Abstract
The mammalian Transient Receptor Potential Vanilloid (TRPV) channels are a family of six tetrameric ion channels localized at the plasma membrane. The group I members of the family, TRPV1 through TRPV4, are heat-activated and exhibit remarkable polymodality. The distal N-termini of group I TRPV channels contain large intrinsically disordered regions (IDRs), ranging from ~ 75 amino acids (TRPV2) to ~ 150 amino acids (TRPV4), the vast majority of which is invisible in the structural models published so far. These IDRs provide important binding sites for cytosolic partners, and their deletion is detrimental to channel activity and regulation. Recently, we reported the NMR backbone assignments of the distal TRPV4 N-terminus and noticed some discrepancies between the extent of disorder predicted solely based on protein sequence and from experimentally determined chemical shifts. Thus, for an analysis of the extent of disorder in the distal N-termini of all group I TRPV channels, we now report the NMR assignments for the human TRPV1, TRPV2 and TRPV3 IDRs.
Keywords: Transient receptor potential, TRP vanilloid, Ion channel, Intrinsically disordered protein, Regulatory domain, Structural dynamics
Biological Context
Transient Receptor Potential (TRP) channels are tetrameric cation channels involved in temperature and pain sensation, nociception and osmoregulation (Ramsey et al. 2006). They are classified into subfamilies primarily based on amino acid sequence and structural characteristics rather than functional properties (Himmel and Cox 2020). Members of the mammalian TRPA (ankyrin), TRPC (canonical), TRPM (melastatin) and TRPV (vanilloid) subfamilies are localized at the plasma membrane and feature extensive N- and C-terminal cytoplasmatic domains. The mammalian TRPML (mucolipin) and TRPP (polycystin) proteins possess a large domain located between the first two transmembrane helices (Hellmich and Gaudet 2014; Samanta et al. 2018; Viet et al. 2019).
The mammalian TRPV subfamily can be further subdivided into group I and group II channels. TRPV1 through TRPV4 (group I) are activated by heat and, together with temperature-sensitive members of other TRP channel subfamilies, constitute the so-called “ThermoTRPs” (Cohen and Moiseenkova-Bell 2014; Islas 2017). Human group I TRPV channels are widely expressed, e.g. in primary sensory neurons, dorsal root ganglia, trigeminal ganglia, testis, spleen, intestine, bladder, brain, heart, liver, endothelia, and many other tissues (Cortright et al. 2001; Toledo Mauriño et al. 2020). In addition to thermosensation, they generally exhibit remarkable polymodality, i.e. the ability to respond to very diverse and seemingly unrelated stimuli such as natural and artificial compounds, pH, ions, small molecules or lipids (Baez-Nieto et al. 2011). Notably, many active ingredients from spices and herbs activate group I TRPV channel members, e.g. capsaicin (from chili) (Caterina et al. 1997), piperine (pepper) (Dong et al. 2019), allicin (garlic) (Macpherson et al. 2005) for TRPV1 or thymol (thyme) and carvacrol (oregano) (Xu et al. 2006) for TRPV3. Group I TRPV channels also play important roles in human health, e.g., in cancer, the immune system, viral infections or neurodegenerative diseases (Prevarskaya et al. 2007; Nilius and Voets 2013; Omar et al. 2017; Bujak et al. 2019; Taga et al. 2022). In contrast to the stimuli-dependent, non-selective group I TRPV calcium channels, the group II members, TRPV5 and TRPV6, are constitutively active and calcium-selective (van Goor et al. 2020).
To date, high-resolution cryo-electron microscopy (cryo-EM) structures have been determined for all six mammalian TRP vanilloid members (e.g. TRPV1 (Liao et al. 2013; Gao et al. 2016), TRPV2 (Huynh et al. 2016; Pumroy et al. 2019), TRPV3 (Singh et al. 2018, 2019; Zubcevic et al. 2018, 2019; Nadezhdin et al. 2021), TRPV4 (Deng et al. 2018), TRPV5 (Hughes et al. 2018; Dang et al. 2019) and TRPV6 (McGoldrick et al. 2018)). These structures demonstrate a significant 3D structural conservation within the transmembrane region of these channels. In addition, all TRPV channels contain an N-terminal ankyrin repeat domain (ARD). However, while TRPV5 and TRPV6 proteins feature an N-terminal α-helix preceding the ARD (e.g. Hughes et al. 2018; McGoldrick et al. 2018), the distal cytosolic N-termini of group I TRPV channels contain intrinsically disordered regions (IDRs) of variable length: ~100 aa for TRPV1, ~ 75 for TRPV2, ~ 120 for TRPV3, and ~ 150 for TRPV4. To date, structural studies could not capture most of these regions critical for protein function and regulation. Nonetheless, the group I TRPV channel IDRs act as important interaction platforms for cytosolic binding partners and post-translational modifications (Voolstra and Huber 2014; Goretzki et al. 2018; Aisenberg et al. 2022) and they play an important role in stimulus-dependent channel activation (e.g. Liao et al. 2013; Zubcevic et al. 2016; Deng et al. 2018; Shimada et al. 2020).
