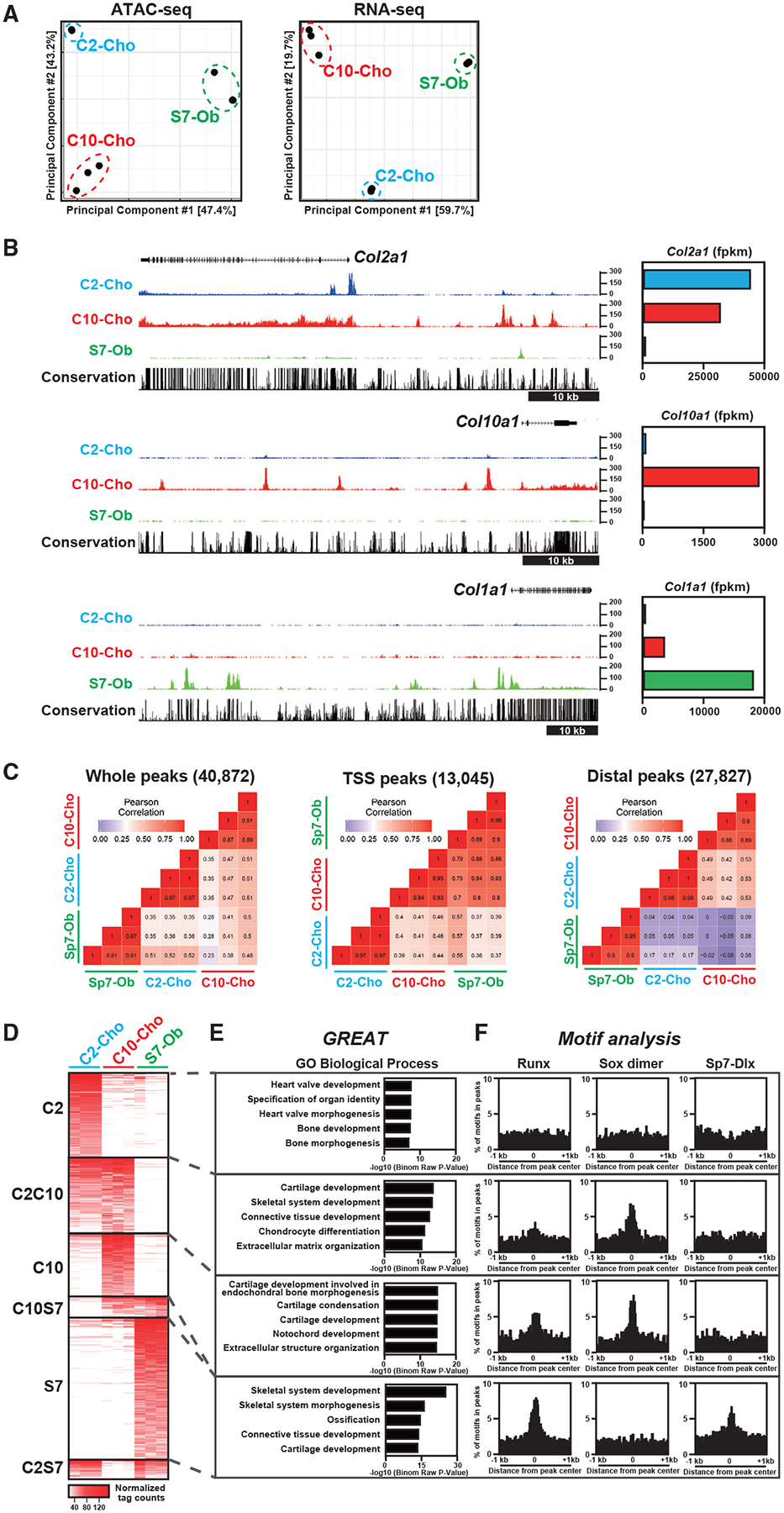
(A) Principal-component analysis (PCA) of ATAC-seq and RNA-seq data. C2-Cho, Col2a1-ECFP-sorted rib chondrocytes; C10-Cho, Col10a1-mCherry-sorted rib chondrocytes; S7-Ob, Sp7-EGFP-sorted calvaria osteoblasts.
(B) CisGenome browser screenshots showing chromatin accessibility around skeletal marker genes (left) and the corresponding gene expression in skeletal cell types (right). Representative data (left) and average of normalized values (right) from biological triplicates are shown.
(C) Correlation analysis of ATAC-seq profiles of the three skeletal cell types. The color indicator represents the Pearson correlation coefficient. TSS peaks, peaks located within 500 bp from the transcription start site (TSS) of the nearest genes; distal peaks, peaks located more than 500 bp from the TSS.
(D) Heatmap showing chromatin accessibility in the cell-type-distinct regions. C2, C2-Cho-distinct peaks; C10, C10-Cho-distinct peaks; S7. S7-Ob-distinct peaks; C2C10, peaks with high intensity in C2-Cho and C10-Cho but not S7-Ob; C10S7, peaks with high intensity in C10-Cho and S7-Ob but not C2-Cho; C2S7, peaks with high intensity in C2-Cho and S7-Ob but not C10-Cho. The Color indicator represents the normalized ATAC-seq peak intensity.
(E) GREAT Gene Ontology (GO) annotations of identified chromatin-accessible regions. The top five enriched terms are shown in each group.
(F) Enrichment of Runx, Sox dimer, and Sp7-Dlx consensus motifs in the identified chromatin-accessible regions in the 2-kb window from the center of the ATAC-seq peaks.
Profiles of biological triplicates were used (A, C, and D). See also Figure S1 and Tables S1 and S2.
To compare chromatin accessibility among skeletal cell types, we analyzed ATAC-seq profiles of each cell population. Distal accessible regions, potential cis-regulatory enhancer regions, better represented the cell-type-distinct signatures of each skeletal cell type than open chromatin-encompassing promoter sites (Figure 1C), consistent with previous studies in different biological contexts (Corces et al., 2016; Gury-BenAri et al., 2016). A comparison of chromatin accessibility profiles highlights the lineage relationship between columnar (i.e., C2-Cho) and hypertrophic chondrocytes (i.e., C10-Cho) in that they are more similar to each other than either is to developing osteoblasts (i.e., S7-Ob) (Figure 1C).
Pairwise comparisons of the distal ATAC-seq peak signals revealed cell-type-distinct peaks (C2-Cho-, C10-Cho-, and S7-Ob-specific) and peaks shared by paired sets (Figures 1D and S1C). Genomic Regions Enrichment of Annotations Tool (GREAT) Gene Ontology (GO) analysis (McLean et al., 2010) highlighted expected cell type signatures; cartilage development terms were associated with C10-Cho-specific and C2C10-shared peaks, whereas S7-specific peaks were associated with skeletal development and ossification (Figure 1E; Table S1).
To examine the relationship between the identified accessible regions and master transcriptional regulators in skeletal development, we performed de novo motif analysis and mapped consensus motifs for Runx, Sox9, and Sp7 (Figure 1F). The Sp7 motif is an Sp7-Dlx-cooperative motif that has been identified previously by osteoblast Sp7 chromatin immunoprecipitation sequencing (ChIP-seq) (Hojo et al., 2016). The Runx motif was slightly enriched in C2C10-shared peaks, moderately enriched in C10-Cho-specific peaks, and highly enriched in S7-Ob-specific peaks. The Sox9 dimer motif (Ohba et al., 2015) was enriched in chondrocyte-related peaks; i.e., C2C10-shared and C10-Cho-specific peaks, whereas the Sp7-Dlx motif was exclusively enriched in S7-Ob-specific peaks.
These results predict the cell-type-distinct and overlapping effects of master regulators on putative enhancer regions in skeletal development. The enrichment of the Runx motif in the accessible regions of hypertrophic chondrocytes and osteoblasts provides genomic support for the direct transcriptional role of Runx factors in each cell type (Håkelien et al., 2014; Meyer et al., 2014; Tai et al., 2017; Wu et al., 2014), consistent with previous studies (Kern et al., 2001; Li et al., 2011), and reinforces its regulatory effects reported in mice (Komori et al., 1997; Yoshida et al., 2004).
Genome-scale analysis of Runx2-DNA associations in osteoblasts and chondrocytes
To determine the relationship of Runx2 with accessible regions, we performed genome-scale profiling of Runx2-DNA association. To this end, we generated a Runx2-Biotin-3xFLAG knockin mouse line using a gene-targeting strategy (Runx2-BioFL mouse; Figure 2; STAR Methods). This approach appended a cassette to the C terminus of the Runx2 protein comprising a biotin (Bio) recognition motif and three copies of the FLAG (FL) epitope to enable biochemical isolation of Runx2 and associated DNA targets or binding partners (Figures 2A–2C). BioFL epitope tagging did not alter the normal characteristics of Runx2. Western blotting detected levels of Runx2-BioFL protein similar to its wild-type counterpart (Figure 2D), and immunohistochemistry of tibial sections also showed an identical temporal and spatial distribution (Figure 2E). Mice homozygous for the Runx2-BioFL allele showed no overt phenotype, were fertile, and had undergone normal skeletal development when analyzed at P1 (Figures 2F and 2G).
Figure 2. Generation of a Runx2 Bio-3xFL knockin mouse line and Runx2 association profiles in osteoblasts and chondrocytes.
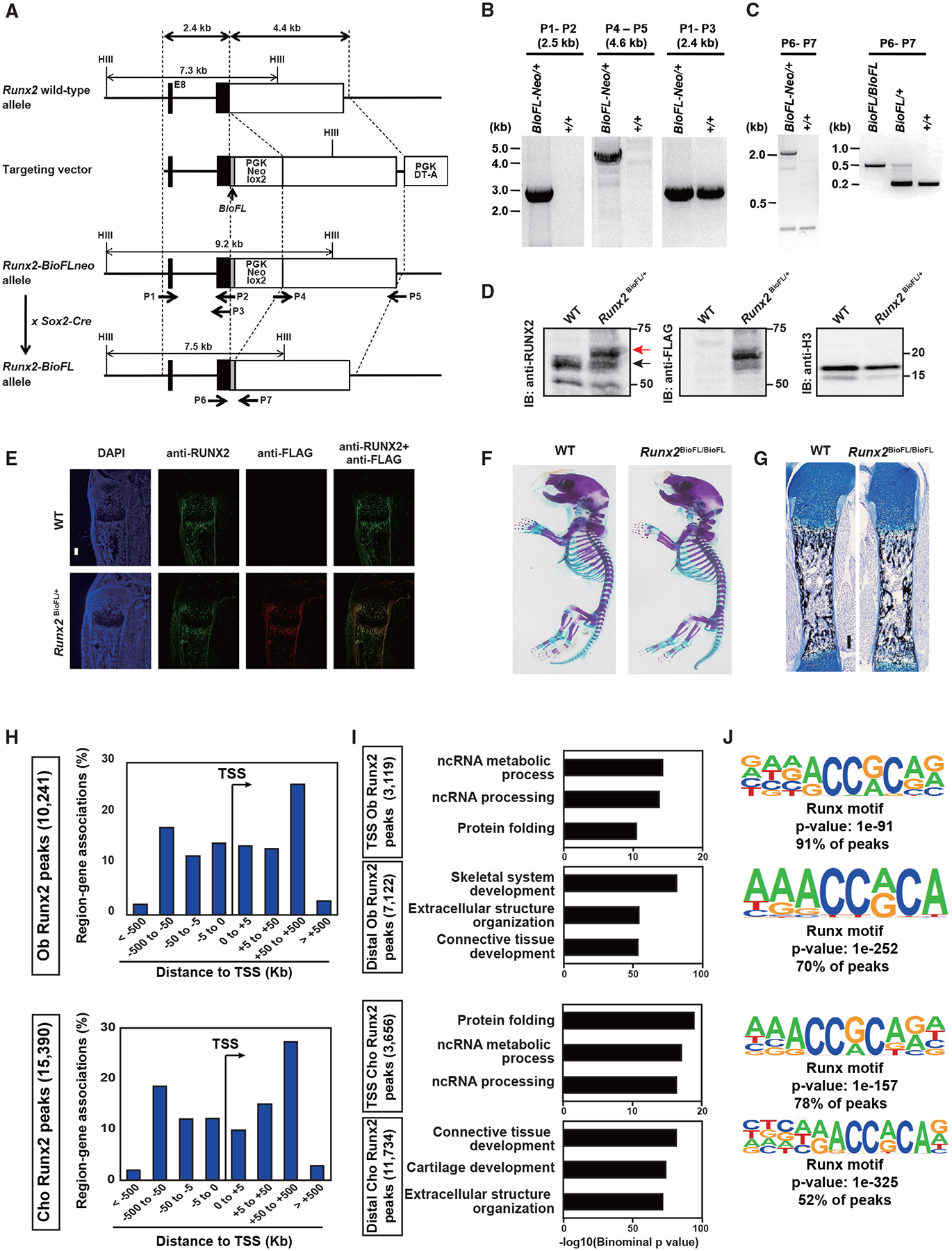
(A) Targeting strategy for the Runx2 Bio-3xFL knockin allele.
(B) Long-range PCR analysis of the Runx2 knockin allele using the primers in (A).
(C) PCR genotyping of Runx2-BioFLneo and Runx2-BioFL mice obtained by crossing Runx2-BioFLneo mice to Sox2-Cre mice; see (A).
(D) Western blot comparison of Runx2 and Runx2-BioFL proteins in primary osteoblasts derived from neonatal calvarial cells. Cells were cultured in an osteogenic medium for 7 days. Red arrow, Runx2-BioFL; black arrow, Runx2. Blotting for histone H3 (H3) was used for the control.
(E) Immunohistochemistry for Runx2 and FL tag detection in tibial sections of P1 Runx2-BioFL pups. Scale bar, 300 μm.
(F) Whole-mount skeleton of P1 pups stained with Alcian blue (cartilage) and alizarin red (mineralized tissue).
(G) Histological analysis of calcified bone matrix stained with von Kossa (black) and cartilage stained with Alcian blue in a P1 femur. Scale bar, 100 μm.
(H) Genome-wide distribution of Runx2-associated regions relative to TSSs in osteoblasts (Obs) and chondrocytes (Chos). The total number of Runx2-associated regions is indicated.
(I) GREAT GO annotations of Runx2-associated regions. The top three enriched terms are shown in each TSS and distal region.
(J) De novo motif analysis of the top 1,000 Runx2-associated regions in Obs and Chos. The p value of the shown motif and the percentage of peaks containing the enriched Runx motif are shown.
Representative images obtained from at least biological duplicates are shown (E–G). See also Table S3.
To compare Runx2 interactions in osteoblasts and chondrocytes, we performed anti-FL antibody-directed ChIP-seq analysis on calvarial osteoblasts and rib chondrocytes from neonatal Runx2-BioFL mice. The intersection of biological replicates identified a stringent set of 10,241 Runx2-BioFL ChIP-seq peaks in osteoblasts and 15,390 peaks in chondrocytes (Figure 2H; Table S3). Peak distribution was similar between the two populations, and Runx2 was associated with the proximal and distal genomic regions (Figure 2H). This pattern is consistent with previous results from Runx2 ChIP-seq in in vitro-cultured osteoblasts (Håkelien et al., 2014; Meyer et al., 2014; Wu et al., 2014). We found that distal Runx2 peaks were highly associated with genes related to osteoblasts or chondrocytes, whereas TSS Runx2 peaks were more associated with general cell activities (Figure 2I). De novo motif analysis showed high enrichment of Runx consensus motifs in transcription start site (TSS) and distal Runx2 peaks (Figure 2J). The enriched motif retains the core sequence of the Runx consensus motif (AACCACA), which was identified by a systematic mutagenesis analysis of the mouse osteocalcin promoter (Ducy and Karsenty, 1995; Geoffroy et al., 1995). Thus, binding of Runx2 to its consensus motif is likely to be the primary mode of action of Runx2 interaction with DNA, independent of the distance of the target region from the TSS of the skeletal cell type, although the possibility that Runx2 may have non-genomic actions critical for its biological roles cannot be ruled out.
Considering the physical and functional interaction between Runx2 and core-binding factor β (Cbfβ) in skeletal cells (Kundu et al., 2002; Yoshida et al., 2002), we investigated the Cbfβ-Runx2 associations with the genome. A Cbfβ ChIP-seq study showed that 64% of the Cbfβ peaks overlapped with Runx2 peaks, which represented 13% of all Runx2 peaks in chondrocytes (Figure S1D). The motif analysis showed that a Runx consensus motif was most enriched in TSS and distal Cbfβ peaks (Figure S1E). These data suggest that Runx2 may pair with Cbfβ in chondrocytes on only a small subset of its consensus motifs, although the Runx2-Cbfβ interaction may be underestimated in the data because of technical limitations of ChIP experiments, including a difference in ChIP efficiency between different antibodies and potentially limited detection of indirect interactions between DNA and cofactors.
