Abstract
The overlapping and opposing promoter elements for the Escherichia coli fepDGC operon and the ybdA gene (encoding a 43-kDa cytoplasmic membrane protein) within the enterobactin gene cluster were investigated by measuring the effects of site-specific mutations on transcript levels and on expression of reporter genes in a bidirectional transcriptional fusion vector. Primary promoter structures for the opposing transcripts overlapped extensively such that their −10 sequences were almost directly opposed on the two strands of the DNA helix and their +1 transcription start sites were only 23 bp apart. Relative to the E. coli consensus sequence, both promoters were poorly conserved at the −35 position and mutations which strengthened the −35 element of either promoter significantly enhanced its transcription, decreased that of the opposing promoter, and dramatically altered iron-mediated regulation of expression. Both the fepD and ybdA primary promoters were shown to require a 5′-TGn-3′ upstream extension of their −10 elements for optimal activities. Secondary promoters were identified for both fepD and ybdA, and their contributions to the overall expression levels were evaluated in these dual expression vector constructs. The data provided strong evidence that the architecture of the regulatory elements within the overlapping fepD and ybdA promoters is configured such that there is a direct competition for binding RNA polymerase and that the expression levels at these promoters are influenced not only by the activity of the opposing promoters but also by additional promoter sequence elements and perhaps accessory regulatory factors. Iron-mediated regulation of these promoters through the repressor protein Fur is a consequence of the relative promoter strengths and the position of an operator site that consists of two overlapping Fur-binding sequences in this compact regulatory region.
Delicate regulatory balances play an essential role in the survival of microorganisms. To that end, microbes have evolved specialized systems to allow adaptation to changes in environmental growth conditions and maintenance of balanced metabolic processes. Since iron is an essential cofactor for important metabolic activities, rapid and efficient responses to iron-limiting environments are required for the growth and pathogenicity of microbial species. Mediated by iron-binding proteins or specific siderophores, the microbial response to iron limitation is effective in solubilizing and transporting nutritional iron sources. However, the intracellular levels of iron must be carefully regulated, since free iron can serve as a reactant in the generation of cell-damaging reactive oxygen species (26). The sensitive mechanisms involved in assimilating an adequate internal iron supply while protecting the cell against the damaging effects of iron-related oxygen radicals are controlled by the global regulatory protein Fur (4). Through interaction with transcriptional control regions (9, 14, 17, 25, 29, 50), Fur regulates the expression of numerous iron-regulated chromosomal genes, including those for iron uptake and a subset of genes with detoxifying activity toward cell-damaging hydroxyl radicals and superoxides (13, 51, 52). It is generally accepted that for most of the iron-regulated promoters examined, Fur repressor protein functions by direct competition with RNA polymerase for access to the promoter-operator region (5, 14, 20, 22, 56).
Escherichia coli strains produce enterobactin as their principal iron chelator and maintain multiple membrane transport systems for internalization of a variety of exogenously produced microbial iron-binding compounds (40). The enterobactin gene cluster includes 14 genes tightly organized into six operons originating from three Fur-controlled bidirectional promoter-operator regions. These three control regions possess distinct regulatory architectures (9, 29, 50), suggesting that control by the Fur repressor is manifest through different regulatory strategies.
Many bidirectional promoter regions have been described in prokaryotes (6, 22, 38), and these offer several effective regulatory configurations and evolutionary advantages. These promoters have been divided into three classes of divergent transcription units: back-to-back, overlapping, and face-to-face (6). The back-to-back variety, e.g., araC-araBAD and malE-malK, is characterized by transcriptional start sites for the opposing mRNAs separated by 75 to 513 bp. The back-to-back promoters may be individually regulated and transcriptionally independent, in most instances dependent on the distance between the 5′ termini of the transcripts expressed. The overlapping, divergent transcription regions are the most compact, with opposing −10 and −35 promoter elements overlapping to various degrees and 5′ ends of transcripts separated by distances ranging from 16 bp (tetR-tetA on Tn10) to 62 bp (nahR-nahG of Pseudomonas plasmid NAH7) (6). Because the promoter elements occupy the same sequence regions on opposing template strands, their influence on each other is a significant component of the regulatory process. In the face-to-face control regions, the promoters are again separated but express opposing transcripts with 5′ ends that are complementary to various extents, e.g., from 10 bp in the bioA-bioBFCD transcripts to 182 bp in the argE-argCBH transcripts (6). Expression levels are thus influenced not only by the likelihood of collisions between converging transcription complexes but also by the presence of antisense mRNA populations which can dictate transcript stability or translation capability.
Among the three enterobactin bidirectional control regions, the promoters for fepB and the entCEBA ybdB operon are situated back to back, with 103 bp separating the opposing transcript start sites (9). These two promoters are independently expressed, each controlled by Fur from distinct operator sites that have at best minimal influence on the opposing promoter. The remaining two control regions, one between fepA and fes and the other between fepD and ybdA, are classified as overlapping, with opposing promoter elements occupying the same DNA regions. There are 48 bp separating the 5′ start sites of the overlapping fepA and fes transcripts, with well-conserved promoter elements in direct opposition (29). Fur strategically occupies a single central operator region situated between the −10 and −35 elements of both promoters, thus providing direct competition to RNA polymerase occupancy of either promoter. Evidence suggests that Fur simultaneously regulates expression of both transcripts (22, 29).
In this study, the promoter elements controlling transcription of the fepDGC operon and the ybdA gene were examined in detail. The fepDGC operon encodes the proteins of the cytoplasmic membrane permease responsible for ferric enterobactin transport (11, 50). The ybdA gene, previously referred to as P43 (50) or orf43 (11), encodes an integral cytoplasmic membrane protein with significant homology to several proton motive force-dependent membrane transporters. Its function remains unknown.
The architecture of the fepD-ybdA bidirectional promoter region is similar to yet significantly different from the design of the fepA-fes components. Initial data indicated that only 23 bp separate the start sites of the fepD and ybdA transcripts (50). The present study used site-directed mutagenesis with reporter gene and transcript analyses to examine in detail the promoter elements involved in expression of the fepDGC and ybdA transcripts. Promoters responsible for fepD and ybdA expression overlap substantially at their −10 regions. These promoters are significantly weaker in consensus than the fepB-entC or the fepA-fes elements and rely on extended promoter elements for maximal activity. Because of their relative positions, mutations which alter the strength of either opposing promoter have a significant influence on the expression level of the divergent promoter. Furthermore, iron-regulated control of promoter activity through the Fur repressor is shown to be dependent upon the relative promoter strength.
MATERIALS AND METHODS
Bacterial strains, plasmids, bacteriophages, and primers.
Escherichia coli strain CC117 (relevant genotype, lacZ phoA) and its recA1 derivative CC118 (37) were used as hosts for all fusion plasmids. CJ236 (relevant genotype, dut-1 ung-1) (30) was used for the isolation of uracil-containing M13 templates. The M13mp19 vector was used in site-directed mutagenesis (41) and nucleotide sequencing procedures, with JM101 (41) and NM522 (24) as host strains for propagation of M13mp19 derivatives.
The bidirectional reporter plasmid, pUJ10, was a gift from V. de Lorenzo (15), and all pFD43 derivatives carrying the fepD-ybdA control region were constructed in this vector (Fig. 1). Plasmid pUJ10 was derived from pCB267, a pBR322 descendant with a copy number of approximately 10. RNA probes used in RNase protection experiments were generated by in vitro transcription of pCAC/FD43-1, which carries the 327-bp BamHI-PstI fragment of pFD43-1 in pGEM4Z (Promega Corp., Madison, Wis.).
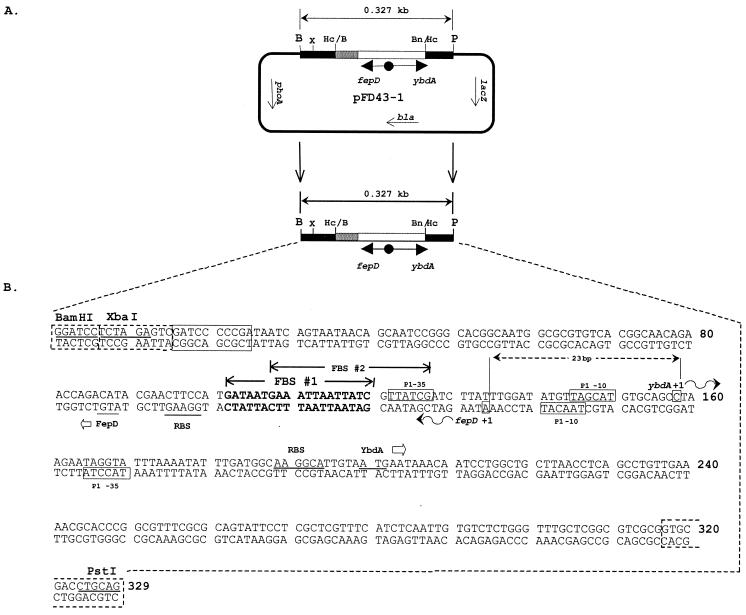
FIG. 1.
