Abstract
The development of clinically effective drugs that could complement existing vaccines is urgently needed to reduce the morbidity and mortality associated with COVID-19. Drug-metabolizing enzymes, membrane-associated drug transporters, and inflammatory responses can partly determine the safety and efficacy of COVID-19 drugs by controlling their concentrations in both the systemic circulation and in peripheral tissues. It is still unknown how these factors affect how well COVID-19 drugs work in the clinic. We explore how drug metabolism and transport, as well as SARS-CoV-2-associated inflammatory response at disease target sites, may affect the clinical outcomes of COVID-19 drugs. In addition, we provide expert opinion on potential strategies for overcoming the clinical pharmacology and pathophysiological obstacles to improve COVID-19 drug effectiveness.
Keywords: clinical pharmacology; pharmacokinetics, pharmacodynamics; drug-metabolizing enzymes; membrane-associated drug transporters; SARS-CoV-2-associated inflammatory response; COVID-19
Unlocking the optimal therapeutic benefits of COVID-19 drugs
Although several vaccines are now available to protect against severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), full protection from SARS-CoV-2 and its variants is not guaranteed for multiple reasons including evasion of neutralizing antibody by some variants of SARS-CoV-2 [1,2], upper respiratory tract liability to viral infection [3,4], and delay in reaching herd immunityi. Therefore, a combination of vaccines and effective viral eradicating drugs, as well as adjunctive therapies will be necessary to reduce the morbidity and mortality associated with coronavirus disease 2019 (COVID-19). At the beginning of the COVID-19 pandemic, drug repurposing (see Glossary) was deployed to accelerate drug development and save cost. However, drug repurposing has not fully delivered its promise partly because drug developers may have overlooked the importance of clinical pharmacology (Box 1 ) in drug development. For instance, it may have been initially anticipated that the pharmacokinetic (PK) and pharmacodynamic (PD) profiles of pre-existing drugs for other diseases such as HIV would function similarly for COVID-19 [5]. Nonetheless, it is presumptuous to think that the original clinical pharmacology profile (Box 1) of pre-existing drugs will apply to COVID-19 owing to anatomical and physiological differences between disease target sites. The failure of many COVID-19 drug-repurposing programs, as well as of approved repurposed COVID-19 drugs such as remdesivir whose optimal therapeutic benefits have yet to be fully uncovered, can be partly attributed to suboptimal therapeutic concentrations at disease target sites such as the lung, brain, and testes due to potential drug interactions with drug-metabolizing enzymes and membrane-associated drug transporters (Box 1) at those sites (Figure 1 , Key figure) coupled with the confounding effect of the SARS-CoV-2-associated inflammatory response (Figure 2).
Box 1. Clinical pharmacology: drug metabolism and transport.
Clinical pharmacology is the study of the PK and PD parameters that inform the safety and efficacy profile of drugs. PK evaluates the overall disposition of drugs including absorption, distribution, metabolism, and excretion processes with a graphical representation of drug concentration versus time curve, whereas PD examines drug action with emphasis on potency and efficacy represented by the dose–response curve. Drug-metabolizing enzymes and membrane-associated drug transporters play a significant role in modulating the PK/PD profile of drugs.
Drug-metabolizing enzymes are enzymes that biotransform drugs to generate metabolites with the goal of improving solubility for elimination processes and/or inactivation of drug action [7]. There are two main phases of drug metabolism – phase I (the addition or exposure of polar functional groups such as alcohols and carboxylic acids on drug molecules) and phase II (primarily conjugation reactions, including sulfation and glucuronidation, that further increase drug solubility; methylation reactions may also reduce solubility and inactivate drug action) [7]. Examples of phase I enzymes are cytochromes P450 (e.g., CYP1A2 and CYP3A4) whereas phase II enzymes include UDP-glucuronosyl transferases (e.g., UGT1A1) and sulfotransferases (e.g., SULT1A1) [29].
Drug transport entails the intrinsic ability of drug molecules to move across biological membranes through passive or facilitated diffusion as well as by carrier-mediated transport processes. Carrier-mediated transporters transfer drug molecules across biological membranes and may need direct or indirect use of energy. When a carrier-mediated transporter uses membrane-associated drug transporters and energy to enable the uphill (against a concentration gradient) transport of drugs and other molecules through biological membranes, it is known as active transport. Active transport is mainly divided into two types depending on the source of energy: primary active transport in which ATP hydrolysis is linked to drug uphill transport [e.g., P-glycoprotein (P-gp) and breast cancer resistance protein (BCRP)], and secondary active transport in which energy from an electrochemical gradient is used to move drugs against their concentration gradient (e.g., sodium/glucose cotransporters SGLT1 and SGLT2). ATP-binding cassette (ABC) and solute carrier (SLC) transporters are primary and secondary active transporters respectively, and are also the major membrane-associated drug transporter superfamilies involved in the cellular influx and efflux of drugs in mammalian systems [7]. Members of the ABC superfamily include P-gp, BCRP, and multidrug resistance-associated proteins (MRPs), whereas SLC members include organic anion transporting polypeptides (OATPs), equilibrative nucleoside transporters (ENTs), and concentrative nucleoside transporters. Alone or in synergy, these membrane-associated drug transporters and drug-metabolizing enzymes determine drug concentrations in the systemic circulation and peripheral tissues. To avoid costly clinical development failures, it is important to investigate potential drug-metabolizing enzymes and membrane-associated drug transporter interactions with investigational new drugs during the early development phase following US FDA guidance [37].
Alt-text: Box 1
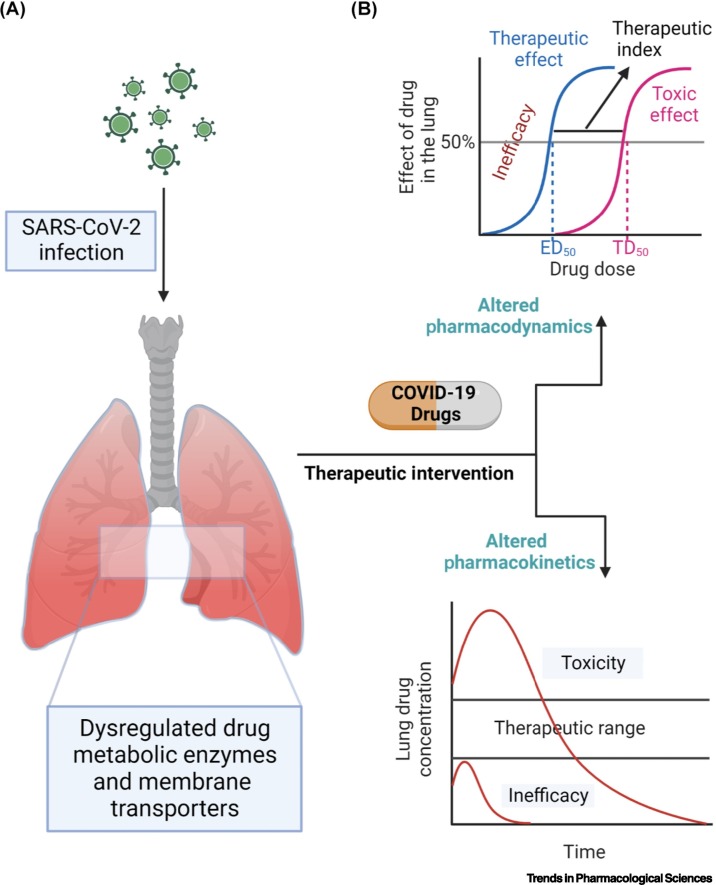
Figure 1.
