Abstract
The ratio of the major monensin analogs produced by Streptomyces cinnamonensis is dependent upon the relative levels of the biosynthetic precursors methylmalonyl-coenzyme A (CoA) (monensin A and monensin B) and ethylmalonyl-CoA (monensin A). The meaA gene of this organism was cloned and sequenced and was shown to encode a putative 74-kDa protein with significant amino acid sequence identity to methylmalonyl-CoA mutase (MCM) (40%) and isobutyryl-CoA mutase (ICM) large subunit (36%) and small subunit (52%) from the same organism. The predicted C terminus of MeaA contains structural features highly conserved in all coenzyme B12-dependent mutases. Plasmid-based expression of meaA from the ermE∗ promoter in the S. cinnamonensis C730.1 strain resulted in a decreased ratio of monensin A to monensin B, from 1:1 to 1:3. Conversely, this ratio increased to 4:1 in a meaA mutant, S. cinnamonensis WM2 (generated from the C730.1 strain by insertional inactivation of meaA by using the erythromycin resistance gene). In both of these experiments, the overall monensin titers were not significantly affected. Monensin titers, however, did decrease over 90% in an S. cinnamonensis WD2 strain (an icm meaA mutant). Monensin titers in the WD2 strain were restored to at least wild-type levels by plasmid-based expression of the meaA gene or the Amycolatopsis mediterranei mutAB genes (encoding MCM). In contrast, growth of the WD2 strain in the presence of 0.8 M valine led only to a partial restoration (<25%) of monensin titers. These results demonstrate that the meaA gene product is significantly involved in methylmalonyl-CoA production in S. cinnamonensis and that under the tested conditions the presence of both MeaA and ICM is crucial for monensin production in the WD2 strain. These results also indicate that valine degradation, implicated in providing methylmalonyl-CoA precursors for many polyketide biosynthetic processes, does not do so to a significant degree for monensin biosynthesis in the WD2 mutant.
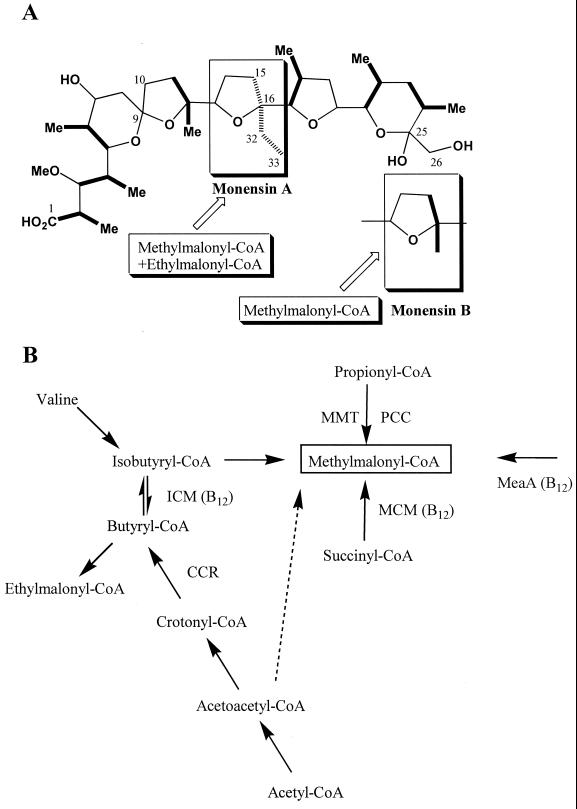
Streptomycetes produce a large number of structurally diverse polyketide antibiotics by a process similar to long-chain fatty acid biosynthesis (20). Polyketide biosynthesis, catalyzed by polyketide synthases, uses carboxylated acyl thioesters, such as malonyl-coenzyme A (CoA), methylmalonyl-CoA, or ethylmalonyl-CoA, as extender units. These precursors form the polyketide carbon backbone and side chains, as seen in the examples of rifamycin (2), erythromycin (11), and monensin (6). Malonyl-CoA and ethylmalonyl-CoA are likely derived from the carboxylation of acetyl-CoA and butyryl-CoA, respectively (16, 35), while methylmalonyl-CoA can be produced from a variety of different pathways (6). As several methylmalonyl-CoA molecules are required to build a single polyketide (six for erythromycin, seven for monensin A, and eight for rifamycin), the levels of methylmalonyl-CoA under certain conditions may represent a limiting factor in production titers. For monensin biosynthesis, either ethylmalonyl-CoA or methylmalonyl-CoA can be used at the same stage of elongation to generate monensin A or monensin B, respectively (14, 24). Thus, in this organism, changes in the levels of methylmalonyl-CoA affect not only the titers but also the ratio of monensin A to monensin B. This feature makes Streptomyces cinnamonensis an excellent organism for probing pathways that contribute in both minor and major ways to generating methylmalonyl-CoA for polyketide biosynthesis.
