Abstract
The ongoing coronavirus disease-19 (COVID-19) pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has drastically changed our way of life and continues to have an unmitigated socioeconomic impact across the globe. Research into potential vaccine design and production is focused on the spike (S) protein of the virus, which is critical for virus entry into host cells. Yet, whether the degree of glycosylation in the S protein is associated with vaccine efficacy remains unclear. Here, we first optimized the expression of the S protein in mammalian cells. While we found no significant discrepancy in purity, homogeneity, or receptor binding ability among S proteins derived from 293F cells (referred to as 293F S-2P), 293S GnTI- cells (defective in N-acetylglucosaminyl transferase I enzyme; 293S S-2P), or TN-5B1-4 insect cells (Bac S-2P), there was significant variation in the glycosylation patterns and thermal stability of the proteins. Compared with the partially glycosylated 293S S-2P or Bac S-2P, the fully glycosylated 293F S-2P exhibited higher binding reactivity to convalescent sera. In addition, 293F S-2P induced higher IgG and neutralizing antibody titres than 293S or Bac S-2P in mice. Furthermore, a prime-boost-boost regimen, using a combined immunization of S-2P proteins with various degrees of glycosylation, elicited a more robust neutralizing antibody response than a single S-2P alone. Collectively, this study provides insight into ways to design a more effective SARS-CoV-2 immunogen.
Keywords: SARS-CoV-2, Spike, 293 cells, Glycosylation, Immunogenicity
1. Introduction
SARS-CoV-2 is the third highly pathogenic betacoronavirus to infect humans in recent times [1], [2]. The ongoing global COVID-19 pandemic has had an inestimable impact on the global economy and public health. As SARS-CoV-2 continues to evolve constantly, leading to the emergence of variants which exhibit different escape mutations associated with diverse transmissibility or pathogenicity such as the WHO defined variants of concerns: B.1.1.7 lineage (Alpha), B.1.135 (Beta), P.1 (Gamma), B.1.167.2 (Delta), and B.1.1.529 (Omicron) [3], [4], [5], [6], [7], threaten the efficacy of current vaccines and therapeutics [8], [9], [10], [11]. SARS-CoV-2 belongs to the betacoronavirus group and utilizes the angiotensin-converting enzyme 2 (ACE2) cellular receptor to gain entry into host cells [12], [13], [14], [15]. Like other coronaviruses, SARS-CoV-2 cellular entry is mediated by the trimeric transmembrane spike (S) glycoprotein, which is also the dominant target of neutralizing antibodies (nAbs) [16], [17], [18]. As the major antigen on SARS-CoV-2 viral surface, S protein is an ideal target for diagnosis of SARS-CoV-2 infection. Recently, a method named femtomolar-level detection of SARS-CoV-2 spike proteins improved the early diagnosis efficiency of COVID-19 [19]. The mature S protein, which is responsible for receptor binding, virus attachment, and fusion [20], [21], [22], consists of S1 and S2 subunits divided at the furin cleavage site [23], [24]. The receptor binding domain (RBD) on the S1 subunit binds to the receptor ACE2 and subsequently induces conformational changes on the S2 subunit from a prefusion to post-fusion state [25].
The S protein is a highly glycosylated protein, with 22 N-linked glycan sequons on the promoter surface in mammalian cells, 8 of these glycans are the oligomannose-type whereas the other 14 are the fully complexed-type [26], [27], [28]. Glycans on membrane viruses, such as the influenza virus and human immunodeficiency virus (HIV-1), play important roles in receptor binding and host cell entry [29], [30]. N-glycans close to receptor binding sites shield the conserved epitopes and abrogate existing antibody binding, protect this conserved and vulnerable region from immune attack. Some of the glycans are involved in recognition of nAbs, like S309 [31]. The HIV-1 envelope (Env) glycoprotein, which is decorated by N-glycans, surrounds the CD4 receptor binding site (CD4bs) and thus limits B cell recognition and hinders the elicitation of CD4bs-targeting nAbs [32]. Indeed, several previous studies have indicated that gradual N-glycan restoration along with heterologous boosting can elicit effective cross-neutralizing Abs against HIV [32], [33], [34]. The advantage of this strategy is that priming with a low glycosylated antigen helps B cells recognize the exposed glycan-deleted epitopes within the densely glycosylated neutralizing epitopes, while boosting with the fully glycosylated antigen further allows these B cells to gradually recognize the densely glycosylated epitopes to induce potent nAbs [34]. Most of the recombinant COVID-19 vaccine candidates in clinical trials are currently derived from mammalian (CHO, 293F) or baculovirus expression systems [35]. Not surprisingly, S trimers from different expression systems have distinct glycosylation profiles [36]. Based on previous studies in HIV, we hypothesized that different glycosylation profiles on the SARS-CoV-2 S protein will have an impact on immunogenicity; whether the strategy of priming with a partially glycosylated S protein and boosting with a fully glycosylated S protein could improve immunogenicity levels deserves exploring.
In this study, we obtained high-quality SARS-CoV-2 S-2P proteins with different glycosylation profiles from three cell lines: 293F, 293S GnTI- and insect cells; 293S GnTI- cells are defective in N-acetylglucosaminyl transferase I enzyme. As anticipated, the S proteins produced in these systems showed different antigenicity levels: S-2P from the mammalian cell system induced the highest nAb titres in mice, particularly when compared with that from insect cells. In addition, we showed that the immunization in mice using spike protein with varied glycosylation led to the production of higher nAb titres compared to using spike with homogenous glycosylation alone in the prime-boost-boost procedure. Overall, we suggest that a combined prime-boost-boost immunization protocol using SARS-CoV-2 S-2P proteins with different glycosylation profiles could improve the immunization strategy that is currently used in COVID-19 vaccine development.
2. Materials and methods
2.1. Protein expression and purification
The mammalian codon optimized spike gene of Wuhan-Hu-1 strain (GenBank accession no. NC_045512.2), 15–1208 aa, was synthesized (Shenggong Biotech, Shanghai, China). In detail, three versions of the mammalian codon-optimized sequences were designed: 1) the ectodomain (aa. 15–1208) of the wild-type S protein containing the tissue plasminogen activator signal peptide (tPA-S-WT); 2) a two-proline stabilized trimer (S-2P) with the original signal peptide (Ori-S-2P); 3) S-2P with a tPA signal peptide (tPA-S-2P). For all constructs, a T4 fibritin trimerization motif (Fd) was added to the C-terminus to promote trimerization, along with an 8-His tag for purification. For the two S-2P constructs, the two proline substitutions (K986P, V987P) were introduced to enhance the stabilization of S trimer, and the furin-silenced mutation (GSAS) at residues 682–685 were introduced to prevent cleavage between S1 and S2. These constructs were than cloned into the pTT5, pVRC8400 and pcDNA3.1 mammalian expression vectors. Constructs were subsequently transfected into 293F (Invitrogen) and 293S GnTI- cells (ATCC) using linear polyethyleneimine (MW 25000, Polysciences) at a cell density of 1.0 × 106 cells/mL. For each construct, 750 μg of plasmid and 1.5 mg PEI was diluted in 25 mL growth media, separately. and then mixed together, 18 min later, the mixture was added to 450 mL growth media. Five days after transfection, the culture medium was collected and purified through a one-step Ni-Sepharose Excel (Cytiva) purification column. The eluted samples containing the S-2P proteins were collected and further dialyzed in phosphate-buffered saline (pH 7.4). Proteins were then concentrated to 1 mg/ml using the 100-kDa Vivaspin turbo 15 mL tubes (Sartorius). S-2P proteins from the baculovirus-insect cell expression system were expressed and purified as previously described [36].