Key figure. Interactions between human pulmonary drug clinical pharmacology and COVID-19 pathophysiology.
(A) SARS-CoV-2 infects the human lung and elicits an immune response. The inflammatory response could alter the levels of clinically relevant drug-metabolizing enzymes and membrane-associated drug transporters in the lung. (B) Dysregulated pulmonary drug-metabolizing enzymes and membrane transporters could modify the concentration of COVID-19 drugs in the lungs leading to altered drug pharmacokinetic (PK) and pharmacodynamic (PD) profiles. For example, drug concentrations below the pulmonary therapeutic range would cause inefficacy whereas drug concentrations higher than the therapeutic range may result in toxicity. Other SARS-CoV-2 target sites, including the brain and testes, are expected to display similar interactions. Abbreviations: ED50, 50% effective dose; TD50, 50% toxicity dose.
Figure 2.
Dysregulation of pulmonary drug pharmacokinetics by COVID-19-associated inflammatory response.
(A) SARS-CoV-2 infection can cause severe inflammatory response with elevated proinflammatory cytokines such as IL-6, IL-1β, and TNF-α. (B) All these inflammatory effects can dysregulate drug metabolism and transport leading to altered pulmonary drug concentrations. Higher or lower pulmonary drug concentrations can lead to toxicity or inefficacy, respectively. Other SARS-CoV-2 target sites, including the brain and testes, are expected to display similar interactions.
These drug-metabolizing enzymes and membrane-associated drug transporters (Box 1) play a crucial role in determining the safety and efficacy profile of drugs by modulating their PK/PD parameters [6., 7., 8.]. Furthermore, pathophysiological events such as the inflammatory response associated with SARS-CoV-2 infection could alter the clinical pharmacology profile of promising COVID-19 drugs [9] by altering the expression of clinically relevant drug-metabolizing enzymes, membrane-associated drug transporters, plasma and tissue proteins, as well as tissue injuries – including injuries to tissues that may house metabolic enzymes and transporters involved in the disposition of COVID-19 drugs. In this opinion article we highlight some current challenges in COVID-19 drug development, discuss the potential role of drug-metabolizing enzymes and membrane-associated drug transporters in the disposition of promising COVID-19 drugs in the lung, brain, and testes, and assess the involvement of SARS-CoV-2-associated inflammatory response in modulating the safety and efficacy of COVID-19 drugs. We also provide strategies for mitigating the effect of drug-metabolizing enzymes, membrane-associated drug transporters, and SARS-CoV-2-associated inflammatory response on the clinical efficacy and safety profile of COVID-19 drugs.
Challenges in the development of COVID-19 drugs
For COVID-19, the lung is the predominant site of SARS-CoV-2 infection, and the virus-eradicating potential of COVID-19 drugs is partly dependent on the spatial distribution and therapeutic concentrations in the alveolar epithelial cells of the lungs (Figure 1). However, it is challenging to measure COVID-19 drug concentrations in the lungs of COVID-19 patients, and plasma drug concentrations are therefore measured as a surrogate for target site drug levels. The drug levels in the plasma may not correlate with levels in the lung, and total lung drug levels are not an accurate representation of drug concentrations in target cells. For example, in animal studies, the levels of remdesivir and its active metabolite GS-441524 were quantified in lung tissues, but the studies did not determine the spatial distribution or concentration of remdesivir or GS-441524 in type II alveolar epithelial cells – the predominant pulmonary cells infected by SARS-CoV-2 [10., 11., 12.]. It is impossible to assess how the reported total lung remdesivir/GS-441524 concentrations correlate with potential viral load because the investigation was not carried out in SARS-CoV-2-infected animals. Furthermore, the limited efficacy of lopinavir for the treatment of SARS-CoV-2 infection was partly attributed to suboptimal pulmonary drug concentrations that may be too low to sufficiently inhibit SARS-CoV-2 replication in the lungs [13]. This is based on earlier animal research demonstrating that the lung concentration of lopinavir is lower than its plasma level [14]; however, the study did not address the distribution or concentration of lopinavir in different pulmonary cell types. To design an ideal dosing regimen that will improve the clinical efficacy and safety profile of promising COVID-19 drugs, it is crucial to understand the spatial distribution and concentration of COVID-19 drugs in type II alveolar epithelial cells as well as in other cells that are primarily susceptible to SARS-CoV-2 infection.
Surprisingly, studies on the pulmonary spatial distribution and concentration of promising COVID-19 drugs such as remdesivir, molnupiravir, and nirmatrelvir are lacking. The spatial distribution of drugs in the lung is further complicated by its heterogeneous nature as well as by other factors including route of administration, dissolution, deposition, sequestration, clearance, and retention [15]. Many of these factors are controlled by pulmonary drug-metabolizing enzymes and membrane-associated drug transporters [15., 16., 17.]. However, sophisticated tools such as mass spectrometry imaging (MSI) and positron emission tomography (PET) can be used in future COVID-19 drug development efforts to unravel the spatial distribution and concentration of the drugs at SARS-CoV-2 target sites such as the lung, brain, and testes [18]. These techniques have been used to reveal the lung and brain spatial distribution of several drugs [19., 20., 21.]. For instance, MSI has been used to determine the pulmonary localization and retention of the long-acting β-adrenergic receptor agonist salmeterol [19], and PET imaging was used to uncover the lung target occupancy of ipratropium (an inhaled bronchodilator) [21]. These studies are informing research and development strategies for respiratory medicines [15]. The overall goal of using techniques such as MSI in drug development is to perform localized drug disposition studies in target tissues that will uncover how drugs and/or their associated metabolites are spatially distributed and concentrated in select target cells within diseased tissues [18].
COVID-19 drug disposition at SARS-CoV-2 target sites
The therapeutic benefits of promising COVID-19 drugs may be constrained by drug-metabolizing enzymes and membrane-associated drug transporters at SARS-CoV-2 target sites in the body. As a result, their effects should be taken into consideration when designing the ideal clinical dosing regimen. Although the lung is the predominant site of SARS-CoV-2 infection, viral tropism is dependent on the expression of the host anchor proteins – angiotensin-converting enzyme 2 receptor and type 2 transmembrane serine protease – which explains the presence of the virus in the brain and testes as well as in other tissues such as gastrointestinal tract and heart [22., 23., 24., 25., 26., 27.]. SARS-CoV-2 infection of the brain is implicated in the proposed pathogenesis of the COVID-19-associated neurocognitive dysfunction observed in 20–70% of investigated COVID-19 patients in Germany and the UK [24,28]. In addition, the presence of SARS-CoV-2 in the testes has been suggested to potentially contribute to viral transmission through sexual intercourse [22], and chronic lung injuries may be induced by viral persistence in the respiratory system. Therefore, it is important to identify the role of drug-metabolizing enzymes and membrane-associated drug transporters in determining the concentration of promising COVID-19 drugs in the lung, brain, and testes to prevent viral persistence and post-recovery complications.
Pulmonary drug metabolism and transport
Clinically relevant drug-metabolizing enzymes and membrane-associated drug transporters are present in the lungs [16,29,30] and may be involved in regulating the pulmonary concentration of COVID-19 drugs whether administered via oral, inhalation, or intravenous routes.