Next, to directly compare the Runx2 binding signatures in chondrocytes and osteoblasts, we performed a correlation analysis between osteoblast and chondrocyte Runx2 ChIP-seq peaks and found that Runx2 binding signatures were cell type distinct (Figure S2A). In particular, the signature in the distal Runx2-bound regions was well separated between the two cell types, whereas those in the proximal regions were not cell type distinct. Distal Runx2-DNA regions were highly associated with genes related to skeletal cell types (Figure 2I). Thus, we addressed the associations between the Runx2 binding signature and chromatin accessibility in each cell type using the distal Runx2 ChIP-seq peaks. We found that Runx2-bound regions in chondrocytes were associated with higher chromatin accessibility in Col10-positive chondrocytes, whereas Runx2 regions in osteoblasts were highly associated with chromatin accessibility in Sp7-positive osteoblasts (Figures S2B–S2D). These data suggest that Runx2 binding signatures are associated with chromatin accessibility in a cell type-distinct manner.
We next investigated whether Runx2 acts on closed chromatin. We extracted 448 chondrocyte Runx2 peaks at the closed chromatin region in C10-Cho and 87 osteoblasts Runx2 peaks at the closed chromatin region in S7-Ob (Figure S2E; STAR Methods). Motif analysis revealed that not only the Runx consensus motif but also CTCF and REST motifs were highly enriched in chondrocyte Runx2 peaks, whereas the Runx motif was not enriched in osteoblast Runx2 peaks (Figure S2F). GREAT GO analysis showed that chondrocyte Runx2 peaks, having the Runx motif at the closed chromatin, were associated with genes related to skeletal system development (Figure S2G; Table S3). For example, a shared Runx2 peak in chondrocytes and osteoblasts was detected in the closed chromatin region flanking Tgfb2 (Figure S2H). These results suggest that Runx2 may act at a limited number of closed chromatin regions to regulate skeletal development, where Runx2 may bind DNA directly through the Runx consensus motifs or indirectly by interacting with other chromatin regulators, including CTCF and REST.
Actions of Runx2 and its potential interaction with other transcriptional regulators on the genome
We next investigated the cell-type-distinct actions of Runx2. We extracted genomic regions where Runx2 binding and chromatin accessibility were enriched in a cell-type-specific manner (Figure 3A; Table S4; see STAR Methods for details). GREAT GO analysis confirmed that osteoblast- and chondrocyte-distinct regions were enriched in skeletal development- and cartilage-related gene sets, respectively (Figure 3B). In contrast, the small number of regions shared between chondrocytes and osteoblasts were less associated with genes related to skeletal cell features (Figure 3B).
Figure 3. Actions of Runx2 and its potential interaction with other transcriptional regulators on the genome.
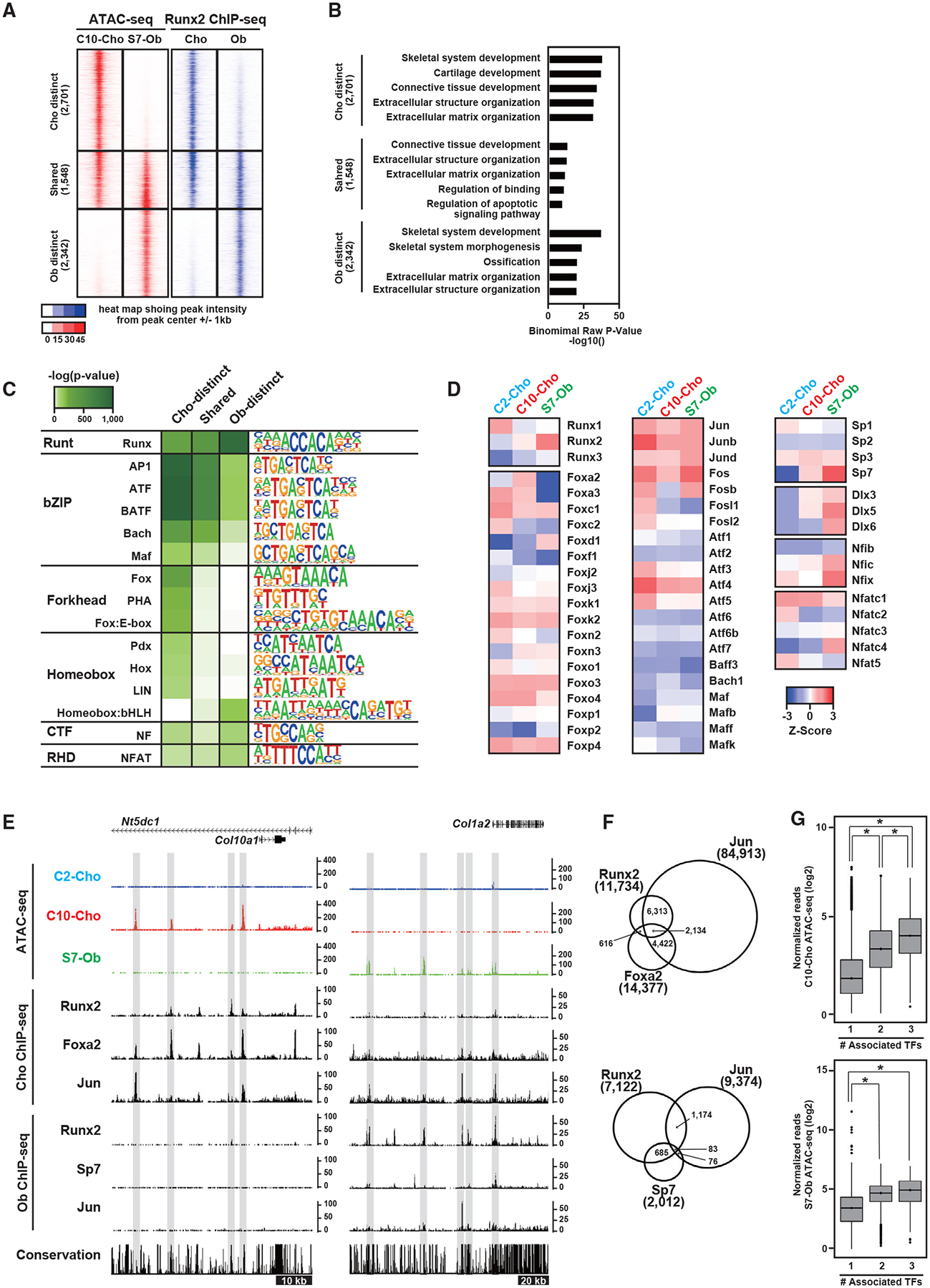
(A) Signal intensities of chromatin accessibility and Runx2-DNA binding in Obs and Chos. The selected regions showing Ob- and Cho-distinct signatures and their shared signatures are shown. The number of selected genomic regions is indicated. Representative data obtained from at least biological duplicates are shown.
(B) GREAT GO annotations of the selected regions, showing the top five enriched terms.
(C and D) Enrichment of motifs in Ob- and Cho-distinct regions and their shared regions. Motifs with p < 1.0 × 10−50 and target sequences with motifs >10% are shown (C). Also shown (D) is the Z score representing the relative gene expression of transcription factors in C2-Cho, C10-Cho, and S7-Ob. Genes were selected from the enriched motifs in (D). Average values obtained from biological triplicates were used. Transcription factors with fragments per kilobase of transcript per million mapped reads (FPKM) >5 are shown.
(E) CisGenome browser screenshots of the flanking regions of Col10a1 and Col1a2 showing chromatin accessibility and the associations of transcription factors in Obs and Chos. Cell-type-distinct regions are highlighted in gray.
(F) Venn diagram showing the overlap of ChIP-seq peaks. Top: Runx2, Foxa2, and Jun in Chos. Bottom: Runx2, Sp7, and Jun in Obs. ChIP-seq peaks distally located more than 500 bp from the nearest TSS were used. Peak numbers for each ChIP-seq peak are shown.
(G) Boxplots showing the normalized reads of ATAC-seq at regions associated with the indicated number of transcription factors. Top: C10-Cho ATAC-seq at regions occupied by Runx2, Foxa2, and/or Jun in Chos. Bottom: S7-Ob ATAC-seq at the regions occupied by Runx2, Sp7, and/or Jun in Obs. The y axis value is log2 (normalized reads + 1). The values were averaged from biological triplicates. *p < 0.0001 versus normalized reads with the one factor-associated site (Tukey-HSD analysis).
See also Figures S2 and S3 and Table S4.
To predict cell-type-distinct transcriptional regulators that may work together with Runx2 in osteoblasts and chondrocytes, we performed de novo motif analysis on peak sets (Figure 3C). In addition to the consensus Runx motif, activator protein-1 (AP-1)-related motifs were highly enriched in both cell types. Additionally, although weaker predictions, CAAT box-binding transcription factor/nuclear factor (NF) and NF of activated T-cells (NFAT) motifs were enriched in both cell types. In contrast, a forkhead motif was specifically enriched in chondrocyte-specific regions, whereas a homeobox:basic-helix-loop-helix (bHLH) motif was enriched in osteoblast-specific ones, suggesting that cell-type-distinct regulatory factors act with Runx2 on enhancers that regulate cell-type-specific gene expression. Because the homeobox domain in the homeobox:bHLH motif is similar to the Sp7-Dlx motif (Hojo et al., 2016), the transcriptional complex containing Sp7 and Dlx is likely to be associated with the motif.
To narrow down the candidate transcription factors that potentially act with Runx2 on cell-type-specific enhancers, we compared the expression of the members of the transcription factors associated with the enriched motifs in C2-Cho, C10-Cho, and S7-Ob populations (Figure 3D). We observed the variable expression of multiple family members, suggesting a complex regulatory relationship among similarly acting family members in each skeletal cell type. For example, the overlapping roles of Foxa2 and Foxa3 have been documented in chondrogenesis (Ionescu et al., 2012) and those of Dlx5 and Dlx6 in skeletal development (Robledo et al., 2002). It is also notable that osteoblasts and chondrocytes highly express ATF4 and show ATF motif enrichment (Figures 3C and 3D), which is consistent with the requirements of ATF transcription factors for skeletogenesis (Wang et al., 2009; Yang et al., 2004). These results suggest that Runx2 may act with these transcription factors on cell-type-specific enhancers to drive the common and distinct aspects of the gene-regulatory program of each cell.
To investigate the associations between Runx2 and selected transcriptional regulators in chromatin accessible regions, we integrated additional ChIP-seq data with Runx2-DNA binding and chromatin accessibility profiles. Based on motif enrichment (Figure 3C), gene expression (Figure 3D), and biological functions reported in the literature (Karsenty, 2008; Kozhemyakina et al., 2015), Jun was identified as a potential co-regulator of Runx2 in chondrocytes and osteoblasts, Foxa2 as a chondrocyte-specific co-regulator, and Sp7 as an osteoblast specific co-regulator. We performed ChIP-seq for chondrocyte Foxa2 and osteoblast Jun (Figure S3) and used publicly available ChIP-seq datasets for chondrocyte Jun and osteoblast Sp7 ChIP-seq (He et al., 2016; Hojo et al., 2016). As shown in Figure 3E, Runx2, Jun, and Foxa2 co-occupied genomic regions flanking Col10a1 in chondrocytes that were accessible in C10-Cho. In osteoblasts, Runx2, Jun, and Sp7 showed DNA associations at S7-Ob-specific accessible regions flanking Col1a2. Interestingly, regions co-occupied by more transcription factors show overall enhanced accessibility in chondrocytes and osteoblasts (Figures 3F and 3G).
Requirement of Runx2 for establishment of osteoblast-distinct chromatin accessibility
To determine whether Runx2 is required for establishment of cell-type-distinct chromatin accessibility, we generated Sp7-EGFP:Cre;Runx2fl/fl;R26RtdTomato compound mutant mice and investigated the effect of Runx2 ablation on mutant osteoblasts. CRE activity under Sp7 regulation conditionally removed Runx2 activity while genetically marking CRE-producing cells and their descendants through recombination-mediated activation of tdTomato. Because Runx2 is genetically upstream of Sp7 (Nakashima et al., 2002), Sp7 activation and expression are expected in early osteoblast progenitors with subsequent loss of Runx2 expression and Runx2 activity in Sp7-positive osteoblasts (Figure 4A). We confirmed that skeletal formation was severely impaired in conditional homozygous Runx2 knockout mice (Runx2 cHomo KO) at embryonic day 18.5 (E18.5), as reported previously (Takarada et al., 2016; Figure 4B); it was partially impaired in the littermates of conditional heterozygous Runx2 KO mice (Runx2 cHet KO) (Figure 4B). Histological analysis did not detect SP7 protein in tdTomato-positive cells in Runx2 cHomo KO calvariae at E18.5 Figure 4C). Considering the expression of key osteoblast marker genes, we confirmed that Runx2 cHomo KO cells did not display a mature osteoblast phenotype (Figure S4A).
Figure 4. Effects of Runx2 ablation on establishment of Ob chromatin accessibility.
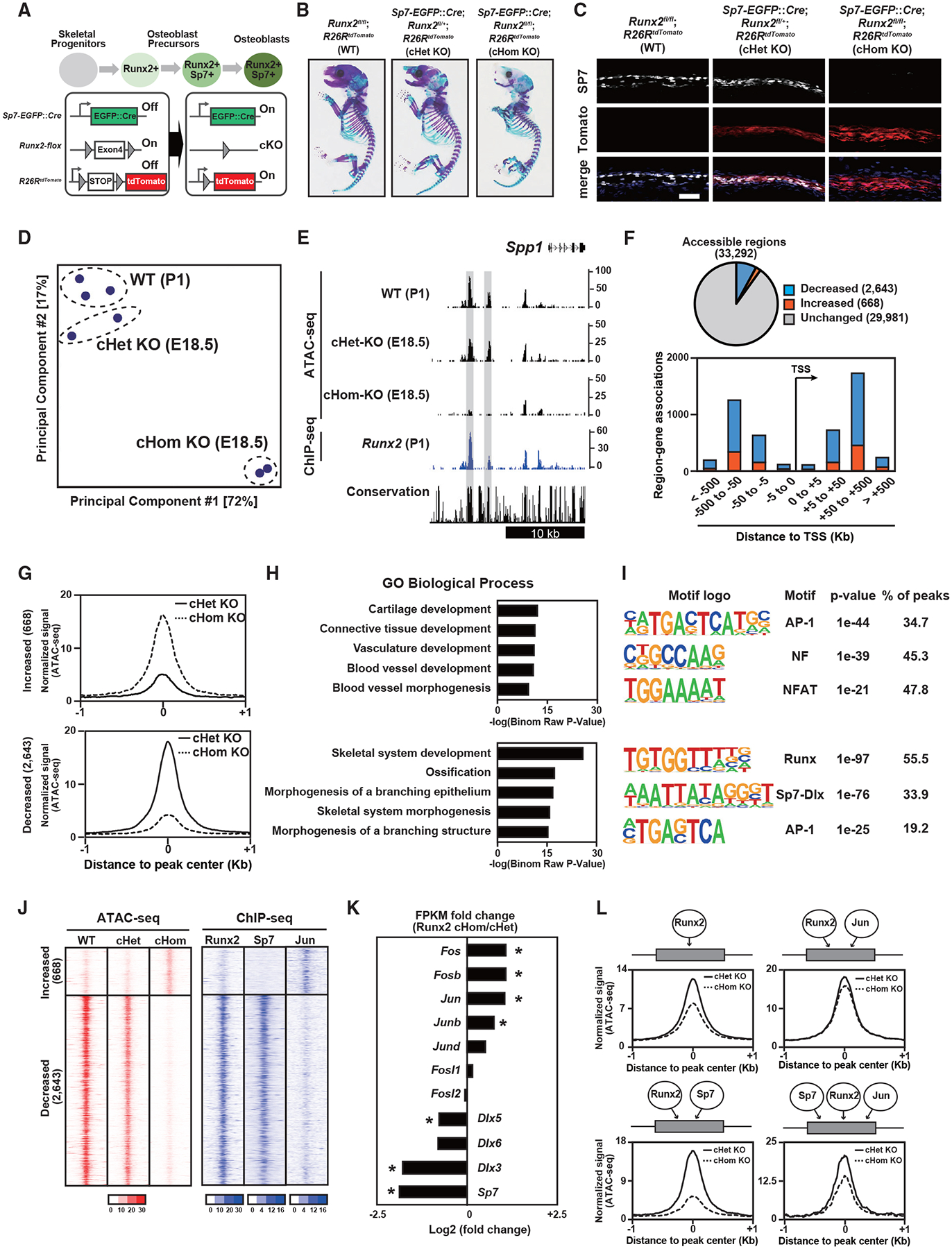
(A) Schematic of Ob differentiation and the genetic strategy.