Given the general lack of structural data available for the distal N-terminus in the majority of group I TRPV channel structures, many questions regarding the structural coupling of these regions to the channel pore, and the conformational propagation of peripheral ligand binding remain unanswered. We have previously described the NMR backbone assignments of the human and chicken TRPV4 IDR with 147 and 133 amino acids, respectively (Goretzki et al. 2022). Now, to extend the structural and dynamic analysis of the IDRs to the remaining members of the human group I TRPV channels, we report the assignments of the N-terminal IDRs of the human TRPV1 (UniProtKB: Q8NER1-1, residues K2-D100, 99 amino acids), TRPV2 (UniProtKB: Q9Y5S1-1, residues T2-R73, 72 aa), and TRPV3 (UniProtKB: Q8NET8-1, residues K2-L119, 118 aa) ion channels.
Methods and experiments
Protein expression and purification
The DNA sequences encoding the H. sapiens N-terminal TRPV1, TRPV2 and TRPV3-IDRs (hsTRPV1-IDR, hsTRPV2-IDR and hsTRPV3-IDR, respectively) with an N-terminal His6-SUMO-tag cloned into a pET11a vector were obtained from Genescript (NJ, USA).
To obtain uniformly 13C, 15N-labeled hsTRPV1-IDR, hsTRPV2-IDR and hsTRPV3-IDR, transformed Escherichia coli BL21-Gold(DE3) cells (Agilent Technologies) were grown in M9 minimal medium supplemented with 0.1 mg/mL Ampicillin, 1 g/l 15NH4Cl and 2 g/l 13C6-labeled glucose as the sole nitrogen and carbon sources. Gene expression was induced at an OD600nm of 0.8-1.0 by adding 1 mM IPTG (isopropyl-1-β-d-galactopyranoside). Cells were grown overnight at 16 °C, harvested via centrifugation (4500xg, 20 min, 4 °C) and stored at -80 °C until further use.
For protein purification, cells were resuspended in lysis buffer (50 mM NaPi pH 7.5, 300 mM NaCl, 5 mM imidazole, 1 mM PMSF, 1 mM benzamidine, lysozyme, DNAse, RNAse, and protease inhibitor (cOmplete Mini, Roche Diagnostics GmbH)). For hsTRPV3-IDR, DTT was added to a final concentration of 1 mM in all purification buffers. Resuspended cells were sonicated on ice (Bandelin SONOPULS) and cell debris was removed by centrifugation (20000xg, 20 min, 4 °C). The supernatants were loaded onto pre-equilibrated gravity flow Ni-NTA (Qiagen) affinity columns at 4 °C. After washing twice (50 mM NaPi pH 7.5, 300 mM NaCl, 5 mM imidazole, then 10 mM imidazole), proteins were eluted in one column volume fractions with 50 mM NaPi pH 7.5, 300 mM NaCl, 250 mM imidazole. Protein containing fractions were pooled and dialyzed overnight (50 mM NaPi pH 7.5, 300 mM NaCl, 1 mM DTT) in the presence of 5 mol% Ulp-1 protease at 4 °C. Reverse Ni-NTA affinity chromatography was employed for separation of cleaved proteins from His6-SUMO-tag containing constructs.
Subsequently, size exclusion chromatography (HiLoad prep grade 10/300 Superdex75 column (GE Healthcare)) using 10 mM Tris-HCl pH 7, 100 mM NaCl (hsTRPV1-IDR, hsTRPV2-IDR) or 10 mM NaPi pH 6.2, 300 mM NaCl, 1 mM DTT (hsTRPV3-IDR) was carried out. Sample purity was verified by SDS-PAGE. The purified, tag-free proteins were frozen in liquid N2 and stored at -80 °C until further use.
Of note, the first residue (M1) was missing in the final constructs used for NMR experiments. Thus, the constructs comprise residues K2-D100 in hsTRPV1-IDR, T2-R73 in hsTRPV2-IDR and K2-L119 in hsTRPV3-IDR, respectively.
NMR spectroscopy
All NMR spectra of human TRPV1-IDR, TRPV2-IDR and TRPV3-IDR were recorded at 25 °C on 600 MHz Bruker AvanceIII HD NMR spectrometer systems equipped with cryogenic triple resonance probes (Bruker Biospin GmbH, Rheinstetten, Germany). The spectrometers were locked on D2O.
The 1H chemical shifts of the 13C,15N-labelled proteins were directly referenced to 3-(trimethylsilyl)propane-1-sulfonate (DSS). 13C and 15N chemical shifts were referenced indirectly to the 1H DSS standard by the magnetogyric ratio (Wishart et al. 1995).
Backbone and side chain chemical shift resonances were assigned with a set of band-selective excitation short-transient (BEST) (Schanda et al. 2006) transverse relaxation-optimized spectroscopy (TROSY)-based (Pervushin et al. 1997) experiments: [1H,15N]-TROSY, HNCO, HN(CA)CO, HNCA, HNCACB (Schulte-Herbrüggen and Sørensen 2000; Lescop et al. 2007; Favier and Brutscher 2011; Solyom et al. 2013). In addition, HBHA(CO)NH spectra (Grzesiek and Bax 1993; Muhandiram and Kay 1994) were recorded for proton resonance assignments of hsTRPV1-IDR and hsTRPV2-IDR. For hsTRPV3-IDR, 13C and 1H side chain chemical shift information was obtained from (H)CC(CO)NH (Montelione et al. 1992; Grzesiek et al. 1993) and [1H,15N]-TOCSY-HSQC spectra (Marion et al. 1989).