2.2. SDS-PAGE and western blotting
Collected protein samples were mixed with 6 × reducing loading buffer (50 mM Tris, pH 6.8, 2 % SDS, 5 % 2-mercaptoethanol, 0.01 % bromophenol blue, 8 % glycerol), boiled for 10 min, and then loaded onto 8 % SDS-PAGE gels. Proteins were electrophoresed at 80 V for 90 min in a BioRad MINI-PROTEIN Tetra system (BioRad Laboratories, CA, USA). The gels were stained with Coomassie Brilliant Blue R-250 (Bio-Rad) for 30 min and then decolorized with eluent overnight. For western blotting, gels were soaked in trans-Blot Turbo 5 × Transfer Buffer (SDS), and then transferred onto nitrocellulose membranes (Whatman, Dassel, Germany) using the trans-Blot Turbo transfer system (Bio-Rad). Membranes were then blocked with 5 % skim milk for 1 h, and then incubated with diluted human sera (1:500 dilution) or anti-His-tag monoclonal antibody (1:5000 dilution) for 1 h. Membranes were then washed thrice with tris-buffered saline (TBS) and then incubated with HRP-conjugated goat anti-mouse or goat anti-human IgG (Abcam) for 45 min. Membranes were washed again, and then incubated with SuperSignal ELISA Pico Chemiluminescent Substrate Kit (Thermo Fisher Scientific). All steps and incubations were performed at room temperature.
2.3. High-performance size-exclusion chromatography (HPSEC)
Purified WT or S-2P proteins were analysed on an HPSEC system (Waters; Milford, MA) using a TSK Gel PW5000xl 7.8 × 300 mm column (TOSOH, Japan) pre-equilibrated with PBS, pH 7.4. Samples were loaded at a flow rate of 0.5 mL/min and detected at 280 nm.
2.4. Negative-stain electron microscopy (EM)
For negative-stain EM, protein samples (0.1 mg/ml) were applied to carbon-coated grids and incubated for 5 min. Excess liquid was removed using filter paper, and samples were immediately stained with 5 % phosphotungstic acid for 2 min. Grids was dried thoroughly and then examined on an FEI Tecnai G2 Spirit electron microscope operating at 120 keV.
2.5. Differential scanning fluorimetry (DSC)
The thermal denaturation midpoints (Tm) of purified S-2P proteins were evaluated by differential scanning calorimetry (VP-Capillary DSC, GE Healthcare), as previously described [37]. Protein samples at 0.3–0.5 mg/mL were dissolved in phosphate-buffered saline (PBS, pH7.4). Samples (300 μL protein) and reference buffer were loaded into the cells. A scan rate of 90℃/h was used to detect the melting temperature. All data processing procedures were performed using Origin 7.0 software.
2.6. PNGase-F and endo-H digestion
Deglycosylation with endo-H (NEB, MA, US) or PNGase-F (NEB) was performed according to manufacturer’s protocols: 20 μg S-2P was mixed with denaturation buffer, 2 μL deglycosylated enzymes, and H2O to a final volume of 20 μL, and incubated at 37℃ overnight. Treated samples were loaded onto SDS-PAGE gels, and detected by western blot using an anti-His antibody.
2.7. Enzyme-linked immunosorbent assay (ELISA)
S-2P protein samples were loaded into the wells of 96-well microtiter plates at 100 ng/well in 100 μL PBS, and incubated overnight at 4℃. Samples were then blocked with 1 × enzyme dilution buffer (PBS + 0.25 % casein + 1 % gelatin + 0.05 % proclin-300) at 37℃ for 2 h. Samples were then incubated with 3-fold serially diluted mAbs (three RBD-targeting, two NTD-targeting and two S2- targeting antibodies isolated and purified in mice or rabbit from our laboratory), SARS-CoV-2 (Wuhan strain) sera (reported in our previous study) from convalescent patients in the First Affiliated Hospital of Xiamen University (China) [36], or biotinylated receptor protein human ACE2 for 1 h, and then washed three times with TBS. Serum samples were diluted from a starting dilution of 1:100. Samples were then incubated with horseradish peroxidase (HRP)-conjugated goat anti-human antibody (Abcam), goat anti-mouse antibody (Abcam), or goat anti-rabbit antibody (Abcam) (all 1:5000 dilution) for 45 min. Streptavidin-HRP (ThermoFisher) was used to detect ACE2 binding. Finally, the wells were incubated with O-phenylenediamine (OPD) substrate for 10 min, with the reactions quenched by the addition of 1 M sulfuric acid. The absorbance was measured at 450 nm on a microplate reader (TECAN, Männedorf, Switzerland). The half-effective titres (ET50) and half-effective concentrations (EC50) were calculated by sigmoid trend fitting using GraphPad Prism software (GraphPad Software, CA, US).
2.8. Mice immunization
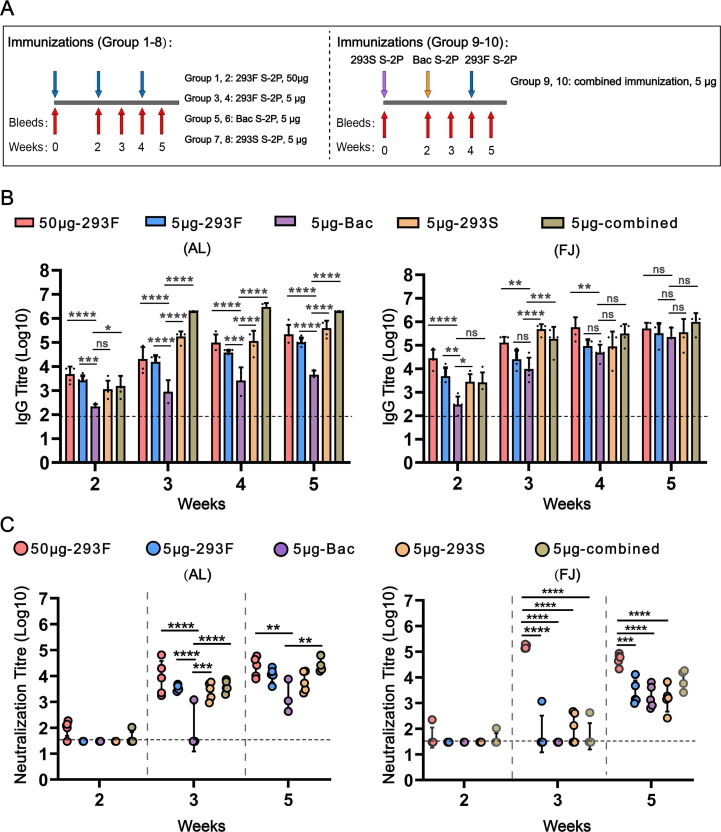
All mouse experiments were performed strictly according to the animal ethics guidelines and protocols set by the Xiamen University Laboratory Animal Management Ethics Committee. Six-week-old female BALB/c mice were purchased from Shanghai Slake Laboratory Animal Co. ltd (Shanghai, China). Mice were immunized via an intramuscular injection into the left or right posterior limb at weeks 0, 2, and 4. Two adjuvants were used: aluminium adjuvant (AL) and Freund’s adjuvant (FJ). S-2P proteins were diluted in saline and formulated with an equal volume of aluminium adjuvant (AL) or Freund’s adjuvant (FJ), respectively (totally 150 μL). Mice were separated into 10 groups (n = 5, per group) and immunized according to the schedule presented in Fig. 4A and Table S1. Briefly, group 1–2 were immunized with 293F S-2P at a high dose (50 μg) adjuvanted with AL or FJ, respectively; group 3–8 received low dose (5 μg) cognate S-2P immunization for each immunization. In which group 3, 5, 7 were immunized with 5 μg 293F S-2P, 5 μg Bac S-2P, 5 μg 293S S-2P formulated with AL separately for every immunization; Group 4, 6, 8 were immunized with 5 μg 293F S-2P, 5 μg Bac S-2P, 5 μg 293S S-2P formulated with FJ seperately; Group 9 and 10 received a combined immunization protocol, with 293S S-2P (5 μg) prime at week 0, Bac S-2P (5 μg) at week 2, and 293F S-2P (5 μg) at week 4, adjuvanted with AL or FJ, respectively; Serum samples were collected at weeks 0, 2, 3, 4, and 5, centrifuged at 13,000 × g for 10 min, and stored at − 20 °C for ELISA and neutralization assays.