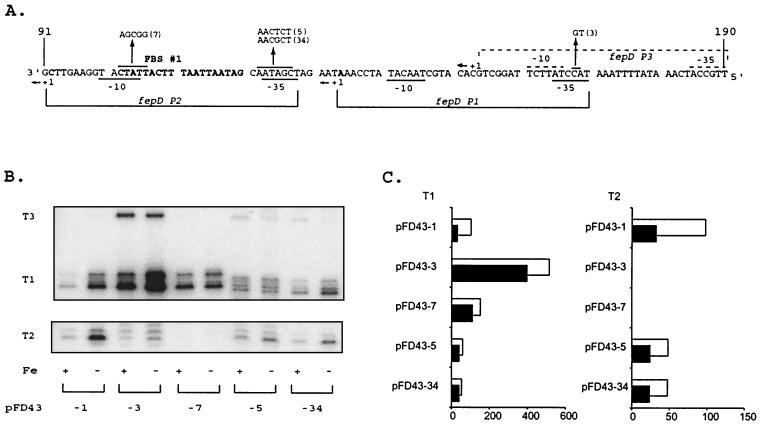
The fepD-ybdA bidirectional transcription fusion vector and sequence of the transcriptional control region. (A) Structure of the wild-type promoter construct, pFD43-1, with fepD fused to phoA and ybdA to lacZ. Abbreviations for the restriction enzymes used: B, BamHI; Bn/Hc, BanI-HincII hybrid site; Hc/B, HincII-BamHI hybrid site; P, PstI; X, XbaI. Plasmid reporter genes are lacZ (β-galactosidase) and phoA (alkaline phosphatase); bla is the selectable β-lactamase gene and serves as an internal standard for transcript quantitation. Arrows within constructs designate the direction of the transcripts. (B) Sequence of the 329-bp region containing the promoter cassette used for mutagenesis, nucleotide sequencing, and reporter gene analysis. The BamHI and PstI cloning sites are noted. Boxed sequences at the termini originate from the vectors used in construction steps (see Materials and Methods). The boxes with dashed edges are from the M13mp19 polylinker, and the solid box near the left end of the sequence is from the pCON4 vector. These sequences are represented by the black and gray boxes, respectively, in the vector map in panel A (not to scale). Predicted overlapping primary 19-bp Fur-binding core sequences are identified and labeled FBS #1 and FBS #2. Putative primary promoter elements (P1) for the fepD and ybdA genes are represented and labeled. The +1 primary transcriptional start sites for ybdA and fepD are identified, and the curved arrows denote the direction of transcription. The proposed ATG translational start sites for both the FepD and YbdA proteins and their predicted corresponding ribosome binding sequences are underlined. Nucleotides are numbered 5′ to 3′ from the BamHI site to the PstI site.
Oligonucleotide primers were synthesized by the University of Missouri DNA Core Facility. The primers used for site-directed mutagenesis and primer extension are listed in Table 1. Mismatched bases resulting in mutations are underlined, and nucleotide sequence positions indicated are relative to those in Fig. 1. For standardization of RNA loads in primer extension experiments, primer M8 (29) was used to generate a 110-bp product from the plasmid-encoded β-lactamase gene, and levels of fepD and ybdA transcripts from the same plasmids were quantitated relative to this standard. This corrects for any potential minor differences in plasmid copy number when cells are grown under different conditions.
TABLE 1.
Primers used in this study
| Primera | Gene | Sequencee | Location (5′→3′)b |
|---|---|---|---|
| m2 | fepD | 5′-GAT CTT ATT TGG ATG TGT TAG CAT GTG C-3′ | −31→−4 |
| m3 | fepD | 5′-GCC TAA GAA TAG CAA TTT AAA ATA TTT G-3′ | −2→+26 |
| m4 | P43 | 5′-GGA TAT GTT AGC ACG TGC AGC CTA AG-3′ | −21→+5 |
| m5 | P43 | 5′-ATT AAT TAT CGT TGA CGA TCT TAT TTG G-3′ | −47→−20 |
| m6 | fepD-P43 | 5′-CTT ATT TGG ATA TGT TGG CAT GTG CAG CCT-3′ | −28→+2 |
| m7 | FBS 1 | 5′-ACG AAC TTC CAT TCG CCT GAA ATT AAT TAT CG-3′ | −68→−37 |
| m30 | P43 | 5′-GGA TAT ATT AGC ATG TGC-3′ | −21→−4 |
| m31 | fepD | 5′-GGA TAT GTT AGT ATG TGC AGC-3′ | −21→−1 |
| m32 | fepD | 5′-CGA ACT TCC CCG ATA ATG-3′ | −67→−50 |
| m33c | fepD | 5′-TAT CGT TAT GGG TCT TAT TTG GAT GTG TTA GCA TG-3′ | −41→−7 |
| m34 | P43 | 5′-GAA ATT AAT TAT CGT TGA CGA TCT TAT TTG G-3′ | −50→−20 |
| pe1 | fepD | 5′-GGC GCG TGT CAC GGC AAC-3′ | −98→−81 |
| pe2 | fepD | 5′-CAG TAA TAA CAG CAA TCC G-3′ | −128→−110 |
| pe3 | P43 | 5′-CAA CAG GCT GAG GTT AAG-3′ | −104→−87d |
| pe4 | P43 | 5′-CAG CCA GGA TTG TTT ATT C-3′ | −86→−68d |
| M8 | bla | 5′-TGG GTG AGC AAA AAC AGG AAG G-3′ |
m designates the primers used for site-directed mutagenesis; pe represents the primers used for primer extension analysis; M8 represents the primer used to detect the bla control transcript (29).
The location of all of the primers relative to the P43 transcription start (Fig. 1) is given except where noted.
Bases changed at the fepD P2 −35 element with primer m33 are also located in FBS 2. This primer also creates the same change in the fepD P1 promoter −10 region as primer m2.
Primers designed for P43 transcript analysis are complementary to the P43 mRNA and are numbered relative to the fepD transcription start.
Mismatched bases are underlined.
Media, growth conditions, chemicals, and enzymes.
Luria-Bertani medium (39) was used for growth of all E. coli strains. 2× YT (49) was used for propagation of M13mp19 derivatives. Iron-poor media were prepared by addition of 200 μM 2,2′-dipyridyl, and iron-replete conditions were maintained with the addition of 20 μM FeSO4 and 10 mM sodium citrate (9, 29, 50). Ampicillin at a final concentration of 100 μg/ml, chloramphenicol at 30 μg/ml, and kanamycin at 40 μg/ml were used. Restriction and modifying enzymes and molecular biology reagents were purchased from Boehringer Mannheim (Indianapolis, Ind.), United States Biochemical Corp. (Cleveland, Ohio), New England Biolabs (Beverly, Mass.), or Fisher Scientific (St. Louis, Mo.). o-Nitrophenyl phosphate and o-nitrophenyl-β-d-galactoside were purchased from Sigma Chemical Co. (St. Louis, Mo.).
General genetic methods and nucleotide sequencing.
Isolation of plasmid and single-stranded M13 DNA for nucleotide sequencing of both single- and double-stranded templates has been described (42, 44). Plasmid DNA was also isolated using the Magic/Wizard DNA Preparation Kit (Promega). All cloning procedures and bacterial transformations followed standard molecular biology protocols (49). Sequencing, primer extension, and RNase protection reactions were performed as described previously (29, 42, 44) and were analyzed on 6% or 8% polyacrylamide–urea sequencing gels and were exposed to Kodak XRP-5 film at −70°C without drying.
Generation of the iron-regulated fepD′-′phoA and ybdA′-′lacZ promoter constructs.