All spectra were processed using Bruker Topspin 3.6.2 or 4.1.1 and analyzed using CARA (www.cara.nmr.ch) or CcpNmr Analysis (Vranken et al. 2005) v2.4 (locally installed) or v2.5 (within the NMRbox virtual environment (Maciejewski et al. 2017)).
Disorder prediction
To predict the structural disorder of the N-terminal regions of the human TRPV1, TRPV2 and TRPV3 channels based on their primary structures, we used the ODiNPred web server (https://st-protein.chem.au.dk/odinpred) (Nielsen and Mulder 2019; Dass et al. 2020).
Further, the POTENCI tool ( https://st-protein02.chem.au.dk/potenci) (Nielsen and Mulder 2018) was used to calculate the random coil chemical shifts for hsTRPV1-IDR, hsTRPV2-IDR, and hsTRPV3-IDR based on their respective amino acid sequence at our experimental conditions (temperature, pH value and ionic strength). Using the backbone NMR assignments, we additionally applied the SSP program (Marsh et al. 2006) to assess the amount of secondary structure formation within the IDRs of the three TRPV channels.
Extent of assignment and data deposition
Although detailed structural information on TRPV channels is available from X-ray and cryo-EM studies, in most cases the distal N-terminal region preceding the ARD escaped description at an atomic level, either because it was purposefully deleted or because it remained unresolved due to its inherent flexibility (Goretzki et al. 2021). With a spectroscopic approach, we recently showed that the N-terminal regions of human and chicken TRPV4 channels are almost completely disordered (Goretzki et al. 2018, 2022). Interestingly, this was somewhat contradicted by sequence-based disorder prediction web tools that indicated the presence of a significant amount of secondary structure propensity in the TRPV4 IDRs (Goretzki et al. 2022).
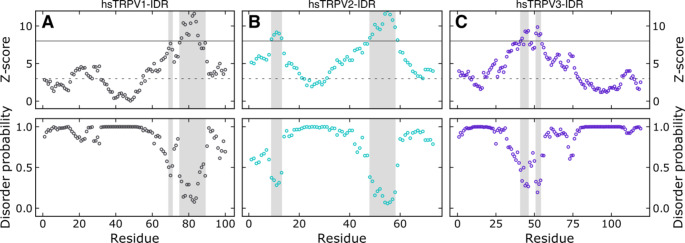
Here, to estimate the potential flexibility and disorder of the N-termini of the three remaining group I human TRPV channels and to compare this to NMR spectroscopic data, we used the sequence-based web server ODiNPred (Dass et al. 2020) (Fig. 1). ODiNPred predicts disorder probabilities larger than 0.5 for 86% of residues within TRPV1-IDR, 78% of residues within TRPV2-IDR and 91% of residues within TRPV3-IDR, which is indicative of highly disordered proteins. Nonetheless, on the C-terminal end of the TRPV1- and TRPV2-IDRs, lower disorder probabilities (< 0.5) are predicted for continuous stretches of amino acids in 14 of 100 and 16 of 72 residues, respectively. Such values are indicative of transient structural order within ensembles sampling the potential conformational space. In addition, in the N-terminal part of hsTRPV2-IDR, a second short region with low per-residue disorder probability is predicted. Notably, in hsTRPV3-IDR only 10 of 109 residues show a disorder probability lower than 0.5.
Fig. 1.
The sequence-based ODiNPred analysis predicts a significant amount of disorder in the distal N-termini of the human TRPV1, TRPV2 and TRPV3 channels. Regions with low predicted disorder propensities (shaded in light grey), as seen in particular for TRPV1 and TRPV2, could indicate the transient formation of structural order. The circles show the residue-specific Z-score (upper panel) and disorder probability (lower panel). A residue specific Z-score larger than 8 (solid line) indicates structural order while a Z-score below three (dashed line) predicts full disorder. Z-scores between three and eight reflect transient local structure propensity. The Z-score and disorder probability were calculated using the ODiNPred webserver (Dass et al. 2020)
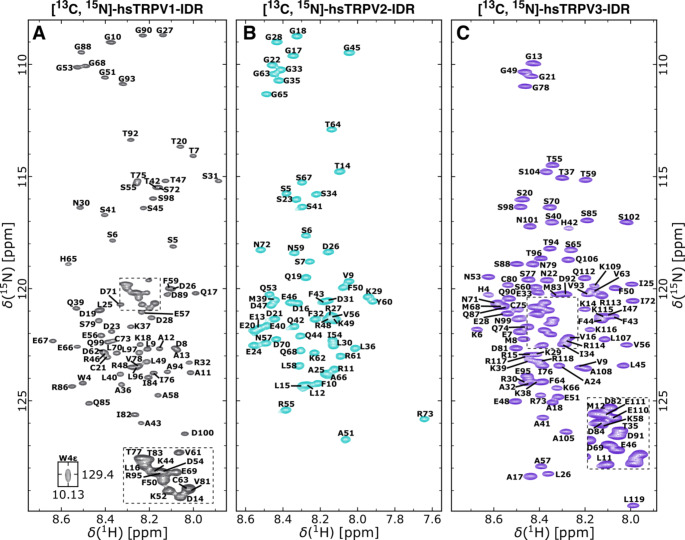
To experimentally characterize the N-terminal regions of the human TRPV1, TRPV2 and TRPV3 channels in solution, we used NMR spectroscopy. In agreement with a predicted low overall secondary structure content, the [1H, 15N]-TROSY-HSQC-spectra of human TRPV1-IDR, hsTRPV2 and TRPV3-IDR show limited signal dispersion in the 1HN dimension (Fig. 2 A-C), indicating a similar chemical environment of all 1HN nuclei and an inherent lack of stable structural elements. By using a set of two- and three-dimensional NMR experiments, the sequence specific resonance assignments for nearly all backbone 1H, 13C and 15N spins could be obtained. In summary, 91.3%, 92.6% and 91.1% of the HN, N´, C´, Cα, Cβ, Hα and Hβ resonances of hsTRPV1-, hsTRPV2- and hsTRPV3-IDR could be assigned, respectively.