In comparison to the relatively limited number of cell types in the liver, the lung contains ~40 different cell types with an unequal distribution of drug-metabolizing enzymes and membrane-associated drug transporters of lower expression and activity [15,29]. Club cells in the bronchioles have been identified as the principal pulmonary source of drug-metabolizing enzymes (i.e., cytochromes P450, CYPs) [29,31]. Within the lung tissue, cells are grouped into epithelial, endothelial, immune, and mesenchymal cell types [29]. The basal, ciliated, and alveolar type I and II cells of the pulmonary epithelium, as well as the immune and endothelial cell types, comprising macrophages and vascular endothelial cells, respectively, play a significant role in the metabolism of drugs in the lung [29]. Phase I metabolic enzymes (Box 1), including CYP1A2, CYP2A6, CYP2B6, CYP2E1, and CYP3A5, as well as phase II enzymes (Box 1) such as glutathione-S-transferases (GSTA1 and GSTA2), N-acetyl transferase (NAT1), sulfotransferase (SULT1A1), and UDP-glucuronosyl transferase (UGT2A1), have been identified in human lung tissues [15,29,31]. Similarly, membrane-associated drug transporters including influx transporters such as organic cation transporters (OCT1, OCT2, OCT3, and OCTN2), efflux transporters such as P-glycoprotein (P-gp), breast cancer resistance protein (BCRP), multidrug resistance-associated protein (MRP1), and multidrug and toxin extrusion (MATE1) are also found in respiratory tract cells such as tracheal/bronchial ciliated cells, basal cells, airway epithelium, bronchial epithelium, alveolar macrophages, alveolar epithelium, and others [30,32,33].
Most of these cells are implicated in the pathophysiology of COVID-19. For example, SARS-CoV-2 appears to target numerous cell types in the proximal airways as well as alveolar type II cells in the distal lung gas-exchange zone, and these cells are significantly involved in drug metabolism and transport [25,34., 35., 36.]. Similarly, macrophage recruitment is an important immune response to SARS-CoV-2 infection of the lungs, and these cells also express clinically relevant drug-metabolizing enzymes and membrane-associated drug transporters that are recommended for routine investigation by the US FDA [37] (Table 1 ). Therefore, alteration of these pulmonary cells due to SARS-CoV-2 infection could lead to dysregulation of drug metabolism and transport.
Table 1.
Localization of selected clinically relevant drug-metabolizing enzymes and membrane-associated drug transporters in human lung, brain, and testisa
| Parameters |
Protein |
Tissue localization |
Refs |
||
|---|---|---|---|---|---|
| Lung | Brain | Testis | |||
| Drug-metabolizing enzymes | CYP1A2 | Ciliated columnar epithelial cells, type I and II pneumocytes | Basal ganglia, substantia nigra, frontal cortex, striatum, pons, hippocampus, cerebellum | ? | [31,48] |
| CYP2B6 | Bronchiolar epithelium, Clara cells, alveolar epithelium | Basal ganglia, frontal cortex, striatum, pons, and cerebellum | ? | [31,48] | |
| CYP2C8 | Bronchiolar epithelium | ? | ? | [31] | |
| CYP2C9 | ? | Basal ganglia, frontal cortex, hippocampus, cerebellum | ? | [48] | |
| CYP2C19 | ? | Basal ganglia, frontal cortex, hippocampus, cerebellum | ? | [48] | |
| CYP2D6 | ? | Thalamus, hypothalamus, substantia nigra, frontal cortex, striatum, pons, hippocampus, cerebellum | Throughout testicular tissue, stronger localization at the seminiferous epithelium and testicular endothelium | [48,54] | |
| CYP3A4 | ? | Thalamus, hypothalamus, and basal ganglia | Throughout testicular tissue | [48,54] | |
| Membrane-associated drug transporters | P-gp | Alveolar macrophages, alveolar epithelium, serous cells of the bronchial mucosa, bronchial capillaries |
Luminal membranes of brain microvessel endothelial cells, apical plasma membrane of choroid plexus epithelial cells, astrocytes, microglia, neurons |
Seminiferous epithelium, interstitial space | [30,47,54] |
| BCRP | Bronchial epithelial cells and seromucinous glands, small endothelial capillaries of the lung, alveolar pneumocytes |
Luminal membrane of microvessel endothelial cells, astrocytes, and microglia | Basal and apical side of the seminiferous epithelium, testicular endothelium | [30,47,54] | |
| OCT2 | Basal cells | Luminal side of brain microvessel endothelial cells | Peritubular myoid cells, Leydig cells | [30,47,52] | |
| OATP1B1 | ? | ? | Basal membrane of Sertoli cells |
[52] | |
| OATP1B3 | ? | ? | Basal membrane of Sertoli cells |
[52] | |
| MATE1 | Apical side of bronchial and bronchiolar epithelial cells, alveolar macrophages | ? | Adluminal compartment of the seminiferous tubules, peritubular myoid cells, Leydig cells | [32,52] | |
| MATE2-K | ? | ? | ? | ||
| OAT1 | ? | ? | Basal membrane of Sertoli cells |
[52] | |
?, presence is unknown.
Many of the repurposed COVID-19 drugs are substrates for the drug-metabolizing enzymes and membrane-associated drug transporters that are present at disease target sites such as human lung, brain, and testes, and may trigger metabolic enzyme/transporter-mediated drug–drug interactions (Table 2 ). For example, remdesivir is a substrate for the metabolic enzymes CYP3A4 and CES1 (carboxyl esterase 1), as well as for the transporters organic anion transporting polypeptide (OATP1B1; influx) and P-gp (efflux) [38]. These pulmonary drug-metabolizing enzymes and membrane-associated drug transporters work together to regulate the levels of active and/or inactive drug (via biotransformation processes) and intracellular drug concentrations (via transepithelial transport), resulting in a distinct local and systemic drug PK/PD profile. There is also a potential for an interaction between drug-metabolizing enzymes and membrane-associated drug transporters where a high pulmonary drug transport turnaround time could limit metabolic processes and vice versa [30]. It has been reported that the pulmonary exposure of inhaled umeclidinium bromide and vilanterol (drugs used for the treatment of chronic obstructive pulmonary disease) increased by ~40% in humans because of inhibition of P-gp-mediated efflux by verapamil, a calcium channel inhibitor [39]. In addition, the bioavailability of intranasally administered drugs may be limited due to interactions with drug-metabolizing enzymes and membrane-associated drug transporters in the nasal cavity [40]. This may have severe consequences for COVID-19 drugs that are being developed for administration through the inhalation route.
Table 2.