(B) Whole-mount skeleton of E18.5 pups stained with Alcian blue and alizarin red.
(C and D) Immunohistochemistry for SP7 expression and native tdTomato expression of E18.5 calvariae. Nuclei were stained by DAPI (blue) (C). Scale bar, 100 μm. Also shown (D) is a PCA plot of ATAC-seq. Profiles of biological duplicates and triplicates were plotted.
(E) CisGenome browser screenshot of the flanking region of Spp1 showing chromatin accessibility in calvaria cells of the indicated genotypes and Runx2-DNA binding in Runx2-BioFL calvaria cells. Runx2-dependent chromatin accessible regions are highlighted in gray.
(F) Pie chart showing the change in chromatin accessibility in tdTomato-sorted Runx2 cHomo KO calvaria cells compared with that in Runx2 cHom KO cells (top). Genome-wide distribution of genomic regions shows altered accessibility because of Runx2 ablation (bottom).
(G) Histogram showing the average of normalized ATAC-seq signals in genomic regions with altered accessibility by Runx2 ablation. 668 significantly increased chromatin-accessible regions (top) and 2,643 significantly decreased chromatin-accessible regions (bottom) in Runx2 cHom KO calvaria cells compared with those in Runx2 cHet KO cells are shown. Average values from biological duplicates were used.
(H) GREAT GO annotations of significantly changed chromatin-accessible regions in (G).
(I) De novo motif analysis of the top 500 significantly changed chromatin-accessible regions in (G). The top three most enriched motifs with p value and percentage of peaks are shown.
(J) Heatmap showing the signal intensities of ATAC-seq in E18.5 calvaria cells with the indicated genotypes and those of ChIP-seq for the indicated transcription factors in wild-type (WT) P1 Obs.
(K) Relative gene expression of selected transcription factors in Runx2 cHom KO and cHet KO calvaria cells at E18.5. The values were normalized from biological duplicates. *p < 0.05.
(L) Histogram showing the average of normalized ATAC-seq signals in Runx2 cHet KO and cHom KO calvaria cells at E18.5. The following distal regions were selected based on their associations with transcription factors: regions associated with Runx2 only (top left, 5,180 regions), those associated with Runx2 and Jun (top right, 7,012), those associated with Runx2 and Sp7 (bottom left, 685), and those associated with Runx2, Jun, and Sp7 (bottom right, 83). The values were normalized from biological duplicates.
Representative images obtained from at least biological duplicates are shown (B and C). See also Figure S4 and Table S5.
To address chromatin accessibility after Runx2 removal, we performed ATAC-seq on tdTomato-positive calvarial cells purified from Runx2 cHomo KO and Runx2 cHet KO littermates using S7-Ob ATAC-seq data as a Runx2 wild-type control. PCA showed that the chromatin accessibility pattern in Runx2 cHomo KO cells was markedly different from that in wild-type and Runx2 cHet KO cells (Figure 4D). For example, Spp1-flanking regions were accessible in wild-type and Runx2 cHet KO cells but not in Runx2 cHomo KO cells (Figure 4E). Runx2-dependent accessible regions were bound by Runx2 in Runx2-BioFL osteoblasts (Figure 4E). We found 3,311 significantly changed accessible regions in Runx2 cHomo KO compared with Runx2 cHet KO cells. Upon Runx2 removal, chromatin accessibility increased in 669 regions and decreased in 2,643 regions (Figures 4F and 4G). Most of the differentially accessible regions were located in distal regions, potential enhancers, and not in the TSS (Figure 4F). Runx2-dependent regions with decreased accessibility after Runx2 removal were highly associated with genes related to skeletal development and ossification (Figure 4H). Regions with decreased accessibility contained a known Runx2 enhancer (Kawane et al., 2014) as well as enhancers around osteoblast marker genes (Figures 4E and S4B; Table S5). De novo motif analysis revealed motifs for AP-1, NF, and NFAT enriched in regions with increased accessibility after Runx2 removal, whereas Runx, Sp7-Dlx, and AP-1 motifs were highly enriched in regions that lost chromatin accessibility (Figure 4I). These results suggest that direct Runx2 binding is required for establishment of chromatin accessibility associated with several osteogenesis-related genes and reveal a potential interplay with other transcription factors, including Sp7 and AP-1.
To investigate the involvement of Runx2 and its co-regulator candidates, we mapped ChIP-seq signals for Sp7, AP-1, and Runx2 in the differentially accessible regions. Runx2 and Sp7 were highly associated with regions requiring Runx2 for accessibility, whereas Jun was associated with regions where Runx2 is associated with suppression of chromatin accessibility (Figure 4J). GREAT GO analysis showed that Sp7 binding regions were highly associated with skeletal development, whereas AP1 binding regions were associated with angiogenesis (Figures S4B–S4D). Transcriptional profiling revealed that expression of Sp7 and Dlx members is decreased, whereas that of AP-1 family members is increased after Runx2 ablation (Figure 4K).
Thus, we hypothesized that binding of co-regulatory factors normally acting with Runx2 may explain the alteration in the chromatin profiles observed after Runx2 removal. To verify this, we investigated the effect of Runx2 ablation on ATAC-seq profiles at the intersected peak regions for Runx2, AP-1, and Sp7. We found an association between Runx2 ablation, heightened chromatin accessibility, and binding of co-regulatory transcriptional partners at target enhancers (Figure 4L). Chromatin accessibility was decreased by Runx2 ablation in putative enhancers sharing Runx2 and Sp7 binding (Runx2-Sp7 shared regions), whereas it was intermediately decreased in regions sharing Runx2, Sp7, and Jun (Runx2-Sp7-Jun shared regions). This suggests a compensatory mechanism to keep chromatin accessibility upon Runx2 ablation by increasing Jun expression. Chromatin accessibility in osteoblasts may be maintained by the cooperative actions of Runx2 and its potential co-regulatory factors, including Sp7 and AP-1, on enhancer modules that regulate osteoblast development.
Pioneering action of Runx2 to establish chromatin accessibility
“Pioneer factors” have been identified that facilitate opening of closed chromatin sites (Iwafuchi-Doi and Zaret, 2016). To address whether Runx2 exhibits pioneer factor properties, we overexpressed Runx2-BioFL in NIH3T3 fibroblasts in the presence of bone morphogenetic protein 2 (BMP2), generating osteoblast-like cells (Ohba et al., 2007). Two days after Runx2-BioFL transduction, cells were cultured in an osteogenic medium in the presence of BMP2. ATAC-seq and Runx2-FL ChIP-seq were performed on day 0 and day 3 after BMP2 addition and osteogenic induction (Figure 5A). Alkaline phosphatase (ALP) staining confirmed Runx2/BMP-dependent osteoblast differentiation (Figure 5B). We selected Runx2-and osteogenic induction-dependent chromatin-accessible regions (STAR Methods; Table S6). Runx2 binding to these regions was observed as early as day 0, but in a closed chromatin conformation. In contrast, by day 3, these regions exhibited enhanced chromatin accessibility and Runx2 binding (Figures 5C and 5D). The “Runx2-responsive” accessible regions were mainly located distal to the TSS (Figure 5E). GREAT GO analysis of Runx2-responsive regions identified a strong association of targets with skeletal development (Figure 5F). De novo motif analysis confirmed a Runx consensus motif highly enriched within these regions, and approximately 80% of the Runx2-responsive regions were predicted to encode a Runx2-binding motif (Figure 5G). These results indicate that Runx2 may act as a pioneer factor for distal enhancers of a certain set of skeletal genes by direct binding to the Runx consensus motif. Figure 5H shows an example of a Runx2-responsive region associated with the osteogenic factor-encoding gene Wnt3a (Tu et al., 2007). An ATAC-seq peak was not detected until day 3, whereas a Runx2 ChIP-seq peak was detected on day 0.
Figure 5. Effects of Runx2 gain of function on acquisition of chromatin accessibility in fibroblasts.
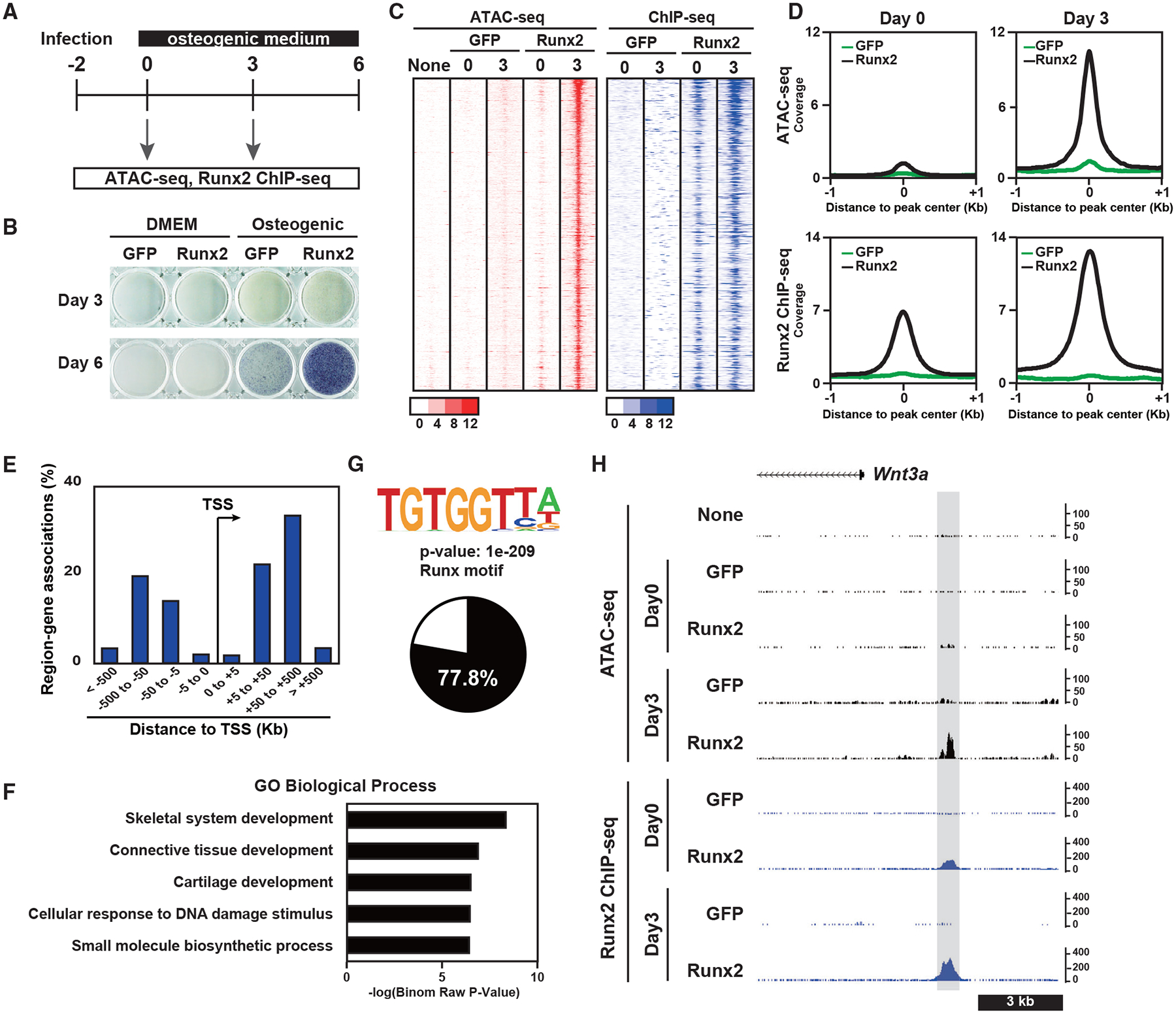
(A) Schematic of the experiment.
(B) ALP staining of NIH 3T3 fibroblasts overexpressing GFP or Runx2-BioFL. Cells were cultured with DMEM or an osteogenic medium.
(C) Heatmap showing the signal intensities of ATAC-seq and Runx2 ChIP-seq of NIH 3T3 cells overexpressing GFP or Runx2-BioFL for the indicated culture periods. We selected 731 genomic regions representing the pioneer action of Runx2. Cells were cultured in an osteogenic medium.
(D) Histogram showing the average of normalized signals in ATAC-seq (top) and Runx2 ChIP-seq (bottom) on day 0 (left) and day 3 (right) in a 2-kb window from the center of the peaks. The values were normalized from biological duplicates.
(E) Genome-wide distribution of Runx2-responsive chromatin-accessible regions relative to TSSs.
(F) GREAT GO annotations of genomic regions representing the pioneer action of Runx2.
(G) Motif logo of de novo motif analysis of 731 genomic regions representing the pioneer action of Runx2 (top) and a pie chart showing the percentage of peaks containing the Runx consensus motif in these genomic regions (bottom).
(H) CisGenome browser screenshot of the flanking region of Wnt3a showing chromatin accessibility and Runx2-DNA binding in NIH 3T3 cells overexpressing GFP or Runx2-BioFL. A genomic region representing the pioneer action of Runx2 is highlighted in gray.
On day3, Runx2 mRNA expression was not altered upon osteogenic induction with BMP2, whereas Runx2 protein expression increased (Figure S5). This is consistent with previous reports showing that BMP2 enhances Runx2 expression and activity through post-transcriptional regulation (Jeon et al., 2006; Ohba et al., 2007). Thus, enhanced binding of Runx2 at day 3 may reflect the enhanced activity of Runx2 for opening of chromatin revealing new sites or cooperative interactions with other factors enabled by opening of chromatin.
Identifying functional osteoblast enhancers targeted by Runx2
To directly address whether Runx2 binding identifies enhancer regions, we performed a CROP-seq assay (Datlinger et al., 2017), where CRISPR-based enhancer KO was combined with single-cell RNA-seq for simultaneous detection of the genotype (from the target guide RNA sequence) and the phenotype (from mRNA levels). We designed single guide RNAs (sgRNAs) for nine putative enhancers as Runx2 targets; the sgRNA targeted the Runx consensus motif located at the center of each enhancer (Table S7). We also prepared sgRNAs for the Runx2 gene as a positive control and for the non-targeting sequence as a negative control (Table S7). The sgRNA library was transduced into the MC3T3-E1 osteoblast cell line, which was engineered to stably express Cas9 (Figures S6A and S6B). We analyzed single-cell RNA-seq (scRNA-seq) data from 5,136 cells in which one sgRNA was detected. Gene expression analysis identified significantly decreased expression of 47 genes, including the known Runx2 targets Bglap, Ibsp, and Spp1, in cells carrying Runx2 sgRNA (Figure 6A). We defined the 47 genes as Runx2-dependent genes in the analysis. GO analysis confirmed that the 47 genes were highly associated with ossification and skeletal development (Figure 6B), validating the general detection approach. Comparative analysis of cells expressing sgRNAs for putative enhancers revealed that expression of Runx2-dependent genes was significantly decreased in cells expressing the sgRNA for Sp7 enhancer-1 (Figures 6C and 6D), whereas that of the housekeeping genes Actb and Gapdh, were not significantly altered (Figure 6D), implicating the Sp7 enhancer-1 as a functional enhancer mediating Runx2 transcriptional regulation of Sp7-directed osteoblast differentiation.
Figure 6. Screening for putative enhancers targeted by Runx2 in Obs.
