Abstract
Introduction
Ankylosing spondylitis (AS) is a chronic progressive inflammatory disease of the spine and its affiliated tissues. AS mainly affects the axial bone, sacroiliac joint, hip joint, spinal facet, and adjacent ligaments. We used machine learning (ML) methods to construct diagnostic models based on blood routine examination, liver function test, and kidney function test of patients with AS. This method will help clinicians enhance diagnostic efficiency and allow patients to receive systematic treatment as soon as possible.
Methods
We consecutively screened 348 patients with AS through complete blood routine examination, liver function test, and kidney function test at the First Affiliated Hospital of Guangxi Medical University according to the modified New York criteria (diagnostic criteria for AS). By using random sampling, the patients were randomly divided into training and validation cohorts. The training cohort included 258 patients with AS and 247 patients without AS, and the validation cohort included 90 patients with AS and 113 patients without AS. We used three ML methods (LASSO, random forest, and support vector machine recursive feature elimination) to screen feature variables and then took the intersection to obtain the prediction model. In addition, we used the prediction model on the validation cohort.
Results
Seven factors—erythrocyte sedimentation rate (ESR), red blood cell count (RBC), mean platelet volume (MPV), albumin (ALB), aspartate aminotransferase (AST), and creatinine (Cr)—were selected to construct a nomogram diagnostic model through ML. In the training cohort, the C value and area under the curve (AUC) value of this nomogram was 0.878 and 0.8779462, respectively. The C value and AUC value of the nomogram in the validation cohort was 0.823 and 0.8232055, respectively. Calibration curves in the training and validation cohorts showed satisfactory agreement between nomogram predictions and actual probabilities. The decision curve analysis showed that the nonadherence nomogram was clinically useful when intervention was decided at the nonadherence possibility threshold of 1%.
Conclusion
Our ML model can satisfactorily predict patients with AS. This nomogram can help orthopedic surgeons devise more personalized and rational clinical strategies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40744-022-00481-6.
Keywords: Ankylosing spondylitis, Machine learning algorithms, Prediction model, Nomogram, Diagnosis
Plain Language Summary
AS is a chronic progressive inflammatory disease of the spine and its affiliated tissues. AS starts gradually, and its early symptoms are mild. Some hospitals lack HLA-B27 and related imaging instruments to assist in the diagnosis of AS. There are relatively few studies on liver function and kidney function of patients with AS. We used ML methods to construct diagnostic models. Our model can satisfactorily predict patients with AS. This diagnostic model can help orthopedic surgeons devise more personalized and rational clinical strategies.
Supplementary Information
The online version contains supplementary material available at 10.1007/s40744-022-00481-6.
Key Summary Points
| We consecutively screened 348 patients with AS through complete blood routine examinations, liver function tests, and kidney function tests. |
| We used three ML methods [LASSO, random forest, and support vector machine recursive feature elimination (SVM-RFE)] to screen feature variables and then took the intersection to obtain the prediction model. In addition, we used the prediction model on the validation cohort. |
| Our diagnostic models can help orthopedic surgeons devise more personalized and rational clinical strategies. |
Introduction
Ankylosing spondylitis (AS) is a chronic progressive inflammatory disease of the spine and its affiliated tissues [1]. AS mainly affects the axial bone, sacroiliac joint, hip joint, spinal facet, and adjacent ligaments [2]. The main clinical manifestations are pain in the waist, sacroiliac joints, and hip and progressive joint stiffness, resulting in joint mobility limitation and joint deformity [3].
Millions of people are diagnosed with AS each year, but the cause of AS remains unknown. The ratio of male to female patients with AS is approximately 3:1 [4], and the incidence rate among relatives of patients is 20 times higher than that in the general population [5]. However, the genetic heritability of AS is influenced by variations in the MHC region, accounting for 40–50% of the total genetic risk of developing the disease, specifically with positivity for HLA-B27 [3].
Machine learning (ML) is a scientific discipline focusing on how computers learn using data. It is the intersection of statistics, which learns relationships from data, and computer science, which emphasizes efficient computational algorithms. ML is now widely used in the study of clinically relevant data [6, 7]. Liang et al. used LASSO regression to find that the platelet-to-lymphocyte ratio could be an independent factor in diagnosing AS [8]. Zhang et al. used machine learning to predict the volumetric response of patients with acute kidney injury [9].
AS starts gradually, and its early symptoms are mild. Patients with AS often have nephropathy and cardiovascular disease [10, 11]. Some hospitals lack HLA-B27 and related imaging instruments to assist in the diagnosis of AS. There are relatively few studies on liver function and kidney function of patients with AS. We used ML methods to construct diagnostic models based on blood routine examination, liver function test, and kidney function test of patients with AS to help clinicians enhance diagnostic efficiency and allow patients to receive systematic treatment as soon as possible.
Methods
Patients
Subjects volunteering for the study signed informed consent forms. The Ethics Committee of the First Affiliated Hospital of Guangxi Medical University approved this study, which adhered to the tenets of the Helsinki Declaration of 1964.
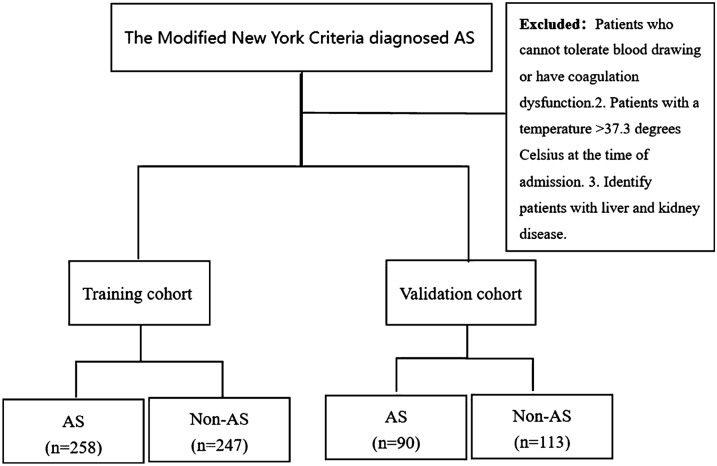
From 2012 to 2021, we consecutively screened 348 patients with AS through complete blood routine examination, liver function test, and kidney function test at the First Affiliated Hospital of Guangxi Medical University according to the modified New York criteria (diagnostic criteria for AS) [12]. Inclusion criteria: (1) patients with AS who met the Modified New York Criteria; (2) patients who had good compliance and no serious cardiovascular and cerebrovascular diseases; (3) patients who voluntarily accepted blood routine examinations, liver function tests, and kidney function tests. Exclusion criteria: (1) patients who cannot tolerate blood drawing or have coagulation dysfunction; (2) patients with a temperature > 37.3 °C at admission; (3) patients with liver and kidney disease .
A total of 360 patients without AS were recruited from among all the inpatients diagnosed with the other disease to complete the blood drawing test. Inclusion criteria: (1) patients clearly diagnosed with non-AS; (2) patients who had good compliance and no serious cardiovascular and cerebrovascular diseases; (3) patients who voluntarily accepted blood routine examinations, liver function tests, and kidney function tests. Exclusion criteria: (1) patients who cannot tolerate blood drawing or have coagulation dysfunction; (2) patients with a temperature > 37.3 °C at admission; (3) patients with liver and kidney disease .
By using random sampling, the patients were randomly divided into training and validation cohorts. The training cohort included 258 patients with AS and 247 patients without AS, whereas the validation cohort included 90 patients with AS and 113 patients without AS (Fig. 1).
Fig. 1.