A 1.0-kb NruI-HpaI DNA fragment from pITS24 (43), spanning the fepD-ybdA intergenic region, was cloned in both orientations into the SmaI site of the pCON4 operon expression vector (16); both constructs produced iron-regulated fusion transcripts. With fepD fused to lacZ (pCON410), 452 Miller units of β-galactosidase (39) was produced under high-iron growth conditions and 1,353 Miller units was produced when iron was limiting. The ybdA′-′lacZ fusion construct (pCON411) produced 636 Miller units under high-iron conditions and 1,671 Miller units under low-iron conditions. Subsequently, a 0.3-kb BamHI-BanI fragment from pCON410, containing the fepD-ybdA bidirectional promoter region, was end filled and ligated with HincII-digested M13mp19 replicative-form DNA for production of single-stranded DNA for site-directed mutagenesis. After confirmation by nucleotide sequencing, wild-type and mutagenized DNA fragments were subcloned into the bidirectional reporter plasmid pUJ10 (15) using the flanking BamHI and PstI sites and were used to transform either CC118 or CC117. The final wild-type construct, pFD43-1 (Fig. 1), resulted in transcriptional fusions to the promoterless alkaline phosphatase (fepD′-′phoA) and β-galactosidase (ybdA′-′lacZ) genes.
Site-directed mutagenesis.
Mutations were generated in the cloned fepD-ybdA promoter region using the uracil-containing template method (34) and the MutaGene M13 in vitro mutagenesis kit (Bio-Rad, Richmond, Calif.). After passage of the recombinant M13mp19 derivatives in CJ236, single-stranded uracil-containing DNA templates were isolated and annealed with one of the mutagenic primers (Table 1); second-strand synthesis, transfection, and propagation of M13 derivatives have been described (9, 29).
Enzyme assays.
Alkaline phosphatase (8) and β-galactosidase (39) activity assays were performed on CC118 cells harboring the various fusion plasmids and grown in low-iron or high-iron media, as described previously (29). The data presented are averages obtained from multiple assays of fusion constructs performed on the same day under identical conditions.
RNA isolation and transcript analyses.
RNA isolation, phosphorylation of primers, and primer extensions followed methods described previously (29, 44). RNA was isolated from CC118 or CC117 cells harboring pFD43-1 or one of its mutant derivatives, grown under iron-poor and iron-replete conditions. A fepD- or ybdA-specific primer (Table 1) was end labeled with [γ-32P]dATP (3,000 Ci/mmol; New England Nuclear, Boston, Mass.), and 250 fmol was annealed to 40 μg of total cellular RNA. Extension reactions were carried out using 10 to 22 U of avian myeloblastosis virus reverse transcriptase (United States Biochemical) per reaction. Quantitative comparison of the resultant extension products was made possible by generating a primer extension product off the bla transcript from the same plasmid using 250 fmol of the M8 primer (29) either in the same reactions or, in some instances, separate but identical reactions. RNase protection assays to detect chromosomally expressed fepD and ybdA transcripts were performed as described elsewhere (29) using the Ambion RPAII kit (Ambion, Inc., Austin, Tex.).
Comparative quantitation of transcripts with a PhosphorImager.
Undried polyacrylamide-urea gels were wrapped and exposed to storage phosphor screens (Eastman Kodak, Rochester, N.Y.). The relative intensities of the individual bands were determined with ImageQuant version 3.0 software and a model 400A PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.). Each RNA sample was normalized using the quantitated value of the internal bla standard. The normalized wild-type-induced value for either the fepD or ybdA transcript was arbitrarily set at 100%. All other transcript levels were then expressed as the percentage of this value.
RESULTS
Identification of fepD and ybdA transcripts.
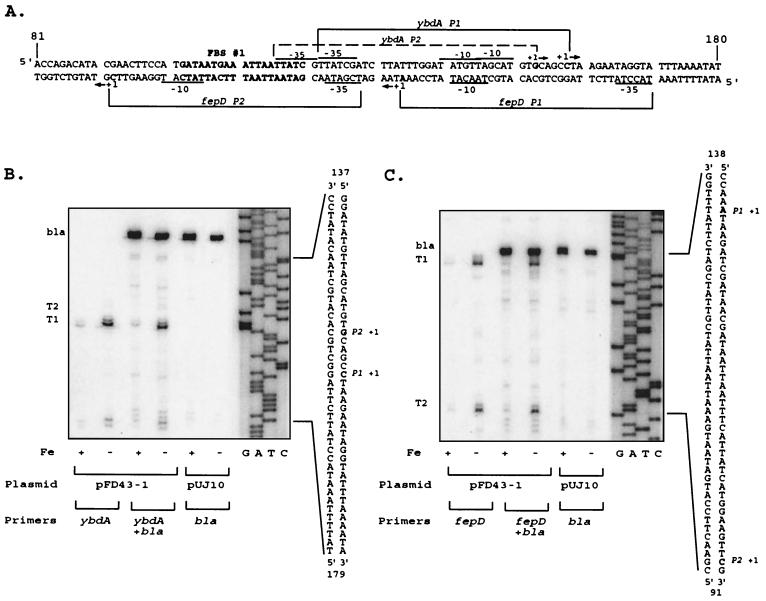
Primer extension analysis of the transcripts encoding ferric enterobactin transport proteins revealed that the major transcription start sites for the divergent fepDGC and ybdA promoters (50), now designated fepD P1 and ybdA P1, are separated by only 23 bp (Fig. 1), significantly more overlapping than the two other bidirectional enterobactin promoter regions. The predicted promoter elements for fepD P1 and ybdA P1 are shown in Fig. 1 and were assigned based on primer extension data (Fig. 2) and spacing considerations from the known geometry of ς70 promoters (27). The ybdA P1 promoter exhibits a ς70-like −10 region, 5′-TAgcAT-3′, that is conserved at four important positions of the hexamer (conserved bases shown in capital letters). However, its proposed −35 region, 5′-TTatCg-3′, has only weak similarity to the consensus. The sequence representing the fepD P1 −10 element, 5′-TAacAT-3′, is also conserved at four key sequence positions and overlaps the ybdA P1 −10 element at 2 nucleotides (nt), while the fepD P1 −35 region, 5′-TacctA-3′, bears almost no resemblance to the consensus sequence. The poor sequence conservation illustrated by both −35 ς70 recognition elements suggests that these promoters are weaker than the consensus E. coli ς70 promoter and may require additional factors for maximal expression (27, 47).
FIG. 2.
Transcripts originating from the fepD-ybdA bidirectional promoter region. (A) Sequence of the fepD-ybdA promoter region (extending from bp 81 to 180 shown in Fig. 1). The predicted “iron box” sequence (50) is boldfaced and designated as FBS #1. Brackets delineate the extent of the designated promoter regions, with corresponding −10 and −35 elements noted for each. The −10 TGn extensions for both P1 promoters are not designated. The dominant transcriptional start sites are shown in boldface and are denoted by +1, and the direction of transcription is indicated by the arrows alongside. (B) ybdA primer extension. Oligonucleotide pe4 (Table 1) was end labeled and used to map the ybdA transcription start sites as described in Materials and Methods. T1 and T2 represent the transcripts corresponding to the ybdA P1 and P2 promoters, respectively; bla represents the bla control transcript generated from the same RNA population with primer M8 (Table 1). The sequence ladder was generated with the pe4 primer on double-stranded pFD43-1 DNA template and was run alongside the extension reactions to identify the 5′ ends of the transcripts, which are noted on the corresponding sequence alongside. Conditions for cell growth prior to RNA isolation, the plasmid template for RNA expression, and primers used are given below the extension autoradiograms. Fe +, high iron; Fe −, low iron; pFD43-1, wild-type fepD-P43 promoter region; pUJ10, vector control. Primers (Table 1): ybdA, pe4 primer; ybdA + bla, pe4 and M8 primers; bla, M8 primer alone. (C) fepD primer extension. Oligonucleotide pe2 (Table 1) was used to map the fepD transcription start sites and generate the sequence markers. T1 and T2 represent the transcripts corresponding to the fepD P1 and P2 promoters, respectively, and bla represents the bla control transcript generated on the same RNA with primer M8 (Table 1). Conditions and designations are as described for the ybdA data in panel B.
Both the ybdA and the fepDGC transcripts were regulated by iron availability (Fig. 2). The regulation was mediated by the iron-binding Fur repressor protein, since there was no difference in transcript levels in a fur null mutant grown under low- or high-iron conditions (data not shown). Based on the consensus 19-bp Fur-binding sequence (17), a strong “iron box” (FBS 1), which might control iron-regulated expression of both fepD P1 and ybdA P1 (Fig. 2), was identified just upstream of the ybdA −35 element (50). A second, conserved, core Fur-binding sequence (FBS 2) overlaps FBS 1 by 13 nt and is thus positioned a half-turn of the helix downstream. By analogy to recent observations (21), this configuration could be viewed as four contiguous units of the hexameric Fur-binding motif 5′-NAT(A/T)AT-3′.