Three generally accepted routes to methylmalonyl-CoA are (i) the isomerization of succinyl-CoA, catalyzed by the coenzyme B12-dependent methylmalonyl-CoA mutase (MCM) (28, 43); (ii) carboxylation of propionyl-CoA, catalyzed by either propionyl-CoA carboxylase (PCC) (7, 35) or methylmalonyl-CoA transcarboxylase (MMT) (22); and (iii) a multistep oxidation of isobutyryl-CoA (34) (Fig. 1). The mutAB genes encoding MCM involved in the first of these pathways have been cloned from monensin A-producing S. cinnamonensis (6), as well as from rifamycin SV-producing Amycolatopsis mediterranei U32 (49) and other prokaryotic and mammalian sources (27, 47). A mutAB disruption has no effect on the monensins A and B total production and ratio in S. cinnamonensis cells (44). Plasmid-based overexpression of the mutAB genes in S. cinnamonensis, on the other hand, has been reported to yield both a slight increase in total monensin production and a decreased ratio of monensin A to monensin B (49). These observations suggest that under these conditions, methylmalonyl-CoA is a limiting factor in monensin biosynthesis and that the natural levels of MCM activity generated from the mutAB genes do not contribute significantly to this process. Genes encoding either PCC or MMT, enzymes capable of generating methylmalonyl-CoA from propionyl-CoA, have not yet been cloned from S. cinnamonensis, and thus their role in providing this precursor through the second pathway has not been evaluated. A PCC encoded by the pcc gene has been cloned and characterized for Streptomyces coelicolor A3(2) (7, 35) and erythromycin-producing Saccharopolyspora erythraea (12). Disruption of the pcc gene in S. erythraea has no effect on erythromycin production (12). However, even in this organism, the role of carboxylation of propionyl-CoA in methylmalonyl-CoA formation is not clear, as there are probably an MMT and additional acyl-CoA (or propionyl-CoA) carboxylases (6, 7, 18, 35).
FIG. 1.
(A) The structures of monensins A and B. Methylmalonyl-CoA positions in both monensins A and B are marked in bold, and the ethylmalonyl-CoA position in monensin A is hatched. (B) Proposed pathways for methylmalonyl-CoA formation in S. cinnamonensis. The dotted arrow indicates a pathway that does not require the meaA, icm, or mutAB genes cloned from this organism. The substrate for MeaA is unknown. Ethylmalonyl-CoA is used for monensin A biosynthesis, while methylmalonyl-CoA is used for both monensin A and B biosynthesis. B12 indicates known or putative coenzyme B12-dependent mutase. MCM, methylmalonyl-CoA mutase; ICM, isobutyryl-CoA mutase; MMT, methylmalonyl-CoA transcarboxylase; PPC, propionyl-CoA carboxylase; CCR, crotonyl-CoA reductase.
The third pathway from methylmalonyl-CoA, clearly established from numerous biosynthetic studies with monensin (34) and tylosin (30) and other polyketide antibiotic producing organisms, is oxidation of isobutyryl-CoA (Fig. 1). Isobutyryl-CoA can be formed either from valine catabolism or from butyryl-CoA by carbon skeleton rearrangement catalyzed by coenzyme B12-dependent isobutyryl-CoA mutase (ICM) (34). The icm genes encoding S. cinnamonensis ICM were recently cloned and sequenced, and insertional inactivation of icm has been shown to have no detectable effect on monensin production (33, 44, 48). Thus, it appears that either ICM or MCM activities can be removed from S. cinnamonensis without significantly affecting the pools of methylmalonyl-CoA for monensin biosynthesis. Methylmalonyl-CoA in these strains might thus be obtained directly from the oxidation of valine-derived isobutyryl-CoA (44) or from the carboxylation of propionyl-CoA. A third possibility was raised by the unexpected observation that [1,3-13C2]acetoacetyl-CoA can be converted intact into [1,2-13C2]methylmalonyl-CoA in S. cinnamonensis in the absence of either ICM or MCM. There are no other known pathways beyond that involving ICM which would explain such an intact interconversion, and these data suggested the presence of another coenzyme B12-dependent mutase and/or unidentified pathway(s) involved in methylmalonyl-CoA formation (44).