Fig. 4.
Immunogenicity of SARS-CoV-2 spike (S) proteins. (A) Mice immunization schedules. Group 1, 3, 5, 7, 9 received immunizations formulated with aluminium adjuvant. Group 2, 4, 6, 8, 10 received immunizations formulated with Freund’s adjuvant. (B) Antigen-specific IgG antibody titres induced by ten immunized groups. (C) Neutralizing antibody titres against SARS-CoV-2 pseudovirus (Wuhan-Hu-1 strain). The dotted line indicates the limit of detection for the assay. Statistical analysis was performed by one-way ANOVA. Significant differences were determined using the Holm-Sidak pairwise multiple comparisons in Graphpad Prism 8.0 (ns = non-significant; *P < 0.1, **P < 0.01, ***P < 0.001, ****P < 0.0001). (A) Groups statistically different from the Bac S-2P group are indicated. (B) Groups statistically different from each other are indicated. An absence of symbols indicates no statistical difference between the groups.
2.9. Neutralization assays
NAbs in mice sera were quantified using a VSV-based neutralization assay, as described previously [38]. In brief, the S gene minus the last 18-aa was codon-optimized for expression in human cells and cloned into the pCAG plasmid to generate pCAG-nCoVSdel18 construct. The pCAG-nCoVSdel18 plasmid was then transfected into Vero-E6 cells. Meanwhile to produce VSV-G particles with enhanced green fluorescent protein (EGFP) reporter gene (VSVdG-EGFP-G virus), the VSVdG-EGFP-G construct (Addgene, 31842) was transfected into 293 T cells (kindly gifted by Dr Jiahuai Han), 48 h later, the VSVdG-EGFP-G viruses were harvested and then inoculated into cells expressing the SARS-CoV-2 Sdel18 truncated protein. After 1 h incubation, the supernatant that containing VSV pseudotyped viruses bearing SARS-CoV-2 spike protein and glycoprotein (G) was collected. and was then incubated with anti-VSV-G rat serum to abrogate the infectivity of pseudovirus mediated by the G protein on VSVdG-EGFP-G viruses. This supernatant was collected for SARS-CoV-2 infection experiments. The viral titer of the SARS-CoV-2 pseudovirus supernatant was determined by counting the number of GFP-expressing cells after BHK21-hACE2 cell (BHK21 cells (ATCC, CCL-10) stably overexpressing human ACE2) infection with gradient diluted supernatants. For neutralization, immunization serum samples were gradient diluted and incubated with 0.05 multiplicity of infection (MOI) VSV-SARS-CoV-2-Sdel18 virus for 1 h. Subsequently, BHK-21-hACE2 cells were seeded into the wells of a 96-well plate (5 × 104 cells/well) and then infected with virus-serum samples for 12 h. The infected cells with GFP fluorescence (GFP expressing) was imaged by Opera Phenix or Operetta CLS equipment (PerkinElmer) and analyzed using the Columbus system (PerkinElmer). In our assay, we controled the number of GFP-expressing cells at around 3000–5000 in the no serum added wells. The neutralizing potency of the sera was defined as the reduction (%) in the number of GFP-positive cells following mAb/serum treatment as compared with the no serum added negative-controlls. The neutralization titer of each sample was expressed as the maximum dilution fold required to achieve infection inhibition by 50 %(ID50). The calculation of ID50 is based on the non-linear fitting (variable slope) using GraphPad Prism 8.
3. Results
3.1. Enhanced expression of the spike protein in mammalian cells
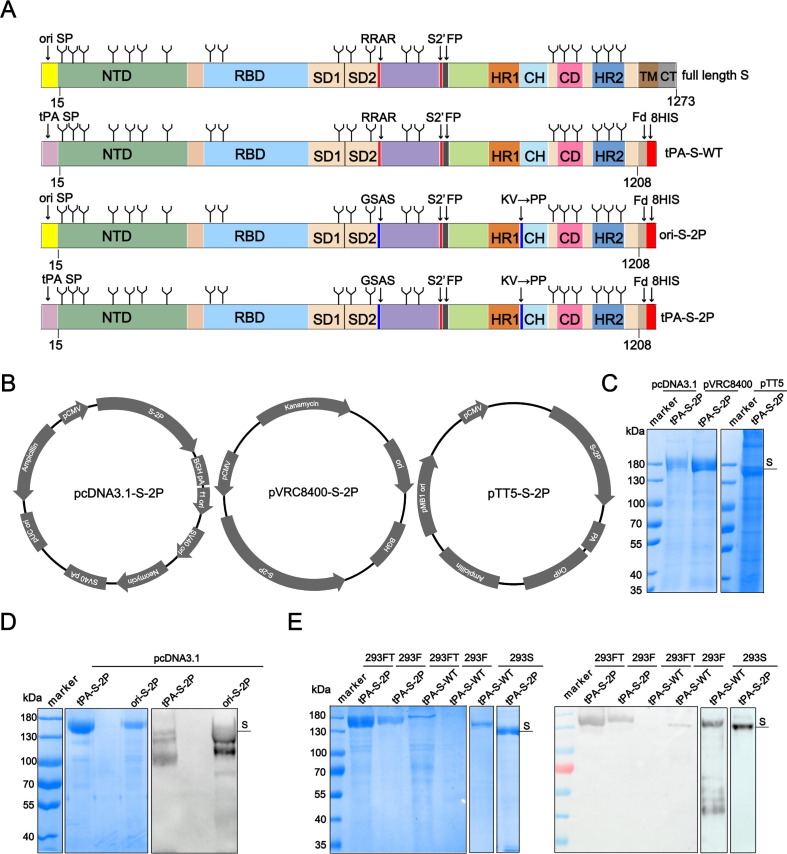
The full-length SARS-CoV-2 S protein of the Wuhan-Hu-1 strain (Genbank accession no. NC_045512.2) contains 1,273 amino acids (aa) ( Fig. 1 A) [39]. To facilitate the expression of soluble S protein, tPA-S-WT, Ori-S-2P and tPA-S-2P constructs with only the ectodomain (1208 aa) of S were designed ( Fig. 1 A). To enhance the expression of the S protein in 293F cells in vitro, we first compared the expression level of tPA-S-2P using three different vectors: pcDNA3.1, pVRC8400, and pTT5 ( Fig. 1 B). The S-2P was resolved at a molecular weight (m.w.) of ∼ 180 kDa in SDS-PAGE ( Fig. 1 C). The yield of tPA-S-2P (2.5 mg/L) using the pCDNA3.1 vector was higher than that expressed using the other two vectors, and showed about 90 % purity after Ni-metal affinity chromatography ( Fig. 1 C). For pVRC8400 and pTT5 vectors, tPA-S-2P yield was much lower (0.5 mg/L), with lower purity. We thus used pcDNA3.1 to compare the signal peptide in terms of the secretion of the S protein in subsequent assays. We found the yield of tPA-S-2P to be 5-times higher than that of ori-S-2P, suggesting that the tPA signal peptide more effectively promoted the secretion of the protein than the original signal peptide ( Fig. 1 D). Taken together, tPA-S-2P coupled with the pcDNA3.1 vector showed the best performance for S protein production in this study.