Metabolic enzyme and transporter-mediated interactions with selected repurposed COVID-19 drugsa
| Drug category | Drug | Metabolic enzymes | Membrane transporters | Membrane transporter inhibition | Metabolic enzyme inhibition | Refs |
|---|---|---|---|---|---|---|
| Inhibition of virus replication | Azithromycin | CYP3A4 | P-gp, MRP2, and OATP | P-gp | [67,75] | |
| Favipiravir | Aldehyde oxidase and xanthine oxidase | OAT1, OAT3, P-gp | CYP2C8 | [67,75]; DrugBankv |
||
| Ivermectin | CYP3A4, CYP2D6, CYP2E1 | P-gp, BCRP, MRP1, MRP2, MRP3 | CYP2C9, CYP2C19, CYP2D6, CYP3A4 | [67]; DynaMed Plusvi |
||
| Lopinavir | CYP3A | P-gp, MRP1, MRP2, OATP1A2, OATP1B1 | P-gp, BCRP, OATP1B1, OATP1B3, OATP2B1, MATE1, BSEP | CYP3A4 | [67,75,76]; DrugBankv |
|
| Molnupiravir | ENT1, ENT2 | [53] | ||||
| Nitazoxanide | Deacetylase, UGT | [67] | ||||
| Nirmatrelvir | CYP3A4 | P-gp | P-gp, OATP1B1 | CYP3A4 | Product monographii | |
| Remdesivir | CES1, cathepsin A, CYP2C8, CYP2D6, and CYP3A4 | P-gp, OATP1B1, ENT1, ENT2 | OATP1B1, OATP1B3, BSEP, MRP4, NTCP, MATE1, OCT1, ENT1, ENT2 | CYP3A4 | [53,67,75,76]; DynaMed Plusvi |
|
| Ritonavir | CYP1A2, CYP2C8, CYP2C9, CYP2C19, CYP3A, CYP2D6 | P-gp, MRP1, MRP2 | P-gp, MRP1, BCRP, OATP1A2, OATP2B1, OATP1B1, OATP1B3, OCT1, MATE1, BSEP | CYP3A4, CYP2D6 | [67,75,76]; DynaMed Plusvi |
|
| Inhibition of virus entry | Chloroquine | CYP2C8, CYP3A4, CYP2D6, CYP3A5, CYP1A1 | OATP1A2 | P-gp, OCT1, MATE1, MATE2 | [67,75]; DrugBankv |
|
| Hydroxychloroquine | CYP2C8, CYP3A4, CYP2D6 | P-gp, OATP1A2, OCT1, MATE1, MATE2 | [67,75] | |||
| Umifenovir | CYP3A4, UGT1A9, UGT2B7 | P-gp, OATP2B1, OATP1B1, OATP1B3, OCT1, MATE1 | [67,75]; DrugBankv |
|||
| Anti-inflammatory | Dexamethasone | CYP3A4 | P-gp, MRP2 | [67] | ||
| Baricitinib | CYP3A4 | P-gp, BCRP, OAT3, MATE2-K | OAT1, OAT2, OAT3, OCT1, OCT2, OATP1B3, BCRP, MATE1, MATE2-K | [67]; DynaMed Plusvi |
||
| Colchicine | CYP3A4 | P-gp | DynaMed Plusvi | |||
| Methylprednisolone | 11β-Hydroxysteroid dehydrogenases and 20-ketosteroid reductases | DrugBankv | ||||
| Ruxolitinib | CYP1A2, CYP2B6, CYP2C9, CYP3A4 | OATP1B1, OCT1, NTCP, | P-gp, BCRP, OATP2B1 | [67,75] | ||
| Fluvoxamine | CYP1A2, CYP2C19, CYP2D6, CYP3A4 | CYP1A2, CYP2C9, CYP3A4, CYP2C19, CYP2D6 | [67]; DynaMed Plusvi |
Abbreviations: BSEP, bile salt export pump; NTCP, sodium-dependent uptake transporter.
Drug metabolism and transport in the brain
Similarly to the lung, cerebral drug metabolism and transport could also limit the concentration of promising COVID-19 drugs in the brain. The blood–brain barrier (BBB), a physiological barrier that separates the brain parenchyma from the systemic circulation and is made up of brain microvascular endothelial cells that are sealed by tight junction proteins, plays a major role in modulating drug concentrations in the brain. This barrier regulates the trafficking of molecules into and out of the brain through various transport pathways [41]. Although this selective permeation is aimed at protecting the brain from neurotoxicants, it inevitably excludes neuroprotective drugs from reaching optimal therapeutic concentrations in the central nervous system (CNS). Although the BBB is the major roadblock to brain drug delivery owing to its large surface area and impermeability, the blood–cerebrospinal fluid barrier (BCSFB) is another barrier located at the choroid plexus and arachnoid membrane that can also limit drug penetration into the brain [41].
Drugs primarily have access to the brain parenchyma through paracellular transport, passive diffusion, and carrier- or receptor-mediated transport processes (Box 1) [41]. Conversely, efflux transporters expressed at the BBB are responsible for extruding drugs out of the brain. Drug physicochemical properties (such as lipophilicity, hydrophilicity, and molecular weight), cerebral blood flow, metabolism, degradation, systemic clearance, and protein binding all influence how much drug crosses the BBB [42].
Drug-metabolizing enzymes and membrane-associated drug efflux transporters in the brain parenchyma, as well as at the BBB and BCSFB, play a key role in limiting drug bioavailability in the CNS, potentially contributing to the formation of CNS viral sanctuary sites for infectious diseases such as HIV [41,43,44]. From a pharmacological perspective, these drug–metabolic enzyme/transporter interactions could modify the overall CNS drug PK profile because of altered brain-to-plasma drug concentration ratios, as exemplified by several antiretroviral drugs [41]. A similar phenomenon is anticipated in the context of CNS SARS-CoV-2 sanctuary site formation and limited brain penetration of COVID-19 drugs. Phase I (CYP3A4 and CYP2D6) and phase II (UGT1A6 and UGT2B7) drug-metabolizing enzymes, as well as influx (OCT1 and OCT2) and efflux transporters (P-gp and BCRP) have been identified in the human brain (Table 1) [43,45., 46., 47., 48.] and are involved in the metabolism and transport of promising COVID-19 drugs (Table 2). Several studies have uncovered the role of membrane-associated drug transporters and drug-metabolizing enzymes in the disposition of antiretroviral drugs used for the treatment of HIV [44]. Our group has shown that P-gp and BCRP limit the penetration of the HIV integrase strand-transfer inhibitor, raltegravir, in an in vitro BBB model [49]. Beyond concentrations, drug distribution in the brain is also important in determining the safety and efficacy profile of drugs. Therefore, evidence-based studies to delineate the concentration and spatial distribution profile of promising COVID-19 drugs in the brain are urgently required.
Testicular drug metabolism and transport
Owing to the restricted penetration of antiretroviral drugs across the blood–testis barrier (BTB), the male genital tract has been identified as a sanctuary site for persistent viral infections such as HIV [44,50]. Given the presence of SARS-CoV-2 in the male genital tract [22,51], a similar occurrence is anticipated in COVID-19. The role of the BTB is to protect developing germ cells from xenobiotics including drugs from entering the abluminal compartment through transepithelial transport processes regulated by efflux and influx transporters [50].
Efflux transporters such as P-gp and BCRP, as well as influx transporters (equilibrative nucleoside transporters, ENT1 and ENT2), have been identified at the BTB and appear to play a significant role in drug disposition in this tissue (Table 1) [50,52]. Our group has shown that P-gp and BCRP limit the penetration of raltegravir in an in vitro BTB model [49]. Conversely, uptake transporters such as ENT1 and ENT2 are also involved in the transport of nucleoside/nucleotide analog drugs because of their structural similarity to endogenous transporter substrates such as adenosine [53], creating an exceptional uptake pathway for increasing drug concentrations. We also found that drug-metabolizing enzymes including CYP3A4, CYP2D6, and UGT1A1 are expressed in human testicular tissues and T cell subsets isolated from testes (Table 1) [54,55]. Furthermore, we and others have found that the antiretroviral drugs efavirenz and darunavir had significantly lower testicular concentrations (below the therapeutic range) compared to plasma concentrations, whereas the other antiretroviral drugs – emtricitabine, lamivudine, and tenofovir – had higher testicular concentrations in HIV-infected tissues [54,55]. This suggests that an alternative transport pathway, potentially mediated by ENTs, may drive the testicular influx of these drugs because they are nucleoside/nucleotide analogs. Interestingly, promising COVID-19 drugs such as remdesivir and EIDD-1931 (the active metabolite of molnupiravir), but not nirmatrelvir, have recently been identified as substrates for ENT1 and ENT2 in in vitro models [53,56].