Fig. 2.
[1H, 15N]-TROSY-HSQC spectra of 13C, 15N-labeled human TRPV1-IDR (residues 2-100) (A), TRPV2-IDR (residues 2–73) (B) and TRPV3-IDR (residues 2-119) (C) recorded at 600 MHz and 25 °C. Human TRPV1-IDR and TRPV2-IDR were measured in 10 mM Tris-HCl pH 7, 100 mM NaCl and hsTRPV3-IDR was measured in 10 mM NaPi pH 6.2, 300 mM NaCl, 1 mM DTT. 10% D2O for field-frequency locking and 0.1 mM DSS for internal 1H chemical shift referencing was added to all samples. Assigned residues are annotated in single letter amino acid code according to the human TRPV1 (UniProtKB: Q8NER1-1), TRPV2 (UniProtKB: Q9Y5S1-1), and TRPV3 (UniProtKB: Q8NET8-1) protein sequences.
All three proteins contain a large number of proline residues, i.e., 14 in TRPV1-IDR, 7 in TRPV2-IDR and 17 in TRPV3-IDR. These do not group in extended proline rich regions as in the N-terminus of TRPV4, but nonetheless frequently cluster in pairs of two or three. For the human TRPV1-IDR, the C´, Cα, Cβ resonances for 12 out of 14 proline residues could be assigned with mean Cβ values of 32.17 ± 0.18 ppm. This leaves only the consecutive proline residues in the triple proline repeat P33*-P34*-P35 unassigned (marked with *).
Likewise, backbone carbon chemical shifts for 6 out of 7 proline residues in hsTRPV2-IDR were assigned with mean Cβ values of 32.12 ± 0.06 ppm, leaving residue 37 in the P37*-P38 double proline motif unassigned.
For the hsTRPV3-IDR, the C´, Caα, Cβ as well as Cγ resonances for 16 of 17 proline residues could be assigned, leaving only P61 in the P61*-P62 motif without chemical shift information. All assigned hsTRPV3-IDR prolines show 13Cβ and 13Cγ values in the range of 32.17 ± 0.08 ppm and 27.47 ± 0.16 ppm, respectively. The mean difference of the proline 13Cβ and 13Cγ chemical shifts is 4.69 ± 0.20 ppm.
Based on the chemical shift values for Cβ and Cγ, it can be assumed that all assigned proline residues in hsTRPV1-IDR, hsTRPV2-IDR and hsTRPV3-IDR are in the trans configuration (Schubert et al. 2002; Shen and Bax 2010). It remains to be seen whether these residues can adopt a stable cis configuration in the presence of ligands, as was observed for TRPV4 (Goretzki et al. 2018).
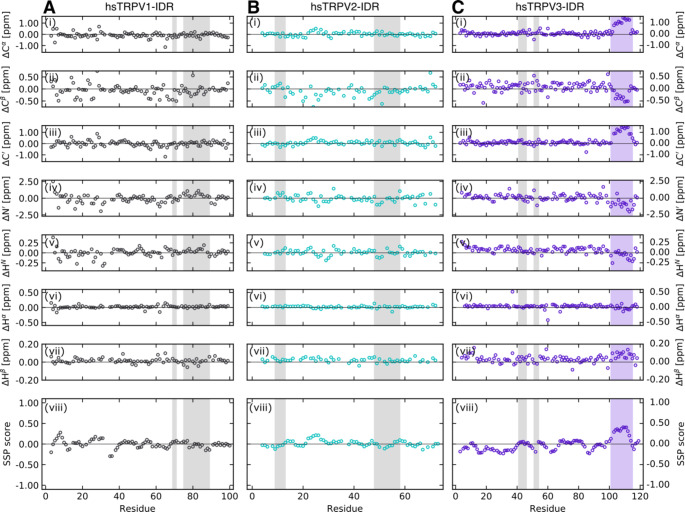
The experimentally obtained chemical shifts of the distal N-termini of hsTRPV1, hsTRPV2 and hsTRPV3 were also used for an initial secondary chemical shift-based structural analysis. The POTENCI web server (Nielsen and Mulder 2018) was used for the prediction of random coil chemical shifts at our experimental conditions. The predicted chemical shifts were compared with those experimentally obtained to reveal potential regions of structural order. For all three TRPV channel constructs, the measured and predicted Cα, Cβ, C´, N´, HN, Hα, and Hβ chemical shift values agree remarkably well (Fig. 3 A-C, i-vii). The mean differences between the experimental and POTENCI-predicted random coil chemical shift values for hsTRPV1-IDR are: ΔCα = -0.08 ± 0.24 ppm, ΔCβ = -0.08 ± 0.27 ppm, ΔC´ = 0.00 ± 0.26 ppm, ΔN´ = -0.03 ± 0.64 ppm, ΔHN = 0.01 ± 0.10 ppm, ΔHα = 0.02 ± 0.05 ppm, and ΔHβ = 0.02 ± 0.03 ppm. Likewise, the mean differences for hsTRPV2-IDR are: ΔCα = -0.02 ± 0.17 ppm, ΔCβ = -0.09 ± 0.22 ppm, ΔC´ = 0.06 ± 0.18 ppm, ΔN´ = -0.03 ± 0.51 ppm, ΔHN = 0.01 ± 0.07 ppm, ΔHα = 0.02 ± 0.04 ppm, and ΔHβ = 0.02 ± 0.02 ppm. For hsTRPV3-IDR the mean differences between experimental and predicted chemical shifts are: ΔCα = 0.18 ± 0.36 ppm, ΔCβ = 0.06 ± 0.21 ppm, ΔC´ = 0.20 ± 0.38 ppm, ΔN´ = 0.00 ± 0.61 ppm, ΔHN = 0.06 ± 0.09 ppm, ΔHα = 0.03 ± 0.11 ppm, and ΔHβ = 0.04 ± 0.04 ppm.