Fig. 1.
Construct design and expression optimization of the SARS CoV-2 Spike (S) protein. (A) Linear representation of the S protein primary structure and construct design. Full-length (FL) S protein, extracellular S protein with tPA signal peptide (tPA-S-WT) and extracellular S-2P protein with tPA (tPA-S-2P) or the original signal peptide (Ori-S-2P). NTD, N-terminal domain; RBD, receptor binding domain; SD1, subdomain 1, SD2, subdomain 2; FP Fusion peptide, HR1, heptad repeat 1, CH, central helix, CD, connector domain, HR2, heptad repeat 2; TM, transmembrane domain; tPA, tissue plasminogen activator; CT, cytoplasmic tail. (B) Construct map of pcDNA3.1, pVRC8400 and pTT5 vectors that were inserted with the tPA-S-2P gene at the multiple cloning site. (C) SDS-PAGE analysis of Ni-Sepharose excel-purified S-2P protein produced with different vectors. (D) SDS-PAGE and western blot analysis of purified S-2P from two constructs with different signal peptides. (E) SDS-PAGE and western blot analysis of purified tPA-S-2P and tPA-S-WT produced via three types of 293 cell lines. To make the western blot image more clearly, we down-adjusted the Gray coefficient of exposure parameter. Human convalescent serum was used for immunoblotting.
We next used this vector/signal peptide combination to express tPA-S-2P and tPA-S-WT in three cell types: 293FT, 293F and 293S GnTI-. The tPA-S-2P yields produced with 293FT and 293S cells were lower than that produced with 293F cells. Unexpectedly, the tPA-S-WT protein yield was too low to obtain high-purity protein from the 293FT and 293F cell lines, but a small amount of pure tPA-S-WT could be harvested from 293S GnTI- cells (0.2 mg/L) ( Fig. 1 E). In SDS-PAGE, we noted that the m.w. of tPA-S-2P from 293S GnTI- cells was lower than that from 293F cells; this difference might be due to the defective glycosylation modification of the cell line [40] ( Fig. 1 E). Considering these findings, we finally chose to express tPA-S-2P in 293F (293F S-2P) and 293S (293S S-2P) cells in subsequent studies.
3.2. Physiochemical properties of SARS-CoV-2 spike proteins
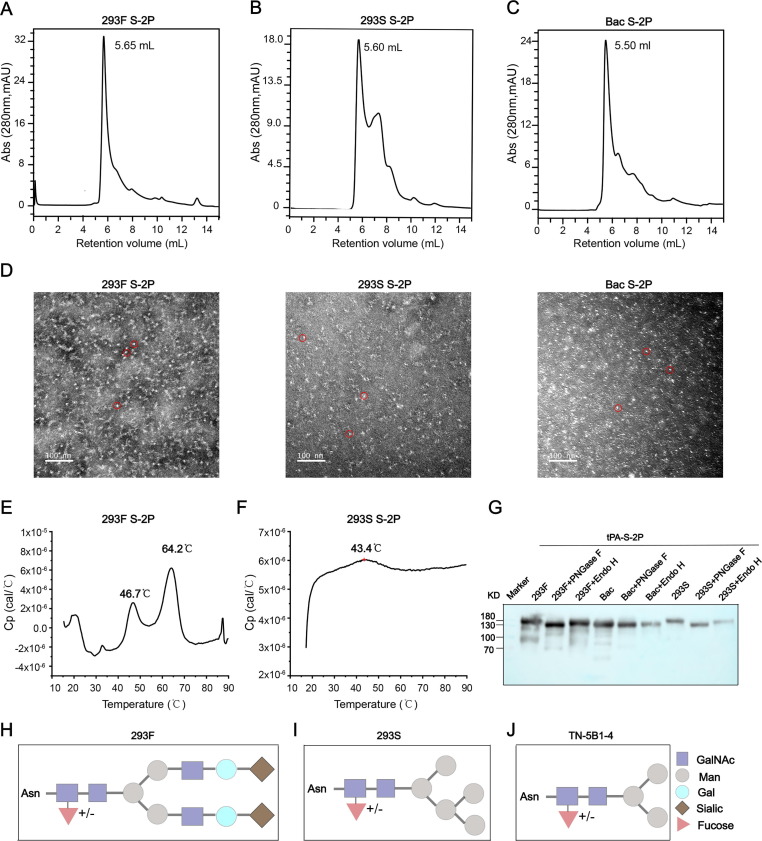
We next investigated the physiochemical properties of the recombinant S proteins from the two 293 cell lines, and compared the expression profiles with that of S-2P from insect cells (Bac S-2P) described in our previous study [36]. First, purified S-2P proteins were applied to high-pressure size-exclusion chromatography (HPSEC) analysis to evaluate their homogeneity. 293F S-2P and 293S S-2P showed major peaks in the HPSEC profile at elution volumes of 5.65 mL or 5.60 mL, respectively ( Fig. 2 A, 2B); these values were similar to Bac S-2P, which eluted at 5.5 mL ( Fig. 2 C). We previously verified Bac S-2P to assume a trimer morphology [36], and the same morphology was confirmed for these S-2P proteins using negative-stain electron microscopy (EM) imaging ( Fig. 2 D). The thermostability of the three proteins was then analyzed using differential scanning calorimetry (DSC). The DSC profile of 293F S-2P showed two peaks at the thermal denaturation midpoints (Tm) of 44.5℃ and 64.2℃ ( Fig. 2 E). The main peak at 64.2℃ was nearly consistent with that reported for Bac S-2P (64.1℃) [36], whereas a wide peak at 43.4℃ was detected for 293S S-2P ( Fig. 2 F).These results indicate that Bac S-2P and 293F S-2P exhibit higher thermostabilities than 293S S-2P.