To circumvent the potential effects of drug-metabolizing enzymes and membrane-associated drug transporters in reaching optimal therapeutic drug concentrations at disease target sites such as lung, brain, and testes, an effective route of administration should be adopted. For example, previous studies have shown that drugs administered intranasally have a high CNS bioavailability compared to their intravenous counterparts [57,58]. Compared to systemic administration, inhaled salmeterol had a higher targeting in the lung subepithelial and epithelial regions [59]. This suggests that low doses of inhaled COVID-19 drugs may achieve optimal therapeutic concentrations and distribution in human lung and brain tissues with minimal systemic exposures. Drug bioavailability can be improved using prodrug approaches, drug delivery bypass systems [29], and CYP450 inhibitors – ritonavir can be coadministered with nirmatrelvir to improve the therapeutic concentrations of nirmatrelvir through ritonavir-mediated inhibition of CYP3A4ii. Drugs that are structurally similar to endogenous substrates of influx transporters including the ENTs and concentrative nucleoside transporters are more likely to achieve optimal therapeutic concentrations at target sites and should be investigated in COVID-19 drug development programs. However, it is unknown how counteracting efflux transporters and/or COVID-19 pathophysiology would impact on these influx transport pathways.
Involvement of SARS-CoV-2-associated inflammatory response in therapeutic efficacy and safety
Pathophysiological events such as the inflammatory response associated with SARS-CoV-2 infection could alter the clinical pharmacology profile of promising COVID-19 drugs [9]. For example, variability in clinical response to remdesivir has been reported in several clinical trials [60., 61., 62., 63., 64.]. Because the extent of inflammatory response may vary among COVID-19 patients, the variability in drug response may be related to its effect on drug-metabolizing enzymes and membrane-associated drug transporters (Figure 2 ). Proinflammatory cytokines such as tumor necrosis factor α (TNF-α), interleukin 6 (IL-6), and interleukin 1β (IL-1β) are overproduced in a typical hospitalized COVID-19 patient and may be responsible for acute respiratory distress syndrome, lung injuries, and multiple-organ damages observed in some COVID-19 patients at the severe stage of the disease [65,66]. The overproduction of proinflammatory cytokines ('cytokine storm') is a consequence of the hyperactivation of immune cells that attempt to clear SARS-CoV-2 virus [25]. Although this inflammatory response may start in the lung and cause multiple lung tissue damage, it may eventually reach other tissues such as the brain and male genital tract through the systemic circulation [65]. Inflammatory response is a well-known regulator of drug-metabolizing enzymes and membrane-associated drug transporters, and frequently results in a rise or decrease in the expression of some clinically important drug-metabolic enzymes and transporters [9].
Several proinflammatory cytokines (e.g., IL-6, IL-1β, TNF-α) dysregulate the expression of drug-metabolizing enzymes (e.g., CYP enzymes) and membrane-associated drug transporters (e.g., efflux transporters) through transcription factors (e.g., constitutive androstane receptor, pregnane X receptor, and nuclear factor κB) that regulate their transcription and translation [9]. Consequently, the PK/PD profile of labile drugs may change due to the alteration of the corresponding absorption, distribution, metabolism, and excretion processes at both the systemic and peripheral levels (Figure 2). Altered drug PK/PD profile mediated by inflammatory response may be distinct between systemic circulation and peripheral tissues depending on the concentrations of clinically-relevant proinflammatory cytokines at both sites. This variability in the drug PK/PD profile can cause intra- and inter-individual differences in drug response among patient populations and may be the underlying reason for differences in clinical outcomes for some repurposed COVID-19 drugs such as remdesivir.
Multiple disease–drug interactions have been suggested for repurposed COVID-19 drugs and should be carefully considered (Table 2) [67]. For example, remdesivir is a substrate of CYP3A4 and P-gp, both of which are known to be dysregulated under inflammatory conditions [9]. Previous clinical studies have shown that CYP3A4, CYP2D6, and P-gp levels are altered in HIV+ individuals and had severe consequences for drug disposition [68,69]. In addition, damage to tissues (mediated by the hyperinflammatory response) housing clinically relevant drug-metabolizing enzymes and membrane-associated drug transporters can also alter the drug PK/PD profile. This is anticipated in COVID-19 because multiple types of organ damage, specifically lung injuries, may affect pulmonary cells expressing drug-metabolizing enzymes and membrane-associated drug transporters. For example, diffuse alveolar damage is present in some COVID-19 patients, and this may affect the integrity of alveolar epithelial cells – a significant source of pulmonary drug-metabolizing enzymes and membrane-associated drug transporters [29,30,70]. Furthermore, the downregulation of ENT1 and ENT2 in the context of COVID-19 is most likely due to the clinical manifestation of acute lung injuries and hypoxia [71]. In addition, competitive inhibition of ENTs-mediated uptake processes caused by elevated extracellular adenosine levels (associated with acute lung injuries) may affect drug delivery [71]. Both of these pathophysiological events may limit the penetration of remdesivir and EIDD-1931 (the active metabolite of molnupiravir). The recruitment of pulmonary macrophage cells in response to SARS-CoV-2 infection could also increase resident drug-metabolizing enzymes and membrane-associated drug transporters, thereby altering drug PK/PD profile. It remains unknown how COVID-19 pathophysiology would affect tissue and plasma protein (albumin and α1 acid glycoprotein) levels because their alteration could also dysregulate the extent of drug tissue and/or plasma protein binding and ultimately their PK/PD profile.
To navigate therapeutic efficacy and safety against a background of COVID-19 pathophysiology, dosing should be individualized based on the above-discussed pathophysiological markers, including clinical measurement of the levels of systemic and peripheral inflammatory markers, markers of tissue damage, the concentration of relevant plasma and tissue proteins, and the expression of clinically relevant drug-metabolizing enzymes and membrane-associated drug transporters. In addition, the administration of COVID-19 drugs should be stratified based on disease stage and viral burden. For instance, if a COVID-19 patient has a high replicating SARS-CoV-2 viral load and a hyperinflammatory response, then anti-inflammatory drugs should be initially considered to dampen the ongoing inflammation and prevent further tissue damage before administering antiviral drugs. Stratifying therapeutic administration in this manner can help to mitigate or prevent disease–drug interactions that could limit the clinical safety and efficacy of promising COVID-19 drugs. Examples of these classes of COVID-19 drugs include anti-inflammatory drugs (dexamethasone and baricitinib) and antiviral drugs (remdesivir, molnupiravir, and nirmatrelvir). Drugs such as sirolimus (immunosuppressant) and metformin (antidiabetic drug) which target the mechanistic target of rapamycin (mTOR) pathway may have great potential for both inhibiting viral replication and dampening the inflammatory response, as well as reducing the associated cellular damages [72., 73., 74.]. These mTOR inhibitor drugs are undergoing several clinical trials (clinical trial identifiers NCT04948203, NCT04461340, and NCT04604678) to determine their safety and efficacy profile in the treatment of COVID-19. If clinically effective, this class of drugs could mitigate or prevent any possible disease–drug interactions because of their potential to offer both anti-inflammatory and antiviral effects.