Recruitment and screening
All clinical data were obtained from the Information System of the First Affiliated Hospital of Guangxi Medical University. The information of patients was identified by their ID number. Age, diagnosis, erythrocyte sedimentation rate (ESR), high-sensitivity C-reactive protein (hs-CRP), blood routine examination, liver function examination, and kidney function examination of all the patients were collected and statistically analyzed. Blood routine examination included white blood cell (WBC) count, red blood cell count (RBC), hemoglobin (HGB), hematocrit value (HCT), mean corpuscular volume (MCV), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), blood platelet count (BPC), mean platelet volume (MPV), platelet distribution width (PDW), absolute value of neutrophil (NEUT#), percentage of neutrophils (NEUT), absolute value of lymphocytes (LYM#), percentage of lymphocytes (LYM), monocyte absolute value (MONO#), percentage of monocytes (MONO), absolute value of eosinophils (ESO#), percentage of eosinophils (ESO), absolute value of basophils (BASO#), percentage of basophils (BASO), red cell distribution width (RDW), and thrombocytocrit (PCT). The liver function examination included total bilirubin (TBil), direct bilirubin (DBil), indirect bilirubin (IBil), DBil/IBil, total protein (TP), albumin (ALB), globulin (GLB), ALB/GLB ratio, gamma-glutamyl transpeptidase (GGT), total bile acid (TBA), aspartate aminotransferase (AST), alanine aminotransferase (ALT), AST/ALT, A-alkaline phosphatase (ALP), prealbumin (PAB), and cholinesterase (ChE). The kidney function examination included blood urea nitrogen (BUN), creatinine (Cr), uric acid (UA), bicarbonate radical (HCO), creatinine clearance rate (Ccr), and cysteine C (Cys-C). We tried to keep the data as complete as possible, and we excluded the very few patients who were missing before we did the statistical analysis.
Statistical Analysis
We used IBM SPSS Statistics 23 and R software (version 4.1.3; https://www.R-project.org) for data analysis. Student t-test was used to compare the mean of continuous variables between the two cohorts (i.e., patients with AS and patients without AS). t-Test data are normally distributed and have homogeneous variances. We verified the data calculation several times. hs-CRP was analyzed using chi-square test. A two-sided probability value of less than 0.05 was considered to be statistically significant for all analyses.
The nomogram survival model was constructed to predict AS by using the “rms” package [13]. The “rms” package was also used to calculate the C value and multifactor logistic regression [14, 15] to verify nomogram prediction ability. The area under the curve (AUC) of the receiver operating characteristic curve (ROC) curve and Harrell’s concordance index were used to evaluate the performance of nomogram predictions. Harrell’s concordance index was calculated to assess nomogram discrimination by using a bootstrap method with 1000 samples [16]. The “corrplot” package was used to analyze the correlation of the independent variables [17]. Decision curve analysis was conducted to determine the clinical usefulness of the nonadherence nomogram by quantifying the net benefits at different threshold probabilities in patients with AS [18]. The net benefit was calculated by subtracting the proportion of all patients who were false-positive cases from the proportion of the patients who were true-positive cases and by weighing the relative harm of forgoing interventions compared with the negative consequences of an unnecessary intervention [19]. In this study, the “rms” and “rmda” packages were used to obtain the thresholds and visualize them.
If all variables are included, machine learning operations become very difficult. So, for the training cohort, we first screened all variables with P < 0.05 using the SPSS software and then continued screening using three ML methods.
Random Forest
The random forest model uses the “randomforest” package in R software to screen out variables and calculate and visualize the relative importance of variables [20]. “%IncMSE” means an increase in the mean squared error. Values are randomly assigned to each prediction variable; if the prediction variable is more important, the model prediction error increases after its value is randomly replaced [21]. Therefore, the greater the value, the greater the importance of the variable. “IncNodePurity” denotes an increase in node purity, which is measured as the sum of the squares of residual errors and represents the impact of each variable on the heterogeneity of observed values at each node of the classification tree. The greater the value, the greater the importance of the variable [22]. Either “%IncMSE” or “IncNodePurity” was chosen as an indicator to judge the importance of the prediction variable. The most important quantity was obtained as the most suitable predictive variable through five iterations of tenfold cross-validation.
Lasso Regression
The LASSO regression model is a contraction method that actively selects from a large, potentially multicollinear set of variables in the regression to screen out risk factors and optimal predictive characteristics from the data of patients with AS. The dependent variables with P < 0.05 after calculation using Student’s t-test were used, and the “glmnet” package in R software was used for LASSO regression analysis and visualization [23, 24].
Support Vector Machine Recursive Feature Elimination
Support vector machine (SVM) recursive feature elimination (SVM-RFE) is a powerful tool, often grouped under ML. The SVM-RFE model was constructed to predict AS by using the “rms” package. In this study, tenfold cross-validation was performed on the data, the output vector characteristic index was obtained, and the variables were sorted from “most useful” to “least useful.” The smaller the AvgRank value, the greater the influence of the dependent variable on the independent variable. After sorting, we generalized error estimation for the entire data and screened the variable with the lowest common diagnosis error rate [25, 26].
Results
Data feature
Tables 1, 2, 3 and 4 show the differences in sex, age, ESR, blood routine examination, liver function and kidney function, and hypersensitive C-reactive protein between patients with AS and patients without AS in the training and validation cohorts. In the training cohort, the majority of patients with AS were male (Tables 1 and 2). The mean age of patients with AS was smaller than that of patients without AS, but the difference was not statistically significant. The proportion of ESR and hs-CRP > 10 in patients with AS was higher than that in patients without AS, and the proportion of hs-CRP < 0.8 was significantly lower than that in patients without AS. WBC, RBC, HGB, HCT, BPC, PDW, NEUT#, NEUT%, and MONO# were higher in patients with AS than in patients without AS, whereas MCHC, MPV, LYM%, and ESO# were lower in patients with AS (Tables 1 and 4). There was no significant difference in MCV, MCH, LYM#, MONO%, ESO%, BASO#, BASO%, RDW, and PCT on blood routine examination. Cr and Ccr of patients with AS were higher than those of patients without AS, whereas BUN and Cys-C were lower than those of patients without AS (Table 3). There was no statistical difference in UA and HCO between patients with AS and those without AS. Furthermore, the liver function test showed no significant differences in IBil, GGT, ALT, or PAB (Table 2). Moreover, DBil/TBil, TP, ALB, GLB, ALB/GLB, and ChE of patients with AS were higher than those of patients without AS, whereas TBil, DBil, TBA, AST, AST/ALT, and ALP were lower.