Fig. 3.
Secondary chemical shifts analysis reveals that the distal N-termini of the human TRPV1, TRPV2 and TRPV3 channels are highly disordered. (i-vii) Secondary chemical shifts are calculated as the differences between the experimentally determined and predicted Cα, Cβ, C´, N´, HN, Hα, Hβ chemical shifts. POTENCI (Nielsen and Mulder 2018) was used for sequence-based prediction of random coil chemical shifts. (viii) Secondary structure propensity (SSP) prediction based on chemical shifts using the SSP script (Marsh et al. 2006). A positive and negative SSP score reflects α-helix and β-sheet propensities, respectively. A SSP value of 1 reflects a fully formed helical-structure and a value of -1 a fully formed βstrand, respectively. Only 13Cα, 13Cβ and 1Hα chemical shifts of non-proline preceding residues were used when running the SSP script. For better comparison with Fig. 1, the regions with low predicted disorder propensities based on ODiNPred are shaded in light grey. The stretch of residues (P103-R113) in the C-terminal region of the hsTRPV3-IDR potentially forming an α-helical element is highlighted in light purple. Of note, ODiNPred does not predict secondary structure formation for this region in TRPV3 (Fig. 1 C)
The notion that the human TRPV1, TRPV2 and TRPV3 distal N-termini are highly disordered is further supported by the sequence-specific secondary structure propensity method (Marsh et al. 2006). As recommended for intrinsically disordered proteins, we used the SSP method to combine Cα, Cβ and Hα chemical shift values into single residue specific scores (Fig. 3 A-C, viii). In contrast to the analysis with the ODiNPred server, which indicated that hsTRPV1-IDR and hsTRPV2-IDR but not hsTRPV3-IDR contain extended regions able to form transient structures (Fig. 1), the SSP scores predict both the TRPV1 and TRPV2-IDR sequences to be highly disordered (mean SSP scores of 0.000 ± 0.097 and 0.015 ± 0.075 for hsTRPV1-IDR and hsTRPV2-IDR, respectively). Globally, the mean SSP score of the hsTRPV3-IDR is also close to 0 (-0.009 ± 0.156). However, a stretch of residues (P103-R113) in the C-terminal region of the hsTRPV3-IDR shows a significantly higher mean SSP score (0.355 ± 0.041), suggesting the potential formation of a helical structural element. The presence of an α-helical structure in this region is supported by recent cryo-EM structures of human TRPV3 (e.g. Zubcevic et al. 2019). Interestingly, AlphaFold (Jumper et al. 2021) also predicts a helical structure for this region.
By averaging the calculated SSP scores, an overall secondary structure content of only 7.7% for hsTRPV1-IDR, 6.0% for hsTRPV2-IDR and 12.0% for hsTRPV3-IDR can be estimated.
Although partial structural information on the N-terminal IDRs of group I TRPV channel members has been reported (e.g. Zubcevic et al. 2019; Pumroy et al. 2019; Nadezhdin et al. 2021), for the most part these regions are missing from structural studies. NMR spectroscopy is an optimal tool to investigate highly flexible and intrinsically disordered proteins. Together with our previous study on TRPV4 (Goretzki et al. 2022) we supplement the available structural information on group I TRPV channels with a detailed view on the extent of intrinsic disorder in their distal N-termini at atomic resolution. Interestingly, while there is some agreement with sequence-based structure predictions, the disorder determined in vitro tends to be more extensive than what is seen in silico. The inherent flexibility of the N-termini of group I TRPV channels is likely a conserved molecular feature underpinning their importance for channel regulation but making them challenging targets for structural studies.