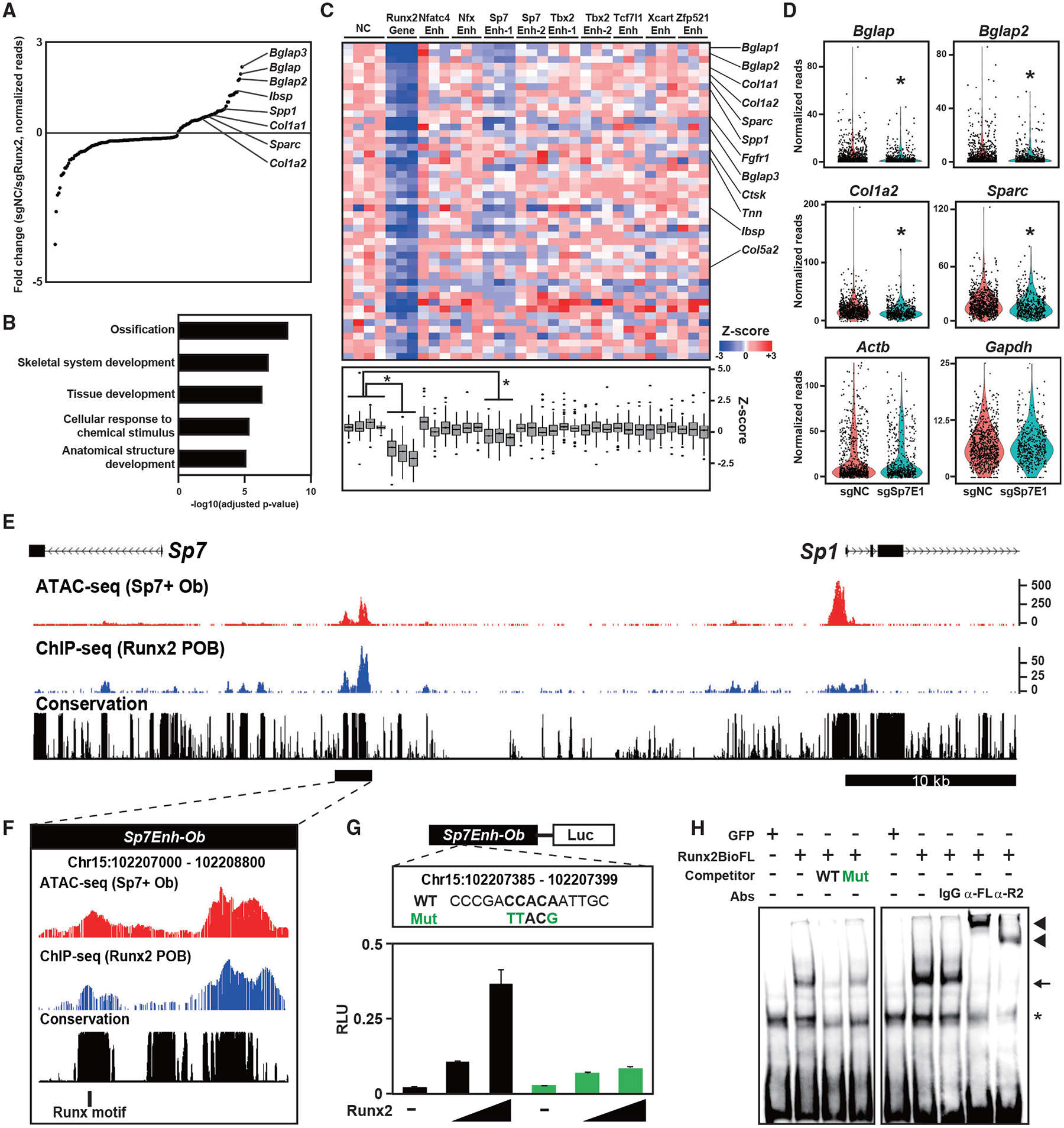
(A) Dot plots showing genes with significantly altered expression induced by sgRunx2 expression. Ob marker genes are highlighted. Normalized values in cells expressing sgRNAs for negative control sequences (sgNC, n = 820) and those for Runx2 (sgRunx2, n = 312) were used.
(B) g:Profiler GO annotations of biological processes in genes with significantly decreased expression induced by sgRunx2 expression.
(C) Heatmap showing Z scores representing relative gene expression in cells expressing the indicated sgRNAs (top) and boxplot of Z scores (bottom). Genes with significantly decreased expression induced by sgRunx2 were selected. Ob-related genes are indicated on the right. *p < 0.05 versus cells expressing sgNC in the one factor-associated site (Tukey-HSD analysis). Normalized values in cells expressing each gRNA (n = 87–368) were used.
(D) Violin plot showing normalized reads of the indicated genes in cells expressing sgNC or Sp7 Enhancer 1 (sgSp7E1). Each dot represents a cell. *p < 5.0 × 10−4 versus cells expressing sgNC. Normalized values from sgNC (n = 820) and sgSp7E1 (n = 498) were used.
(E and F) CisGenome browser screenshot of the Sp7Enh-Ob region showing chromatin accessibility and Runx2-DNA binding in Obs. The region indicated by the black bar in (E) is enlarged in (F), indicating a Runx motif targeted by sgSp7E1.
(G) Schematic of the luciferase reporter driven by Sp7Enh-Ob (top) and luciferase reporter assay in 293T cells transfected with WT and mutated (Mut) reporter constructs together with different amounts of Runx2-expressing plasmids (bottom). WT and Mut DNA sequences of the Runx motif targeted by sgSp7E1 (top) are shown. Data are presented as the means ± SD of triplicate experiments.
(H) EMSA of Runx2-BioFL using a probe containing the Runx motif targeted by sgSp7E1. WT and Mut probes, whose sequences are shown in (G), were used for the competitor assay. Anti-FL (α-FL) and anti-Runx2 antibodies (α-R2) and an immunoglobulin G (IgG) negative control were used for the supershift assay. Arrow, RUNX2-DNA complex; arrowhead, RUNX2-DNA-antibody complex; asterisk, non-specific signal.
See also Figures S6 and S7 and Table S6.
Sp7 enhancer-1 is located in a conserved block between Sp7 and Sp1, exhibiting Runx2 binding and Runx2-dependent chromatin accessibility in osteoblasts (Figures 6E and 6F; Figure S7). To verify the effect of Runx2 on Sp7 enhancer-1, we cloned the 1.8-kb putative enhancer region (chromosome 15 [chr15]: 102 207 000–102 208 800) containing Sp7 enhancer-1 (Sp7Enh-Ob; Figure 6F). We examined the effect of Runx2 expression on enhancer-driven luciferase activity. We found that luciferase reporter activity was elevated in a Runx2-dependent manner and that its activity was attenuated by mutagenesis in the Runx motif that was targeted by sgRNA in the CROP-seq assay (Figure 6G). An electrophoretic mobility shift assay (EMSA) revealed direct binding of Runx2 to the Runx motif (Figure 6H). These results suggest that Runx2 directly binds to the distal Sp7 enhancer and regulates Sp7 transcription.
A luciferase assay showed that the combination of Runx2, constitutively active Alk6, and Smad1 synergistically induced Sp7 enhancer activity (Figure S7C). Considering that Smad1 binding to the Sp7 enhancer was detected in embryonic limb bud cells (Salazar et al., 2019), the result suggests that the interaction of Runx2 and BMP-Smad signaling regulates Sp7 transcription through the identified enhancer.
Verification of the Sp7 enhancer activity in vivo
To investigate the activity of Sp7Enh-Ob in vivo, we generated a transgenic reporter mouse line expressing a lacZ-GFP fusion cassette (Peterson et al., 2012) under the control of Sp7Enh-Ob. Whole-mount β-galactosidase staining showed reporter activities specific to the developing skeletal tissues (Figure 7A). Immunohistochemistry revealed that reporter activity is restricted to a subset of SP7-positive osteoblasts in calvariae and long bones (Figure 7B). Although endogenous SP7 was expressed in osteoblasts and pre-hypertrophic chondrocytes in the long bone, enhancer-driven reporter activity was only detected in osteoblasts (Figure 7B).
Figure 7. In vivo Sp7 enhancer activity and the contribution of the enhancer activity to skeletal development.
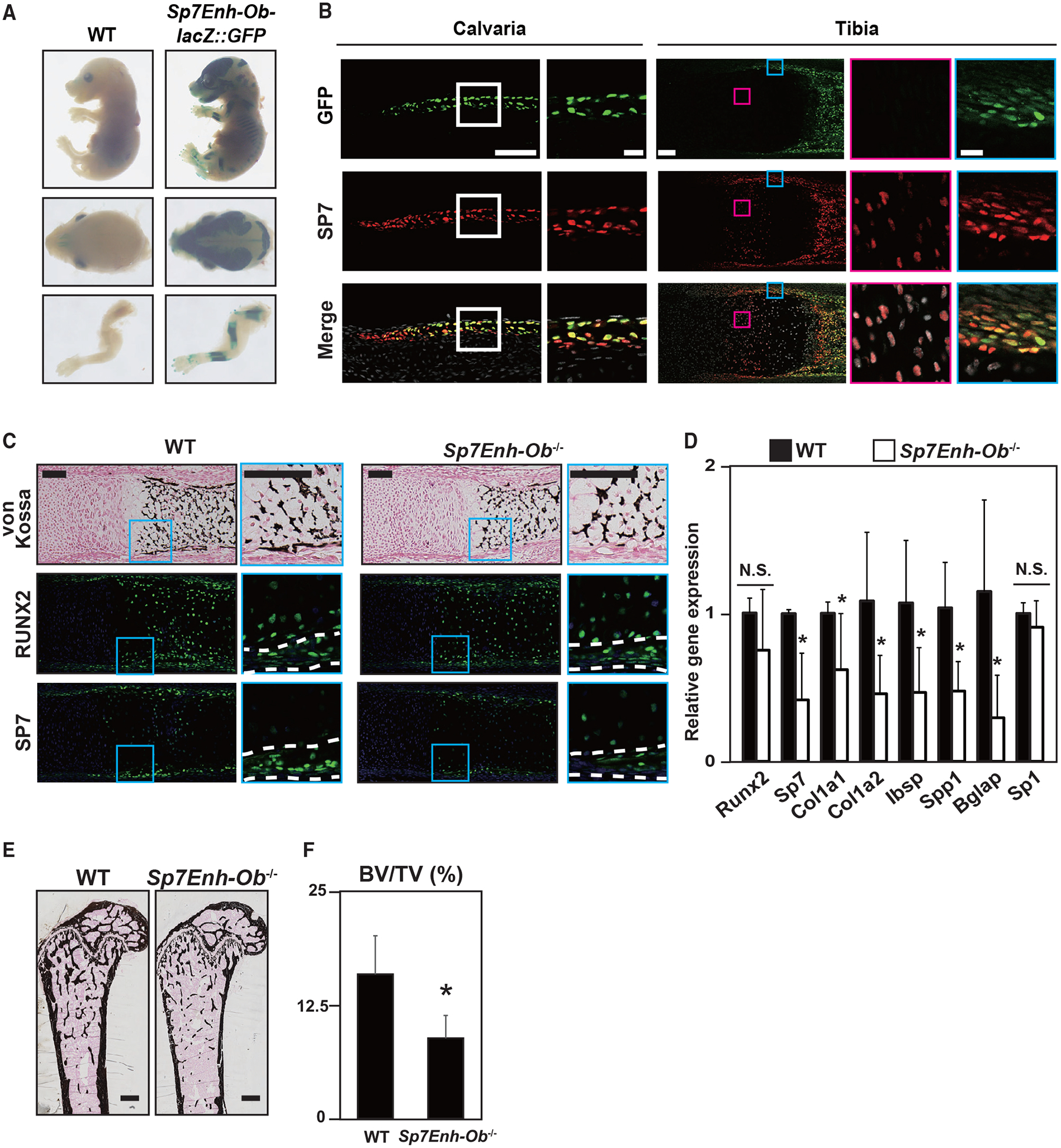
(A) β-Galactosidase staining to visualize Sp7Enh-Ob-driven lacZ:GFP reporter activity in E16.5 transgenic mice. Similar reporter activities were detected in 9 of 13 independent transgenic lines. Views of whole bodies, heads, and forelimbs are shown.
(B) Immunohistochemistry for SP7 and GFP signal of Sp7Enh-Ob-driven lacZ:GFP reporter activity in E17.5 calvariae and tibiae of transgenic reporter mice. Boxed regions are enlarged. Blue box, perichondrium containing Obs; pink box, prehypertrophic chondrocytes. Scale bars, 100 μm in the whole view and 20 μm in the enlarged view.
(C) von Kossa staining and immunohistochemistry for RUNX2 and SP7 in WT and Sp7Enh-Ob−/−metatarsals at E17.5. Regions between white broken lines indicate the perichondrium. Scale bar, 100 μm.
(D) qRT-PCR analysis of Sp1, Sp7, and osteogenic markers in primary Obs derived from the calvariae of WT and Sp7Enh-Ob−/−at E17.5. Cells were cultured with an osteogenic medium for 10 days. Data are presented as the mean ± SD of biological duplicate experiments with technical triplicates. *p < 0.05; N.S., not significant.
(E) von Kossa staining of 10-week-old femora in WT and Sp7Enh-Ob−/−. Scale bar, 500 μm.
(F) Microcomputed tomography (μCT) analysis for BV/TV (bone volume/tissue volume) in 10-week-old femora of Sp7Enh-Ob−/−male mice. Data are presented as the means ± SD. n = 6, *p < 0.05. Representative images obtained from at least biological duplicates are shown (B–D).
To examine the contribution of Sp7Enh-Ob to skeletal development, we generated a KO of Sp7Enh-Ob (Sp7Enh-Ob−/−) (Figure S7). Histological analysis revealed impaired bone collar formation in Sp7Enh-Ob−/−mice at E17.5, whereas cartilage mineralization was normal (Figure 7C). Compared with wild-type skeletal elements, decreased SP7 expression in the perichondrium and periosteum regions of Sp7Enh-Ob−/−was observed at E17.5 We observed significantly decreased expression of Sp7 and osteoblast marker genes in Sp7Enh-Ob−/−primary osteoblasts relative to wild-type osteoblasts (Figure 7D), whereas the closely linked Sp1 gene was not significantly different between the two genotypes (Figure 7D). Similar results were observed in Sp7Enh-Ob+/− MC3T3-E1 cells and their wild type (Figures S6C and S6D). In adult mice, bone mineral density was decreased in Sp7Enh-Ob−/−compared with the wild type (Figures 7E and 7F). The distal Sp7 enhancer is essential for normal osteoblast-specific activation of Sp7 (located downstream of Runx2) and for the normal programming of Sp7-driven osteoblast differentiation.
DISCUSSION
Characterization of the gene-regulatory landscape is key to understanding how the genome is linked to developmental processes. Chromatin accessibility is determined by the cooperative actions of transcription factors on gene-regulatory elements and is an essential step in establishing cellular identity (Long et al., 2016). This study shows that chondrocytes and osteoblasts are characterized by different repertoires of distal chromatin-accessible regions. Distinct combinations of transcription factors, which are likely associated with Runx2, contribute to enhancer activity. Motif analysis integrated with gene expression profiles suggests complex and redundant associations among multiple transcription factors in the regulatory regions. For example, the integrity of biological functions of key transcription regulators, including Foxa2, Foxa3, and Foxc1 for chondrogenesis (Ionescu et al., 2012; Tan et al., 2018; Xu et al., 2021; Yoshida et al., 2015) and Sp7 and Dlx members for osteogenesis (Hojo et al., 2016), has been confirmed. The common enrichment of AP-1 and ATF transcription factors in osteoblasts and chondrocytes is consistent with the requirement of AP-1 and ATF4 for both cell types (Bozec et al., 2010; He et al., 2016; Karreth et al., 2004; Wang et al., 2009; Yang et al., 2004). Considering that AP-1 is also highly associated with active enhancers in many cell types and ubiquitously expressed in throughout the body, AP-1 is likely to act as a general enhancer component (Andersson et al., 2014; Madrigal and Alasoo, 2018).
Integrative analysis with ChIP-seq analysis for Runx2 and selected transcriptional regulators indicated that a number of transcriptional regulators associated with genomic regions are correlated with the degree of chromatin accessibility. These findings suggest that Runx2 and the transcriptional regulators form so-called “hotspots” to maximize chromatin accessibility and gene expression (Long et al., 2016; Rickels and Shilatifard, 2018). Combinatorial gain- or loss-of-function analyses are needed to assess the requirements of combinatorial transcription factors in chromatin accessibility in the future.
Our findings support the hypothesis that Runx2 acts as a pioneer factor. Pioneer factors are defined as a specific class of proteins that (1) bind to target DNA sequences in heterochromatin, (2) implement epigenetic stability of the open chromatin state, (3) permit binding with other transcription factors, and (4) initiate chromatin remodeling (Mayran and Drouin, 2018). Runx2 meets the first and second criteria in the study. The interactions of Runx2 with chromatin remodelers, including histone deacetylases and Mediator MED23 (Liu et al., 2016; Stein et al., 2004), suggest that Runx2 meets the fourth criterion. However, extensive genome-scale analysis of Runx2 interaction with other transcriptional regulators is necessary to determine whether Runx2 meets the full definition of a pioneer factor. Considering that other Runx transcription factors have also been reported to have pioneer activity in different cell types (Hass et al., 2021; Lee et al., 2019; Lichtinger et al., 2012; Wang et al., 2018), further comparative analysis of the members will provide a better understanding of the universal or Runx member-specific pioneer actions across cell types.