Table 1.
Differences on blood routine examination
| Type | Training cohort | Validation cohort | ||||
|---|---|---|---|---|---|---|
| AS | NC | P-value | AS | NC | P-value | |
| (N = 258) | (N = 247) | (N = 258) | (N = 247) | |||
| Sex | ||||||
| Male | 224 | 120 | < 0.001 | 77 | 64 | < 0.001 |
| Female | 34 | 127 | 13 | 49 | ||
| Age | ||||||
| Mean (median) | 34.1 (33) | 37.05 (38) | 0.058 | 34.61 (34) | 41.31 (36) | 0.002 |
| ESR | ||||||
| Mean (median) | 33.09 (30) | 26.11 (14) | 0.002 | 37.93 (32.5) | 22.86 (16) | < 0.001 |
| WBC | ||||||
| Mean (median) | 8.43 (8.17) | 7.81 (7.23) | 0.008 | 8.32 (7.93) | 7.81 (7.46) | 0.15 |
| RBC | ||||||
| Mean (median) | 4.95 (4.88) | 4.52 (4.53) | < 0.001 | 4.82 (4.82) | 4.81 (4.70) | 0.925 |
| HGB | ||||||
| Mean (median) | 132.36 (134.45) | 124.15 (126) | < 0.001 | 134.81 (136.95) | 132.22 (132.2) | 0.238 |
| HCT | ||||||
| Mean (median) | 0.407 (0.413) | 0.378 (0.38) | < 0.001 | 0.406 (0.406) | 0.404 (0.402) | 0.685 |
| MCV | ||||||
| Mean (median) | 82.84 (85) | 84.12 (85.72) | 0.125 | 84.44 (85.2) | 84.37 (86.6) | 0.945 |
| MCH | ||||||
| Mean (median) | 27.01 (27.75) | 27.62 (28.32) | 0.057 | 28.06 (28.3) | 27.67 (28.43) | 0.353 |
| MCHC | ||||||
| Mean (median) | 325.06 (327) | 327.76 (327.9) | 0.013 | 332.05 (329.95) | 327.33 (328) | 0.004 |
| BPC | ||||||
| Mean (median) | 335.44 (319.2) | 301.87 (282) | < 0.001 | 319.6 (305.64) | 297.3 (279) | 0.098 |
| MPV | ||||||
| Mean (median) | 8.22 (7.9) | 8.86 (8.9) | < 0.001 | 8.04 (7.85) | 8.4 (8.14) | 0.033 |
| PDW | ||||||
| Mean (median) | 0.154 (0.16) | 0.138 (0.16) | < 0.001 | 0.1499 (0.16) | 0.1596 (0.16) | 0.005 |
| NEUTa | ||||||
| Mean (median) | 5.45 (5.1) | 4.80 (4.06) | 0.002 | 5.41 (5.2) | 4.94 (4.44) | 0.144 |
| NEUT% | ||||||
| Mean (median) | 0.635 (0.64) | 0.591 (0.593) | < 0.001 | 0.641 (0.65) | 0.612 (0.62) | 0.05 |
| LYMa | ||||||
| Mean (median) | 2.09 (2.07) | 2.07 (2.08) | 0.508 | 2.02 (1.98) | 2.07 (1.97) | 0.636 |
| LYM% | ||||||
| Mean (median) | 0.259 (0.254) | 0.297 (0.289) | < 0.001 | 0.252 (0.244) | 0.283 (0.28) | 0.019 |
| MONOa | ||||||
| Mean (median) | 0.644 (0.6) | 0.591 (0.55) | 0.013 | 0.66 (0.64) | 0.566 (0.51) | 0.006 |
| MONO% | ||||||
| Mean (median) | 0.078 (0.073) | 0.078 (0.076) | 0.927 | 0.0787 (0.078) | 0.0737 (0.073) | 0.051 |
| ESOa | ||||||
| Mean (median) | 0.19 (0.12) | 0.202 (0.14) | 0.34 | 0.178 (0.125) | 0.198 (0.12) | 0.518 |
| ESO% | ||||||
| Mean (median) | 0.0232 (0.018) | 0.0292 (0.02) | 0.078 | 0.023 (0.017) | 0.0267 (0.018) | 0.314 |
| BASOa | ||||||
| Mean (median) | 0.037 (0.04) | 0.037 (0.03) | 0.615 | 0.178 (0.03) | 0.198 (0.03) | 0.493 |
| BASO% | ||||||
| Mean (median) | 0.0049 (0.004) | 0.00496 (0.004) | 0.93 | 0.0048 (0.004) | 0.00466 (0.005) | 0.681 |
| RDW | ||||||
| Mean (median) | 0.144 (0.14) | 0.141 (0.13) | 0.203 | 0.138 (0.13) | 0.138 (0.13) | 0.91 |
| PCT | ||||||
| Mean (median) | 0.272 (0.256) | 0.261 (0.25) | 0.163 | 0.259 (0.248) | 0.25 (0.234) | 0.222 |
ESR erythrocyte sedimentation rate, hs-CRP high-sensitivity C-reactive protein, WBC white blood cell count, RBC red blood cell count, HGB hemoglobin, HCT hematocrit value, MCV mean corpuscular volume, MCH mean corpuscular hemoglobin, MCHC mean corpuscular hemoglobin concentration, BPC blood platelet count, MPV mean platelet volume, PDW platelet distribution width, NEUT neutrophil percentage, LYM lymphocyte percentage, MONO monocyte percentage, ESO eosinophil percentage, BASO basophil percentage, RDW red cell distribution width, PCT thrombocytocrit
aDenotes absolute value
The bold text means that the P value was < 0.05
Table 2.
Differences on liver function examination
| Type | Training cohort | Validation cohort | ||||
|---|---|---|---|---|---|---|
| AS | NC | P-value | AS | NC | P-value | |
| (N = 258) | (N = 247) | (N = 90) | (N = 113) | |||
| TBil | ||||||
| Mean (median) | 8.59 (7.25) | 10.55 (9.4) | < 0.001 | 8.96 (7.85) | 9.58 (8.1) | 0.384 |
| DBil | ||||||
| Mean (median) | 2.81 (2.5) | 3.36 (3.2) | < 0.001 | 2.85 (2.7) | 3.05 (2.6) | 0.511 |
| IBil | ||||||
| Mean (median) | 5.77 (5.0) | 7.18 (6.1) | 0.315 | 6.11 (5.45) | 6.54 (5.8) | 0.399 |
| DBil/IBil | ||||||
| Mean (median) | 0.343 (0.3) | 0.333 (0.32) | < 0.001 | 0.33 (0.3) | 0.32 (0.3) | 0.663 |
| TP | ||||||
| Mean (median) | 76.7 (76) | 70.34 (70) | < 0.001 | 77.3 (77.85) | 71.6 (72.9) | < 0.001 |
| ALB | ||||||
| Mean (median) | 42.78 (43.2) | 38.38 (38.9) | < 0.001 | 43.63 (44.15) | 41.63 (42.5) | 0.002 |
| GLB | ||||||
| Mean (median) | 33.92 (33.15) | 31.97 (31.2) | 0.007 | 33.69 (33.1) | 29.95 (29.3) | < 0.001 |
| ALB/GLB | ||||||
| Mean (median) | 1.33 (1.3) | 1.28 (1.3) | < 0.001 | 1.33 (1.3) | 1.43 (1.5) | 0.018 |
| GGT | ||||||
| Mean (median) | 35.11 (25) | 29.63 (19) | 0.093 | 48.74 (27) | 41.57 (23) | 0.52 |
| TBA | ||||||
| Mean (median) | 4.92 (3.5) | 7.696 (5.0) | < 0.001 | 4.49 (3.1) | 5.8 (4.2) | 0.056 |
| AST | ||||||
| Mean (median) | 21.31 (20) | 28.66 (25) | < 0.001 | 22.6 (22) | 27.6 (21) | 0.062 |
| ALT | ||||||
| Mean (median) | 21.85 (17) | 22.19 (15) | 0.834 | 25.17 (19) | 28.87 (21) | 0.452 |
| AST/ALT | ||||||
| Mean (median) | 1.24 (1.1) | 1.73 (1.6) | < 0.001 | 1.14 (1.1) | 1.21 (1.6) | 0.354 |
| ALP | ||||||
| Mean (median) | 102.3 (92) | 113.46 (86) | 0.036 | 108.64 (97.5) | 101.79 (80) | 0.412 |
| PAB | ||||||
| Mean (median) | 225.64 (224.45) | 225.08 (222.5) | 0.934 | 230.71 (215) | 239.3 (238.5) | 0.393 |
| ChE | ||||||
| Mean (median) | 8349.3 (8248.5) | 8210.1 (8121) | 0.37 | 8545.73 (8587) | 8331.71 (8277) | 0.386 |
TBil total bilirubin, DBil direct bilirubin, IBil indirect bilirubin, TP total protein, ALB albumin, GLB globulin, GGT gamma-glutamyl transpeptidase, TBA total bile acid, AST aspartate aminotransferase, ALT alanine aminotransferase, ALP a alkaline phosphatase, PAB prealbumin, ChE cholinesterase
The bold text means that the P value was < 0.05
Table 3.
Differences on kidney function examination
| Type | Training cohort | Validation cohort | ||||
|---|---|---|---|---|---|---|
| AS | NC | P-value | AS | NC | P-value | |
| (N = 258) | (N = 247) | (N = 90) | (N = 113) | |||
| BUN | ||||||
| Mean (median) | 4.47 (4.32) | 5.05 (4.47) | 0.001 | 4.40 (4.21) | 4.61 (4.56) | 0.267 |
| Cr | ||||||
| Mean (median) | 69.22 (68.5) | 59.75 (56) | 0.008 | 68.32 (67) | 64.36 (62) | 0.113 |
| UA | ||||||
| Mean (median) | 337.51 (331.5) | 322.63 (307) | 0.105 | 355.16 (351) | 300.49 (290) | < 0.001 |
| HCO | ||||||
| Mean (median) | 26.12 (26.4) | 24.19 (24.3) | 0.111 | 26.57 (26.2) | 25.55 (26.1) | 0.009 |
| Ccr | ||||||
| Mean (median) | 101.54 (99) | 97.84 (97.4) | 0.005 | 111.8 (113) | 100.3 (97) | 0.006 |
| Cys-C | ||||||
| Mean (median) | 0.79 (0.77) | 0.866 (0.79) | < 0.001 | 0.74 (0.683) | 0.82 (0.79) | 0.005 |
BUN blood urea nitrogen, Cr creatinine, UA uric acid, HCO bicarbonate radical, Ccr creatinine clearance rate, Cys-C cysteine C
The bold text means that the P value was < 0.05
Table 4.