Bidirectional transcriptional fusion constructs.
Transcription regulation within this promoter region was examined by generating mutations in the proposed control elements and analyzing their effects in two independent assays: (i) reporter gene expression following ligation of this region into the dual reporter and transcriptional fusion vector pUJ10 (15) and (ii) direct measurement of transcript levels by primer extension (Fig. 2). The plasmid pFD43-1 expressed appreciable alkaline phosphatase activity from the fepD′-′phoA fusion gene but low levels of β-galactosidase from the ybdA′-′lacZ fusion gene, and both promoters were iron repressible (Table 2). In a Fur− background, expression of either reporter enzyme was indistinguishable after low- or high-iron growth (data not shown). When the control region was reversed (pFD43-1R) and the fepD promoter was now fused to lacZ, there were again low levels of β-galactosidase activity, but the ybdA promoter (now fused to phoA) produced significant levels of alkaline phosphatase. These data suggested that both promoters were functional but that production of β-galactosidase from either fusion transcript was inefficient in this vector system. This difficulty may be attributable to the vector transcript sequence just upstream of the lacZ start codon (present in all promoter fusions using this vector). It is predicted to have the potential of forming a stem-loop structure which (with some transcripts) may effectively sequester the lacZ ribosome binding site, leading to poor translation initiation (data not shown). Several other studies using this same and related vectors have reported low levels of enzyme activity (3, 12, 19, 53). However, despite the low β-galactosidase reporter activity in these constructs, sequence modifications to the potential control elements led to changes in these activity levels that were consistent with changes in the actual transcript levels measured by primer extension.
TABLE 2.
Reporter gene activities from dual transcriptional fusion vector
| Plasmida | Mutationb | Alkaline phosphatase activityc
|
β-Galactosidase activityc
|
||
|---|---|---|---|---|---|
| + Fe | − Fe | + Fe | − Fe | ||
| pUJ10d | No insert | 7 | 10 | 0 | 0 |
| pFD43-1e | Wild type | 722 | 1,898 | 3 | 11 |
| pFD43-2 | fepD P1 −10 | 331 | 996 | 3 | 14 |
| pFD43-4 | ybdA P1 −10 | 1,044 | 2,411 | 0 | 0 |
| pFD43-6 | Both −10 | 275 | 915 | 0 | 0 |
| pFD43-30 | ybdA P1 “extended −10” | 1,052 | 1,883 | 1 | 1 |
| pFD43-31 | fepD P1 “extended −10” | 341 | 793 | 5 | 20 |
| pFD43-32 | fepD P2 −10 | 1,402 | 2,518 | 4 | 11 |
| pFD43-33 | fepD P1 −10 and P2 −35 | 98 | 143 | 2 | 4 |
| pFD43-3 | fepD P1 −35 (up) | 6,162 | 8,773 | 6 | 11 |
| pFD43-5 | ybdA P1 −35 (up) | 1,028 | 1,398 | 30 | 34 |
| PFD43-34 | ybdA P1 −35 (up) | 1,222 | 1,644 | 36 | 57 |
| pFD43-7 | FBS 1 | 2,730 | 3,347 | 15 | 28 |
| pFD43-1Rf | Wild type | 582 | 1,550 | 5 | 21 |
| pFD43-2R | ybdA P1 −10 | 23 | 43 | 6 | 27 |
| pFD43-6R | Both −10 | 20 | 32 | 1 | 6 |
Plasmids designated pFD43 contain the 327-bp fepD-ybdA control region in the vector pUJ10, with fepD fused to phoA and ybdA fused to lacZ. The suffix -1 designates the wild-type control sequence. All other suffixes represent the corresponding mutants described in Fig. 3 to 5. The last three plasmids represent the corresponding promoter fragments in the reverse orientation in pUJ10, with -1R containing the wild-type sequence, -2R containing the -2 mutant sequence, and -6R containing the -6 mutant sequence.
Activity is measured in Miller units.
There was no promoter for pUJ10 when alkaline phosphatase and β-galactosidase activity was measured.
For this group of plasmids, the promoter when alkaline phosphatase activity was measured was fepD. The promoter when β-galactosidase activity was measured was ybdA.
For this group, the alkaline phosphatase and β-galactosidase promoters were ybdA and fepD, respectively.
The primary transcripts from pFD43-1 detected by primer extension (Fig. 2) corresponded to those identified previously (50) for both the fepDGC operon and the ybdA gene. There were additional extension products apparent from these reactions, some of which represent transcripts from alternative secondary promoters (ybdA P2 and fepD P2) that maintain expression levels when the primary promoters are inactivated by mutations. Primer extension and RNase protection reactions detected the secondary transcripts expressed from the chromosomal fepD and ybdA genes (data not shown), but they were generally very weak, making it difficult to assess the contributions of these potential promoters to the overall expression of these genes.
Promoter elements controlling the ybdA gene.
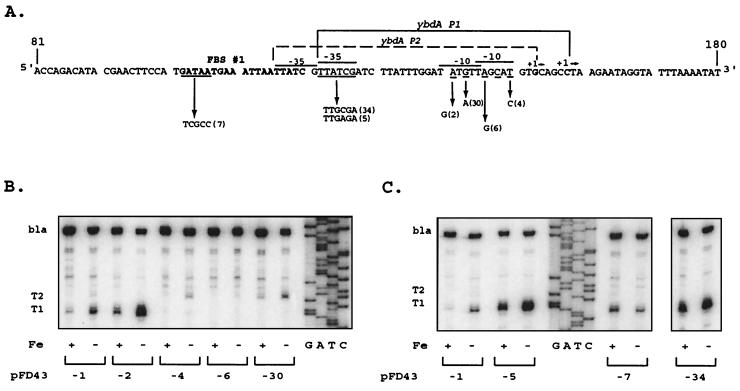
Mutations that altered the −10 region of the ybdA P1 promoter (Fig. 3A, pFD43-4 and pFD43-6) eliminated expression of the T1 transcript (Fig. 3B), confirming the role of this sequence in ybdA expression. In the fusion vector (Table 2), ybdA′-′lacZ activity was eliminated by these mutations, and in the opposite orientation, ybdA′-′phoA activity was reduced to <5% (pFD43-2R and pFD43-6R). A second transcript, T2, was weakly detected in RNA from pFD43-1 and became more prominent in pFD43-4 (Fig. 3B). The transcriptional start site for the ybdA P2 promoter is 5 bp upstream from that of ybdA P1 and correlates with a reasonable −10 element (5′-TATgtT-3′) but again with a poor −35 sequence (5′-TTatCg-3′). The minor T2 transcript was detected at 1 to 5% of the level of T1 in these constructs, which correlates with the low level of alkaline phosphatase expression when the two mutant promoters were fused to phoA in pFD43-2R and pFD43-6R (Table 2). The mutation in pFD43-2 (Fig. 3A) simultaneously eliminated ybdA P2 expression (Fig. 3B) and that of the opposing fepD P1 promoter (see below). The activity of ybdA P1 was enhanced significantly (Fig. 3B).
FIG. 3.
Primer extension of ybdA promoter mutants. (A) Nucleotide sequence of wild-type and mutant ybdA promoters (only the relevant strand is shown). Promoters are identified as in Fig. 2. Nucleotides underlined represent those altered by site-directed mutagenesis using “m” primers (Table 1). The corresponding arrow points from the wild-type bases to the altered bases, followed by the identification number in parentheses, which corresponds to the number after “pFD43” in panels B and C and Table 2. The number in parentheses corresponds as well to the primer number in Table 1. (B and C) Primer extension analysis of mRNA expressed from wild-type and mutant promoters. Primer pe4 was used in extension reactions with mRNA from pFD43-1 and from the designated mutant derivatives, as described in Materials and Methods. Only the relevant portions of the autoradiograms are shown. The sequencing ladder was generated with primer pe4 and pFD43-1 as templates. RNA preparations from high-iron (+) and low-iron (−) cultures are designated as in Fig. 2. As shown in panel C, extension products for pFD43-34 were analyzed on the same gel but were separated from the other preparations shown.