Acknowledgements
BG acknowledges a PhD fellowship by the Max Planck Graduate Center (MPGC). ZNM acknowledges a fellowship of the German-American Fulbright Commission. We thank the Centre of Biomolecular Magnetic Resonance (BMRZ) at the Goethe University Frankfurt funded by the state of Hesse for support. Funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy - EXC 2051 - Project ID 390713860.
Author contributions
C.W., B.G., Z.N.M., F.T. and P.S. optimized protocols, expressed and purified proteins; C.W. and B.G. recorded data; C.W., B.G. and Z.N.M. analyzed data; U.A.H. conceptualized and supervised the study; C.W., B.G. and U.A.H. wrote the manuscript; all authors reviewed the manuscript.
Funding
Max Planck Graduate Center PhD Fellowship (to BG), German-American Fulbright Commission (to ZNM), BMRZ funded by the State of Hesse, DFG EXC 2051 – Project ID 390713860.
Open Access funding enabled and organized by Projekt DEAL.
Availability of data and material
The backbone assignments of the wildtype human TRPV1, TRPV2 and TRPV3 N-terminal intrinsically disordered regions have been deposited in the BioMagResBank (https://bmrb.io) under the accession numbers 51353 (TRPV1-IDR, residues 2-100), 51354 (TRPV2-IDR, residues 2–73) and 51355 (TRPV3-IDR, residues 2-119).
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Aisenberg WH, McCray BA, Sullivan JM et al (2022) Multi-ubiquitination of TRPV4 reduces channel activity independent of surface localization. J Biol Chem 101826. 10.1016/j.jbc.2022.101826 [DOI] [PMC free article] [PubMed]
- Baez-Nieto D, Castillo JP, Dragicevic C, et al. Thermo-TRP channels: biophysics of polymodal receptors. Adv Exp Med Biol. 2011;704:469–490. doi: 10.1007/978-94-007-0265-3_26. [DOI] [PubMed] [Google Scholar]
- Bujak JK, Kosmala D, Szopa IM et al (2019) Inflammation, Cancer and Immunity—Implication of TRPV1 Channel. Front Oncol 9. 10.3389/fonc.2019.01087 [DOI] [PMC free article] [PubMed]
- Caterina MJ, Schumacher MA, Tominaga M, et al. The capsaicin receptor: a heat-activated ion channel in the pain pathway. Nature. 1997;389:816–824. doi: 10.1038/39807. [DOI] [PubMed] [Google Scholar]
- Cohen MR, Moiseenkova-Bell VY. Structure of thermally activated TRP channels. Curr Top Membr. 2014;74:181–211. doi: 10.1016/B978-0-12-800181-3.00007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortright DN, Crandall M, Sanchez JF, et al. The tissue distribution and functional characterization of human VR1. Biochem Biophys Res Commun. 2001;281:1183–1189. doi: 10.1006/bbrc.2001.4482. [DOI] [PubMed] [Google Scholar]
- Dang S, van Goor MK, Asarnow D, et al. Structural insight into TRPV5 channel function and modulation. Proc Natl Acad Sci. 2019;116:8869–8878. doi: 10.1073/pnas.1820323116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dass R, Mulder FAA, Nielsen JT. ODiNPred: comprehensive prediction of protein order and disorder. Sci Rep. 2020;10:14780. doi: 10.1038/s41598-020-71716-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Paknejad N, Maksaev G, et al. Cryo-EM and X-ray structures of TRPV4 reveal insight into ion permeation and gating mechanisms. Nat Struct Mol Biol. 2018;25:252–260. doi: 10.1038/s41594-018-0037-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong Y, Yin Y, Vu S, et al. A distinct structural mechanism underlies TRPV1 activation by piperine. Biochem Biophys Res Commun. 2019;516:365–372. doi: 10.1016/j.bbrc.2019.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Favier A, Brutscher B. Recovering lost magnetization: polarization enhancement in biomolecular NMR. J Biomol NMR. 2011;49:9–15. doi: 10.1007/s10858-010-9461-5. [DOI] [PubMed] [Google Scholar]
- Gao Y, Cao E, Julius D, Cheng Y. TRPV1 structures in nanodiscs reveal mechanisms of ligand and lipid action. Nature. 2016;534:347–351. doi: 10.1038/nature17964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretzki B, Glogowski NA, Diehl E, et al. Structural Basis of TRPV4 N Terminus Interaction with Syndapin/PACSIN1-3 and PIP2. Structure. 2018;26:1583–1593e5. doi: 10.1016/j.str.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goretzki B, Guhl C, Tebbe F, et al. Unstructural Biology of TRP Ion Channels: The Role of Intrinsically Disordered Regions in Channel Function and Regulation. J Mol Biol. 2021;433:166931. doi: 10.1016/j.jmb.2021.166931. [DOI] [PubMed] [Google Scholar]
- Goretzki B, Tebbe F, Mitrovic S-A, Hellmich UA. Backbone NMR assignments of the extensive human and chicken TRPV4 N-terminal intrinsically disordered regions as important players in ion channel regulation. Biomol NMR Assign. 2022 doi: 10.1007/s12104-022-10080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grzesiek S, Bax A. Amino acid type determination in the sequential assignment procedure of uniformly 13 C/15 N-enriched proteins. J Biomol NMR. 1993;3:185–204. doi: 10.1007/BF00178261. [DOI] [PubMed] [Google Scholar]