Differences in hypersensitive C-reactive protein
| hs-CRP | |||||
|---|---|---|---|---|---|
| Type | < 0.8 | 0.8–10 | > 10 | Overall | P-value |
| AS | 49(17.8%) | 85 (30.8%) | 142 (51.4%) | 276 | < 0.001 |
| NC | 83 (31.2%) | 109 (40.9%) | 74 (27.9%) | 266 | |
hs-CRP high-sensitivity C-reactive protein
The bold text means that the P value was < 0.05
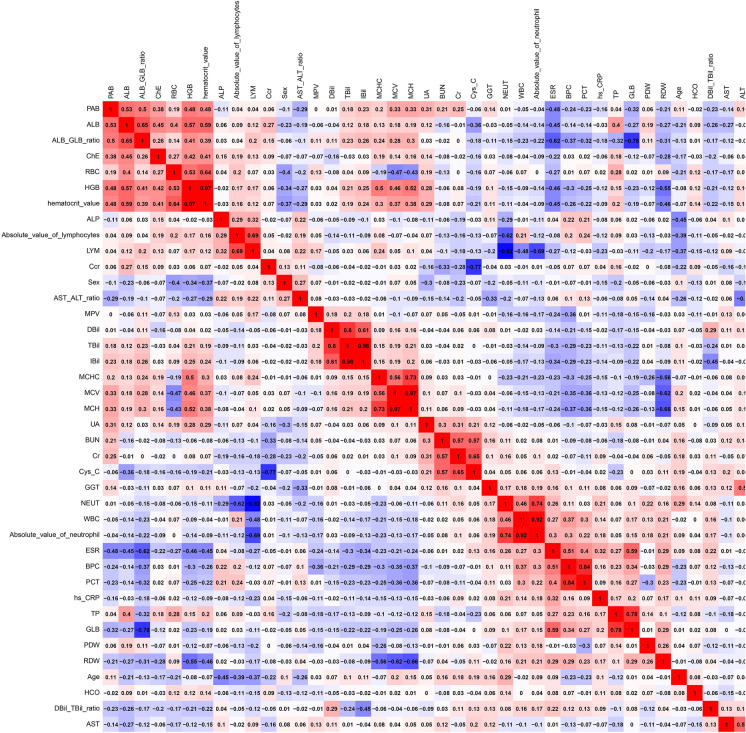
The correlation heat map (Fig. 2) shows the correlations between all the variables. Positive correlation was noted between HGB and HCT (hematocrit value), TBil and DBil, TBil and IBil, MCV and MCH, WBC and absolute value of neutrophils, BPC and PCT, TP and GLB, EO and absolute value of eosinophils, and BASO and absolute value of basophils. In contrast, a negative correlation was noted between LYM and NEUT and between Ccr and Cys-C (Fig. 3).
Fig. 2.
Heat map of the correlations between all the variables
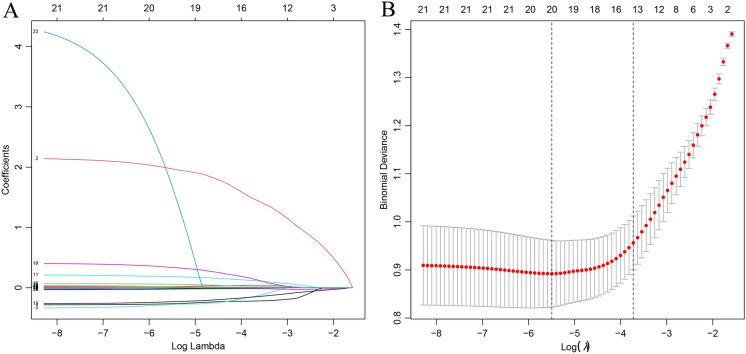
Fig. 3.
LASSO coefficient profiles of the factors, using cross-validation to the optimal penalty parameter lambda. A The results of the LASSO regression analysis of dependent variables. B The 20 factors that exhibited significant differences between the patients with AS and those without AS
Machine Learning
In the training cohort, 30 factors were included in ML with P < 0.05 after t-test screening.
Random Forest
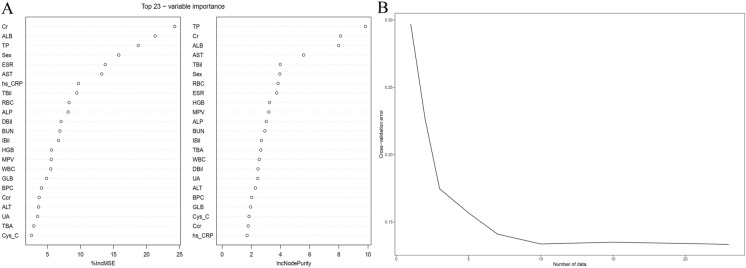
Figure 4A shows the 23 most important factors calculated using the two random forest algorithms “%IncMSE” and “IncNodePurity.” Fig. 4B shows that the ideal regression effect can be obtained by retaining the ten most important factors after tenfold cross-validation. Table 6 lists the ten important factors for the final selection of random forest regression.
Fig. 4.
Random forest screening variables. A The 23 most important factors calculated using the two random forest algorithms “%IncMSE” and “IncNodePurity.” B The ideal regression effect can be obtained by retaining the ten most important factors after tenfold cross-validation
Table 6.
The final selection of random forest regression
| Type | %IncMSE | IncNodePurity |
|---|---|---|
| TP | 18.747674 | 9.819856 |
| Cr | 24.262817 | 8.123714 |
| ALB | 21.292089 | 7.991552 |
| AST | 13.215127 | 5.586151 |
| TBil | 9.453452 | 3.981226 |
| Sex | 15.800084 | 3.950362 |
| RBC | 8.282169 | 3.834942 |
| ESR | 13.748547 | 3.737694 |
| HGB | 5.604174 | 3.250661 |
| MPV | 5.577898 | 3.205734 |
TP total protein, Cr creatinine, ALB albumin, AST aspartate aminotransferase, TBil total bilirubin, RBC red blood cell count, ESR erythrocyte sedimentation rate, HGB hemoglobin, MPV mean platelet volume
Lasso Regression
Figure 3A shows the results of the LASSO regression analysis of dependent variables. Figure 3B shows the 20 factors that exhibited significant differences between the patients with AS and those without AS. Table 5 presents the factors screened by LASSO regression (Table 6).
Table 5.
LASSO regression screened variables
| hs-CRP | Sex | ESR | WBC | RBC |
|---|---|---|---|---|
| HGB | BPC | MPV | IBil | TP |
| ALB | TBA | AST | ALT | ALP |
| BUN | Cr | UA | Ccr | Cys-C |
hs-CRP high-sensitivity C-reactive protein, ESR erythrocyte sedimentation rate, WBC white blood cell count, RBC red blood cell count, HGB hemoglobin, BPC blood platelet count, MPV mean platelet volume, IBil indirect bilirubin, TP total protein, ALB albumin, TBA total bile acid, AST aspartate aminotransferase, ALT alanine aminotransferase, ALP a alkaline phosphatase, BUN blood urea nitrogen, Cr creatinine, UA uric acid, Ccr creatinine clearance rate, Cys-C cysteine C
Svm-Rfe
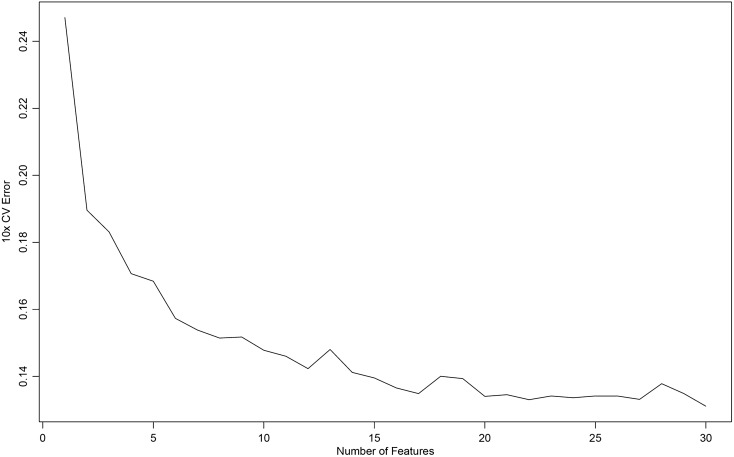
Figure 5 shows that, when 30 factors were selected as diagnostic models after SVM-RFE calculation, the error rate was the lowest, and all the factors included were meaningful for diagnosis. Table 7 shows the order of importance of the 30 factors in SVM-RFE. The smaller the AvgRank value, the greater the influence of the dependent variable on the independent variable.