Since the ybdA P1 −35 element is poorly conserved, alternative control factors may be required for maximal ybdA P1 expression. A mutagenic primer (Table 1, m5), designed to make this promoter element more consensus, introduced the desired base substitutions but simultaneously resulted in deletion of the C residue at position 126. A second primer (Table 1, m34) introduced the desired G substitution at position 124 but deleted the T residue at 125. In both mutations, the −35 region was made more consensus (Fig. 3A, pFD43-5 and pFD43-34), although the spacing between the −10 and −35 elements was reduced to 16 nt. However, ybdA T1 transcript levels were increased 7-fold when iron was limiting and 12-fold under repressing conditions (Fig. 3C). Expression of the ybdA′-′lacZ fusion gene (Table 2) reflected these changes, with a 3- to 5-fold increase in β-galactosidase activity under low-iron conditions and a >10-fold increase when iron was sufficient. This increase in expression under derepressing (low-iron) conditions suggested an increased promoter affinity for RNA polymerase. However, the >10-fold increase in expression seen under normally repressing (high-iron) conditions suggested that these mutations also resulted in a decreased ability of Fur to control this promoter. Fur affinity measurements supported this conclusion (Christoffersen and McIntosh, unpublished data). An additional mutation (pFD43-7), which disrupts 5 nt within the first hexanucleotide motif in FBS 1(Fig. 3A), deregulated the expression of the T1 transcript (Fig. 3C) and ybdA′-′lacZ expression (Table 2) and also enhanced promoter activity, although not to the levels seen with mutations 5 and 34.
An additional feature of the ybdA P1 promoter: evidence for an extended −10 binding element.
A subset of ς70-dependent promoters with poor −35 recognition regions is characterized by a −10 region that is extended immediately upstream by a 5′-TGn-3′ motif as an additional sequence element required for maximal promoter activity (31, 33, 55). In effect, the TGn motif provides an additional contact for RNA polymerase (10, 31) and serves as a replacement element for the absent or poor −35 recognition region. This class of promoters may also require a positive activator (31, 33, 45). It has been suggested that the positive activator stimulates closer contact between RNA polymerase, the TGn motif, and the −10 promoter element, thus eliminating the need for the initial −35 recognition region (10, 33).
The mutation in pFD43-30 altered the TGn sequence of the ybdA P1 promoter and almost eliminated T1 transcript levels (Fig. 3B) as well as β-galactosidase activity from the ybdA′-′lacZ fusion (Table 2). The ybdA P2 promoter was strengthened by the alteration of this TGn element (Fig. 3A), and T2 transcript levels increased (Fig. 3B).
Promoter elements controlling expression of the fepDGC operon.
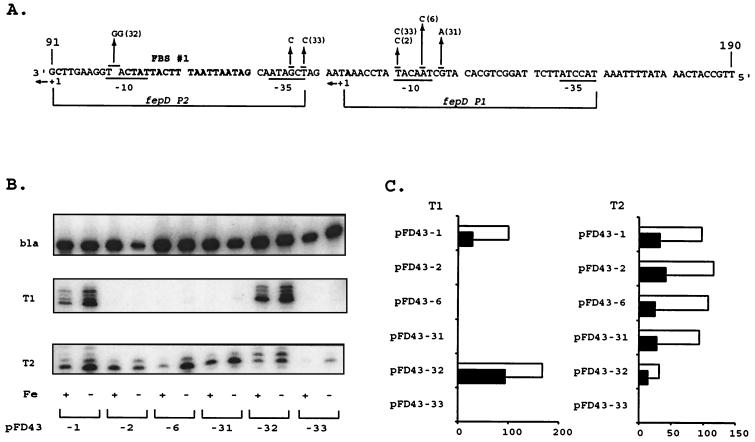
The mutation in pFD43-2 was targeted to the −10 region of the fepD P1 promoter, while that in pFD43-6 simultaneously changed the −10 regions of both the fepD and ybdA P1 promoters (Fig. 4A). In both mutations, the fepD T1 transcript was not detected (Fig. 4B), but fepD′-′phoA expression was reduced by only 50% (Table 2). A second transcript, fepD T2, which was expressed at levels equivalent to those for T1 in the wild-type construct pFD43-1 (Fig. 4B), was not affected by these mutations. The 5′ end of the fepD T2 transcript is 42 bp downstream of the fepD T1 +1, with a promoter that is represented by 5′-TATcAT-3′ as the −10 element and 5′-TcGAtA-3′ as the −35 region (Fig. 4A). To examine the contribution of the T2 transcript to expression of fepDGC, a mutation that changed the −10 region of the fepD P2 promoter (pFD43-32) was constructed (Fig. 4A) and resulted in a fivefold decrease of T2 transcript expression (Fig. 4B and C). However, alkaline phosphatase levels from fepD′-′phoA increased (Table 2) due to a significant rise in fepD T1 transcript levels (Fig. 4B and C); under high-iron conditions the effect was more pronounced, resulting in a decrease in iron regulation. The sequences changed with the pFD43-32 mutation are adjacent to the predicted Fur-binding region and may have reduced its binding affinity, causing a reduction in repression capability (Christoffersen and McIntosh, unpublished). Another mutation (pFD43-33), constructed to change both the fepD P2 −35 and fepD P1 −10 elements, resulted in severe reduction of fepD′-′phoA expression (Table 2) and of both T1 and T2 transcripts (Fig. 4B and C).
FIG. 4.
Primer extension analysis of the fepD promoter mutants. (A) Nucleotide sequence of the fepD promoter region (note that the sequence polarity is 5′ → 3′ from the right). The fepD P1 and P2 promoters are designated as in Fig. 2. Mutational alterations are designated as described in Fig. 3. (B) Primer extension of mRNA expressed from wild-type and mutant promoters. Primer pe2 was used in extension reactions with mRNA from pFD43-1 and from designated mutant derivatives, as described in Materials and Methods. Only the regions of the autoradiograms around T1 and T2 are shown. The fepD transcript mapping was performed independently of the bla control since the extension product with the bla-specific primer was similar in size to the fepD T1 transcripts. bla control transcripts were identified from separate reactions using the same mRNA populations, were separated on the same gel, and are presented above the extension reactions. RNA preparations from high-iron (+) and low-iron (−) cultures are designated below each lane. (C) Comparative effects of the mutations on both fepD transcripts. The bar graph depicts the relative contributions from the two fepD transcripts (T1 and T2), quantitated from primer extension gels by PhosphorImager analysis (as described in Materials and Methods) relative to the normalized bla transcript control from the same RNA samples. Data are presented for each designated clone as percentages of the induced levels of the wild-type fepD T1 transcript, arbitrarily defined as 100%. T1 and T2 represent the fepD transcripts derived from the fepD P1 and P2 promoters, respectively. Black bars represent repressed high iron (+ Fe) levels, while empty bars represent the induced (−Fe) condition.
The fepD P1 promoter is also characterized by a TGn motif immediately upstream of its −10 element. When this sequence was changed (pFD43-31), fepD′-′phoA expression was reduced by 50%, reflecting a significant decrease in fepD T1 transcript levels, with no obvious effects on fepD T2 transcript levels (Fig. 4B and C).
As with the ybdA P1 promoter, a mutation (pFD43-3) was generated in the fepD P1 promoter −35 element to increase the consensus of this region (Fig. 5A). The mutation strongly enhanced expression of the fepD T1 transcript (Fig. 5B and C) and resulted in a sharp increase in fepD′-′phoA levels (Table 2). The fepD T2 transcript was repressed (Fig. 5B and C), probably a direct result of the increased fepD P1 promoter strength. Expression from the pFD43-3 promoter also was deregulated, even though the fepD P1 −35 sequence changed is some distance from the Fur-binding operator region. The Fur operator mutation (pFD43-7) deregulated expression of the T1 transcript (Fig. 5B and C) and of phoA (Table 2) but also eliminated expression from fepD P2, since it altered its −10 promoter element (Fig. 5A).
FIG. 5.
Transcript analysis of the fepD promoter-operator mutants. All components are configured and labeled as described in the legend to Fig. 4. The promoter elements for the fepD P3 promoter (A), which were mapped according to the newly identified T3 transcript from pFD43-3 (B), are denoted by dotted overlines, and the relevant + 1 site is identified. (C) T1 and T2 represent the fepD transcripts derived from the fepD P1 and P2 promoters, respectively. Black bars represent repressed high iron levels, while empty bars represent the induced (− Fe) condition.