Concluding remarks and future directions
Over the past 2 years significant progress has been made in vaccine and drug development to reduce the morbidity and mortality associated with COVID-19. For example, the US FDA has authorized four vaccinesiii and two drugsiv for the prevention and treatment of SARS-CoV-2 infection, respectively. However, these vaccines may not offer full protection against SARS-CoV-2 and its variants because breakthrough infections are still observed in some vaccinated individuals. Likewise, there is limited clinical evidence for effective eradication of SARS-CoV-2 and its variants by existing COVID-19 drugs. It is possible that many repurposed drugs – both approved and failed – may have untapped therapeutic potential for the treatment of COVID-19, and unlocking optimal therapeutic benefits will require an optimized dosing regimen that achieves sufficient drug concentrations to eradicate SARS-CoV-2 (and variants) at the disease target sites. Therefore, evidence-based and mechanistic studies will be necessary to answer the most pressing questions concerning COVID-19 drug development programs (see Outstanding questions). For instance, research in SARS-CoV-2 animal models must show time-dependent changes in the pulmonary, CNS, and testicular concentrations of promising COVID-19 drugs (remdesivir, molnupiravir, and nirmatrelvir) and the associated changes in viral burden. Furthermore, sophisticated analytical platforms such as MSI and PET should be used to investigate the time-dependent spatial distribution of promising COVID-19 drugs in SARS-CoV-2 animal models at target sites. Such studies should also evaluate the impact of different routes of drug administration on the tissue PK and distribution profiles. In developing inhaled COVID-19 drugs, drug-metabolizing enzymes and membrane-associated drug transporters resident in the upper respiratory tract including the nasal epithelial cells must be considered and their impact on drug disposition quantified accordingly. Data generated from these studies should be used to predict optimal dosing regimens by applying physiologically based pharmacokinetic (PBPK) modeling.
Outstanding questions.
Do COVID-19 drugs reach optimal concentrations in the lung, brain, and testes of infected patients?
Are promising COVID-19 drugs well-distributed at the predominant sites of infected cells within target tissues?
Are target site membrane-associated drug transporters and/or drug-metabolizing enzymes responsible for the limited efficacy of some investigational and approved COVID-19 drugs?
Could the optimal route of administration be dependent on the stage of SARS-CoV-2 infection?
What is the relationship between disease target site (lung, brain, and testes) expression of membrane-associated drug transporters and drug-metabolizing enzymes, and different stages of SARS-CoV-2 infection?
Is COVID-19 drug dosing adjustment necessary to achieve clinically meaningful efficacy and safety outcomes given the potential for target site transporter and/or enzyme-mediated drug interactions?
Alt-text: Outstanding questions
Acknowledgments
Acknowledgments
This work is supported by grants from the Ontario HIV Treatment Network (fund 506657) and the Canadian Institutes of Health Research (fund 51794) to R.B. C.K.N. is the recipient of a Canadian Institutes of Health Research Doctoral Scholarship, Ontario Graduate Scholarship, Pfizer Canada Graduate Fellowship, and Leslie Dan Faculty of Pharmacy Dean's Fellowship. BioRender was used to create the figures.
Declaration of interests
The authors declare no competing interests
Glossary
- Bioavailability
the fraction of an administered drug that reaches the systemic circulation.
- Brain parenchyma
the nervous tissue of the brain that comprises two main cell types, neurons and glial cells, that are responsible for the normal functions of the brain.
- COVID-19-associated neurocognitive dysfunction
neuropsychiatric symptoms and neurocognitive deficits that are observed in some COVID-19 patients, such as anosmia, inability to focus and pay attention, new-onset anxiety, depression, psychosis, seizures, and even suicidal ideation.
- Disease–drug interactions
these occur when an underlying disease condition alters the pharmacokinetic/pharmacodynamic (PK/PD) profile of drugs by modifying their pharmacological profile (for example, metabolism and transport, protein binding, and others) leading to clinically relevant safety and efficacy issues.
- Drug–drug interactions
these occur when two or more drugs are coadministered, one of which (the perpetrator) alters the PK/PD profile of the other drug (victim) by modifying its pharmacology (for example, metabolism and transport, protein binding, other) leading to clinically relevant safety and efficacy issues.
- Drug repurposing
a strategy for developing drugs by identifying pre-existing drugs that could be used to treat other diseases.
- Hyperinflammatory response
overactivation of immune responses to pathogens (i.e., bacteria, viruses, toxins), trauma, and other external factors that may affect the normal physiology of the body.
- Mass spectrometry imaging (MSI)
a label-free analytical technique that allows the direct localization and quantification of drugs, metabolites, and biomarkers in biological tissues.
- Mechanistic target of rapamycin (mTOR)
a protein kinase that controls several metabolic processes in response to environmental factors including nutrient levels, growth factors, and stress.
- Paracellular transport
the transport of molecules including drugs and biomolecules through intercellular spaces between adjacent cells.
- Physiologically based pharmacokinetic modeling (PBPK)
a computational method used to predict pharmacokinetic parameters that represent the absorption, distribution, metabolism, and excretion profile of a drug.
- Positron emission tomography (PET)
a molecular imaging technique that uses radioisotopes to visualize and quantify changes in biomolecular processes and drug biodistribution in mammalian systems.
- Proinflammatory cytokines
secreted proteins such as IL-6 and TNF-α that play a key role in initiating inflammation in response to pathogen invasion or other external factors that alter normal body physiology.
- SARS-CoV-2 sanctuary
disease target sites where SARS-CoV-2 evades the effects of antiviral drugs, thus allowing its survival and persistence.
Resources
ihttps://ourworldindata.org/covid-vaccinationsiihttps://covid-vaccine.canada.ca/info/pdf/paxlovid-pm-en.pdfiiihttps://www.fda.gov/emergency-preparedness-and-response/coronavirus-disease-2019-covid-19/covid-19-vaccinesivhttps://www.fda.gov/drugs/emergency-preparedness-drugs/coronavirus-covid-19-drugsvhttps://go.drugbank.com/vihttps://www.dynamed.com/References
- 1.Garcia-Beltran W.F., et al. Multiple SARS-CoV-2 variants escape neutralization by vaccine-induced humoral immunity. Cell. 2021;184:2372–2383. doi: 10.1016/j.cell.2021.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hoffmann M., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184:2384–2393. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krammer F. SARS-CoV-2 vaccines in development. Nature. 2020;586:516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 4.Su F., et al. Induction of mucosal immunity through systemic immunization: Phantom or reality? Hum. Vaccines Immunother. 2016;12:1070–1079. doi: 10.1080/21645515.2015.1114195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatakrishnan K., et al. Challenges in drug development posed by the COVID-19 pandemic: an opportunity for clinical pharmacology. Clin. Pharmacol. Ther. 2020;108:699–702. doi: 10.1002/cpt.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nwabufo C.K., et al. Employing in vitro metabolism to guide design of F-labelled PET probes of novel α-synuclein binding bifunctional compounds. Xenobiotica. 2021;51:885–900. doi: 10.1080/00498254.2021.1943566. [DOI] [PubMed] [Google Scholar]