- Grzesiek S, Anglister J, Bax A. Correlation of Backbone Amide and Aliphatic Side-Chain Resonances in 13 C/15 N-Enriched Proteins by Isotropic Mixing of 13 C Magnetization. J Magn Reson Ser B. 1993;101:114–119. doi: 10.1006/jmrb.1993.1019. [DOI] [Google Scholar]
- Hellmich UA, Gaudet R. Structural biology of TRP channels. Handb Exp Pharmacol. 2014;223:963–990. doi: 10.1007/978-3-319-05161-1_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel NJ, Cox DN. Transient receptor potential channels: current perspectives on evolution, structure, function and nomenclature. Proc R Soc B Biol Sci. 2020;287:20201309. doi: 10.1098/rspb.2020.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes TET, Lodowski DT, Huynh KW, et al. Structural basis of TRPV5 channel inhibition by econazole revealed by cryo-EM. Nat Struct Mol Biol. 2018;25:53–60. doi: 10.1038/s41594-017-0009-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh KW, Cohen MR, Jiang J, et al. Structure of the full-length TRPV2 channel by cryo-EM. Nat Commun. 2016;7:11130. doi: 10.1038/ncomms11130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islas LD (2017) Molecular Mechanisms of Temperature Gating in TRP Channels. In: Neurobiology of TRP Channels. CRC Press, Boca Raton: CRC Press, 2017., pp 11–25 [PubMed]
- Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596:583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescop E, Schanda P, Brutscher B. A set of BEST triple-resonance experiments for time-optimized protein resonance assignment. J Magn Reson. 2007;187:163–169. doi: 10.1016/j.jmr.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Liao M, Cao E, Julius D, Cheng Y. Structure of the TRPV1 ion channel determined by electron cryo-microscopy. Nature. 2013;504:107–112. doi: 10.1038/nature12822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewski MW, Schuyler AD, Gryk MR, et al. NMRbox: A Resource for Biomolecular NMR Computation. Biophys J. 2017;112:1529–1534. doi: 10.1016/j.bpj.2017.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson LJ, Geierstanger BH, Viswanath V, et al. The Pungency of Garlic: Activation of TRPA1 and TRPV1 in Response to Allicin. Curr Biol. 2005;15:929–934. doi: 10.1016/j.cub.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Marion D, Driscoll PC, Kay LE, et al. Overcoming the overlap problem in the assignment of proton NMR spectra of larger proteins by use of three-dimensional heteronuclear proton-nitrogen-15 Hartmann-Hahn-multiple quantum coherence and nuclear Overhauser-multiple quantum coherence spectroscopy. Biochemistry. 1989;28:6150–6156. doi: 10.1021/bi00441a004. [DOI] [PubMed] [Google Scholar]
- Marsh JA, Singh VK, Jia Z, Forman-Kay JD. Sensitivity of secondary structure propensities to sequence differences between α- and γ-synuclein: Implications for fibrillation. Protein Sci. 2006;15:2795–2804. doi: 10.1110/ps.062465306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGoldrick LL, Singh AK, Saotome K, et al. Opening of the human epithelial calcium channel TRPV6. Nature. 2018;553:233–237. doi: 10.1038/nature25182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montelione GT, Lyons BA, Emerson SD, Tashiro M. An efficient triple resonance experiment using carbon-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically-enriched proteins. J Am Chem Soc. 1992;114:10974–10975. doi: 10.1021/ja00053a051. [DOI] [Google Scholar]
- Muhandiram DR, Kay LE. Gradient-Enhanced Triple-Resonance Three-Dimensional NMR Experiments with Improved Sensitivity. J Magn Reson Ser B. 1994;103:203–216. doi: 10.1006/jmrb.1994.1032. [DOI] [Google Scholar]
- Nadezhdin KD, Neuberger A, Trofimov YA, et al. Structural mechanism of heat-induced opening of a temperature-sensitive TRP channel. Nat Struct Mol Biol. 2021;28:564–572. doi: 10.1038/s41594-021-00615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen JT, Mulder FAA. POTENCI: prediction of temperature, neighbor and pH-corrected chemical shifts for intrinsically disordered proteins. J Biomol NMR. 2018;70:141–165. doi: 10.1007/s10858-018-0166-5. [DOI] [PubMed] [Google Scholar]
- Nielsen JT, Mulder FAA. Quality and bias of protein disorder predictors. Sci Rep. 2019;9:5137. doi: 10.1038/s41598-019-41644-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilius B, Voets T. The puzzle of TRPV4 channelopathies. EMBO Rep. 2013;14:152–163. doi: 10.1038/embor.2012.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omar S, Clarke R, Abdullah H, et al. Respiratory virus infection up-regulates TRPV1, TRPA1 and ASICS3 receptors on airway cells. PLoS ONE. 2017;12:e0171681. doi: 10.1371/journal.pone.0171681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. Proc Natl Acad Sci. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prevarskaya N, Zhang L, Barritt G. TRP channels in cancer. Biochim Biophys Acta - Mol Basis Dis. 2007;1772:937–946. doi: 10.1016/j.bbadis.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Pumroy RA, Samanta A, Liu Y et al (2019) Molecular mechanism of TRPV2 channel modulation by cannabidiol. Elife 8. 10.7554/eLife.48792 [DOI] [PMC free article] [PubMed]