The pFD43-3-enhanced P1 promoter mutation resulted in increased expression of a new transcript, T3, located 21 bp upstream of the fepD T1 transcript start site. The T3 transcript was weakly detected in primer extension reactions with the wild-type template (Fig. 2C and 5B). The predicted P3 promoter elements are well conserved (−10, 5′-TATtcT-3′; and −35, 5′-TTGcCA-3′). The strength of the P3 promoter elements suggested that the limited expression of a T3 transcript from the wild-type promoter sequences reflected a repression of the potential fepD P3 promoter by elements of the P1 promoter. Since the wild-type fepD P1 −35 promoter element is very weak, an alternative activator binding in this region might explain the repression of P3. This mutation also creates an extended −10 motif for the P3 promoter. Thus, although the P3 promoter already has strong similarity to ς70 consensus determinants, this new TGn extension might account for increased P3 transcriptional activity by allowing it to better compete for RNA polymerase.
The mutations in pFD43-5 and pFD43-34, which enhanced the ybdA P1 −35 element, simultaneously changed the fepD P2 −35 element (Fig. 5A). However, neither mutation made this hexameric sequence any less consensus. Primer extension products (Fig. 5B and C) for fepD P1 (which reflect the 1-nt deletion generated in these mutations) and P2, as well as reporter gene activity (Table 2), suggest a slight reduction in transcriptional expression, probably due to the competition with the stronger ybdA P1 promoter. Under these conditions, the transcript from the fepD P3 promoter is more obvious. Regulation is also affected because these mutations alter Fur-binding affinity (Christoffersen and McIntosh, unpublished).
DISCUSSION
The ferric enterobactin gene island incorporates three different bidirectional promoter structures and regulatory strategies to link iron-responsive control by the Fur repressor to the variable expression of six transcripts encoding various transport or siderophore biosynthesis gene groups. The back-to-back promoters between fepB and entC are sufficiently separated such that each is controlled more or less independently by its interactions with Fur and the transcribing polymerase (9). The promoter for fes and the primary promoter for fepA are positioned such that the −35 element for one promoter slightly overlaps the −10 element of the opposing promoter, and Fur then occupies the sequences between both elements of both promoters, allowing coincidental repression of both promoters by competition between Fur and RNA polymerase for access to these sequences (29). Mutations which reduced the strength of fepA P1 increased expression of the fes promoter by 10 to 20% under both low- and high-iron conditions but did not alter the regulation pattern. When both fepA promoters were inactivated, however, there was no increase in fes expression, and Fur exhibited a stronger repression of the fes promoter (29). In contrast, when the fes promoter was eliminated, transcription from fepA P1 was strongly enhanced (T. Morris, unpublished data) and no longer responded to iron regulation (29).
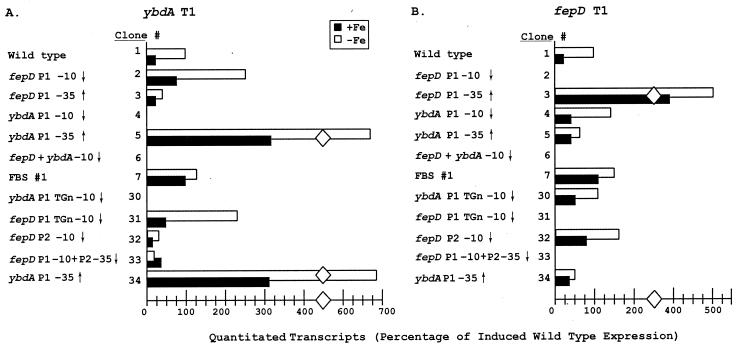
The tightly overlapping promoters between fepD and ybdA are influenced significantly by the expression levels of the opposing promoters, and iron regulation is imposed by the Fur repressor interacting at a site that is not symmetrically positioned relative to the primary promoters. For each of the promoter mutations generated in the compact fepD-ybdA control region, transcripts from the opposing promoter were analyzed to assess the regulatory effects on expression levels. Mutations at either primary P1 promoter generally had a reciprocal effect on the opposing primary P1 promoter (Fig. 6). Promoter down-mutations at the fepD P1 promoter (pFD43-2 and pFD43-31) or at the ybdA P1 promoter (pFD43-4 and pFD43-30) resulted in increased expression from the opposing P1 promoter. Transcript levels were repressed from either P1 promoter when the opposing promoter was strengthened by mutation (e.g., pFD43-5 and pFD43-34 at ybdA P1 and pFD43-3 at fepD P1). These data indicate that RNA polymerase occupancy of or activity at the opposing promoter is a significant component of the regulatory mechanism for both fepD P1 and ybdA P1 promoters. The Fur operator mutation (pFD43-7) removes a repressing effect to both P1 promoters. The result was a fourfold increase in transcript levels under high-iron conditions, when Fur-Fe(II) would frequently occupy its binding site, and a 25 to 50% increase in expression under iron-limited conditions, when Fur-Fe(II) would only rarely occupy this site.
FIG. 6.
Comparative effects of promoter mutations on the opposing transcripts, with quantitative comparison between the ybdA and fepD T1 transcripts expressed from the wild-type and mutant promoter-operator regions. Quantitation was achieved by comparison to the normalized bla transcript within each RNA preparation (as described in Materials and Methods). Data are presented as the percentage of induced wild-type expression of the ybdA T1 transcript (A) or the fepD T1 transcript (B). Black bars represent high-iron conditions, while empty bars represent low-iron culture conditions. In both cases, the diamond is used to represent a 150-U break in the bar graphs. The different clones, no. 1 to 7 and 30 to 34, are presented under the column “Clone #.” The ↑ and ↓ designate the mutant as one in which the consensus of the corresponding promoter region was increased and decreased, respectively.
While these data support the interpretation that the opposing promoters are competitive, the mutations may also have affected proximal sequences in this compact control region to produce unexpected changes in promoter activity (23). For example, although both mutations at the ybdA P1 −10 region (pFD43-4 and pFD43-30) led to increased fepD P1 expression, there was a significantly smaller increase in expression with pFD43-30. This may be explained by the fact that although ybdA P1 was eliminated, pFD43-30 inadvertently strengthened the ybdA P2 promoter, which is also positioned to compete directly with fepD P1.
The mutational analysis in these experiments was complicated by the observation that both the fepDGC operon and ybdA can be expressed from either of two promoters, an observation based on multicopy reporter gene analyses and primer extension from plasmid-encoded mRNAs. The roles of these secondary promoters for expression from the single-copy chromosomal genes and the growth conditions under which they may be optionally expressed have not been defined. Analysis of the chromosomally encoded fepD and ybdA transcripts by primer extension and RNase protection proved difficult due to their weak expression; the corresponding mRNA levels are significantly lower than those for the other four enterobactin transcripts. Similarly low expression levels have been described for other hydrophobic cytoplasmic membrane-associated proteins within analogous permease systems (2). For the ybdA gene, detection of the T2 transcript was very weak among various RNA preparations, suggesting that it at best represents only a minor component of ybdA gene expression. RNase protection analysis (data not shown) consistently detected both fepD transcripts, although in chromosome-encoded RNA preparations, fepD T1 was more strongly expressed than fepD T2. The almost equal expression levels of these two transcripts in plasmid-derived mRNA populations may indicate that the fepD P2 promoter is more competitive in the multicopy situation (54).
The fepD P2 promoter contributes significantly to fepD transcript expression in these experiments, but it overlaps the ybdA P1 promoter only minimally. Mutations which alter fepD P2 had variable effects on expression of ybdA. When the fepD P2 −10 was inactivated (pFD43-32), expression from the upstream fepD P1 promoter was enhanced and somewhat deregulated (Fig. 6). There was a corresponding fourfold decrease in ybdA P1 transcript levels; this could be interpreted to result from increased competition from the opposing fepD P1 promoter. However, this mutation also enhanced ybdA P2 expression levels by four- to fivefold and resulted in normal β-galactosidase levels from the fusion construct (Table 2). Since ybdA P2 also strongly overlaps fepD P1, it is not clear how it could escape the repressive effects of an enhanced fepD P1. The variable effects on the two tandem ybdA promoters is reminiscent of the regulatory configuration of the gal P1 and P2 promoters, where cyclic AMP-catabolite gene activator protein coordinately represses P2 and activates P1 (1). In such a comparison, the site that the pFD43-32 mutation altered would be required for full ybdA P1 expression through the binding of an activator and would simultaneously repress ybdA P2. Coincidentally, the gal promoters also belong to the “extended −10” class (45).