- 7.Nwabufo C.K. Relevance of ABC transporters in drug development. Curr. Drug Metab. 2022;23:434–446. doi: 10.2174/1389200223666220621113524. [DOI] [PubMed] [Google Scholar]
- 8.Nwabufo C.K., Aigbogun O.P. Diagnostic and therapeutic agents that target alpha-synuclein in Parkinson’s disease. J. Neurol. 2022;269:5762–5786. doi: 10.1007/s00415-022-11267-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunvald A.-C.D., et al. Clinical and molecular perspectives on inflammation-mediated regulation of drug metabolism and transport. Clin. Pharmacol. Ther. 2022;112:277–290. doi: 10.1002/cpt.2432. [DOI] [PubMed] [Google Scholar]
- 10.Sahakijpijarn S., et al. In vivo pharmacokinetic study of remdesivir dry powder for inhalation in hamsters. Int. J. Pharm. X. 2021;3 doi: 10.1016/j.ijpx.2021.100073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu W., et al. Pharmacokinetics and tissue distribution of remdesivir and its metabolites nucleotide monophosphate, nucleotide triphosphate, and nucleoside in mice. Acta Pharmacol. Sin. 2021;42:1195–1200. doi: 10.1038/s41401-020-00537-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu H., et al. High expression of ACE2 receptor of 2019-nCoV on the epithelial cells of oral mucosa. Int. J. Oral Sci. 2020;12:8. doi: 10.1038/s41368-020-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang Y., Chen L. Lung tissue distribution of drugs as a key factor for COVID-19 treatment. Br. J. Pharmacol. 2020;177:4995–4996. doi: 10.1111/bph.15102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumar G.N., et al. Metabolism and disposition of the HIV-1 protease inhibitor lopinavir (ABT-378) given in combination with ritonavir in rats, dogs, and humans. Pharm. Res. 2004;21:1622–1630. doi: 10.1023/b:pham.0000041457.64638.8d. [DOI] [PubMed] [Google Scholar]
- 15.Bäckström E., Fridén M. In: Inhaled Medicines. Kassinos S., et al., editors. Elsevier; 2021. Drug distribution in lung tissue; pp. 301–318. [Google Scholar]
- 16.Bosquillon C. Drug transporters in the lung--do they play a role in the biopharmaceutics of inhaled drugs? J. Pharm. Sci. 2010;99:2240–2255. doi: 10.1002/jps.21995. [DOI] [PubMed] [Google Scholar]
- 17.Ehrhardt C., et al. Current progress toward a better understanding of drug disposition within the lungs: summary proceedings of the First Workshop on Drug Transporters in the Lungs. J. Pharm. Sci. 2017;106:2234–2244. doi: 10.1016/j.xphs.2017.04.011. [DOI] [PubMed] [Google Scholar]
- 18.Nwabufo C.K., Aigbogun O.P. Potential application of mass spectrometry imaging in pharmacokinetic studies. Xenobiotica. 2022 doi: 10.1080/00498254.2022.2119900. Published online September 1, 2022. [DOI] [PubMed] [Google Scholar]
- 19.Hamm G.R., et al. Revealing the regional localization and differential lung retention of inhaled compounds by mass spectrometry imaging. J. Aerosol Med. Pulm. Drug Deliv. 2020;33:43–53. doi: 10.1089/jamp.2019.1536. [DOI] [PubMed] [Google Scholar]
- 20.Vallianatou T., et al. A mass spectrometry imaging approach for investigating how drug-drug interactions influence drug blood–brain barrier permeability. Neuroimage. 2018;172:808–816. doi: 10.1016/j.neuroimage.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Schou M., et al. Pulmonary PET imaging confirms preferential lung target occupancy of an inhaled bronchodilator. EJNMMI Res. 2019;9:9. doi: 10.1186/s13550-019-0479-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D., et al. Clinical characteristics and results of semen tests among men with coronavirus disease 2019. JAMA Netw. Open. 2020;3 doi: 10.1001/jamanetworkopen.2020.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazza M.G., et al. Persistent psychopathology and neurocognitive impairment in COVID-19 survivors: effect of inflammatory biomarkers at three-month follow-up. Brain Behav. Immun. 2021;94:138–147. doi: 10.1016/j.bbi.2021.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boldrini M., et al. How COVID-19 affects the brain. JAMA Psychiatry. 2021;78:682. doi: 10.1001/jamapsychiatry.2021.0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rendeiro A.F., et al. The spatial landscape of lung pathology during COVID-19 progression. Nature. 2021;593:564–569. doi: 10.1038/s41586-021-03475-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwabenland M., et al. Deep spatial profiling of human COVID-19 brains reveals neuroinflammation with distinct microanatomical microglia–T-cell interactions. Immunity. 2021;54:1594–1610. doi: 10.1016/j.immuni.2021.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trypsteen W., et al. On the whereabouts of SARS-CoV-2 in the human body: a systematic review. PLoS Pathog. 2020;16 doi: 10.1371/journal.ppat.1009037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo M.S., et al. Frequent neurocognitive deficits after recovery from mild COVID-19. Brain Commun. 2020;2:fcaa205. doi: 10.1093/braincomms/fcaa205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enlo-Scott Z., et al. Drug metabolism in the lungs: opportunities for optimising inhaled medicines. Expert Opin. Drug Metab. Toxicol. 2021;17:611–625. doi: 10.1080/17425255.2021.1908262. [DOI] [PubMed] [Google Scholar]
- 30.Enlo-Scott Z., et al. In: Inhaled Medicines. Kassinos S., et al., editors. Elsevier; 2021. Epithelial permeability and drug absorption in the lungs; pp. 267–299. [Google Scholar]
- 31.Oesch F., et al. Xenobiotica-metabolizing enzymes in the lung of experimental animals, man and in human lung models. Arch. Toxicol. 2019;93:3419–3489. doi: 10.1007/s00204-019-02602-7. [DOI] [PubMed] [Google Scholar]
- 32.Berg T., et al. Expression of MATE1, P-gp, OCTN1 and OCTN2, in epithelial and immune cells in the lung of COPD and healthy individuals. Respir. Res. 2018;19:68. doi: 10.1186/s12931-018-0760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gustavsson L., et al. In: Drug Transporters Vol 1: Role and Importance in ADME and Drug Development. Nicholls G., Youdim K., editors. Royal Society of Chemistry; 2016. Drug transporters in the lung: expression and potential impact on pulmonary drug disposition; pp. 184–228. [Google Scholar]
- 34.Ziegler C.G.K., et al. SARS-CoV-2 receptor ACE2 is an interferon-stimulated gene in human airway epithelial cells and is detected in specific cell subsets across tissues. Cell. 2020;181:1016–1035. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoffmann M., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou Y.J., et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182:429–446. doi: 10.1016/j.cell.2020.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Center for Drug Evaluation and Research . FDA; 2020. In Vitro Drug Interaction Studies – Cytochrome P450 Enzyme- and Transporter-Mediated Drug Interactions. Guidance for Industry. [Google Scholar]
- 38.Deb S., et al. ADME and pharmacokinetic properties of remdesivir: its drug interaction potential. Pharmaceuticals (Basel) 2021;14:655. doi: 10.3390/ph14070655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mehta R., et al. Effect of verapamil on systemic exposure and safety of umeclidinium and vilanterol: a randomized and open-label study. Int. J. Chron. Obstruct. Pulmon. Dis. 2013;8:159–167. doi: 10.2147/COPD.S40859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oliveira P., et al. Drug-metabolizing enzymes and efflux transporters in nasal epithelium: influence on the bioavailability of intranasally administered drugs. Curr. Drug Metab. 2016;17:628–647. doi: 10.2174/1389200217666160406120509. [DOI] [PubMed] [Google Scholar]
- 41.Osborne O., et al. The paradox of HIV blood–brain barrier penetrance and antiretroviral drug delivery deficiencies. Trends Neurosci. 2020;43:695–708. doi: 10.1016/j.tins.2020.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Banks W.A. From blood–brain barrier to blood–brain interface: new opportunities for CNS drug delivery. Nat. Rev. Drug Discov. 2016;15:275–292. doi: 10.1038/nrd.2015.21. [DOI] [PubMed] [Google Scholar]