Fig. 2.
Characterization of purified SARS-CoV-2 spike (S) protein. (A-C) 293F S-2P, 293S S-2P, and Bac S-2P proteins were analysed by high-pressure size-exclusion chromatography (HPSEC). (D) Negative-stain EM micrographs of purified 293F S-2P, 293S S-2P, and Bac S-2P proteins. (E-F) Differential scanning calorimetry (DSC) profiles of 293F S-2P and 293S S-2P. Two thermal denaturation midpoints (Tm) are shown for 293F S-2P at 46.7℃ (Tm1) and 64.2℃ (Tm2), but just one weak peak at 43.4℃ for 293S S-2P. (G) Western blot of 293F-S-2P, 293S-S-2P, Bac-S-2P proteins following treatment with endo-H or PNGase F. Untreated samples act as the control. (H-J) The glycosylation anatomy of proteins produced by 293F cell, 293S GnTI- and Tn-5B1-4 cells (insect cell). Different glycan types are labelled with different colours and shapes.
Deglycosylation analysis was next used to investigate the extent of glycosylation on these spike proteins. Before deglycosylation treatment, we noted a discrepancy in the m.w.: 293F S-2P, 293S S-2P, and Bac S-2P measured at ∼ 200 kDa, ∼180 kDa, and ∼ 170 kDa, respectively ( Fig. 2 G). Proteins were digested with PNGase F or endo-H. PNGase F treatment led to a discernible reduction in the m.w. of all three proteins, with decreases of ∼ 50 kDa, ∼40 kDa, ∼30 kDa for 293F S-2P, 293S S-2P, and Bac S-2P, respectively. In contrast, Endo H treatment led to m.w. reductions of ∼ 20 kDa, ∼30 kDa, ∼10 kDa for 293F S-2P, 293S S-2P, and Bac S-2P, respectively ( Fig. 2 G).
PNGase F cleaves between the innermost GlcNAc and Asn residues and releases almost all the N-glycans on the glycoprotein, this result indicated high levels of glycosylation in 293F S-2P, moderate levels in 293S S-2P, and lower levels in Bac S-2P. In contrast, Endo H treatment led to m.w. reductions of ∼ 20 kDa, ∼30 kDa, ∼10 kDa for 293F S-2P, 293S S-2P, and Bac S-2P, respectively ( Fig. 2 G). The lower m.w. reductions of 293F S-2P than 293S S-2P might because endo-H cleaves between two N-acetylglucosamine residues of high-mannose and some hybrid-type but not complex-type N-glycans, resulting in one N-acetylglucosamine residue on the Asn [41]. The m.w. reduction is higher treated by PNGase F but lower treated by Endo H for 293F S-2P compared to 293S S-2P, suggested 293F S-2P possess higher level of complex-type N-glycans than 293S S-2P. These results were consistent with the glycosylation patterns of these cell lines. Specifically, 293F cells produce proteins with fully, complexed glycans ( Fig. 2 H), whereas simple Man5GlcNAc2Asn intermediate-type glycans are present on those produced by 293S GnTI- cells ( Fig. 2 I); this difference is likely due to a defect in the activity of N-acetylglucosaminyl transferase I, a critical enzyme for hybrid and complexed N-linked glycosylation [40], [42]. In contrast, insect cells (TN-5B1-4) appear to produce proteins with terminal mannose glycans but without terminal sialylated N-glycans ( Fig. 2 J) [43], [44], [45], [46].
3.3. Antigenicity and receptor binding of SARS-CoV-2 spike protein
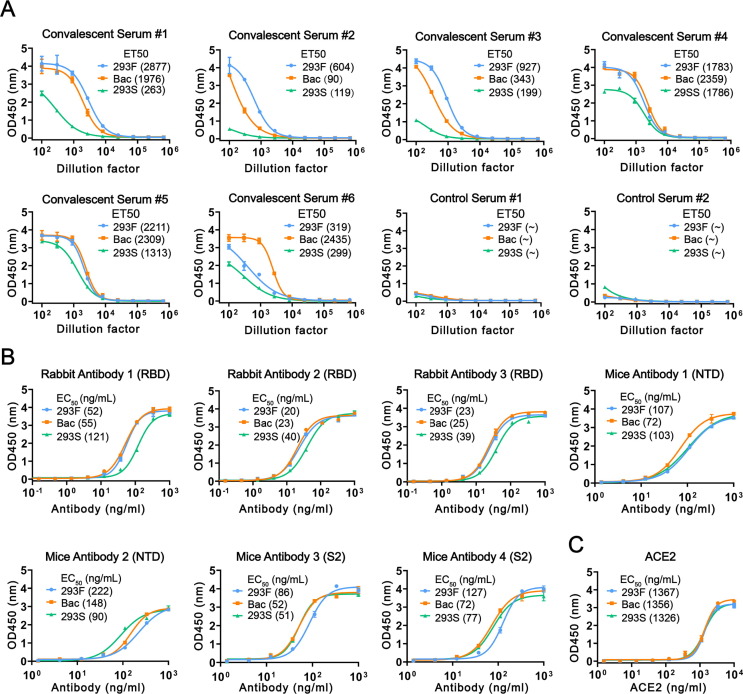
We next evaluated the antigenicity of these S-2P proteins with ELISA using a panel of six COVID-19 convalescent human sera and antibodies that target different domains of the S protein. The reaction titres are represented as half-effective titres (ET50). 293F S-2P showed high reactivity against five of the six sera samples (#1, #2, #3, #4, #5) ( Fig. 3 A). In contrast, Bac S-2P showed high reactivity against only three sera (#4, # 5, #6) and slightly lower reactivity for the other three samples (#1, #2, #3). Notably, 293S S-2P showed poor reactivity to all six sera. There were no detectable reactions for the healthy individual’s sera (Control, Fig. 3 A). We further tested the binding reactivities of these three proteins using rabbit or mouse monoclonal antibodies targeting the receptor-binding domain (RBD), the N-terminal domain (NTD), or the S2 subunit. These mAbs all targeted the epitopes on monomer. Three RBD-targeting rabbit antibodies showed comparable reactivities toward 293F S-2P and Bac S-2P, but slightly lower reactivity toward 293S S-2P. Regarding the mouse monoclonal antibodies, one of the NTD-targeting antibodies showed higher reactivity toward 293S S-2P over 293F and Bac S-2P, while there was no significant difference in binding for the two S2-targeted mouse antibodies among the 3 proteins ( Fig. 3 B). We surmised that these differences in reactivities toward convalescent sera and antibodies might be contributed by different glycans on the RBD and the NTD.
Fig. 3.
Antigenicity and receptor binding of S-2P proteins. (A) Reactivities of S-2P proteins against COVID-19 convalescent human sera (#1-#6) and control sera (#1-#2), ∼ denotes not detected. (B) Reactivities of S-2P proteins against three rabbit monoclonal antibodies targeting the receptor-binding domain (RBD), two mouse monoclonal antibodies targeting the N-terminal domain (NTD) and two antibodies targeting the S2 subunit. (C) Reactivities of S-2P proteins to the ACE2 receptor.