In addition to its pioneering action, we demonstrated that Runx2 is required for establishment of osteoblast chromatin accessibility. Chromatin accessibility was dynamically altered upon Runx2 removal, and we speculate that the degree of chromatin alteration is dependent on co-occupancy of other transcription factors in Runx2-mediated enhancer modules. The interplay between Runx2 and other transcription factors may account for the plasticity of the cellular identity between osteoblasts and chondrocytes. Removal of osteogenic inputs, including hedgehog activity (Long et al., 2004), Wnt/β-catenin activity (Day et al., 2005; Hill et al., 2005; Rodda and McMahon, 2006), and Sp7 (Nakashima et al., 2002), in skeletal progenitors results in ectopic chondrocyte formation in the periosteum.
Our data add insight and definition to Runx2-Sp7 interactions revealed through epistasis analysis of genetic mutants. Runx2 mediates transactivation of an Sp7 cis-regulatory element located approximately 11 kb distal to the Sp7 TSS. the enhancer was activated exclusively in osteoblasts, whereas Sp7 is expressed in osteoblasts and pre-hypertrophic chondrocytes. However, considering that the osteoblast phenotype was weaker for the Sp7 enhancer KO mouse compared with Sp7 gene KO (Nakashima et al., 2002), additional regulatory inputs are predicted to govern Sp7 osteoblast expression. Multiple enhancers may have redundant roles, as reported for different skeletal genes (Osterwalder et al., 2018; Will et al., 2017).
Our findings provide insights into the regulatory landscape encompassing the cell-type-distinct enhancer network in mammalian skeletal programming. Runx2 plays a critical role in initiating osteoblast development and chondrocyte maturation, likely by facilitating chromatin accessibility and directing cell-type-specific gene-regulatory programs. The study lays a foundation for a more detailed exploration of gene-regulatory circuits in mammalian skeletal development.
Limitations of the study
In this study, we performed ChIP-seq for multiple transcription factors using chondrocytes and osteoblasts. Although these cells were enriched, contamination of other cell types was not completely ruled out. Future studies should employ FACS followed by ChIP-seq or single-cell ChIP-seq to examine cell-type-specific datasets, which will help us to better understand the stage-specific actions of transcription factors in skeletal development.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Hironori Hojo (hojo@g.ecc.u-tokyo.ac.jp).
Materials availability
Newly generated materials from this study are available upon request from the lead contact, Hironori Hojo (hojo@g.ecc.u-tokyo.ac.jp).
Data and code availability
ATAC-seq, ChIP-seq, and RNA-seq data have been deposited at Gene Expression Omnibus (GEO: GSE178293). scRNA-seq data have been deposited at the DDBJ BioProject database (DDBJ BioProject database: PRJDB8387).
This paper does not report original code.
Any additional information required to reanalyze the data presented in this work is available from the lead contact upon request.
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Mice
To construct Runx2-BioFL-targeting vectors, homologous arms were subcloned by PCR using C57BL/6J mouse genomic DNA and Platinum Pfx DNA Polymerase (11708–013; Invitrogen, Carlsbad, CA). The 5′ homologous arm (chr17:44744857–44747419) carrying Aat II and Nde I sites at its C-terminus was cloned into the Sac II and Not I sites, which lies upstream of the PGKneo cassette flanked by two loxP sites, of the pPGKneobpAlox2PGKDTA vector (generously provided by P. Soriano, Mount Sinai School of Medicine, NY, USA). Biotin-3xFLAG (5′-GGGTCCGGCCTGAACGACATCTTCGAGGCTCAGAAAATCGAATGGCACGAAGGCGCGCCGAGCTCGAGGGACTACAAAGACCATGACGGTGATTATAAAGATCATGACATCGACTACAAGGATGACGATGACAAG-3′) was cloned into the Aat II and Nde I sites of the 5′ homologous arm with a stop codon. The 3′ homologous arm (chr17:44740547–44744853) was cloned into the Sal I site, which lies downstream of the PGKneo cassette, of the pPGKneobpAlox2PGKDTA vector. The targeting vector was linearized with Sac II and introduced by electroporation into 129/Sv 3 C57BL/6J F1 hybrid ES cells (v6.5). After the expansion of the G418-resistant clones, homologous recombination was screened using long-range PCR. The primer sequences used in the screening were as follows: P1, 5′-tgacttctttccgttttaaattaccc-3′ (in 5′ external region); P2, 5′-atctttataatcaccgtcatggtctt-3′ (Biotin-3xFLAG sequence); P3, 5′-tggttaggcaaatttggatttaatag-3′ (5′ homologous arm); P4, 5′-gaggattgggaagacaatagcag −3′ (Neo sequence); and P5, 5′-atattcatcttgaaagcgctacaaac-3′ (3′ external region). Correctly targeted ES cells were injected into B6D2F1 3 C57BL/6 blastocysts to generate chimeric mice. Runx2BioFL–Neo/+ mice were then crossed with Sox2-Cre transgenic mice (Hayashi et al., 2002) to remove the PGKneo cassette. The following primers were used for PCR genotyping of Runx2BioFL/+ (Figure 2C): P6, 5′-ttgcaagatcatgactagggatt-3′; and P7, 5′-ccccaactgttttgaattctagc-3′. Runx2BioFL/BioFL mice were maintained on a C57BL/6J background. Postnatal day 1 (P1) males and females of the mice were used in this study.
To generate Sp7 enhancer transgenic reporter mice (Sp7Enh-Ob-lacZ∷GFP), the Sp7 enhancer region (chr15:102207000–102208800, mm9) was cloned into the Hsp68-lacZ∷nGFP reporter construct (Peterson et al., 2012). The entire reporter cassette was purified by electroelution. Transgenic founders were generated by pronuclear injection of the transgene DNA into 100 fertilized zygotes and subsequent transfer of the zygotes into the oviducts of pseudopregnant wild-type ICR mice. For the G0 transgenic analysis, 200 injected zygotes were harvested. After the F0 animals were genotyped and the transgenic animals were bred to confirm germline transmission, the Sp7Enh-Ob-lacZ∷GFP transgenic reporter mice were maintained on a C57BL/6J background. E16.5 and E17.5 males and females of the mice were used in this study.
To generate the Sp7 enhancer knockout (Sp7Enh-Ob+/) mouse line, Cas9-mediated removal of the Sp7 enhancer (chr15:102207224–102208686) was performed. The following single guide RNA (sgRNA)-target sequences flanking the enhancer were identified using the CRISPR Design Tool (crispr.mit.edu): 5′ sgRNA-1: ggcatctccaccgcttacgctgg; 5′ sgRNA-2: ccaccgcttacgctggccacct; 3′ sgRNA-1: ccaagaagttagacggggatggg; and 3′ sgRNA-2: gggtggtatcccgttatttttgg. Cas9 protein (Guide-it recombinant Cas9 protein, TaKaRa) and sgRNA (Fasmac) were delivered by electroporation to C57BL/6J embryos at the pronuclear stage as described previously with slight modifications (Hashimoto and Takemoto, 2015). Briefly, the embryos were washed three times with Opti-MEM I (Thermo Fisher Scientific) supplemented with 0.1% polyvinylalchol (PVA) and once with 0.1% PVA-Opti MEM I containing Cas9 protein (100 ng/μL) and four sgRNAs (25 ng/μL each). The embryos were then placed in a line in the gap of an electrode (LF501PT1–10, BEX, Tokyo, Japan) filled with RNA containing 0.1% PVA-Opti-MEM I (total volume: 5 μL), and electroporation was performed using a CUY21EDIT II electroporator (BEX). After electroporation, the embryos were cultured in mWM and transferred into the oviducts of 0.5-day-post-coitum ICR recipients. After the F0 animals were genotyped and the knockout animals were bred to confirm germline transmission, Sp7Enh-Ob+/− mice were maintained on a C57BL/6J background. To minimize the off-target effects of sgRNAs, Sp7Enh-Ob+/− mice were used for experiments after five generations. The following primers were used for the PCR genotyping of Sp7Enh-Ob+/− (Figure S7): P1, 5′-ggcatgcaggttttaaagttcagtg-3′; P2, 5′-gcttgtcatcgtcatccttgtagtc-3′; and P3, 5′-ttcatgaatgtctgtgtgagggtgt-3′. Sp7Enh-Ob+/− mice were maintained on a C57BL/6J background. E17.5 males and females and 10-week-old males of the mice were used in this study.
The R26RtdTomato reporter mouse line (Madisen et al., 2010) was obtained from the Jackson Laboratory. The R26RtdTomato reporter and Sp7-EGFP:Cre BAC transgenic reporter (Rodda and McMahon, 2006) mouse lines were maintained on a C57BL/6J background. Runx2-flox mice were generated by Dr. Taku Saito at the University of Tokyo (Nagata K et al., unpublished data). E18.5 males and females of the compound mice, Sp7-EGFP∷Cre; R26RtdTomato;Runx2-flox, were used in this study.
Transgenic reporter mouse lines of Col2a1-ECFP (Chokalingam et al., 2009) and Col10a1-mCherry (Maye et al., 2011) were maintained in ICR mice. P1 males and females of the mice were used in this study.
All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Southern California, Los Angeles, CA (IACUC #11830 and #11892) and the Animal Care and Use Committee of the University of Tokyo, Tokyo, Japan (approval number P17–037). All mouse experiments in this study were carried out in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, USA, and the Animal Care and Use Committee of the University of Tokyo, Japan.
Cloning and culture of cells in vitro
NIH3T3 (CRL-1658, ATCC), MC3T3-E1 (CRL-2593, ATCC), HEK293T (CRL-3216, ATCC), Plat-E (RV-101, Cell Biolabs), and COS-7 (CRL-1651, ATCC) were used in this study. NIH3T3, HEK293T, Plat-E, and COS-7 cells were cultured in high-glucose Dulbecco’s modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. MC3T3-E1 cells were cultured in alpha-MEM (A10490–01, Life Technologies) containing 10% FBS and 1% penicillin/streptomycin (10% FBS/alpha-MEM). For osteoblast differentiation, cells were cultured in 10% FBS/DMEM supplemented with 50 μg/mL ascorbic acid phosphate, 10 mM β-glycerophosphate, and 100 ng/mL recombinant human bone morphogenetic protein (BMP)2 (rhBMP2, 355-BM, R&D). All cells were grown at 37°C in a humidified incubator with 5% CO2.
Primary osteoblasts were isolated from osteoblast-enriched partial calvarias consisting of the frontal bone and a rostral part of the parietal bone (Hojo et al., 2016). The partial calvaria was digested in five changes of 0.1% collagenase D (Roche Applied Science) and 0.2% Dispase (383–02281, Fujifilm) for 10 min each at 37°C. Cells were cultured in the osteogenic medium containing 10% FBS/DMEM supplemented with 50 μg/mL ascorbic acid phosphate, 10 mM β-glycerophosphate, and 100 ng/mL rhBMP-2.
For the Runx2 gain-of-function analysis, NIH3T3cells were transduced with retroviruses produced by Plat-E retrovirus packaging cells transfected with either pMx-GFP or pMX-Runx2BioFL. Blasticidin selection was performed after transduction at 10 mg/mL for 2 days. Cells were then cultured in 10% FBS/DMEM supplemented with 50 μg/mL ascorbic acid phosphate, 10 mM β-glycerophosphate, and 100 ng/mL rhBMP-2. Alkaline phosphatase (ALP) staining was performed as previously described (Hojo et al., 2012).
To establish a stable Cas9-expressing osteoblast cell line, MC3T3-E1 cells were transduced with lentiviruses produced by the transfection of pKLV2-EF1aBsd2ACas9-W (Tzelepis et al., 2016), psPAX2 (Addgene Plasmid #12260, Cambridge, MA), and pMD2.G (Addgene Plasmid #12259) using Fugene HD transfection reagent (E2311, Promega) in 293T cells. Blasticidin selection was performed 2 days after transduction at 10 μg/mL for 3 days. By limited dilution, single clones were plated in each well of 96-well plates and expanded. A positive clone was selected based on Cas9 expression and normal osteoblast differentiation in an osteogenic medium. Clone #76 was used in this study (Figures S6A and S6B).
To establish Sp7Enh-Ob knockout cells, we used a dual gRNA expression lentiviral vector called pKLV2.2-mU6gRNA5(SapI)-hU6gRNA5(BbsI)-PGKpuroBFP-W (Addgene Plasmid #72667) (Tzelepis et al., 2016). The following gRNA sequences were inserted into the vector: gRNA1, GGCATCTCCACCGCTTACGC and gRNA2, GCAAGAAGTTAGACGGGGAT (chr15:102207224–102208686). The lentivirus carrying these gRNAs was transduced into clone #76 MC3T3-E1 cells stably expressing Cas9. Puromycin selection was performed 2 days after transduction at 4 μg/mL for 3 days. By limited dilution, single clones were plated in each well of 96-well plates and expanded. Three Sp7Enh-Ob+/− clones were selected based on Sanger sequencing with genomic DNA and used for differentiation assays (Figures S6C and S6D).
METHOD DETAILS
Cell extraction for FACS-sorting and ChIP-seq
To isolate rib chondrocytes, we manually dissected rib cartilage to include columnar cartilage and part of mineralized cartilage, but to exclude the vascularized bone marrow. For FACS-sorting, the ribs were initially digested in 2 mg/mL Pronase (53702, EMD Millipore Corporation) for 30 min at 37°C, washed three times with phosphate buffered saline (PBS), and then digested in 0.2% collagenase D (1088858, Roche Applied Science) for 3 h at 37°C. For ChIP-seq experiments, the ribs were digested in 2 mg/mL Pronase for 1 h at 37°C, washed three times with PBS, and then digested in 0.67 units/mL Liberase TM (5401119001, Sigma-Aldrich) for 2 h at 37°C. To isolate calvarial osteoblasts for both FACS-sorting and ChIP-seq experiments, we manually dissected osteoblast-enriched partial calvarias consisting of the frontal bone and a rostral part of the parietal bone (Hojo et al., 2016). The partial calvarias were digested in five changes of 0.1% collagenase D and 0.2% dispase for 10 min each at 37°C. For FACS, cells were resuspended in PBS containing 2% FBS and then filtered through a 40-mm nylon cell strainer (352340, BD Falcon). FACSAria II and FACSMelody (BD Biosciences, Franklin Lakes, NJ) were used for FACS analysis.
Skeletal staining, histology, and radiological analysis
Whole mount skeletons were stained with 0.05% alizarin red (A5533, Sigma-Aldrich) and 0.15% alcian blue 8GX (A5268, Sigma-Aldrich), and then transferred with glycerol/KOH solutions from 20% glycerol/1% KOH to 80% glycerol/1% KOH. For whole-mount lacZ staining, reporter mice were fixed in 1% formaldehyde and 0.2% glutaraldehyde in PBS for 90 min, stained in X-gal for 2 h at 37°C, and cleared in 80% glycerol/PBS. For histological analysis with alcian blue and von Kossa staining, sections were stained with 1% aqueous silver nitrate (196-00831, Fujifilm) solution and 1% alcian blue (pH 2.5, 8GX, Sigma-Aldrich) solutions. Unreacted silver was removed using 5% sodium thiosulfate. Counterstaining was performed using nuclear fast red staining. Micro-computed tomography (μCT) scanning of the harvested femurs was performed using a microfocus X-ray CT system SMX-90CT (Shimadzu, Kyoto, Japan); the three-dimensional construction software package TRI/3D-BON (Ratoc System Engineering, Tokyo) was used for quantitative analysis.