A novel meaA gene encoding an MCM-like protein with unknown function has been found in both Streptomyces collinus (15) and Methylobacterium extorquens AM1 (9, 37). The predicted MeaA amino acid sequence contains the distinctive coenzyme B12-binding domain and exhibits high end-to-end homology to the large subunit of MCM and both subunits of ICM. The meaA gene has been shown to be involved in acetate assimilation in both of these organisms (9, 15, 37). In both S. collinus and S. coelicolor, the meaA gene was found to be 20 to 40 bp downstream of the crotonyl-CoA reductase (CCR)-encoding gene ccr, with the same transcriptional orientation. CCR plays a key role in the catalysis of the last reductive step in the biosynthesis of butyryl-CoA from acetyl-CoA in S. collinus (15) and a significant role in providing butyryl-CoA from monensin A biosynthesis in S. cinnamonensis (24). The genetic organization of meaA and ccr implies a possible role of meaA in the butyryl-CoA or methylmalonyl-CoA pathways. Our interest in the clarification of the role of various methylmalonyl-CoA formation pathways in polyketide-producing streptomycetes prompted us to further examine this novel mutase gene.
In this study, we report the cloning and sequencing of meaA from monensin-producing S. cinnamonensis cells. Gene disruption, gene overexpression, and labeling studies clearly demonstrate that MeaA is involved in methylmalonyl-CoA formation, but that this process does not involve an acetoacetyl-CoA–butyryl-CoA intermediate. Furthermore, the generation and analysis of an meaA icm mutant of S. cinnamonensis has clearly demonstrated that under the tested conditions, the expression of these two genes is crucial for providing the majority of this methylmalonyl-CoA used for monensins A and B production. Surprisingly, valine catabolism does not contribute significantly to providing methylmalonyl-CoA in this mutant.
MATERIALS AND METHODS
Chemicals.
Chemicals were purchased from Fisher Scientific (Pittsburgh, Pa.). Ethyl [3,4-13C2]acetoacetate and perdeuterated valine were from Cambridge Isotope Laboratories Inc. (Andover, Md.). Monensin A standard was from Sigma Co. (St. Louis, Mo.). The S. cinnamonensis C730.1 strain was provided by Eli Lilly & Company.
Bacterial strains, plasmids, and cultural conditions.
Table 1 contains a list of the strains and plasmids used in this work. Escherichia coli XL1-Blue, DH5α, and ET12567 were grown at 37°C in Luria-Bertani medium supplemented with either ampicillin (Amp; 100 μg/ml) or apramycin (50 μg/ml) when necessary (18). Cultures of S. cinnamonensis were grown in YEME medium (19) at 30°C for the isolation of genomic and plasmid DNAs, preparation of protoplasts, and fatty acid analysis. R2YE medium (19) was used for the preparation of Streptomyces spore suspensions and for the regeneration of protoplasts after transformation. For the dl-perdeuterated valine feeding experiments, the following liquid medium was used: yeast extract (0.3%)–malt extract (0.5%)–peptone (0.3%)–glucose (1.0%), pH 7.2.
TABLE 1.
Strains, plasmids, and cosmids used in this studya
| Strain or plasmid | Description | References or source |
|---|---|---|
| E. coli | ||
| XL1-Blue | supE44 recA1 hsdR17 endA1 gyrA46 thi relA1 lacF′(proAB+ lacIq lacZΔM15 Tn10Tetr) | 8 |
| DH5α | supE44 DlacU169 (φ80lacZΔM15) hsdR17 recAl endA1 gyrA96 thi-1 relA1 | 17 |
| ET12567 | F−dam-13::Tn9 dcm-6 | 25 |
| S. cinnamonensis | ||
| C730.1 | Monensin overproducer (wild-type strain for this study) | 34 |
| icm mutant | icmA::hyg | 44 |
| mcm mutant | mutB::hyg | 44 |
| WM2 | meaA::ermE | This study |
| WD2 | icmA::hyg, meaA::ermE | This study |
| L1 | ccr::hyg | 24 |
| Major plasmids and cosmids | ||
| pBluescript | Ampr | 1 |
| SuperCos I | Ampr | Stratagene |
| pKC1139 | Streptomyces-E. coli shuttle vector, Amr | 5 |
| pSE34 | pWHM3 with ermE* promoter, Thior | Pfizer Inc. |
| pIJ4026 | pUC18 carrying ermE; Ampr | Pfizer Inc. |
| pHL1 | pUC119 with 5.7-kb Pstl insert containing S. cinnamonensis ccr, Ampr | 24 |
| pME291 | meaA-positive cosmid clone, Ampr | This study |
| pME291K | pBluescript with 5.2-kb BglII/KpnI insert containing complete meaA, Ampr | This study |
| pZR8 | pSE34 with complete meaA, Thior | This study |
| pZR22 | pKC1139 with XbaI/HindIII 5.6-kb meaA::ermE fragment, Eryr Amr | This study |
| pZC32 | pSE34 with MCM-encoding genes mutAB from A. mediterranei, Thior | 49, 50 |
Abbreviations: Ampr, ampicillin resistant; Amr, apramycin resistant; Eryr, erythromycin resistant; Thior, thiostrepton resistant; hyg, hygromycin resistance gene; ermE, erythromycin resistance gene.