Because glycans can play a critical role in the specific recognition of virus and receptors, we further evaluated the binding ability of three S-2P proteins to the ACE2 receptor using ELISA. The fully glycosylated 293F S-2P showed a similar reactivity to ACE2 as the partially glycosylated Bac S-2P, with EC50 values ranging from ∼ 1,326–1,367 ng/mL ( Fig. 3 C). This suggests that the reactivity of ACE2 to the three proteins might not be affected by glycosylation near the receptor-binding motif (RBM).
3.4. Immunogenicity of glycosylated SARS CoV-2 spike proteins in mice
We next sought to evaluate the immunogenicity of the differentially glycosylated S-2P proteins in BALB/C mice. In addition, the efficiency of different adjuvants (aluminium and Freund’s adjuvant), dosage (5 μg and 50 μg) to stimulating the immune responses were evaluated in our study. The overall immunization schedule and animal groupings were presented in the methods and Table S1 ( Fig. 4 A). End-point ELISA and cell-based vesicular stomatitis virus (VSV) pseudovirus-type neutralization assays were used to determine the specific IgG titres and nAb titres [38]. For the Bac S-2P groups, the IgG titres with FJ (4–5-log) was higher than that with AL (3–3.5-log). In both the AL and FJ combined groups, the IgG titres increased dramatically at week 3 following the second immunization, with IgG titres ranging from 5-log to 6.5-log which was higher than the other S-2P immunized groups (group 3–8). In addition, there was no significant increase in IgG antibody titres for 293F S-2P at the higher (50 μg) dosage ( Fig. 4 B). Seroconversion of the nAbs occurred at one week after the first vaccination (week 2) in the high-dosage 293F S-2P groups (group 1–2) and in the combined immunization groups (group 9–10). For 293 S-2P with AL, the nAbs titres were significantly increased to 3–4.5-log after second immunization (week 3), which was maintained until after the third immunization (week 5) ( Fig. 4 C). For Bac S-2P with AL (Group 5), the nAbs titres climbed from 2-log to 3.5-log after the third immunization, similar to our previous results [36]. The combined S-2P immunization protocol (Group 9–10) elicited the slightly higher nAbs titres, with up to 4.5-log in week 5. For all proteins at the low dosage, nAb titres were lower in FJ than in AL, indicating that AL S-2P is immunogenic in a prime-boost-boost immunization regimen. We also found that the nAb titres were dose-dependent, with a dramatic increase in titres to 5-log in the 50 μg 293F S-2P group with FJ (Group2) at week 3 ( Fig. 4 C). Comparatively, the neutralizing antibody responses induced by the combined immunization formulated with AL or FJ (GMT[AL] = 31,347; GMT[FJ] = 8,184) were 2.5 ∼ 10-times more potent than the convalescent sera in our previous study (GMT = 2,962) [38]. These results suggest that the trimer-stabilized conformation of S-2P from 293 cells may provide higher immunogenicity than that from insect cells; meanwhile, the combined immunization regiment with spike proteins with distinct glycosylation extent might provide an effective strategy to induce higher level of nAbs against SARS-CoV-2.
4. Discussion
The S protein is the principal target for the development of vaccines and/or other therapeutic drugs against SARS-CoV-2 and the ensuing COVID-19 disease. Potent nAbs induced by the S protein can be exploited in the treatment or prevention of SARS-CoV-2 infection. Optimizing the preparation of the recombinant S protein is a critical step in vaccine production. Previous studies have shown the S protein to be highly glycosylated [26], and that it may display functional epitopes. However, sometimes dense glycan presentation can mask the immunogenic epitopes and endow the virus with an opportunity to evade the immune system [47]. While deglycosylation steps or the use of different cells to express the S protein are ways to address this obstacle, it is essential to balance those factors that affect the presentation of native epitopes and those that elicit neutralizing or non-neutralizing antibodies.
COVID-19 vaccine research has explored the use of recombinant proteins derived from different cells; for instance, S-trimer expressed in mammalian cells by Clover Biopharmaceuticals and NVX-CoV2373 prepared in insect cells by Novavax [48]. Here, we explored several factors—signal peptides, vectors, and host cells—that could be used to enhance S-2P protein expression. Remarkably, we found that a codon-optimized S-2P cloned into pcDNA3.1 vector with the tPA signal peptide could improve the yield by up to 5-fold in 293 cells as compared with the original construct design in this study. Furthermore, we showed that S proteins produced in different systems have vastly different properties: 293F S-2P showed the highest molecular weight, followed by 293S S-2P, and finally, Bac S-2P, indicating significant differences in glycosylation modifications on the S-2P protein. In addition, a small peak around the 7.5 mL retention volume of 293S S-2P was shown might be due to the lower thermostabilities revealed by the wide peak of Tm. Thus, the choice of host cell for recombinant S protein production should be carefully considered for yield and degree of glycosylation.
Interestingly, these proteins react differently to various monoclonal antibodies targeting different domains. Most of the serums showed weak reactivity to the partially glycosylated 293S S-2P and Bac S-2P proteins as compared with 293F S-2P; this may indicate that some of the antibodies raised after infection are glycan-dependent, or that some native epitopes might be absent in the partially glycosylated trimers. Therefore, these S-2P proteins with various glycosylation profiles could be used to detect or screen for antibodies that target glycans. Furthermore, the findings from our immunogenicity analysis suggested that S-2P proteins from mammalian expression systems induced a more potent nAb response than did proteins produced from the baculovirus-insect cell expression system. Yet, it is worth noting that whereas the 293S S-2P protein showed poor reactivity with patient sera, it induced a more potent nAb response in mice than did Bac S-2P. TN-5B1-4 cells are known to produce proteins with fucosylated N-glycans but no human-type sialylated N-glycans; this might influence the immunogenicity of Bac-S-2P. Other points to consider could be dosage optimization or the use of more effective adjuvants to elicit a higher immune response in future studies.
The persistent spread of SARS-CoV-2 around the world has led to a rapid increase in the appearance of variant strains, such as the B.1.1.7 (Alpha), B.1.135 (Beta), P.1 (Gamma) and B.1.167.2 (Delta) [3], [5], [6], [7], [10]. These variants contain mutations in different domains of the S protein, particularly within the RBD [5]. These mutations have also resulted in a decline in the efficacy of the current suite of vaccines and nAbs [3], [10]. Additional mutations will likely continue to affect vaccine efficacy. We found that priming with 293S S-2P and then boosting with Bac S-2P and then 293F S-2P can elicit a higher nAb response. We propose that this immunization procedure—important in the production of nAbs targeting the conserved glycosylated epitopes in HIV [32], [34]—could be a potential strategy to reduce the impact of virus mutations in COVID-19 vaccine development. Meanwhile, there are limitations in this study. As we know, many aspects of mammalian biological systems, such as the immune system, are species-specific. There are still differences in the immune responses between mice and human, and our finding in naïve mice should be exercised when transplanting into humans. Whether immunization strategies with different glycosylation forms can reach the same conclusions in human experiments remains a challenge.