- 43.Alam C., et al. Role and modulation of drug transporters in HIV-1 therapy. Adv. Drug Deliv. Rev. 2016;103:121–143. doi: 10.1016/j.addr.2016.05.001. [DOI] [PubMed] [Google Scholar]
- 44.Whyte-Allman S.-K., Bendayan R. HIV-1 sanctuary sites – the role of membrane-associated drug transporters and drug metabolic enzymes. AAPS J. 2020;22:118. doi: 10.1208/s12248-020-00498-1. [DOI] [PubMed] [Google Scholar]
- 45.Declèves X., et al. Drug metabolism at the blood–brain and blood–CSF barriers. AAPS Adv. Pharm. Sci. Ser. 2014;10:101–124. [Google Scholar]
- 46.Dickens D., et al. In: Drug Transporters (Vol. 1): Role and Importance in ADME and Drug Development. Nicholls G., Youdim K., editors. Royal Society of Chemistry; 2016. Drug transporters at the blood–brain barrier; pp. 151–183. [Google Scholar]
- 47.Liu L., Liu X. In: Drug Transporters in Drug Disposition, Effects and Toxicity. Liu X., Pan G., editors. Springer; 2019. Contributions of drug transporters to blood–brain barriers; pp. 407–466. [Google Scholar]
- 48.Kuban W., Daniel W.A. Cytochrome P450 expression and regulation in the brain. Drug Metab. Rev. 2021;53:1–29. doi: 10.1080/03602532.2020.1858856. [DOI] [PubMed] [Google Scholar]
- 49.Hoque M.T., et al. Raltegravir permeability across blood–tissue barriers and the potential role of drug efflux transporters. Antimicrob. Agents Chemother. 2015;59:2572–2582. doi: 10.1128/AAC.04594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Miller S.R., Cherrington N.J. Transepithelial transport across the blood–testis barrier. Reproduction. 2018;156:R187–R194. doi: 10.1530/REP-18-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costa G.M.J., et al. SARS-CoV-2 infects, replicates, elevates angiotensin II and activates immune cells in human testes. MedRxiv. 2022 doi: 10.1101/2022.02.05.22270327. Published online February 8, 2022. [DOI] [Google Scholar]
- 52.Hau R.K., et al. Localization of xenobiotic transporters expressed at the human blood–testis barrier. Drug Metab. Dispos. 2022;50:770–780. doi: 10.1124/dmd.121.000748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Miller S.R., et al. Remdesivir and EIDD-1931 interact with human equilibrative nucleoside transporters 1 and 2: implications for reaching SARS-CoV-2 viral sanctuary sites. Mol. Pharmacol. 2021;100:548–557. doi: 10.1124/molpharm.121.000333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang Y., et al. Antiretroviral drug transporters and metabolic enzymes in human testicular tissue: potential contribution to HIV-1 sanctuary site. J. Antimicrob. Chemother. 2016;71:1954–1965. doi: 10.1093/jac/dkw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Whyte-Allman S.K., et al. Drug efflux transporters and metabolic enzymes in human circulating and testicular T-cell subsets: relevance to HIV pharmacotherapy. AIDS. 2020;34:1439–1449. doi: 10.1097/QAD.0000000000002548. [DOI] [PubMed] [Google Scholar]
- 56.Hau R.K., et al. PF-07321332 (nirmatrelvir) does not interact with human ENT1 or ENT2 : implications for COVID-19 patients. Clin. Transl. Sci. 2022;15:1599–1605. doi: 10.1111/cts.13292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rassy D., et al. Intranasal methylprednisolone effectively reduces neuroinflammation in mice with experimental autoimmune encephalitis. J. Neuropathol. Exp. Neurol. 2020;79:226–237. doi: 10.1093/jnen/nlz128. [DOI] [PubMed] [Google Scholar]
- 58.Espinosa A., et al. Intranasal dexamethasone reduces mortality and brain damage in a mouse experimental ischemic stroke model. Neurotherapeutics. 2020;17:1907–1918. doi: 10.1007/s13311-020-00884-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bäckström E., et al. Uncovering the regional localization of inhaled salmeterol retention in the lung. Drug Deliv. 2018;25:838–845. doi: 10.1080/10717544.2018.1455762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang R., Mylonakis E. In inpatients with COVID-19, none of remdesivir, hydroxychloroquine, lopinavir, or interferon β-1a differed from standard care for in-hospital mortality. Ann. Intern. Med. 2021;174:JC17. doi: 10.7326/ACPJ202102160-017. [DOI] [PubMed] [Google Scholar]
- 61.Beigel J.H., et al. Remdesivir for the treatment of COVID-19 – final report. N. Engl. J. Med. 2020;383:1813–1826. doi: 10.1056/NEJMoa2007764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Goldman J.D., et al. Remdesivir for 5 or 10 days in patients with severe COVID-19. N. Engl. J. Med. 2020;383:1827–1837. doi: 10.1056/NEJMoa2015301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Y., et al. Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled, multicentre trial. Lancet. 2020;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Spinner C.D., et al. Effect of remdesivir vs standard care on clinical status at 11 days in patients with moderate COVID-19: a randomized clinical trial. JAMA. 2020;324:1048. doi: 10.1001/jama.2020.16349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pilla Reddy V., et al. Pharmacokinetics under the COVID-19 storm. Br. J. Clin. Pharmacol. 2020 doi: 10.1111/bcp.14668. Published online November 23, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Aziz M., et al. Elevated interleukin-6 and severe COVID-19: a meta-analysis. J. Med. Virol. 2020;92:2283–2285. doi: 10.1002/jmv.25948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar D., Trivedi N. Disease–drug and drug–drug interaction in COVID-19: risk and assessment. Biomed. Pharmacother. 2021;139 doi: 10.1016/j.biopha.2021.111642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jones A.E., et al. Variability in drug metabolizing enzyme activity in HIV-infected patients. Eur. J. Clin. Pharmacol. 2010;66:475–485. doi: 10.1007/s00228-009-0777-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jetter A., et al. Do activities of cytochrome P450 (CYP)3A, CYP2D6 and P-glycoprotein differ between healthy volunteers and HIV-infected patients? Antivir. Ther. 2010;15:975–983. doi: 10.3851/IMP1648. [DOI] [PubMed] [Google Scholar]
- 70.Konopka K.E., et al. Diffuse alveolar damage (DAD) resulting from coronavirus disease 2019 infection is morphologically indistinguishable from other causes of DAD. Histopathology. 2020;77:570–578. doi: 10.1111/his.14180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller S.R., et al. Response to comments on 'remdesivir and EIDD-1931 interact with human equilibrative nucleoside transporters 1 and 2: implications for reaching SARS-CoV-2 viral sanctuary sites.'. Mol. Pharmacol. 2022;101:121–122. doi: 10.1124/molpharm.121.000448. [DOI] [PubMed] [Google Scholar]
- 72.Fattahi S., et al. PI3K/Akt/mTOR pathway: a potential target for anti-SARS-CoV-2 therapy. Immunol. Res. 2022;70:269–275. doi: 10.1007/s12026-022-09268-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Patocka J., et al. Rapamycin: drug repurposing in SARS-CoV-2 infection. Pharmaceuticals. 2021;14:217. doi: 10.3390/ph14030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ventura-López C., et al. Treatment with metformin glycinate reduces SARS-CoV-2 viral load: an in vitro model and randomized, double-blind, Phase IIb clinical trial. Biomed. Pharmacother. 2022;152 doi: 10.1016/j.biopha.2022.113223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Yee S.W., et al. Drugs in COVID-19 clinical trials: predicting transporter-mediated drug–drug interactions using in vitro assays and real-world data. Clin. Pharmacol. Ther. 2021;110:108–122. doi: 10.1002/cpt.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ambrus C., et al. Interactions of anti-COVID-19 drug candidates with hepatic transporters may cause liver toxicity and affect pharmacokinetics. Sci. Rep. 2021;11:17810. doi: 10.1038/s41598-021-97160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]