Fig. 5.
The 30 factors were selected as diagnostic models after SVM-RFE calculation
Table 7.
The order of importance of the 30 factors in SVM-RFE
| Type | AvgRank | Type | AvgRank | Type | AvgRank |
|---|---|---|---|---|---|
| ALB | 1.6 | hs-CRP | 13.2 | HCT | 21.3 |
| Sex | 2.8 | Cys-C | 13.7 | ALP | 21.5 |
| ESR | 4.9 | Cr | 13.8 | DBil | 21.8 |
| AST | 6.4 | Ccr | 14 | TBA | 23.3 |
| PDW | 6.7 | DBil/TBil | 15.2 | MONO# | 24.3 |
| BUN | 6.7 | WBC | 19.9 | ALB/GLB | 24.3 |
| AST/ALT | 9 | HGB | 20.1 | RBC | 24.7 |
| TP | 9.9 | NEUT# | 20.2 | MPV | 26.6 |
| GLB | 11.5 | NEUT | 20.7 | ||
| BPC | 12.2 | MCHC | 20.8 | ||
| TBil | 12.7 | LYM | 21.2 |
ALB albumin, ESR erythrocyte sedimentation rate, AST aspartate aminotransferase, RDW red cell distribution width, BUN blood urea nitrogen, AST aspartate aminotransferase, TP total protein, GLB globulin, TBil total bilirubin, hs-CRP high-sensitivity C-reactive protein, Cys-C cysteine C, Cr creatinine, Ccr creatinine clearance rate, DBil direct bilirubin, WBC white blood cell count, HGB hemoglobin, NEUT# absolute value of neutrophils, MONO# absolute value of Monocyte, MCHC mean corpuscular hemoglobin concentration, LYM lymphocyte percentage, HCT hematocrit value, ALP alkaline phosphatase, TBA total bile acid, RBC red blood cell count, MPV mean platelet volume
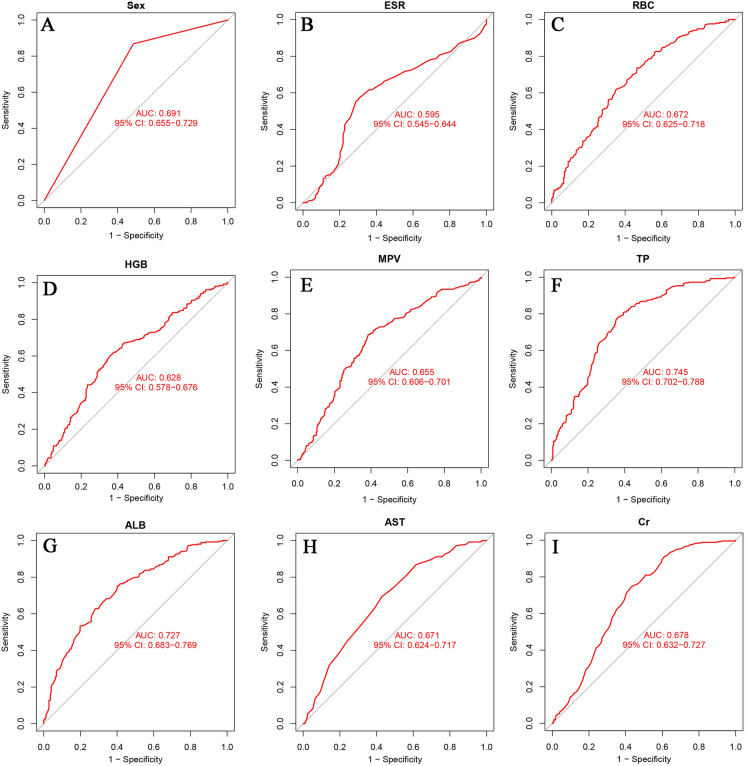
Figure 6 shows the intersection of variables screened using LASSO, random forest, and SVM-RFE; nine variables were finally obtained: sex, ESR, RBC, HGB, MPV, TP, ALB, AST, and Cr. The AUC values for the nine variables are shown in Fig. 7.
Fig. 6.
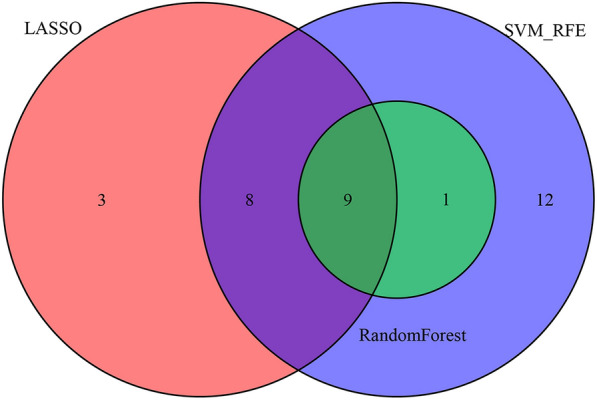
The intersection of variables screened using LASSO, random forest, and SVM-RFE
Fig. 7.
AUCs of the intersection of variables screened
Diagnostic Mode
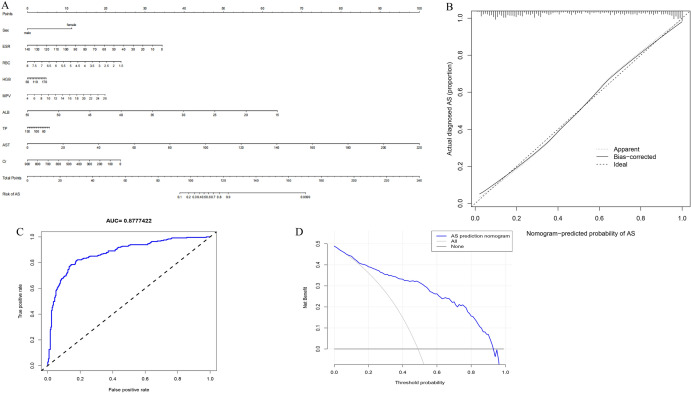
As can be seen in Fig. 8A, nine variables were included in the nomogram model. The optimal cutoff value of this nine-variable nomogram model is 179.459, with a sensitivity of 0.857 (95% CI 0.814–0.899), specificity of 0.806 (95% CI 0.756–0.855), and PPV of 0.822 (95% CI 0.766–0.867). NPV was 0.843 (95% CI 0.797–0.890). The C-index for the prediction nomogram was 0.878. The AUC value of the nomogram curve was 0.8777422 (95% CI 0.847–0.908) (Fig. 8C). In addition, calibration curves exhibited a satisfactory agreement between nomogram predictions and actual probabilities (Fig. 8B). The decision curve (Fig. 8D) showed that if the threshold probability of a patient and a doctor is > 1 and < 92%, respectively, using this nonadherence nomogram to predict AS nonadherence risk is more beneficial than the intervention-in-all-patients scheme or the intervention-in-none scheme[27].
Fig. 8.
The nine factors establish a nomogram for AS. A Nomogram for predicting AS probability. B Calibration curves for predicting AS probability. C AUC of the nomogram based on the nine characteristics. D Decision curve analysis for the nine-characteristic AS prediction nomogram
Simplified Diagnostic Mode
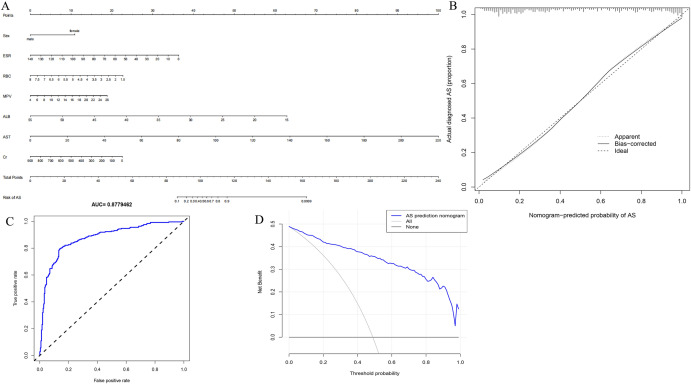
On the basis of the importance of the variables screened by ML and combined with clinical practicability, we attempted to simplify the diagnostic model to achieve high diagnostic efficiency. RBC, HGB, ALB, and TP are partially repeatable, and deletion of HGB and TP does not seriously impact the final diagnostic model, but also makes the model more concise. Finally, we selected seven variables, namely sex, ESR, RBC, MPV, ALB, AST, and CR, for the simplified diagnostic model. The optimal cutoff value of this nine-variable nomogram model is 173.139, with a sensitivity of 0.860 (95% CI 0.818–0.903), specificity of 0.798 (95% CI 0.747–0.848), and PPV of 0.816 (95% CI 0.770–0.862). NPV was 0.845 (95% CI 0.799–0.892). The C-index for the prediction nomogram was 0.878. The AUC value of the nomogram curve was 0.8779462 (95% CI 0.847–0.909) (Fig. 9C). In addition, calibration curves exhibited a satisfactory agreement between nomogram predictions and actual probabilities (Fig. 9B). The decision curve (Fig. 9D) revealed that when the threshold value of the model was set in the range of 1%–100%, the decision curve was above the NONE line and ALL line, thus indicating that the model has clinical usefulness in this range.