Immunohistochemistry
Tissues were fixed in 4% paraformaldehyde (PFA)/PBS for 1 h at 4°C and soaked in 30% sucrose/PBS overnight at 4°C. After embedding in OCT compound, cryosections were cut into 12 μm-thick sections. The sections were blocked with 3% bovine serum albumin (A7960, Sigma-Aldrich) and 1% heat-inactivated sheep serum (S2263, Sigma-Aldrich) in PBS with Tween 20. The sections were then incubated with primary antibodies overnight at 4°C, followed by incubation with secondary antibodies for 1 h at room temperature, and mounted with Immu-Mount (99–904-02, Fisher Scientific). The following antibodies were used in the study: anti-Runx2 (1:1,000 with sc-10758X or 1:50 with sc-10758, Santa Cruz Biotechnology), anti-FLAG M2 (1:500, F1804, Sigma-Aldrich), anti-Sp7 (1:5,000, ab22552, Abcam), Alexa Fluor 488 (1:500, secondary anti-rabbit, Life Technologies), Alexa Fluor 546 (1:500, secondary anti-rabbit; anti-mouse, Life Technologies), and Alexa Fluor 647 (1:500, secondary anti-rabbit, Life Technologies). For immunostaining with anti-mouse antibodies, a Mouse on Mouse (M.O.M.) Basic Kit (BMK-2202, Vector Laboratories) was used.
RT-qPCR analysis
Total RNA was extracted and purified using an RNeasy mini kit (74106, Qiagen) and reverse-transcribed into single-strand complementary DNA using a ReverTra Ace qPCR RT Master Mix kit (FSQ-301, Toyobo) according to the manufacturer’s instructions. qPCR was performed using a FastStart Universal SYBR Green Master kit (4913850, Roche) on a 7500 Fast Real-Time PCR System (Applied Biosystems). The expression of target genes was normalized to that of the reference gene (Actb). The primers used in this study are listed in Table S7.
Western blotting
Whole-cell lysates or nuclear proteins were isolated using RIPA Lysis and Extraction Buffer (89900, Thermo Fisher Scientific) or NEPER nuclear and cytoplasmic extraction reagents (PI78833, Thermo Fisher Scientific), according to the manufacturer’s protocol. Standard protocols were used for western blotting and immunostaining. Immunoblotting images were captured using a LAS-4000 luminescent image analyzer (GE Fujifilm). All antibodies used in this study are listed in the key resources table.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Mouse monoclonal anti-FLAG M2 antibody | Sigma-Aldrich | F1804; RRID:AB_262044 |
| Mouse monoclonal anti-RUNX2 antibody (used for Western blotting and EMSA) | Abcam | ab76956; RRID:AB_1565955 |
| Rabbit polyclonal anti- RUNX2 antibody (used for immunohistochemistry) | Santa Cruz Biotechnology | sc-10758; RRID:AB_2184247 |
| Rabbit polyclonal anti-SP7 antibody | Abcam | ab22552; RRID:AB_2194492 |
| Mouse monoclonal anti-FOXA2 antibody | Merck | 17–10258; RRID:AB_11204693 |
| Rabbit polyclonal anti-C-JUN antibody | Abcam | ab31419; RRID:AB_731605 |
| Rabbit polyclonal anti-CBFb antibody | Abcam | ab195411 |
| Mouse monoclonal anti-CRISPR-Cas9 antibody | Abcam | ab191468; RRID:AB_2692325 |
| Rabbit polyclonal to Histone H3 antibody | Abcam | ab1791; RRID:AB_302613 |
| Alexa Fluor 488 secondary antibody | Invitrogen | A-11034; RRID:AB_2576217 |
| Alexa Fluor 546 secondary antibody | Invitrogen | A-11030; RRID:AB_2534089, A-11035; RRID:AB_2534093 |
| Alexa Fluor 647 secondary antibody | Invitrogen | A-21245; RRID:AB_2535813 |
| Goat Anti-Mouse IgG (H + L), HRP Conjugate | Promega | W402B; RRID:AB_430834 |
| Goat Anti-Rabbit IgG (H + L), HRP Conjugate | Promega | W401B; RRID:AB_430833 |
| Chemicals, peptides, and recombinant proteins | ||
| Recombinant human BMP2 | R&D | 355-BM |
| Critical commercial assays | ||
| Tagment DNA Enzyme 1 | Illumina | 15027865 |
| ThruPLEX®-FD Prep Kit | Rubicon Genomics | R40012 |
| TruSeq Illumina kit | illumina | RS-122-2001 |
| SMARTer Stranded Total RNA-seq Kit v2 Pico Input Mammalian | Takara | 634411 |
| Chromium Single Cell 3′ Library & Gel Bead Kit | 10x Genomics | PN-120237 |
| Chromium Single Cell A Chip Kits | 10x Genomics | PN-120236 |
| Chromium Multiplex Kit | 10x Genomics | PN-120262 |
| Deposited data | ||
| Raw and analyzed ChIP-seq, ATAC-seq and bulk RNA-seq data | This paper | GSE178293 |
| Raw and analyzed scRNA-seq data | This paper | DDBJ BioProject database: PRJDB8387 |
| Mouse osteoblast Sp7 ChIP-seq and Sp7-positive osteoblast RNA-seq data | GEO | GSE76187 |
| Mouse chondrocyte Jun ChIP-seq data | GEO | GSE73372 |
| Experimental models: Cell lines | ||
| MC3T3-E1 | ATCC | CRL-2593 |
| NIH3T3 | ATCC | CRL-1658 |
| HEK293T | ATCC | CRL-3216 |
| COS7 | ATCC | CRL-1651 |
| Plat-E | Cell Biolabs | RV-101 |
| Experimental models: Organisms/strains | ||
| Col2-ECFP transgenic reporter mice | (Chokalingam et al., 2009) | N/A |
| Col10-mCherry transgenic reporter mice | (Maye et al., 2011) | N/A |
| Sp7/Osx-EGFP∷Cre BAC transgenic reporter mice | (Rodda and McMahon, 2006), The Jackson laboratory | Stock No: 006361 |
| R26RtdTomato reporter mice | The Jackson laboratory | Stock No: 007905 |
| Runx2BioFL/+ mice | This paper, The Jackson laboratory | Stock No: 029512 |
| Runx2flox/+ mice | Nagata K et al., unpublished | N/A |
| Sp7Enh-Ob-lacZ∷GFP transgenic reporter mice | This paper | N/A |
| Sp7Enh-Ob+/− mice | This paper | N/A |
| Recombinant DNA | ||
| pMx-GFP | (Hojo et al., 2016) | N/A |
| pMx-Runx2-BioFL | This paper | N/A |
| pKLV2-EF1aBsd2ACas9-W | (Tzelepis et al., 2016) | Addgene 67978 |
| pKLV2.2-mU6gRNA5(SapI)-hU6gRNA5(BbsI)-PGKpuroBFP-W | (Tzelepis et al., 2016) | Addgene 72667 |
| CROPseq-Guide-Puro | (Datlinger et al., 2017) | Addgene 86708 |
| psPAX2 | Unpublished | Addgene 12260 |
| pMD2.G | Unpublished | Addgene 12259 |
| pGL4.23-Sp7Enh-Ob | This paper | N/A |
| pGL4.23- Sp7Enh-Ob-mut | This paper | N/A |
| caALK6 | (Ohba et al., 2008) | N/A |
| Smad1 | (Ohba et al., 2008) | N/A |
| Software and algorithms | ||
| CisGenome software v.2.0 | (Ji et al., 2008) | http://www.biostat.jhsph.edu/~hji/cisgenome/ |
| Trimmomatic v.0.33 | (Bolger et al., 2014) | http://www.usadellab.org/cms/?page=trimmomatic |
| Bowtie v.1.1.2 | (Langmead et al., 2009) | http://bowtie-bio.sourceforge.net/index.shtml |
| Tophat-2 v.2.1.0 | (Trapnell et al., 2009) | https://ccb.jhu.edu/software/tophat/index.shtml |
| Cufflinks v.2.2.1 | (Trapnell et al., 2010) | http://cole-trapnell-lab.github.io/cufflinks/ |
| HOMER v.3 | (Heinz et al., 2010) | http://homer.ucsd.edu/homer/ |
| factoextra v.1.0.7 | (Kassambara and Mundt, 2020) | https://cran.r-project.org/web/packages/factoextra/index.html |
| ggplot2 v.3.3.2 | (Hadley, 2016) | https://ggplot2.tidyverse.org/ |
| Java TreeView | (Saldanha, 2004) | http://jtreeview.sourceforge.net/ |
| GREAT v.3.0.0 | (McLean et al., 2010) | http://great.stanford.edu/public/html/ |
| BEDTools v.2.16.2 | (Quinlan and Hall, 2010) | https://bedtools.readthedocs.io/en/latest/ |
| DiffBind v.2.10.0 | (Ross-Innes et al., 2012) | https://bioconductor.org/packages/release/bioc/html/DiffBind.html |
| Cellranger v.2.0.0 | (Zheng et al., 2017) | https://bioconductor.org/packages/release/bioc/html/DEsingle.html |
| DEsingle | (Miao et al., 2018) | https://bioconductor.org/packages/release/bioc/html/DEsingle.html |
| g:Profiler | (Raudvere et al., 2019) | https://biit.cs.ut.ee/gprofiler/gost |
Luciferase reporter assay
Sp7 enhancer region (chr15:102207000–102208800, mm9, called as Sp7Enh-Ob in this study) was cloned into the pGL4.23 luciferase vector. For site-directed mutagenesis in a Runx motif in the reporter (pGL4.23-Sp7Enh-Ob-mut), PCR with the following primers were used: forward: TTACGATTGCTGGGAGTCGCGTCC; reverse: TCGGGACCGCAGATATGCAGCT. Cells were plated onto 24-well plates, and 0.4 μg of a DNA mixture containing a firefly luciferase reporter driven by tested regulatory elements, a thymidine kinase promoter-renilla luciferase reporter, and effector plasmids were introduced by Fugene HD transfection reagent (E2311, Promega). Forty-eight hours after transfection, luciferase activity was measured using the Dual-Luciferase Reporter Assay System (E1910, Promega) with a GloMax 96 Microplate Luminometer (Promega).
EMSA
Nuclear extracts were prepared from COS7 cells transfected with either GFP or Runx2-BioFL expression vectors were prepared as previously described (Dignam et al., 1983). Nuclear protein extract (1 μg) was used for the binding reaction. The binding mixture contained 50 mM KCl, 25 mM HEPES-KOH (pH 7.9), 5 mM MgCl2, 1 mM DTT, 12% glycerol, 0.1% NP-40, 0.5 μg of Poly d(I-C), and 100 fmol of the Biotin-labeled probe. The binding reaction was performed on ice for 30 min. For competition analysis, unlabeled probes were added at a 100-fold molar excess. For the supershift assay, 1 μg of anti-FLAG M2 antibody (Sigma-Aldrich, F1804), anti-Runx2 antibody (ab76956, Abcam), or non-immune IgG (ab46540, Abcam) were incubated with the nuclear protein extract in the binding mixture on ice for 30 min prior to the addition of probes. Images of the EMSA were captured using an LAS-4000 luminescent image analyzer. The sequences of the probes used in this study are listed in Table S7.
ATAC reaction
ATAC-seq was performed as previously described (Buenrostro et al., 2013). Briefly, 10,000–50,000 cells were collected and lysed using lysis buffer containing 10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% IGEPAL CA-630. Tn5 transposase reaction using the Tagment DNA Enzyme 1 (TDE1) (Illumina, catalog # 15027865) was carried out at 37°C for 30 min. The reacted DNA was purified using a MinElute PCR purification kit (28004, QIAGEN) and amplified for 8–15 cycles to produce libraries for sequencing. The ATAC-seq libraries were sequenced on an Illumina HiSeq X sequencer. Two or three biological replicates were analyzed.
Chromatin immunoprecipitation and DNA sequencing
Protein-DNA complexes were crosslinked with 1% formaldehyde, and the crosslinking was quenched by the addition of 1 M glycine. After washing with PBS three times, cells were lysed in lysis buffer (50 mM HEPES-KOH pH7.5, 140 mM NaCl, 1mM EDTA, 10% glycerol, 0.5% NP40, 0.25% Triton-X 100) at 4°C with agitation for 10 min, then washed with Buffer 2 (200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl pH8.0) for 10 min at room temperature. Chromatin was fragmented by sonication (Branson Digital Sonifier with Microtip) in Buffer 3 (1 mM EDTA, 0.5 mM EGTA, 10 mM Tris-HCl pH8.0) to obtain 100–600 bp of DNA fragments. Lysates were cleared by centrifugation and supplemented with NaCl, NaDeoxycholate, TritonX 100, and Sarkosyl (L9150, Sigma-Aldrich) to final concentrations of 130 mM, 0.13%, 1.3%, and 0.5%, respectively. Dynabeads (Life Technologies) were blocked with a blocking buffer (5 mg/ml BSA in PBS) and incubated with antibodies in the blocking buffer overnight. The unbound antibody was removed by extensive washes with blocking buffer. The bead/antibody complex was incubated with the lysate overnight at 4°C. Beads were washed five times with RIPA buffer (1% NP40, 0.7% NaDeoxycholate, 1 mM EDTA, 50 mM HEPES-KOH pH7.5, and 0.5 M LiCl). DNA was eluted by heating the beads in an elution buffer (50 mM Tris-HCl pH8.0, 10 mM EDTA, and 1% SDS) at 65°C for 15 min, and the eluate was incubated overnight at 65°C for reverse crosslinking. DNA was purified using a MinElute PCR Purification Kit. All buffers were supplemented with a protease inhibitor cocktail mix (04,693,159,693159001, Roche). ChIP-seq libraries were constructed using a ThruPLEX®-FD Prep Kit (R40012, Rubicon Genomics) according to the manufacturer’s instructions. Library quality was validated using a Bioanalyzer (Agilent) before sequencing on Hiseq X and NextSeq500 (Illumina) platforms. Information about the antibodies is listed in the Key Resources Table. We used 5 μg of antibodies for one ChIP experiment.
Bulk RNA-seq
Total RNA was extracted and purified using an RNeasy mini kit (74,106, Qiagen) or RNeasy micro kit (74,004, Qiagen). For RNA-seq with rib chondrocytes sorted from Col2a1-ECFP and Col10a1-mCherry mice at P1, 100 ng of total RNA was subjected to analysis. RNA quality was confirmed by Experion (700–7105, Bio-lad) analysis, and the RNA libraries were constructed using a Tru-Seq Illumina kit (RS-122-2001, Illumina) with some modifications. For RNA-seq with tdTomato-sorted calvarial cells isolated from Sp7-EGFP:Cre;Runx2fl/+;R26RtdTomato and Sp7-EGFP:Cre;Runx2fl/fl;R26RtdTomato at E18.5, 1.7 ng of total RNA was subjected to the analysis. RNA quality was confirmed using a Bioanalyzer with an RNA6000 pico kit (5067–1513, Agilent), and the RNA libraries were constructed using a SMARTer Stranded Total RNA-Seq Kit v2 Pico Input Mammalian (634,411, Takara). Library quality was validated using a Bioanalyzer (Agilent) before sequencing on Hiseq X and NextSeq500 (Illumina) platforms. Two or three biological replicates were analyzed.
CROP-seq analysis
Individual gRNA cassettes were annealed from two oligonucleotides and cloned into lentiviral CROPseq-Guide-Puro vectors (Addgene Plasmid #86708) as previously described (Datlinger et al., 2017). The cloning of gRNAs in CROPseq-Guide-Puro vectors was performed by restriction enzymes, confirmed by Sanger sequencing, and pooled as the gRNA library. The lentiviral library was prepared in 293T cells by transfection of the pooled library plasmid psPAX2 (Addgene Plasmid #12260) and pMD2.G (Addgene Plasmid #12259) using Fugene HD transfection reagent (E2311, Promega). The lentiviral library was transduced into #76 MC3T3-E1 cells stably expressing Cas9 (Figure S6A and S6B). We used the lentivirus at multiplicity of infection 2 in 1.0 × 106 cells. Two days after transduction, cells were cultured in an osteogenic medium containing 50 ng/mL rhBMP2, 10 μg/mL blasticidin, and 5 μg/mL puromycin for 7 days. The single-cell suspension was prepared by enzyme digestion with trypsin-EDTA solution 1X (Wako, 206–17291). Single-cell RNA-seq (scRNA-seq) was performed with chromium platform chromium (10x Genomics) using Chromium Single Cell 3′ Library & Gel Bead Kit v2 (10x Genomics, PN-120237), Chromium Single Cell A Chip Kits (10x Genomics, PN-120236), and Chromium Multiplex Kit (10x Genomics, PN-120262). Library quality was validated using a Bioanalyzer (Agilent) before sequencing on Hiseq2500 (Illumina) platforms. Paired-end sequences of 26 bp and 98 bp were performed. The gRNA sequence, oligo DNA sequence, and targeted putative enhancer coordinates are listed in Table S7.