The mutations in pFD43-33 were constructed to remove both fepD promoters by combining the fepD P1 −10 mutation (same as pFD43-2) with a fepD P2 −35 mutation. Since the pFD43-2 mutation at fepD P1 led to an increase in ybdA P1 activity, it was anticipated that removal of both opposing promoters might enhance ybdA P1 even more. However, its activity was strongly reduced in this mutant (Fig. 6; Table 2). The mutation contains two base substitutions in the ybdA P1 −35 region, which suggests that although this region does not conform to a ς70-type −35 promoter element, it must be important for ybdA P1 expression.
The fepD T2 transcript has its +1 initiation site located between the fepD ribosome binding sequence and its ATG translational start site (Fig. 1 and 2A). While this might suggest reduced translational efficiency of the FepD polypeptide, there are several examples of transcripts which initiate near the translational start codon and do not have typical ribosome binding sequences and have been shown to produce full-length products (35, 36, 48). The possibility that fepD T2 produces a functional FepD translational product remains to be determined.
The most striking characteristic of the fepD and ybdA promoters is their weak resemblance to the ς70 consensus sequences. The varying degrees of consensus encountered with E. coli promoters (well conserved to poorly conserved) have been shown by in vitro expression assays to parallel the initiation frequencies seen at those promoters (28, 31, 38, 46). Although no natural E. coli promoters have been found with a perfect consensus, promoter function is optimized in vivo through the productive interactions between RNA polymerase, regulatory proteins, and the promoter DNA sequences to which they bind (28, 38). With many positively regulated genes, the promoter sequences are not well conserved, yet expression is efficient in the presence of their activators (18, 46, 47). In those promoters which are represented by poor sequence conservation at the −35 element, stability of contacts between promoter elements and RNA polymerase can be provided by productive interactions with a positive regulatory protein or by additional promoter sequence elements, such as an extension of the −10 region. A 5′-TGn-3′ motif immediately upstream of the −10 element has been suggested to provide missing contract points required for productive transcription in the absence of a typical −35 element (31, 46). With a well-conserved −35 promoter element, the TGn motif serves a minor role (31, 33). The significance of the TGn motif has been defined in several promoters (7, 31, 32, 45).
Neither the fepD nor the ybdA promoter is conserved at the −35 element, suggesting that transcription from these regions is inherently weak or must rely on alternate control mechanisms that might include activator proteins (46) or additional promoter elements to optimize activity. Mutational analysis provided strong evidence that both the fepD P1 and ybdA P1 promoters are representatives of the extended −10 class of promoters. Without the TGn motif, expression was strongly reduced from either promoter. When the −35 element from either the fepD P1 or ybdA P1 promoter was made more consensus, expression was five- to sixfold higher than with the wild-type promoter. Although the TGn extension was not removed from these enhanced promoters, it was likely not a factor in the increased activity (31, 33).
Evidence has also suggested that the TGn extension is rarely the sole adjunct to the −10 element when there is poor conservation at the −35 element (31, 33). For the majority of these promoters, an activator has been implicated as essential for adequate transcriptional activity (31, 45). While this study does not directly address whether fepD P1 or ybdA P1 activities are enhanced by such regulators, observations with several of the mutant promoters provide some initial evidence that activation may play a role. Mutations 32 and 33, which inactivated the fepD promoters, were expected to enhance ybdA promoter activity but instead led to repressive effects, an observation consistent with an additional regulatory factor that is affected by these changes. The pFD43-3 mutation created a well-conserved fepD P1 −35 element and resulted in increased fepD expression. However, coincident with this increase was the appearance of a new transcript, T3, which maps to a well-conserved promoter upstream of fepD P1. If it is assumed that, in the absence of a −35 element, an activator protein binds around the −35 region and that the pFD43-3 mutation resulted in the independence of fepD P1 from activator-mediated expression, then the data suggest that fepD T3 expression is normally repressed by the activator required for expression of fepD P1. In the TGn-dependent λPRE promoter, it has been shown that conversion of either of the promoter elements to consensus results in independence from activator function (31). Since fepD P1 in pFD43-3 has a well conserved −35 element, if the requirement for an activator was abolished, the highly competitive promoter elements for fepD P3 might be uncovered.
The detailed investigation of the fepD-ybdA bidirectional promoter region presented in this study has revealed a regulatory architecture unique among the three enterobactin divergent control regions. Promoter sequences are weak renditions of typical E. coli counterparts and rely on additional features for optimal activity. The strength of these promoters has a considerable impact on the ability of Fur to regulate their expression. Furthermore, there is a tight correlation between the position of the Fur-binding sequences within this region and the relative promoter strengths, such that alteration of either the affinity for Fur or of the ability of RNA polymerase to function at these promoters has an important impact on the regulatory responses of these promoters. As was postulated in an earlier study (50) and confirmed by binding measurements (Christoffersen and McIntosh, unpublished), the overall regulation of this divergent promoter-operator region is determined by the strength of the opposing promoters and the location and affinity of the Fur-binding operator sequence.
ACKNOWLEDGMENTS
We thank C. Manoil, V. de Lorenzo, D. Touati, and A. Eisenstark for bacterial strains and plasmids used in this study. We are grateful to J. Lavrrar, M. Hunt, M. Heidari, and C. Lorson for technical advice and helpful discussions during the course of this work.
This study was supported in parts by grants MCB 9201942 (from the National Science Foundation), GM54243 (from the National Institutes of Health), and URB-00-055 McIntosh (from the University of Missouri Research Board). India Hook-Barnard is a predoctoral trainee supported by training grant 5 T32 AI07276 from the National Institutes of Health.
REFERENCES
- 1.Adhya S, Miller W. Modulation of the two promoters of the galactose operon of Escherichia coli. Nature. 1979;279:492–494. doi: 10.1038/279492a0. [DOI] [PubMed] [Google Scholar]
- 2.Ames G F L. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 3.Atlung T, Brondsted L. Role of the transcriptional activator AppY in regulation of the cyx appA operon of Escherichia coli by anaerobiosis, phosphate starvation and growth phase. J Bacteriol. 1994;176:5414–5422. doi: 10.1128/jb.176.17.5414-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bagg A, Neilands J. Molecular mechanism of regulation of siderophore-mediated iron assimilation. Microbiol Rev. 1987;51:509–518. doi: 10.1128/mr.51.4.509-518.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagg A, Neilands J B. Ferric uptake regulation protein acts as a repressor, employing iron(II) as a cofactor to bind the operator of an iron transport operon in Escherichia coli. Biochemistry. 1987;26:5471–5477. doi: 10.1021/bi00391a039. [DOI] [PubMed] [Google Scholar]
- 6.Beck C F, Warren R A J. Divergent promoters, a common form of gene organization. Microbiol Rev. 1988;52:318–326. doi: 10.1128/mr.52.3.318-326.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belyaeva T, Griffiths L, Minchin S, Cole J, Busby S. The E. coli cysG promoter belongs to the ‘extended −10’ class of bacterial promoters. Biochem J. 1993;296:851–857. doi: 10.1042/bj2960851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brickman E, Beckwith J. Analysis of the regulation of Escherichia coli alkaline phosphatase synthesis using deletions and φ80 transducing phages. J Mol Biol. 1975;96:307–316. doi: 10.1016/0022-2836(75)90350-2. [DOI] [PubMed] [Google Scholar]
- 9.Brickman T J, Ozenberger B A, McIntosh M A. Regulation of divergent transcription from the iron-responsive fepB-entC promoter-operator regions in Escherichia coli. J Mol Biol. 1990;212:669–682. doi: 10.1016/0022-2836(90)90229-F. [DOI] [PubMed] [Google Scholar]
- 10.Chan B, Spassky A, Busby S. The organization of open complexes between Escherichia coli RNA polymerase and DNA fragments carrying promoters either with or without consensus −35 region sequences. Biochem J. 1990;270:141–148. doi: 10.1042/bj2700141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chenault S S, Earhart C F. Organization of genes encoding membrane proteins of the Escherichia coli ferrienterobactin permease. Mol Microbiol. 1991;5:1405–1413. doi: 10.1111/j.1365-2958.1991.tb00787.x. [DOI] [PubMed] [Google Scholar]