Currently, mRNA vaccines (Comirnaty, Spikevax) or viral vector vaccines (Vaxzevria) against SARS-CoV-2 have proven effective in controlling the COVID-19 pandemic virus. mRNA or viral vector vaccines introduce genetic material encoding antigenic proteins for host expression. Because these proteins undergo post-translational modifications in the host cell, the glycosylation of viral proteins may be identical or highly similar to that of virion surface proteins. However, it is not clear whether the glycosylation produced by nucleic acid vaccine immunization is related to the expression of virus infection. To achieve proper glycosylation matching, mRNA vaccines may need to be expressed in the same cell type that the virus infects at the time of infection, since the glycosylation of proteins may differ when expressed in different cell types of the same species. This may be due to the highly complex glycosylation pathway involving many different glycosyltransferases, donor sugars, and chaperone proteins, which can exhibit varying degrees of glycosylation modification in different cell types and cellular states [49], [50]. mRNA vaccines can be rapidly produced on a large scale in response to a pandemic, which is more advantageous than recombinant protein vaccines. However, the differences in glycosylation between nucleic acid vaccines and recombinant protein vaccines and the contribution to induce immune responses are still worthy of continued research.
5. Conclusions
The ongoing COVID-19 pandemic continues to pose a serious threat to public health. Effective vaccine and therapeutic options are thus urgently needed. Glycans covered on the surface of the spike protein may be critical in promoting immune escape and may hinder the development of an effective broad-spectrum vaccine. Our results outline the characteristics of the SARS-CoV-2 S protein with different glycosylation modifications and demonstrate the importance of the glycosylation patterns in the antigenicity and immunogenicity of the spike protein. We further suggest that a combined immunization strategy may be beneficial for the development of a more potent vaccine.
Funding sources
This work was supported by grants from the Key Research Program of the Ministry of Science and Technology (Grant No. 2020YFC0842600) and National Natural Science Foundation (Grant No. 82001756, 82041038), The funders had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Ethical statement
All mouse experiments were performed strictly according to the animal ethics guidelines and protocols set by the Xiamen University Laboratory Animal Management Ethics Committee.
Author contribution
Conceptualization, Y.G.; Investigation, T.D., G.C., Y.Z., L.X., Y.L., H.S., H.Z., Q.F., J.H., D.W., S.G., Shaoyong L.; Data analysis, T.D., T.L., Y.W., T.Z., Y.C., Q.Y., Q.Zheng., H.Y., Q.Zhao, J.Z., Shaowei L., N.X. and Y.G.; Writing—original draft preparation, T.D., T.L.; Writing—review and editing, T.D., Y.G. and Shaowei L.; Supervision, Shaowei L., N.X. and Y.G.; Funding acquisition, Shaowei L. and Y.G.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.vaccine.2022.09.057.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Petrosillo N., Viceconte G., Ergonul O., Ippolito G., Petersen E. COVID-19, SARS and MERS: are they closely related? Clin Microbiol Infect. 2020;26(6):729–734. doi: 10.1016/j.cmi.2020.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhuang Z., Lai X., Sun J., et al. Mapping and role of T cell response in SARS-CoV-2-infected mice. J Exp Med. 2021;218(4) doi: 10.1084/jem.20202187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann M., Arora P., Groß R., Seidel A., Hörnich B.F., Hahn A.S., et al. SARS-CoV-2 variants B.1.351 and P.1 escape from neutralizing antibodies. Cell. 2021;184(9):2384–2393.e12. doi: 10.1016/j.cell.2021.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q., Nie J., Wu J., Zhang L., Ding R., Wang H., et al. SARS-CoV-2 501Y.V2 variants lack higher infectivity but do have immune escape. Cell. 2021;184(9):2362–2371.e9. doi: 10.1016/j.cell.2021.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Q., Wu J., Nie J., Zhang L., Hao H., Liu S., et al. The impact of mutations in SARS-CoV-2 spike on viral infectivity and antigenicity. Cell. 2020;182(5):1284–1294.e9. doi: 10.1016/j.cell.2020.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(4):812–827.e19. doi: 10.1016/j.cell.2020.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tegally H., Wilkinson E., Lessells R.J., Giandhari J., Pillay S., Msomi N., et al. Sixteen novel lineages of SARS-CoV-2 in South Africa. Nat Med. 2021;27(3):440–446. doi: 10.1038/s41591-021-01255-3. [DOI] [PubMed] [Google Scholar]
- 8.Lopez Bernal J., Andrews N., Gower C., Gallagher E., Simmons R., Thelwall S., et al. Effectiveness of Covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. 2021;385(7):585–594. doi: 10.1056/NEJMoa2108891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Abu-Raddad L.J., Chemaitelly H., Butt A.A. Effectiveness of the BNT162b2 Covid-19 vaccine against the B.1.1.7 and B.1.351 variants. N Engl J Med. 2021;385(2):187–189. doi: 10.1056/NEJMc2104974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang P., Nair M.S., Liu L., et al. Antibody resistance of SARS-CoV-2 variants B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 11.S. Cele, L. Jackson, D.S. Khoury, et al., Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization, Nature (2021). [DOI] [PMC free article] [PubMed]
- 12.Dai L., Zheng T., Xu K., Han Y., Xu L., Huang E., et al. A universal design of betacoronavirus vaccines against COVID-19, MERS, and SARS. Cell. 2020;182(3):722–733.e11. doi: 10.1016/j.cell.2020.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181(2):271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yan R., Zhang Y., Li Y., Xia L., Guo Y., Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020;367(6485):1444–1448. doi: 10.1126/science.abb2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuhn J.H., Li W., Choe H., et al. Angiotensin-converting enzyme 2: a functional receptor for SARS coronavirus. Cell Mol Life Sci. 2004;61(21):2738–2743. doi: 10.1007/s00018-004-4242-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuchun N., Guangwen W., Xuanling S., Hong Z., Yan Q., Zhongping H.e., et al. Neutralizing antibodies in patients with severe acute respiratory syndrome-associated coronavirus infection. J Infect Dis. 2004;190(6):1119–1126. doi: 10.1086/423286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang B., Liu S., Tan T., et al. Treatment with convalescent plasma for critically ill patients with SARS-CoV-2 infection. Chest. 2020;158(1):e9–e13. doi: 10.1016/j.chest.2020.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drosten C., Günther S., Preiser W., van der Werf S., Brodt H.-R., Becker S., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Engl J Med. 2003;348(20):1967–1976. doi: 10.1056/NEJMoa030747. [DOI] [PubMed] [Google Scholar]