QUANTIFICATION AND STATISTICAL ANALYSIS
General statistical analysis
For statistical analysis, R was used. The number of replicates is indicated in the figure legends. Student’s t test and Tukey-HSD analysis were used as indicated in the figure legends. Statistical details are written in the figure legends.
ATAC-seq, ChIP-seq, and bulk RNA-seq
For ATAC-seq and ChIP-seq, the sequence reads were trimmed using Trimomatic-0.33 (Bolger et al., 2014) and aligned to the mouse genome reference sequence mm9 by Bowtie aligner (Langmead et al., 2009). Peak calling was performed by two-sample analysis using CisGenome software (Ji et al., 2008) with a p-value cutoff of 10−5 compared with the input control. Peaks were incorporated into further analysis, displaying an FDR of <0.01. For RNA-seq, the sequence reads were trimmed by Trimomatic-0.33 (Bolger et al., 2014) and aligned to the mouse genome reference sequence mm9 using Tophat-2.1.0 (Trapnell et al., 2009). Quantification of gene expression and differentially expressed gene analysis were performed using Cufflinks/Cuffdiff 2.2.1 (Trapnell et al., 2010, 2012). The data from Cufflinks/Cuffdiff 2.2.1 were output as fragments per kilobase of transcript per million mapped reads (FPKM) values, which is a normalized count of a transcript’s abundance.
In the analysis in Figure 1A, PCA was performed using the R package factoextra and visualized as plots using ggplot2 (Hadley, 2016). For analyzing ATAC-seq data, we used selected ATAC-seq peaks based on raw fe > 30. For analyzing RNA-seq data, we used selected genes based on fpkm >2. In Figure 1C, the correlation matrix was created by the R function “cor” and the correlation heatmap was generated using the ggplot2 package in R. The correlation matrix was reordered according to the correlation coefficient using hierarchical clustering. The values of the correlation coefficients are shown in the heatmap. All peak regions were unified to obtain ATAC-seq peak intensity. Then, ATAC-seq tag densities within a 150 bp window were calculated using the HOMER package (Heinz et al., 2010) and normalized using the quantile normalized method (Le Martelot et al., 2012). For analysis in Figure 1D, we used selected ATAC-seq peaks with the normalized peak intensity based on the following criteria: raw fe > 30, fold change >5, and p value < 0.05, for cell-type specific regions. Heatmaps were generated using the Homer program (Heinz et al., 2010) and Java TreeView software (Saldanha, 2004). Gene ontology analysis was performed using the online Genomic Regions Enrichment of Annotations Tool (GREAT), version 3.0.0 (McLean et al., 2010) with default settings. To examine the distribution of the consensus motifs in the peaks, we mapped each position weight matrix (PWM) back to the whole mouse genome mm9 using a likelihood ratio (LR) cutoff of 500, compared to a pre-calculated third-order Markov chain background model; peaks are normalized to a 2,000 bp window at the peak center (Figure 1F). Each PWM was obtained from a previous study (Hojo et al., 2016; Ohba et al., 2015). Peak-associated genes were obtained from GREAT analysis with the default setting.
In Figure 2, peak distribution in the genome and gene ontology were performed using GREAT version 3.0.0 (Figures 2H and 2I). TSS peaks and distal peaks were defined as peaks <500 bp from the nearest TSS and >500 bp from the nearest TSS, respectively. In Figure 2J, de novo motif analysis was performed using the HOMER package (Heinz et al., 2010).
In Figure S2, we performed a pairwise comparison of normalized peak intensities between the chondrocyte Runx2 ChIP-seq peaks and osteoblast Runx2 ChIP-seq peaks using the DiffBind package (Ross-Innes et al., 2012). The correlation analysis was performed in the DiffBind with default setting (Figure S2A). The normalized ATAC-seq peak intensity was obtained using Homer annotatePeaks.pl command with the default setting (Figure S2C). To identify the Runx2 bound region at closed chromatin in a cell type, we calculated the normalized peak reads in ATAC-seq and Runx2 ChIP-seq using the DiffBind package (Figure S2E). We extracted chondrocyte Runx2 peaks where the normalized reads in ATAC-seq of C10-Cho were less than 1 and osteoblast Runx2 peaks where the normalized reads in ATAC-seq of S7-Ob were less than 1. Heatmaps were generated using the Homer program (Heinz et al., 2010) and Java TreeView software (Saldanha, 2004).
In Figure 3, to identify the cell-type specific- or shared-genomic regions where both Runx2 binding and chromatin accessibility were enriched, we performed the pairwise comparison using DiffBind. In the analysis, the chondrocyte signatures were obtained by grouping chondrocyte Runx2 ChIP-seq profiles and C10 ATAC-seq profiles, and the osteoblast signatures were obtained by grouping osteoblast Runx2 ChIP-seq profiles and S7-ATAC-seq profiles. DiffBind was applied with the parameters “summits = 150” in the step of “dba.count” to resize peaks. The statistical analysis in the pairwise comparison provided the mean of normalized peak reads in each group, the fold change (FC) of the values in each genomic region between the two groups, and the false discovery rate (FDR) for each region. In the analysis, we used a default setting of DiffBind that used the DESeq2 package for differential peak intensities. We selected genomic regions displaying cell type-specific Runx2-DNA binding and accessible chromatin by the following three criteria: (1) FC > 2, (2) FDR <0.05, and (3) genomic regions called as peaks in all replicates of Runx2 ChIP-seq and ATAC-seq in the cell type but not called in any replicates of ChIP-seq or ATAC-seq in its counterpart cell types. Similarly, the shared Runx2 peaks in both chondrocytes and osteoblasts were selected by the following three criteria: (1) −2 ≤ FC ≤ 2, (2) FDR >0.05, and (3) genomic regions called peaks in all replicates of Runx2 ChIP-seq and ATAC-seq in both chondrocytes and osteoblasts.
Figure 3Cshows motif analysis performed using Homer under default settings. Motifs were selected using the following criteria: “p value < 1.0e-50” and “% of the target sequence with motif >10%” As shown in Figure 3E, transcription factors were selected based on FPKM >5. The Z score was calculated using the log2 transformed FPKM+1 value among the members of transcription factors. Heatmaps were generated using heatmap.2. Figure 3F shows peak intersections performed using BEDTools v.2.16.2 (Quinlan and Hall, 2010). The normalized ATAC-seq peak intensity was obtained using Homer annotatePeaks.pl command with the default setting. As shown in Figure 3G, statistical analysis was performed using one-way ANOVA with post-hoc Tukey-HSD.
In Figure 4, to identify the differential binding regions between genotypes, DiffBind was applied with the following parameters “summits = 150” in the step of “dba.count” to resize peaks. PCA plot was also generated by DiffBind using the normalized peak intensity. To obtain statistically different chromatin-accessible regions, we selected the peaks based on FDR <0.05. Peak distribution in the genome was analyzed using GREAT version 3.0.0. Histograms and heatmaps were generated using Homer within a 2-kb window, and the heatmap was visualized using Java TreeView (Saldanha, 2004). Intersected peaks shown in Figure 3F were used (Figure 4L).
In Figure 5, to identify Runx2-responsive and osteoblast induction-responsive regions, DiffBind was applied with the following parameters “summits = 250” in the step of “dba.count” to resize peaks. Pairwise comparison was performed on ATAC-seq profiles between (1) day 0 GFP-expressing cells vs. day 0 Runx2-expressing cells; (2) No-transduction cells (None) vs. day 0 Runx2-expressing cells. We then selected chromatin regions that showed significant increases in ATAC-seq signals based on the following criteria: FDR <0.05, fold change >4. By further intersections with these selected regions, we obtained shared regions as Runx2-mediated open chromatin regions at day 0. Similarly, we performed a pairwise comparison with ATAC-seq profiles between (1) day 3 GFP-expressing cells vs. day 3 Runx2-expressing cells; (2) None vs. day 3 Runx2-expressing cells. Using the same criteria of FDR <0.05, fold change >4, and following intersection, we selected peaks as Runx2-and osteogenic induction-mediated open chromatin regions on day 3. Finally, to obtain the genomic regions where chromatin was closed at day 0 in Runx2-expressing cells, we performed intersection to obtain Runx2-and osteogenic induction-mediated open chromatin regions on day 3 where chromatin was closed on day 0. Heatmaps and histograms were generated using Homer within a 2-kb window, and the heatmap was visualized using Java TreeView (Saldanha, 2004).
CROP-seq analysis
To map sequence reads, gRNA sequences were aligned to the mm10 reference genome. Cellranger v2.0.0 (Zheng et al., 2017) was used as the reference genome. A gene expression matrix was generated by collecting cells that had one gRNA sequence and was used for further analysis. To identify differentially expressed genes between cells with and without Runx2 sgRNA, DEsingle (Miao et al., 2018) was performed with raw read counts. 47 genes were identified to be significantly downregulated in cells carrying Runx2 sgRNA. To determine the effects of sgRNAs targeting putative enhancers on the expression of the identified 47 genes, fold-change gene expression was calculated with normalized read counts of cells carrying sgRNAs and the negative control. The values were further transferred to z-scores to determine the distribution of the fold change across the genes. Differentially expressed genes were defined as those with an FDR <0.05. Gene ontology analysis was performed using g:Profiler (Raudvere et al., 2019). Violin plots were generated using ggplot2 with normalized read counts. Statistical analysis was performed using Tukey-HSD.
Supplementary Material
Highlights.
Chromatin accessibility is distinct between neonatal osteoblasts and chondrocytes
Runx2-DNA binding is highly associated with distinct chromatin accessibility
Runx2 is essential for establishment of chromatin accessibility in osteoblasts
An Sp7 distal enhancer driven by Runx2 contributes to osteoblast differentiation
ACKNOWLEDGMENTS
We thank Drs. Henry M. Kronenberg, Clifford J. Tabin, Kevin A. Peterson, Yuichi Nishi, Lick Pui Lai, Sean M. Hasso, and Lori L. O’Brien for helpful input; David Butler and Peter Maye for sharing Col2-ECFP and Col10a1-mCherry mice; and Jill McMahon, Charles Nicolet, Selene Tyndale, Helen Truong, Ryoko Kudo, Motoki Goto, Daisuke Tanaka, Asuka Uchida, Chie Kataoka, and Nozomi Nagumo for providing technical assistance. The supercomputing resources were provided by the Human Genome Center at the University of Tokyo. This work utilized the core research facility of the Center for Disease Biology and Integrative Medicine at the Graduate School of Medicine and the One-stop Sharing Facility Center for Future Drug Discoveries at the Graduate School of Pharmaceutical Sciences, The University of Tokyo. This study was funded by Japan Society for the Promotion of Science (JSPS) postdoctoral fellowships for research abroad (to H.H.); grants-in-aid for science research from JSPS (16H06279 [PAGS], 17H05106, 18K19636, and 20H03885 to H.H.; 21H04952 to U.C.; and 17H04403 and 21H03142 to S. Ohba); Rising Star Awards from the American Society for Bone and Mineral Research (to H.H. and S. Ohba); Mochida Memorial Foundation research grants (to H.H. and S.O.*); the Uehara Memorial Foundation Research Grant (to H.H. and S.O.*); the Takeda Science Foundation Research Grant (to H.H. and S.O.*); the Japan Agency for Medical Research and Development (AMED; JP21bm0704071 and JP21zf0127002 to H.H.); the Japan Science and Technology Agency through the Center of Innovation Program (JPMJCE1304 to U.C.); and the National Institutes of Health (DK056246 to A.P.M.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111315.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Akiyama H, Chaboissier MC, Martin JF, Schedl A, and de Crombrugghe B (2002). The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 16, 2813–2828. 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R, Gebhard C, Miguel-Escalada I, Hoof I, Bornholdt J, Boyd M, Chen Y, Zhao X, Schmidl C, Suzuki T, et al. (2014). An atlas of active enhancers across human cell types and tissues. Nature 507, 455–461. 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, and Usadel B (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozec A, Bakiri L, Jimenez M, Schinke T, Amling M, and Wagner EF (2010). Fra-2/AP-1 controls bone formation by regulating osteoblast differentiation and collagen production. J. Cell Biol. 190, 1093–1106. 10.1083/jcb.201002111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro JD, Giresi PG, Zaba LC, Chang HY, and Greenleaf WJ (2013). Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218. 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chokalingam K, Juncosa-Melvin N, Hunter SA, Gooch C, Frede C, Florert J, Bradica G, Wenstrup R, and Butler DL (2009). Tensile stimulation of murine stem cell-collagen sponge constructs increases collagen type I gene expression and linear stiffness. Tissue Eng. Part A 15, 2561–2570. https://doi.org/10.1089/ten.TEA.2008.0451 10.1089/ten.TEA.2008.0451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces MR, Buenrostro JD, Wu B, Greenside PG, Chan SM, Koenig JL, Snyder MP, Pritchard JK, Kundaje A, Greenleaf WJ, et al. (2016). Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat. Genet 48, 1193–1203. 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datlinger P, Rendeiro AF, Schmidl C, Krausgruber T, Traxler P, Klug-hammer J, Schuster LC, Kuchler A, Alpar D, and Bock C (2017). Pooled CRISPR screening with single-cell transcriptome readout. Nat. Methods 14, 297–301. 10.1038/nmeth.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day TF, Guo X, Garrett-Beal L, and Yang Y (2005). Wnt/beta-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev. Cell 8, 739–750. 10.1016/j.devcel.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, and Roeder RG (1983). Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489. 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, and Karsenty G (1995). Two distinct osteoblast-specific cis-acting elements control expression of a mouse osteocalcin gene. Mol. Cell Biol 15, 1858–1869. 10.1128/mcb.15.4.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, and Karsenty G (1997). Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell 89, 747–754. 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- Geoffroy V, Ducy P, and Karsenty G (1995). A PEBP2 alpha/AML-1-related factor increases osteocalcin promoter activity through its binding to an osteoblast-specific cis-acting element. J. Biol. Chem 270, 30973–30979. 10.1074/jbc.270.52.30973. [DOI] [PubMed] [Google Scholar]
- Gury-BenAri M, Thaiss CA, Serafini N, Winter DR, Giladi A, Lara-Astiaso D, Levy M, Salame TM, Weiner A, David E, et al. (2016). The spectrum and regulatory landscape of intestinal innate lymphoid cells are shaped by the microbiome. Cell 166, 1231–1246.e13. 10.1016/j.cell.2016.07.043. [DOI] [PubMed] [Google Scholar]
- Hadley W (2016). Ggplot2 (Springer Science+Business Media, LLC; ). [Google Scholar]
- Håkelien AM, Bryne JC, Harstad KG, Lorenz S, Paulsen J, Sun J, Mikkelsen TS, Myklebost O, and Meza-Zepeda LA (2014). The regulatory landscape of osteogenic differentiation. Stem cells (Dayton, Ohio) 32, 2780–2793. 10.1002/stem.1759. [DOI] [PubMed] [Google Scholar]
- Hashimoto M, and Takemoto T (2015). Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep 5, 11315. 10.1038/srep11315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass MR, Brissette D, Parameswaran S, Pujato M, Donmez O, Kottyan LC, Weirauch MT, and Kopan R (2021). Runx1 shapes the chromatin landscape via a cascade of direct and indirect targets. PLoS Genet. 17, e1009574. 10.1371/journal.pgen.1009574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Lewis P, Pevny L, and McMahon AP (2002). Efficient gene modulation in mouse epiblast using a Sox2Cre transgenic mouse strain. Mech. Dev 119, S97–s101. 10.1016/s0925-4773(03)00099-6. [DOI] [PubMed] [Google Scholar]
- He X, Ohba S, Hojo H, and McMahon AP (2016). AP-1 family members act with Sox9 to promote chondrocyte hypertrophy. Development 143, 3012–3023. 10.1242/dev.134502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, and Glass CK (2010). Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell 38, 576–589. 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill TP, Später D, Taketo MM, Birchmeier W, and Hartmann C (2005). Canonical Wnt/beta-catenin signaling prevents osteoblasts from differentiating into chondrocytes. Dev. Cell 8, 727–738. 10.1016/j.devcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Hojo H, Ohba S, He X, Lai LP, and McMahon AP (2016). Sp7/Osterix is restricted to bone-forming vertebrates where it acts as a Dlx Co-factor in osteoblast specification. Dev. Cell 37, 238–253. 10.1016/j.devcel.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hojo H, Ohba S, Yano F, Saito T, Ikeda T, Nakajima K, Komiyama Y, Nakagata N, Suzuki K, Takato T, et al. (2012). Gli1 protein participates in Hedgehog-mediated specification of osteoblast lineage during endochondral ossification. J. Biol. Chem 287, 17860–17869. 10.1074/jbc.M112.347716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ionescu A, Kozhemyakina E, Nicolae C, Kaestner KH, Olsen BR, and Lassar AB (2012). FoxA family members are crucial regulators of the hypertrophic chondrocyte differentiation program. Dev. Cell 22, 927–939. 10.1016/j.devcel.2012.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, and Zaret KS (2016). Cell fate control by pioneer transcription factors. Development 143, 1833–1837. 10.1242/dev.133900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon EJ, Lee KY, Choi NS, Lee MH, Kim HN, Jin YH, Ryoo HM, Choi JY, Yoshida M, Nishino N, et al. (2006). Bone morphogenetic protein-2 stimulates Runx2 acetylation. J. Biol. Chem 281, 16502–16511. 10.1074/jbc.M512494200. [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, and Wong WH (2008). An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol 26, 1293–1300. 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karreth F, Hoebertz A, Scheuch H, Eferl R, and Wagner EF (2004). The AP1 transcription factor Fra2 is required for efficient cartilage development. Development 131, 5717–5725. 10.1242/dev.01414. [DOI] [PubMed] [Google Scholar]
- Karsenty G (2008). Transcriptional control of skeletogenesis. Annu. Rev. Genomics Hum. Genet 9, 183–196. 10.1146/annurev.genom.9.081307.164437. [DOI] [PubMed] [Google Scholar]
- Kassambara A, and Mundt F (2020). Factoextra: Extract and Visualize the Results of Multivariate Data Analyses. R Package Version 1.0.7. https://CRAN.R-project.org/package=factoextra.