- 12.Clementz T. The gene coding for 3-deoxy-manno-octulosonic acid transferase and the rfaQ gene are transcribed from divergently arranged promoters in Escherichia coli. J Bacteriol. 1992;174:7750–7756. doi: 10.1128/jb.174.23.7750-7756.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Compan I, Touati D. Interaction of six global transcription regulators in expression of manganese superoxide dismutase in Escherichia coli K-12. J Bacteriol. 1993;175:1687–1696. doi: 10.1128/jb.175.6.1687-1696.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Giovannini F, Herrero M, Neilands J B. Metal ion regulation of gene expression. Fur repressor-operator interaction at the promoter region of the aerobactin system of pColV-K30. J Mol Biol. 1988;203:875–884. doi: 10.1016/0022-2836(88)90113-1. [DOI] [PubMed] [Google Scholar]
- 15.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Lorenzo V, Herrero M, Neilands J B. pCON4 and pCON5: improved plasmid vectors to study bacterial promoters. FEMS Microbiol Lett. 1988;50:17–23. [Google Scholar]
- 17.de Lorenzo V, Wee S, Herrero M, Neilands J B. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J Bacteriol. 1987;169:2624–2630. doi: 10.1128/jb.169.6.2624-2630.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deuschle U, Kammerer W, Gentz R, Bujard H. Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures. EMBO J. 1986;5:2987–2994. doi: 10.1002/j.1460-2075.1986.tb04596.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eberl L, Givskov M, Schwab H. The divergent promoters mediating transcription of the par locus of plasmid RP4 are subject to autoregulation. Mol Microbiol. 1992;6:1969–1979. doi: 10.1111/j.1365-2958.1992.tb01370.x. [DOI] [PubMed] [Google Scholar]
- 20.Escolar L, de Lorenzo V, Pérez-Martin J. Metalloregulation in vitro of the aerobactin promoter of Escherichia coli by the Fur (ferric uptake regulation) protein. Mol Microbiol. 1997;26:799–808. doi: 10.1046/j.1365-2958.1997.6211987.x. [DOI] [PubMed] [Google Scholar]
- 21.Escolar L, Pérez-Martin J, de Lorenzo V. Binding of the Fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J Mol Biol. 1998;283:537–547. doi: 10.1006/jmbi.1998.2119. [DOI] [PubMed] [Google Scholar]
- 22.Escolar L, Pérez-Martin J, de Lorenzo V. Coordinated repression in vitro of the divergent fepA-fes promoters of Escherichia coli by the iron uptake regulation (Fur) protein. J Bacteriol. 1998;180:2579–2582. doi: 10.1128/jb.180.9.2579-2582.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodrich J A, McClure W R. Competing promoters in prokaryotic transcription. Trends Biochem Sci. 1991;16:394–397. doi: 10.1016/0968-0004(91)90162-o. [DOI] [PubMed] [Google Scholar]
- 24.Gough J A, Murray N E. Sequence diversity among related genes for recognition of specific targets in DNA molecules. J Mol Biol. 1983;166:1–19. doi: 10.1016/s0022-2836(83)80047-3. [DOI] [PubMed] [Google Scholar]
- 25.Griggs D W, Konisky J. Mechanism for iron-regulated transcription of the Escherichia coli cir gene: metal-dependent binding of Fur protein to the promoter. J Bacteriol. 1989;171:1048–1054. doi: 10.1128/jb.171.2.1048-1054.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haber F, Weiss J. The catalytic decomposition of hydrogen peroxide by iron salts. Proc R Soc Lond A. 1934;147:332–351. [Google Scholar]
- 27.Hawley D K, McClure W R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983;11:2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hoopes B C, McClure W R. Strategies in regulation of transcription initiation. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C.: American Society for Microbiology; 1987. pp. 1231–1240. [Google Scholar]
- 29.Hunt M D, Pettis G S, McIntosh M A. Promoter and operator determinants for Fur-mediated iron regulation in the bidirectional fepA-fes control region of the Escherichia coli enterobactin gene system. J Bacteriol. 1994;176:3944–3955. doi: 10.1128/jb.176.13.3944-3955.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joyce C M, Grindley N D F. Method for determining whether a gene of Escherichia coli is essential: application of the polA gene. J Bacteriol. 1984;158:636–643. doi: 10.1128/jb.158.2.636-643.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 32.Kenney T J, Churchward G. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J Bacteriol. 1996;178:3564–3571. doi: 10.1128/jb.178.12.3564-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar A, Malloch R A, Fujita N, Smillie D A, Ishihama A, Hayward R S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 34.Kunkel T. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci USA. 1985;82:488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lacks S A, Lopez P, Greenberg B, Espinosa M. Identification and analysis of genes for tetracycline resistance and replication functions in the broad-host-range plasmid pLS1. J Mol Biol. 1986;192:753–765. doi: 10.1016/0022-2836(86)90026-4. [DOI] [PubMed] [Google Scholar]
- 36.Lopez P, Martinez S, Diaz A, Espinosa M, Lacks S A. Characterization of the polA genes of Streptococcus pneumoniae and comparison of the DNA polymerase I it encodes to homologous enzymes from E. coli and phage T7. J Biol Chem. 1989;264:4255–4263. [PubMed] [Google Scholar]
- 37.Manoil C. Analysis of protein localization by use of gene fusions with complementary properties. J Bacteriol. 1990;172:1035–1042. doi: 10.1128/jb.172.2.1035-1042.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McClure W R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- 39.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 40.Neilands J B. Microbiol envelope proteins related to iron. Annu Rev Microbiol. 1982;36:285–309. doi: 10.1146/annurev.mi.36.100182.001441. [DOI] [PubMed] [Google Scholar]
- 41.Norrander J, Kempe T, Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983;26:101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- 42.Ozenberger B A, Brickman T J, McIntosh M A. Nucleotide sequence of Escherichia coli isochorismate synthetase gene entC and evolutionary relationship of isochorismate synthetase and other chorismate-utilizing enzymes. J Bacteriol. 1989;171:775–783. doi: 10.1128/jb.171.2.775-783.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ozenberger B A, Nahlik M S, McIntosh M A. Genetic organization of multiple fep genes encoding ferric enterobactin transport functions in Escherichia coli. J Bacteriol. 1987;169:3638–3646. doi: 10.1128/jb.169.8.3638-3646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettis G S, Brickman T J, McIntosh M A. Transcriptional mapping and nucleotide sequence of the Escherichia coli fepA-fes enterobactin region. J Biol Chem. 1988;263:18857–18863. [PubMed] [Google Scholar]
- 45.Ponnambalam S, Webster C, Bingham A, Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of normal −35 region sequences. J Biol Chem. 1986;261:16043–16048. [PubMed] [Google Scholar]
- 46.Raibaud O, Schwartz M. Positive control of transcription initiation in bacteria. Annu Rev Genet. 1984;17:173–206. doi: 10.1146/annurev.ge.18.120184.001133. [DOI] [PubMed] [Google Scholar]
- 47.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 48.Sabelnikov A G, Greenberg B, Lacks S A. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 50.Shea C M, McIntosh M A. Nucleotide sequence and genetic organization of the ferric enterobactin transport system: homology to other periplasmic binding protein-dependent systems in Escherichia coli. Mol Microbiol. 1991;5:1415–1428. doi: 10.1111/j.1365-2958.1991.tb00788.x. [DOI] [PubMed] [Google Scholar]
- 51.Tardat B, Touati D. Iron and oxygen regulation of Escherichia coli MnSOD expression: competition between the global regulators Fur and ArcA for binding to DNA. Mol Microbiol. 1993;9:53–63. doi: 10.1111/j.1365-2958.1993.tb01668.x. [DOI] [PubMed] [Google Scholar]
- 52.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in Δfur mutants of Escherichia coli: protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tsolis R M, Bäumler A J, Stojiljkovic I, Heffron F. Fur regulon of Salmonella typhimurium: identification of new iron-regulated genes. J Bacteriol. 1995;177:4628–4637. doi: 10.1128/jb.177.16.4628-4637.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vidal-Ingigliardi D, Raibaud O. A convenient technique to compare the efficiency of promoters in Escherichia coli. Nucleic Acids Res. 1985;13:5919–5926. doi: 10.1093/nar/13.16.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Voskuil M I, Voepel K, Chambliss G H. The −16 region, a vital sequence for the utilization of a promoter in Bacillus subtilis and Escherichia coli. Mol Microbiol. 1995;17:271–279. doi: 10.1111/j.1365-2958.1995.mmi_17020271.x. [DOI] [PubMed] [Google Scholar]
- 56.Wee S, Neilands J B, Bittner M L, Hemming B C, Haymore B L, Seetharam R. Expression, isolation and properties of Fur (ferric uptake regulation) protein of Escherichia coli K 12. Biol Met. 1988;1:62–68. doi: 10.1007/BF01128019. [DOI] [PubMed] [Google Scholar]