Fig. 9.
The seven factors establish a nomogram for AS. A Nomogram for predicting AS probability. B Calibration curves for predicting AS probability. C AUC of the nomogram based on the seven characteristics. D Decision curve analysis for the seven-characteristic AS prediction nomogram
Validation Cohort
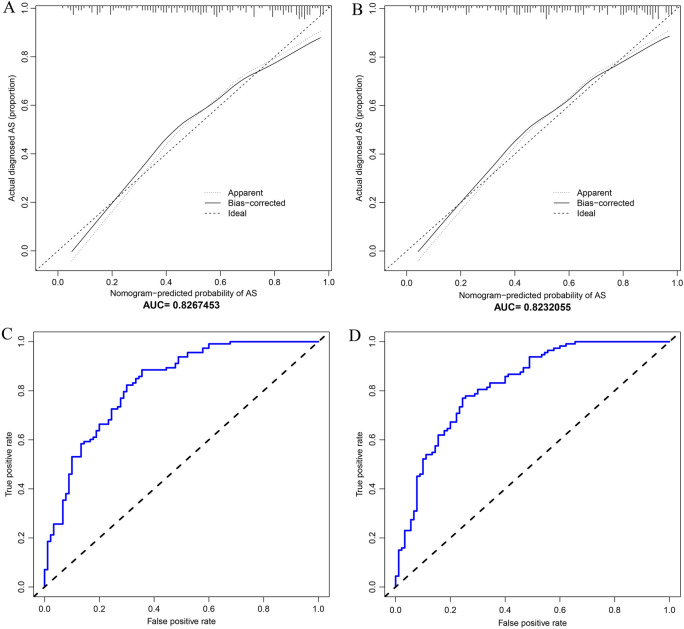
The two diagnostic models were employed for the validation cohort for verification. The C values of this nomogram were 0.827 (nine-factor diagnostic model) and 0.823 (seven-factor diagnostic model). The calibration curves exhibited a satisfactory agreement between nomogram predictions and actual probabilities (Fig. 10A, B). Figures 10C, D show the AUC values of the two diagnostic models were 0.8267453 (nine-factor diagnostic model) and 0.8232055 (seven-factor diagnostic model). We selected the seven-factor diagnostic model for subsequent analyses.
Fig. 10.
Validation cohort. A In validation cohort, calibration curves for nine characteristics predicting AS probability. B In validation cohort, calibration curves for seven characteristics predicting AS probability. C In validation cohort, AUC of the nomogram based on the nine characteristics. D In validation cohort, AUC of the nomogram based on the nine characteristics
Discussion
Using clinically relevant data and ML algorithms, we established a prediction model (Fig. 8A) for AS. The prediction model is based on a series of predictions. In addition, three ML models were used to filter variables and then verified on a validation cohort. This artificial-intelligence-based strategy can be used by clinicians to help them choose easier diagnostic methods [28].
ML has contributed to a paradigm shift in health care wherein computers learn from patient data without employing explicit programming tasks [29]. ML offers the advantages of extensive applicability, objectivity, and repeatability when dealing with large datasets and reliable data [30, 31]. Moreover, it can help improve the quality of early diagnosis, identify disease progression, and increase the likelihood of predicting specific patient outcomes in orthopedic procedures, such as outcome scores, risk of complications, and implant survival [32, 33]. These benefits facilitate decision-making and information sharing between clinicians and patients and facilitate effective planning and rational use of healthcare services [34].
AS is a chronic progressive inflammatory disease of the spine and its affiliated tissues. Through ML screening, we screened a total of nine variables that can be used to predict AS: sex, ESR, RBC, MPV, ALB, AST, and Cr. Most of the patients with AS are male [35], but the prevalence of AS in women is gradually increasing [36]. The proportion of male patients with AS receiving medical treatment is much higher than that of female patients [8]. Male patients with AS are more likely to develop hip and spinal mobility disorders than women [36, 37]. Males score higher in our diagnostic model.
ESR is the distance of the erythrocyte subsidence at the end of the first hour to represent the rate of erythrocyte sedimentation. An increase in ESR is considered an inflammatory reaction or hyperglobulinemia in clinicopathology [38]. AS is a chronic inflammatory disease that can lead to accelerated ESR [39]. Studies have shown that ESR is associated with poor physical activity in patients with AS [40]. In our diagnostic model, ESR is positively correlated with the final score. The higher ESR is, the more likely it is to be diagnosed AS.
The results of the current study revealed that RBC and HGB increased in patients with AS [41]. Ninety percent of RBC is composed of HGB, which is mainly responsible for the transport of oxygen and carbon dioxide in the body. In addition, RBC can clear circulating immune complexes, reduce T-cell proliferation, and promote phagocytosis [42, 43]. The proportion of CD4+ T-cells in patients with AS is reduced, which may be the reason for the increase in RBC and HGB in patients with AS [44]. The higher the RBC, the higher the nomogram score. In clinical practice, MPV is often used to determine the risk of bleeding and changes in bone marrow hematopoietic function. A decrease in the MPV in patients with AS may be caused by bone marrow suppression caused by a chronic inflammatory reaction due to AS [45]. So, the lower the MPV, the higher the score in nomogram. Further research on the mechanism is required.
TP and ALB are often used in the clinical monitoring of a patient’s nutritional status. Our study showed that TP and ALB of patients with AS were higher than those of patients without AS. Elevated AST is often used for the diagnosis of liver diseases, and AST decreases significantly in patients with AS, which has no special clinical significance [46]. Cr is the product of muscle metabolism in the human body, and Cr increases significantly in patients with AS, which may be caused by impaired immune function in patients with AS. The average UA in patients with AS was also higher than that in patients without AS. These studies on AS provide a new direction for further research. In our diagnostic model, ALB and Cr values correlated positively with the final nomogram score, while AST correlated inversely.
The proportion of hs-CRP (Table 4) in patients with AS > 10 was significantly higher than that in patients without AS, and the proportion of hs-CRP < 0.8 was lower than that in patients without AS. In a study by Seulkee et al., CRP was higher in patients with symptoms of AS than in patients without symptoms [6]. WBC and NEUT were elevated in patients with AS, consistent with chronic inflammation. However, ML did not include them in predictive models .
Machine learning is widely used in diagnosing, treating, treating, preventing, and managing AS diseases. Riel et al. used computed tomography (CT) to construct an early diagnosis model using machine learning methods [47]. Samuel et al. used single-cell transcriptome and surface epitope analysis of AS to classify diseases using machine learning methods [48]. Liang et al. used LASSO regression to find that the severity of the platelet-to-lymphocyte ratio was related to the severity of AS, which is helpful for diagnosing and treating physicians [8].
This study aimed to use a dataset of 708 patients to select the best ML model. Our work has several advantages. First, there are few studies on AS using age, ESR, blood routine examination, liver function, and kidney function, and we did not find similar studies. Second, we used three ML methods to filter the data and used the validation cohort for verification. Finally, upon comparison, our model exhibited superior predictive power and ease of usage for clinicians to diagnose AS.
However, there are some limitations to this study. First, the retrospective nature of this study may have led to subjective bias and selection bias. Second, the ML algorithm model we developed is limited to one hospital, which may limit its use in other areas and requires further validation. Third, our study lacks imaging data, which may improve our diagnostic efficacy. Fourth, the predictive performance is average and can be improved further.