- Ramsey IS, Delling M, Clapham DE. AN INTRODUCTION TO TRP CHANNELS. Annu Rev Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- Samanta A, Hughes TET, Moiseenkova-Bell VY (2018) Transient Receptor Potential (TRP) Channels. In: Sub-cellular biochemistry. pp 141–165 [DOI] [PMC free article] [PubMed]
- Schanda P, Van Melckebeke H, Brutscher B. Speeding Up Three-Dimensional Protein NMR Experiments to a Few Minutes. J Am Chem Soc. 2006;128:9042–9043. doi: 10.1021/ja062025p. [DOI] [PubMed] [Google Scholar]
- Schubert M, Labudde D, Oschkinat H, Schmieder P. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13 C chemical shift statistics. J Biomol NMR. 2002;24:149–154. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- Schulte-Herbrüggen T, Sørensen OW. Clean TROSY: Compensation for Relaxation-Induced Artifacts. J Magn Reson. 2000;144:123–128. doi: 10.1006/jmre.2000.2020. [DOI] [PubMed] [Google Scholar]
- Shen Y, Bax A. Prediction of Xaa-Pro peptide bond conformation from sequence and chemical shifts. J Biomol NMR. 2010;46:199–204. doi: 10.1007/s10858-009-9395-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada H, Kusakizako T, Dung Nguyen TH, et al. The structure of lipid nanodisc-reconstituted TRPV3 reveals the gating mechanism. Nat Struct Mol Biol. 2020;27:645–652. doi: 10.1038/s41594-020-0439-z. [DOI] [PubMed] [Google Scholar]
- Singh AK, McGoldrick LL, Sobolevsky AI. Structure and gating mechanism of the transient receptor potential channel TRPV3. Nat Struct Mol Biol. 2018;25:805–813. doi: 10.1038/s41594-018-0108-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh AK, McGoldrick LL, Demirkhanyan L, et al. Structural basis of temperature sensation by the TRP channel TRPV3. Nat Struct Mol Biol. 2019;26:994–998. doi: 10.1038/s41594-019-0318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solyom Z, Schwarten M, Geist L, et al. BEST-TROSY experiments for time-efficient sequential resonance assignment of large disordered proteins. J Biomol NMR. 2013;55:311–321. doi: 10.1007/s10858-013-9715-0. [DOI] [PubMed] [Google Scholar]
- Taga A, Peyton MA, Goretzki B, et al. TRPV4 mutations causing mixed neuropathy and skeletal phenotypes result in severe gain of function. Ann Clin Transl Neurol. 2022 doi: 10.1002/acn3.51523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo Mauriño JJ, Fonseca-Camarillo G, Furuzawa-Carballeda J, et al. TRPV Subfamily (TRPV2, TRPV3, TRPV4, TRPV5, and TRPV6) Gene and Protein Expression in Patients with Ulcerative Colitis. J Immunol Res. 2020;2020:1–11. doi: 10.1155/2020/2906845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Goor MK, de Jager L, Cheng Y, van der Wijst J. High-resolution structures of transient receptor potential vanilloid channels: Unveiling a functionally diverse group of ion channels. Protein Sci. 2020;29:1569–1580. doi: 10.1002/pro.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viet KK, Wagner A, Schwickert K, et al. Structure of the Human TRPML2 Ion Channel Extracytosolic/Lumenal Domain. Structure. 2019;27:1246–1257e5. doi: 10.1016/j.str.2019.04.016. [DOI] [PubMed] [Google Scholar]
- Voolstra O, Huber A. Post-Translational Modifications of TRP Channels. Cells. 2014;3:258–287. doi: 10.3390/cells3020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, et al. The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct Funct Genet. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- Wishart D, Bigam C, Yao J, et al. 1H, 13 C and 15 N chemical shift referencing in biomolecular NMR. J Biomol NMR. 1995;6:135–140. doi: 10.1007/BF00211777. [DOI] [PubMed] [Google Scholar]
- Xu H, Delling M, Jun JC, Clapham DE. Oregano, thyme and clove-derived flavors and skin sensitizers activate specific TRP channels. Nat Neurosci. 2006;9:628–635. doi: 10.1038/nn1692. [DOI] [PubMed] [Google Scholar]
- Zubcevic L, Herzik MA, Chung BC, et al. Cryo-electron microscopy structure of the TRPV2 ion channel. Nat Struct Mol Biol. 2016;23:180–186. doi: 10.1038/nsmb.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubcevic L, Herzik MA, Wu M, et al. Conformational ensemble of the human TRPV3 ion channel. Nat Commun. 2018;9:4773. doi: 10.1038/s41467-018-07117-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubcevic L, Borschel WF, Hsu AL et al (2019) Regulatory switch at the cytoplasmic interface controls TRPV channel gating. Elife 8. 10.7554/eLife.47746 [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The backbone assignments of the wildtype human TRPV1, TRPV2 and TRPV3 N-terminal intrinsically disordered regions have been deposited in the BioMagResBank (https://bmrb.io) under the accession numbers 51353 (TRPV1-IDR, residues 2-100), 51354 (TRPV2-IDR, residues 2–73) and 51355 (TRPV3-IDR, residues 2-119).