- Kawane T, Komori H, Liu W, Moriishi T, Miyazaki T, Mori M, Matsuo Y, Takada Y, Izumi S, Jiang Q, et al. (2014). Dlx5 and mef2 regulate a novel runx2 enhancer for osteoblast-specific expression. J. Bone Miner. Res 29, 1960–1969. 10.1002/jbmr.2240. [DOI] [PubMed] [Google Scholar]
- Kern B, Shen J, Starbuck M, and Karsenty G (2001). Cbfa1 contributes to the osteoblast-specific expression of type I collagen genes. J. Biol. Chem 276, 7101–7107. 10.1074/jbc.M006215200. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al. (1997). Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell 89, 755–764. [DOI] [PubMed] [Google Scholar]
- Kozhemyakina E, Lassar AB, and Zelzer E (2015). A pathway to bone: signaling molecules and transcription factors involved in chondrocyte development and maturation. Development 142, 817–831. 10.1242/dev.105536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu M, Javed A, Jeon JP, Horner A, Shum L, Eckhaus M, Muenke M, Lian JB, Yang Y, Nuckolls GH, et al. (2002). Cbfbeta interacts with Runx2 and has a critical role in bone development. Nat. Genet 32, 639–644. 10.1038/ng1050. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, and Salzberg SL (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25. 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Martelot G, Canella D, Symul L, Migliavacca E, Gilardi F, Liechti R, Martin O, Harshman K, Delorenzi M, Desvergne B, et al. (2012). Genome-wide RNA polymerase II profiles and RNA accumulation reveal kinetics of transcription and associated epigenetic changes during diurnal cycles. PLoS Biol. 10, e1001442. 10.1371/journal.pbio.1001442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JW, Kim DM, Jang JW, Park TG, Song SH, Lee YS, Chi XZ, Park IY, Hyun JW, Ito Y, and Bae SC (2019). RUNX3 regulates cell cycle-dependent chromatin dynamics by functioning as a pioneer factor of the restriction-point. Nat. Commun 10, 1897. 10.1038/s41467-019-09810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F, Lu Y, Ding M, Napierala D, Abbassi S, Chen Y, Duan X, Wang S, Lee B, and Zheng Q (2011). Runx2 contributes to murine Col10a1 gene regulation through direct interaction with its cis-enhancer. J. Bone Miner. Res 26, 2899–2910. 10.1002/jbmr.504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtinger M, Ingram R, Hannah R, Müller D, Clarke D, Assi SA, Lie-ALing M, Noailles L, Vijayabaskar MS, Wu M, et al. (2012). RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J. 31, 4318–4333. 10.1038/emboj.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Yao X, Yan G, Xu Y, Yan J, Zou W, and Wang G (2016). Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nat. Commun 7, 11149. 10.1038/ncomms11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F, Chung UI, Ohba S, McMahon J, Kronenberg HM, and McMahon AP (2004). Ihh signaling is directly required for the osteoblast lineage in the endochondral skeleton. Development 131, 1309–1318. [DOI] [PubMed] [Google Scholar]
- Long HK, Prescott SL, and Wysocka J (2016). Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell 167, 1170–1187. 10.1016/j.cell.2016.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrigal P, and Alasoo K (2018). AP-1 takes centre stage in enhancer chromatin dynamics. Trends Cell Biol. 28, 509–511. 10.1016/j.tcb.2018.04.009. [DOI] [PubMed] [Google Scholar]
- Maye P, Fu Y, Butler DL, Chokalingam K, Liu Y, Floret J, Stover ML, Wenstrup R, Jiang X, Gooch C, and Rowe D (2011). Generation and characterization of Col10a1-mcherry reporter mice. Genesis 49, 410–418. 10.1002/dvg.20733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayran A, and Drouin J (2018). Pioneer transcription factors shape the epigenetic landscape. J. Biol. Chem 293, 13795–13804. 10.1074/jbc.R117.001232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLean CY, Bristor D, Hiller M, Clarke SL, Schaar BT, Lowe CB, Wenger AM, and Bejerano G (2010). GREAT improves functional interpretation of cis-regulatory regions. Nat. Biotechnol 28, 495–501. 10.1038/nbt.1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer MB, Benkusky NA, and Pike JW (2014). The RUNX2 cistrome in osteoblasts: characterization, down-regulation following differentiation, and relationship to gene expression. J. Biol. Chem 289, 16016–16031. 10.1074/jbc.M114.552216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Z, Deng K, Wang X, and Zhang X (2018). DEsingle for detecting three types of differential expression in single-cell RNA-seq data. Bioinformatics 34, 3223–3224. 10.1093/bioinformatics/bty332. [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et al. (1997). Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89, 773–779. 10.1016/s0092-8674(00)80260-3. [DOI] [PubMed] [Google Scholar]
- Nakashima K, Zhou X, Kunkel G, Zhang Z, Deng JM, Behringer RR, and de Crombrugghe B (2002). The novel zinc finger-containing transcription factor osterix is required for osteoblast differentiation and bone formation. Cell 108, 17–29. 10.1016/s0092-8674(01)00622-5. [DOI] [PubMed] [Google Scholar]
- Ohba S, He X, Hojo H, and McMahon AP (2015). Distinct transcriptional programs underlie Sox9 regulation of the mammalian chondrocyte. Cell Rep. 12, 229–243. 10.1016/j.celrep.2015.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba S, Ikeda T, Kugimiya F, Yano F, Lichtler AC, Nakamura K, Takato T, Kawaguchi H, and Chung UI (2007). Identification of a potent combination of osteogenic genes for bone regeneration using embryonic stem (ES) cell-based sensor. FASEB J. 21, 1777–1787. 10.1096/fj.06-7571com. [DOI] [PubMed] [Google Scholar]
- Ohba S, Kawaguchi H, Kugimiya F, Ogasawara T, Kawamura N, Saito T, Ikeda T, Fujii K, Miyajima T, Kuramochi A, et al. (2008). Patched1 haploinsufficiency increases adult bone mass and modulates Gli3 repressor activity. Dev. Cell 14, 689–699. 10.1016/j.devcel.2008.03.007. [DOI] [PubMed] [Google Scholar]
- Olsen BR (2019). Vertebrate Skeletal Development, First edition (Academic Press, an imprint of Elsevier). [Google Scholar]
- Osterwalder M, Barozzi I, Tissières V, Fukuda-Yuzawa Y, Mannion BJ, Afzal SY, Lee EA, Zhu Y, Plajzer-Frick I, Pickle CS, et al. (2018). Enhancer redundancy provides phenotypic robustness in mammalian development. Nature 554, 239–243. 10.1038/nature25461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, et al. (1997). Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell 89, 765–771. 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- Peterson KA, Nishi Y, Ma W, Vedenko A, Shokri L, Zhang X, McFarlane M, Baizabal JM, Junker JP, van Oudenaarden A, et al. (2012). Neural-specific Sox2 input and differential Gli-binding affinity provide context and positional information in Shh-directed neural patterning. Genes Dev. 26, 2802–2816. 10.1101/gad.207142.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudvere U, Kolberg L, Kuzmin I, Arak T, Adler P, Peterson H, and Vilo J (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–w198. 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickels R, and Shilatifard A (2018). Enhancer logic and mechanics in development and Disease. Trends Cell Biol. 28, 608–630. 10.1016/j.tcb.2018.04.003. [DOI] [PubMed] [Google Scholar]
- Robledo RF, Rajan L, Li X, and Lufkin T (2002). The Dlx5 and Dlx6 homeobox genes are essential for craniofacial, axial, and appendicular skeletal development. Genes Dev. 16, 1089–1101. 10.1101/gad.988402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodda SJ, and McMahon AP (2006). Distinct roles for Hedgehog and canonical Wnt signaling in specification, differentiation and maintenance of osteoblast progenitors. Development 133, 3231–3244. 10.1242/dev.02480. [DOI] [PubMed] [Google Scholar]
- Ross-Innes CS, Stark R, Teschendorff AE, Holmes KA, Ali HR, Dunning MJ, Brown GD, Gojis O, Ellis IO, Green AR, et al. (2012). Differential oestrogen receptor binding is associated with clinical outcome in breast cancer. Nature 481, 389–393. 10.1038/nature10730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar VS, Capelo LP, Cantù C, Zimmerli D, Gosalia N, Pregizer S, Cox K, Ohte S, Feigenson M, Gamer L, et al. (2019). Reactivation of a developmental Bmp2 signaling center is required for therapeutic control of the murine periosteal niche. Elife 8, e42386. 10.7554/eLife.42386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ (2004). Java Treeview–extensible visualization of microarray data. Bioinformatics 20, 3246–3248. 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Stein GS, Lian JB, van Wijnen AJ, Stein JL, Montecino M, Javed A, Zaidi SK, Young DW, Choi JY, and Pockwinse SM (2004). Runx2 control of organization, assembly and activity of the regulatory machinery for skeletal gene expression. Oncogene 23, 4315–4329. 10.1038/sj.onc.1207676. [DOI] [PubMed] [Google Scholar]
- Stricker S, Fundele R, Vortkamp A, and Mundlos S (2002). Role of Runx genes in chondrocyte differentiation. Dev. Biol 245, 95–108. 10.1006/dbio.2002.0640. [DOI] [PubMed] [Google Scholar]
- Tai PWL, Wu H, van Wijnen AJ, Stein GS, Stein JL, and Lian JB (2017). Genome-wide DNase hypersensitivity, and occupancy of RUNX2 and CTCF reveal a highly dynamic gene regulome during MC3T3 pre-osteoblast differentiation. PLoS One 12, e0188056. 10.1371/journal.pone.0188056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takarada T, Nakazato R, Tsuchikane A, Fujikawa K, Iezaki T, Yoneda Y, and Hinoi E (2016). Genetic analysis of Runx2 function during intramembranous ossification. Development 143, 211–218. 10.1242/dev.128793. [DOI] [PubMed] [Google Scholar]
- Takeda S, Bonnamy JP, Owen MJ, Ducy P, and Karsenty G (2001). Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice. Genes Dev. 15, 467–481. 10.1101/gad.845101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Z, Niu B, Tsang KY, Melhado IG, Ohba S, He X, Huang Y, Wang C, McMahon AP, Jauch R, et al. (2018). Synergistic co-regulation and competition by a SOX9-GLI-FOXA phasic transcriptional network coordinate chondrocyte differentiation transitions. PLoS Genet. 14, e1007346. 10.1371/journal.pgen.1007346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, and Salzberg SL (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, and Pachter L (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc 7, 562–578. 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, and Pachter L (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol 28, 511–515. 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu X, Joeng KS, Nakayama KI, Nakayama K, Rajagopal J, Carroll TJ, McMahon AP, and Long F (2007). Noncanonical Wnt signaling through G protein-linked PKCdelta activation promotes bone formation. Dev. Cell 12, 113–127. 10.1016/j.devcel.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzelepis K, Koike-Yusa H, De Braekeleer E, Li Y, Metzakopian E, Dovey OM, Mupo A, Grinkevich V, Li M, Mazan M, et al. (2016). A CRISPR dropout screen identifies genetic vulnerabilities and therapeutic targets in acute myeloid leukemia. Cell Rep. 17, 1193–1205. 10.1016/j.celrep.2016.09.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta C, Iwamoto M, Kanatani N, Yoshida C, Liu Y, Enomoto-Iwamoto M, Ohmori T, Enomoto H, Nakata K, Takada K, et al. (2001). Skeletal malformations caused by overexpression of Cbfa1 or its dominant negative form in chondrocytes. J. Cell Biol 153, 87–100. 10.1083/jcb.153.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Diao H, Getzler AJ, Rogal W, Frederick MA, Milner J, Yu B, Crotty S, Goldrath AW, and Pipkin ME (2018). The transcription factor Runx3 establishes chromatin accessibility of cis-regulatory landscapes that drive memory cytotoxic T lymphocyte formation. Immunity 48, 659–674.e6. 10.1016/j.immuni.2018.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Lian N, Li L, Moss HE, Wang W, Perrien DS, Elefteriou F, and Yang X (2009). Atf4 regulates chondrocyte proliferation and differentiation during endochondral ossification by activating Ihh transcription. Development 136, 4143–4153. 10.1242/dev.043281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will AJ, Cova G, Osterwalder M, Chan WL, Wittler L, Brieske N, Heinrich V, de Villartay JP, Vingron M, Klopocki E, et al. (2017). Composition and dosage of a multipartite enhancer cluster control developmental expression of Ihh (Indian hedgehog). Nat. Genet 49, 1539–1545. 10.1038/ng.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Whitfield TW, Gordon JAR, Dobson JR, Tai PWL, van Wijnen AJ, Stein JL, Stein GS, and Lian JB (2014). Genomic occupancy of Runx2 with global expression profiling identifies a novel dimension to control of osteoblastogenesis. Genome Biol. 15, R52. 10.1186/gb-2014-15-3-r52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P, Yu HV, Tseng KC, Flath M, Fabian P, Segil N, and Crump JG (2021). Foxc1 establishes enhancer accessibility for craniofacial cartilage differentiation. Elife 10, e63595. 10.7554/eLife.63595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, et al. (2004). ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology; implication for Coffin-Lowry Syndrome. Cell 117, 387–398. 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- Yoshida CA, Furuichi T, Fujita T, Fukuyama R, Kanatani N, Kobayashi S, Satake M, Takada K, and Komori T (2002). Core-binding factor beta interacts with Runx2 and is required for skeletal development. Nat. Genet 32, 633–638. 10.1038/ng1015. [DOI] [PubMed] [Google Scholar]
- Yoshida CA, Yamamoto H, Fujita T, Furuichi T, Ito K, Inoue K.i., Yamana K, Zanma A, Takada K, Ito Y, and Komori T (2004). Runx2 and Runx3 are essential for chondrocyte maturation, and Runx2 regulates limb growth through induction of Indian hedgehog. Genes Dev. 18, 952–963. 10.1101/gad.1174704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Hata K, Takashima R, Ono K, Nakamura E, Takahata Y, Murakami T, Iseki S, Takano-Yamamoto T, Nishimura R, and Yoneda T (2015). The transcription factor Foxc1 is necessary for Ihh-Gli2-regulated endochondral ossification. Nat. Commun 6, 6653. 10.1038/ncomms7653. [DOI] [PubMed] [Google Scholar]
- Zheng GXY, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, Ziraldo SB, Wheeler TD, McDermott GP, Zhu J, et al. (2017). Massively parallel digital transcriptional profiling of single cells. Nat. Commun 8, 14049. 10.1038/ncomms14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ATAC-seq, ChIP-seq, and RNA-seq data have been deposited at Gene Expression Omnibus (GEO: GSE178293). scRNA-seq data have been deposited at the DDBJ BioProject database (DDBJ BioProject database: PRJDB8387).
This paper does not report original code.
Any additional information required to reanalyze the data presented in this work is available from the lead contact upon request.