Conclusion
We established two prediction models that offer the advantages of good performance, high accuracy, and simplicity of use. We can effectively serve patients with AS and help doctors make a diagnosis by using predictive models. Of course, clinicians always have the final word for interpretation based on their domain expertise. In future studies, we will attempt to cover a wide range of clinical variables so that our diagnostic model can be used more accurately in a wider population.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We are grateful to Dr. Xinli Zhan and Dr. Chong Liu (Spine and Osteopathy Ward, The First Affiliated Hospital of Guangxi Medical University) for their kind assistance in all stages of the present study.
Funding
This work was sponsored by the National Natural Science Foundation of China (81560359); National Natural Science Foundation of China (81860393). Funding bodies had no role in the study design, collection, analysis, and interpretation of the data or in writing the manuscript. The authors funded the journal’s Rapid Service Fee.
Author Contributions
All authors contributed to the study conception and design. Jichong Zhu, Chong Liu and Xinli Zhan designed the study. Qing Lu, Tuo Liang, Jie Jiang, Liyi Chen, Shengsheng Huang, Guobing Deng and Xuhua Sun analyze the data. Wenyong Jiang, Jiarui Chen, Tianyou Chen, Hao Li, Xiang Tao, Binguang Fan and Yuanlin Yao digital visualization. Shaofeng Wu, Shian Liao, Chaojie Yu, Zhen Ye, Hao Guo, Wuhua Chen, Wenkang Chen and Chenxin Zhou collected data on routine blood data. Jichong Zhu wrote and revised the manuscript. Chong Liu and Xinli Zhan revised the manuscript. All authors read and approved the final manuscript.
Disclosures
Jichong Zhu, Qing Lu, Tuo Liang, JieJiang, Hao Li, Chenxin Zhou, Shaofeng Wu, Tianyou Chen, Jiarui Chen, Guobing Deng, Yuanlin Yao, Shian Liao, Chaojie Yu, Shengsheng Huang, Xuhua Sun, Liyi Chen, Wenkang Chen, Zhen Ye, Hao Guo, Wuhua Chen, Wenyong Jiang, Binguang Fan, Xiang Tao, Xinli Zhan and Chong Liu declare that they have no competing interests.
Compliance with Ethics Guidelines
All subjects volunteered for the study and signed informed consent forms. To ensure confidentiality, the names of study participants were not included in the data. Information obtained from the data of the study participants is kept confidential. This study was performed in accordance with the Helsinki Declaration of 1964 and its later amendments. In addition, the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University approved the study (2021-(KY-E-136)).
Data Availability
The original contributions presented in the study are included in the article or the supplementary material files. Further inquiries can be directed to the corresponding authors.
Contributor Information
Xinli Zhan, Email: zhanxinli@stu.gxmu.edu.cn.
Chong Liu, Email: liuchong@stu.gxmu.edu.cn.
References
- 1.Ranganathan V, Gracey E, Brown MA, Inman RD, Haroon N. Pathogenesis of ankylosing spondylitis—recent advances and future directions. Nat Rev Rheumatol. 2017;13(6):359–367. doi: 10.1038/nrrheum.2017.56. [DOI] [PubMed] [Google Scholar]
- 2.Fiorillo MT, Haroon N, Ciccia F, Breban M. Editorial: ankylosing spondylitis and related immune-mediated disorders. Front Immunol. 2019;10:1232. doi: 10.3389/fimmu.2019.01232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mauro D, Thomas R, Guggino G, Lories R, Brown MA, Ciccia F. Ankylosing spondylitis: an autoimmune or autoinflammatory disease? Nat Rev Rheumatol. 2021;17(7):387–404. doi: 10.1038/s41584-021-00625-y. [DOI] [PubMed] [Google Scholar]
- 4.Wright GC, Kaine J, Deodhar A. Understanding differences between men and women with axial spondyloarthritis. Semin Arthritis Rheum. 2020;50(4):687–694. doi: 10.1016/j.semarthrit.2020.05.005. [DOI] [PubMed] [Google Scholar]
- 5.Morin M, Hellgren K, Frisell T. Familial aggregation and heritability of ankylosing spondylitis—a Swedish nested case–control study. Rheumatology (Oxford) 2020;59(7):1695–1702. doi: 10.1093/rheumatology/kez519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S, Kang S, Eun Y, Won HH, Kim H, Lee J, et al. Machine learning-based prediction model for responses of bDMARDs in patients with rheumatoid arthritis and ankylosing spondylitis. Arthritis Res Ther. 2021;23(1):254. doi: 10.1186/s13075-021-02635-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van Calster B, Wynants L. Machine learning in medicine. N Engl J Med. 2019;380(26):2588. doi: 10.1056/NEJMc1906060. [DOI] [PubMed] [Google Scholar]
- 8.Liang T, Chen J, Xu G, Zhang Z, Xue J, Zeng H, et al. Platelet-to-lymphocyte ratio as an independent factor was associated with the severity of ankylosing spondylitis. Front Immunol. 2021;12:760214. doi: 10.3389/fimmu.2021.760214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang Z, Ho KM, Hong Y. Machine learning for the prediction of volume responsiveness in patients with oliguric acute kidney injury in critical care. Crit Care (London, England) 2019;23(1):112. doi: 10.1186/s13054-019-2411-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He D, Wang R, Liang S, Liang D, Xu F, Zeng C, et al. Comparison of secondary IgA nephropathy in patients with ankylosing spondylitis and rheumatoid arthritis. Mod Rheumatol. 2020;30(4):648–656. doi: 10.1080/14397595.2019.1651493. [DOI] [PubMed] [Google Scholar]
- 11.Ding T, Li B, Su R, Su R, Wang Y, Gao C, et al. Elevated Th17 cells are associated with cardiovascular complications in ankylosing spondylitis. Rheumatology (Oxford). 2021 Keab888. [DOI] [PubMed]
- 12.van der Linden S, Valkenburg HA, Cats A. Evaluation of diagnostic criteria for ankylosing spondylitis. A proposal for modification of the New York criteria. Arthritis Rheum. 1984;27(4):361–368. doi: 10.1002/art.1780270401. [DOI] [PubMed] [Google Scholar]
- 13.Pan X, Jin X, Wang J, Hu Q, Dai B. Placenta inflammation is closely associated with gestational diabetes mellitus. Am J Transl Res. 2021;13(5):4068–4079. [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang S, Tong YX, Zhang XH, Zhang YJ, Xu XS, Xiao AT, et al. A novel and validated nomogram to predict overall survival for gastric neuroendocrine neoplasms. J Cancer. 2019;10(24):5944–5954. doi: 10.7150/jca.35785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang R, He S, Sun H, Gong H, Yang X, Cai X, et al. Identifying the risk factors and estimating the prognosis in patients with pelvis and spine ewing sarcoma: a population-based study. Spine. 2021;46(19):1315–1325. doi: 10.1097/BRS.0000000000004022. [DOI] [PubMed] [Google Scholar]
- 16.Wu M, Li X, Zhang T, Liu Z, Zhao Y. Identification of a nine-gene signature and establishment of a prognostic nomogram predicting overall survival of pancreatic cancer. Front Oncol. 2019;9:996. doi: 10.3389/fonc.2019.00996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang H, Liu R, Sun L, Guo W, Ji X, Hu X. Comprehensive analysis of gene expression changes and validation in hepatocellular carcinoma. Onco Targets Ther. 2021;14:1021–1031. doi: 10.2147/OTT.S294500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vickers AJ, Holland F. Decision curve analysis to evaluate the clinical benefit of prediction models. Spine J. 2021;21(10):1643–1648. doi: 10.1016/j.spinee.2021.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang H, Zhang L, Liu Z, Wang X, Geng S, Li J, et al. Predicting medication nonadherence risk in a Chinese inflammatory rheumatic disease population: development and assessment of a new predictive nomogram. Patient Prefer Adherence. 2018;12:1757–1765. doi: 10.2147/PPA.S159293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alderden J, Pepper GA, Wilson A, Whitney JD, Richardson S, Butcher R, et al. Predicting pressure injury in critical care patients: a machine-learning model. Am J Crit Care. 2018;27(6):461–468. doi: 10.4037/ajcc2018525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang H, Wang W, Haggerty J, Schuster T. Predictors of patient satisfaction and outpatient health services in China: evidence from the WHO SAGE survey. Fam Pract. 