- 19.Ahmadivand A., Gerislioglu B., Ramezani Z., Kaushik A., Manickam P., Ghoreishi S.A. Functionalized terahertz plasmonic metasensors: femtomolar-level detection of SARS-CoV-2 spike proteins. Biosens Bioelectron. 2021;177 doi: 10.1016/j.bios.2021.112971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosch B.J., van der Zee R., de Haan C.A.M., Rottier P.J.M. The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol. 2003;77(16):8801–8811. doi: 10.1128/JVI.77.16.8801-8811.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia S., Yan L., Xu W., Agrawal A.S., Algaissi A., Tseng C.-T., et al. A pan-coronavirus fusion inhibitor targeting the HR1 domain of human coronavirus spike. Sci Adv. 2019;5(4):eaav4580. doi: 10.1126/sciadv.aav4580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kirchdoerfer R.N., Cottrell C.A., Wang N., Pallesen J., Yassine H.M., Turner H.L., et al. Pre-fusion structure of a human coronavirus spike protein. Nature. 2016;531(7592):118–121. doi: 10.1038/nature17200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walls A.C., Park Y.-J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181(2):281–292.e6. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Andersen K.G., Rambaut A., Lipkin W.I., Holmes E.C., Garry R.F. The proximal origin of SARS-CoV-2. Nat Med. 2020;26(4):450–452. doi: 10.1038/s41591-020-0820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin X., Xu K., Jiang P., Lian J., Hao S., Yao H., et al. Virus strain from a mild COVID-19 patient in Hangzhou represents a new trend in SARS-CoV-2 evolution potentially related to Furin cleavage site. Emerg Microbes Infect. 2020;9(1):1474–1488. doi: 10.1080/22221751.2020.1781551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalziel M., Crispin M., Scanlan C.N., Zitzmann N., Dwek R.A. Emerging principles for the therapeutic exploitation of glycosylation. Science. 2014;343(6166):37–45. doi: 10.1126/science.1235681. [DOI] [PubMed] [Google Scholar]
- 28.Bagdonaite I., Wandall H.H. Global aspects of viral glycosylation. Glycobiology. 2018;28(7):443–467. doi: 10.1093/glycob/cwy021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kong L., Lee J.H., Doores K.J., Murin C.D., Julien J.-P., McBride R., et al. Supersite of immune vulnerability on the glycosylated face of HIV-1 envelope glycoprotein gp120. Nat Struct Mol Biol. 2013;20(7):796–803. doi: 10.1038/nsmb.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Go E.P., Ding H., Zhang S., Ringe R.P., Nicely N., Hua D., et al. Glycosylation benchmark profile for HIV-1 envelope glycoprotein production based on eleven env trimers. J Virol. 2017;91(9) doi: 10.1128/JVI.02428-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., et al. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583(7815):290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 32.Dubrovskaya V., Guenaga J., de Val N., Wilson R., Feng Y.u., Movsesyan A., et al. Targeted N-glycan deletion at the receptor-binding site retains HIV Env NFL trimer integrity and accelerates the elicited antibody response. PLoS Pathog. 2017;13(9) doi: 10.1371/journal.ppat.1006614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingale J., Stano A., Guenaga J., Sharma S., Nemazee D., Zwick M., et al. High-density array of well-ordered HIV-1 spikes on synthetic liposomal nanoparticles efficiently activate B cells. Cell Rep. 2016;15(9):1986–1999. doi: 10.1016/j.celrep.2016.04.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dubrovskaya V., Tran K., Ozorowski G., Guenaga J., Wilson R., Bale S., et al. Vaccination with glycan-modified HIV NFL envelope trimer-liposomes elicits broadly neutralizing antibodies to multiple sites of vulnerability. Immunity. 2019;51(5):915–929.e7. doi: 10.1016/j.immuni.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung J.Y., Thone M.N., Kwon Y.J. COVID-19 vaccines: the status and perspectives in delivery points of view. Adv Drug Deliv Rev. 2020;170:1–25. doi: 10.1016/j.addr.2020.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li T., Zheng Q., Yu H., Wu D., Xue W., Xiong H., et al. SARS-CoV-2 spike produced in insect cells elicits high neutralization titres in non-human primates. Emerg Microbes Infect. 2020;9(1):2076–2090. doi: 10.1080/22221751.2020.1821583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanders R.W., Derking R., Cupo A., Julien J.-P., Yasmeen A., de Val N., et al. A next-generation cleaved, soluble HIV-1 Env trimer, BG505 SOSIP.664 gp140, expresses multiple epitopes for broadly neutralizing but not non-neutralizing antibodies. PLoS Pathog. 2013;9(9) doi: 10.1371/journal.ppat.1003618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xiong H.-L., Wu Y.-T., Cao J.-L., Yang R., Liu Y.-X., Ma J., et al. Robust neutralization assay based on SARS-CoV-2 S-protein-bearing vesicular stomatitis virus (VSV) pseudovirus and ACE2-overexpressing BHK21 cells. Emerg Microbes Infect. 2020;9(1):2105–2113. doi: 10.1080/22221751.2020.1815589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu F., Zhao S.u., Yu B., Chen Y.-M., Wang W., Song Z.-G., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579(7798):265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reeves P.J., Callewaert N., Contreras R., Khorana H.G. Structure and function in rhodopsin: high-level expression of rhodopsin with restricted and homogeneous N-glycosylation by a tetracycline inducible N-acetylglucosaminyltransferase I-negative HEK293S stable mammalian cell line. PNAS. 2002;99(21):13419–13424. doi: 10.1073/pnas.212519299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tarentino A.L., Gomez C.M., Plummer T.H., Jr. Deglycosylation of asparagine-linked glycans by peptide:N-glycosidase F. Biochemistry. 1985;24(7):4665–4671. doi: 10.1021/bi00338a028. [DOI] [PubMed] [Google Scholar]
- 42.Stanley P., Chaney W. Control of carbohydrate processing the leclA CHO mutation results in partial loss of N-acetylglucosaminyltransferase I activity. Mol Cell Biol 1985;5(6):1204–11. [DOI] [PMC free article] [PubMed]
- 43.Mabashi-Asazuma H., Kuo C.W., Khoo K.H., et al. A novel baculovirus vector for the production of nonfucosylated recombinant glycoproteins in insect cells. Glycobiology. 2014;24(3):325–340. doi: 10.1093/glycob/cwt161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mabashi-Asazuma H., Shi X., Geisler C., et al. Impact of a human CMP-sialic acid transporter on recombinant glycoprotein sialylation in glycoengineered insect cells. Glycobiology. 2013;23(2):199–210. doi: 10.1093/glycob/cws143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi X., Jarvis D.L. Protein N-glycosylation in the baculovirus-insect cell system. Curr Drug Targets. 2007;8(10):1116–1125. doi: 10.2174/138945007782151360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jarvis D.L. Developing baculovirus-insect cell expression systems for humanized recombinant glycoprotein production. Virology. 2003;310(1):7. doi: 10.1016/s0042-6822(03)00120-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ferreira R.C., Grant O.C., Moyo T., et al. Structural rearrangements maintain the glycan shield of an HIV-1 envelope trimer after the loss of a glycan. Sci Rep. 2018;8(1):15031. doi: 10.1038/s41598-018-33390-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang J., Wang W., Chen Z., et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature. 2020;586(7830):572–577. doi: 10.1038/s41586-020-2599-8. [DOI] [PubMed] [Google Scholar]
- 49.Ozdilek A., Avci F.Y. Glycosylation as a key parameter in the design of nucleic acid vaccines. Curr Opin Struct Biol. 2022;73 doi: 10.1016/j.sbi.2022.102348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moremen K.W., Tiemeyer M., Nairn A.V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat Rev Mol Cell Biol. 2012;13(7):448–462. doi: 10.1038/nrm3383. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.