2020;37(4):465–472. doi: 10.1093/fampra/cmaa011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfau M, von der Emde L, Dysli C, Möller PT, Thiele S, Lindner M, et al. Determinants of cone and rod functions in geographic atrophy: AI-based structure–function correlation. Am J Ophthalmol. 2020;217:162–173. doi: 10.1016/j.ajo.2020.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Zhang M, Zhu K, Pu H, Wang Z, Zhao H, Zhang J, et al. An immune-related signature predicts survival in patients with lung adenocarcinoma. Front Oncol. 2019;9:1314. doi: 10.3389/fonc.2019.01314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang S, Su W, Zhong C, Yang T, Chen W, Chen G, et al. An Eight-CircRNA assessment model for predicting biochemical recurrence in prostate cancer. Front Cell Dev Biol. 2020;8:599494. doi: 10.3389/fcell.2020.599494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duan KB, Rajapakse JC, Wang H, Azuaje F. Multiple SVM-RFE for gene selection in cancer classification with expression data. IEEE Trans Nanobiosci. 2005;4(3):228–234. doi: 10.1109/TNB.2005.853657. [DOI] [PubMed] [Google Scholar]
- 26.Zhao E, Xie H, Zhang Y. Predicting diagnostic gene biomarkers associated with immune infiltration in patients with acute myocardial infarction. Front Cardiovasc Med. 2020;7:586871. doi: 10.3389/fcvm.2020.586871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang YQ, Liang CH, He L, Tian J, Liang CS, Chen X, et al. Development and validation of a radiomics nomogram for preoperative prediction of lymph node metastasis in colorectal cancer. J Clin Oncol. 2016;34(18):2157–2164. doi: 10.1200/JCO.2015.65.9128. [DOI] [PubMed] [Google Scholar]
- 28.Sorrentino FS, Jurman G, De Nadai K, Campa C, Furlanello C, Parmeggiani F. Application of artificial intelligence in targeting retinal diseases. Curr Drug Targets. 2020;21(12):1208–1215. doi: 10.2174/1389450121666200708120646. [DOI] [PubMed] [Google Scholar]
- 29.Ngiam KY, Khor IW. Big data and machine learning algorithms for health-care delivery. Lancet Oncol. 2019;20(5):e262–e273. doi: 10.1016/S1470-2045(19)30149-4. [DOI] [PubMed] [Google Scholar]
- 30.Handelman GS, Kok HK, Chandra RV, Razavi AH, Lee MJ, Asadi H. eDoctor: machine learning and the future of medicine. J Intern Med. 2018;284(6):603–619. doi: 10.1111/joim.12822. [DOI] [PubMed] [Google Scholar]
- 31.Thrall JH, Li X, Li Q, Cruz C, Do S, Dreyer K, et al. Artificial intelligence and machine learning in radiology: opportunities, challenges, pitfalls, and criteria for success. J Am Coll Radiol JACR. 2018;15(3 Pt B):504–508. doi: 10.1016/j.jacr.2017.12.026. [DOI] [PubMed] [Google Scholar]
- 32.Herzog NJ, Magoulas GD. Brain asymmetry detection and machine learning classification for diagnosis of early dementia. Sensors (Basel, Switzerland). 2021;21(3):778. doi: 10.3390/s21030778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lynch CM, Abdollahi B, Fuqua JD, de Carlo AR, Bartholomai JA, Balgemann RN, et al. Prediction of lung cancer patient survival via supervised machine learning classification techniques. Int J Med Inf. 2017;108:1–8. doi: 10.1016/j.ijmedinf.2017.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Doupe P, Faghmous J, Basu S. Machine learning for health services researchers. Value Health. 2019;22(7):808–815. doi: 10.1016/j.jval.2019.02.012. [DOI] [PubMed] [Google Scholar]
- 35.Nygaard A, Ljungdalh PS, Iachina M, Nikolov TN, Schiottz-Christensen B. Incidence of ankylosing spondylitis and spondyloarthritis in 2000–2013: a nationwide Danish cohort study. Scand J Rheumatol. 2020;49(1):21–27. doi: 10.1080/03009742.2019.1616324. [DOI] [PubMed] [Google Scholar]
- 36.Crossfield SSR, Marzo-Ortega H, Kingsbury SR, Pujades-Rodriguez M, Conaghan PG. Changes in ankylosing spondylitis incidence, prevalence and time to diagnosis over two decades. RMD Open. 2021;7(3):e001888. doi: 10.1136/rmdopen-2021-001888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ibn Yacoub Y, Amine B, Laatiris A, Hajjaj-Hassouni N. Gender and disease features in Moroccan patients with ankylosing spondylitis. Clin Rheumatol. 2012;31(2):293–297. doi: 10.1007/s10067-011-1819-x. [DOI] [PubMed] [Google Scholar]
- 38.Jiao JB, Huang JC, Chen X, Jin Y. Albumin to globulin ratio, neutrophil to lymphocyte ratio, and globulin levels do not outperform ESR or CRP when diagnosing periprosthetic joint infection. BMC Musculoskelet Disord. 2022;23(1):404. doi: 10.1186/s12891-022-05357-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kang KY, Chung MK, Kim HN, Hong YS, Ju JH, Park SH. Severity of sacroiliitis and erythrocyte sedimentation rate are associated with a low trabecular bone score in young male patients with ankylosing spondylitis. J Rheumatol. 2018;45(3):349–356. doi: 10.3899/jrheum.170079. [DOI] [PubMed] [Google Scholar]
- 40.Chen CH, Chen HA, Liao HT, Liu CH, Tsai CY, Chou CT. The clinical usefulness of ESR, CRP, and disease duration in ankylosing spondylitis: the product of these acute-phase reactants and disease duration is associated with patient's poor physical mobility. Rheumatol Int. 2015;35(7):1263–1267. doi: 10.1007/s00296-015-3214-4. [DOI] [PubMed] [Google Scholar]
- 41.Carson JL, Stanworth SJ, Dennis JA, Trivella M, Roubinian N, Fergusson DA, et al. Transfusion thresholds for guiding red blood cell transfusion. Cochrane Database Syst Rev. 2021;12(12):Cd002042. doi: 10.1002/14651858.CD002042.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sherwood TA, Virella G. The binding of immune complexes to human red cells: complement requirements and fate of the RBC-bound IC after interaction with human phagocytic cells. Clin Exp Immunol. 1986;64(1):195–204. [PMC free article] [PubMed] [Google Scholar]
- 43.Long K, Meier C, Bernard A, Williams D, Davenport D, Woodward J. T-cell suppression by red blood cells is dependent on intact cells and is a consequence of blood bank processing. Transfusion. 2014;54(5):1340–1347. doi: 10.1111/trf.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M, Zhou X, Zhou L, Yu Z, Fu L, Yang P. Meta-analysis of changes in the number and proportion of regulatory T cells in patients with ankylosing spondylitis. BioMed Res Int. 2020;2020:8709804. doi: 10.1155/2020/8709804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kisacik B, Tufan A, Kalyoncu U, Karadag O, Akdogan A, Ozturk MA, et al. Mean platelet volume (MPV) as an inflammatory marker in ankylosing spondylitis and rheumatoid arthritis. Jt Bone Spine. 2008;75(3):291–294. doi: 10.1016/j.jbspin.2007.06.016. [DOI] [PubMed] [Google Scholar]
- 46.Lozano-Paniagua D, Parrón T, Alarcón R, Requena M, López-Guarnido O, Lacasaña M, et al. Evaluation of conventional and non-conventional biomarkers of liver toxicity in greenhouse workers occupationally exposed to pesticides. Food Chem Toxicol. 2021;151:112127. doi: 10.1016/j.fct.2021.112127. [DOI] [PubMed] [Google Scholar]
- 47.Castro-Zunti R, Park EH, Choi Y, Jin GY, Ko SB. Early detection of ankylosing spondylitis using texture features and statistical machine learning, and deep learning, with some patient age analysis. Comput Med Imaging Graph. 2020;82:101718. doi: 10.1016/j.compmedimag.2020.101718. [DOI] [PubMed] [Google Scholar]
- 48.Alber S, Kumar S, Liu J, Huang ZM, Paez D, Hong J, et al. Single cell transcriptome and surface epitope analysis of ankylosing spondylitis facilitates disease classification by machine learning. Front Immunol. 2022;13:838636. doi: 10.3389/fimmu.2022.838636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are included in the article or the supplementary material files. Further inquiries can be directed to the corresponding authors.