Molecular cloning.
Isolation of S. cinnamonensis genomic DNA was performed according to standard methods (19). Genomic DNA was partially digested by Sau3A1 to yield a majority of fragments of around 30 to 40 kb. Sucrose gradient centrifugation was run at 36,000 rpm for 16 h at 4°C in a Sorvall TH-641 Rotor (Sorvall Ultracentrifuge; Du Pont, Wilmington, Del.). Fractions containing 30-to 40-kb fragments were collected and precipitated. Cosmid SuperCos I DNA was prepared according to the instruction manual (Stratagene, La Jolla, Calif.). Twenty microliters of a ligation mixture of genomic DNA (1 μg) and SuperCos I (0.2 μg) was packaged with the Gigapack III XL lambda extract kit (Stratagene) and then transfected into the E. coli XL1- Blue strain. The E. coli XL1-Blue cells competent for transfection were prepared in Luria-Bertani medium with 0.2% (wt/vol) maltose–10 mM MgSO4. About 3,600 recombinant colonies were chosen as a working cosmid library of S. cinnamonensis and subsequently screened using a radiolabeled pHL1 probe that included the ccr gene and 0.5 kb of meaA gene (24). Radioactive DNA labeling was performed with a RTG DNA labeling beads (-dCTP) kit from Pharmacia (Piscataway, N.I.), with [α-32P]dCTP (3,000 Ci/mmol) from Amersham Life Science (Arlington Heights, Ill.). Nonradioactive probing of cosmid subclones with pHL1 was accomplished using digoxigenin DNA labeling and detection kits (Boehringer, Mannheim, Germany). Southern and colony hybridizations were performed according to established methods (36) with a Porablot NY amp nylon membrane (Macherey-Nagel, Duren, Germany). Conditions for prehybridization and hybridization were slightly modified from standard protocols, with final washes for Southern hybridization and colony hybridization being carried out at 68°C in 0.5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Small-scale isolation of high-copy-number plasmid DNA from E. coli was accomplished by the TENS method (51). Large-scale E. coli plasmid preparation was carried out with the Wizard Plus SV Minipreps DNA Purification System (Promega, Madison, Wis.). Low-copy-number cosmid DNA was isolated with Nucleobond AX100 cartridges (Macherey-Nagel). Isolation of DNA fragments from agarose gels was carried out with QIAquick Gel Extraction Kit (Qiagen, Santa Clarita, Calif.). Restriction digestion, phenol extraction, ethanol and isopropanol precipitation, treatment of DNA with the Klenow fragment of DNA polymerase I and alkaline phosphatase, and T4 DNA ligation were accomplished by following standard protocols (36). Preparation and transformation of competent E. coli cells were performed by standard methods (36).
Nucleotide sequence analysis.
Positive cosmids identified with pHL1 were selected, and subclones were prepared with the high-copy-number vector pBluescript SK. DNA sequencing was performed with an ABI PRISM 377 DNA Sequencer (Perkin-Elmer, Foster City, Calif.) according to the conditions recommended by manufacturer. The sequence was analyzed for open reading frames (ORFs) by using CODONPREFERENCE in Genetics Computer Group or FramePlot 2.3∗ software available on the Internet (http://www.nih.go.jp/∼jun/cgi-bin/frameplot.pl). Protein database searches were performed with BLAST and FASTA, protein comparisons were made with COMPARE, and protein sequence alignments were made with LINEUP and PILEUP (all from the University of Wisconsin Genetics Computer Group Package).
Protein overexpression.
Streptomyces protoplasts were transformed in the presence of 25% polyethylene glycol 1000 (19). For overexpression of the meaA gene in S. cinnamonensis, plasmid pZR8 was constructed by PCR with two synthetic oligonucleotides as primers, using cosmid DNA pME291 as a template. Primer 1 (5′-ATACTCTAGAGGAACATGACAGAGC-3′) and primer 2 (5′-ATCTAAGCTTACGAAGGCGGTTGATGG-3′) contained XbaI and HindIII sites (underlined), respectively. The PCR product was purified directly from the gels (Qiagen), digested, and subcloned into pSE34 to produce pZR8. S. cinnamonensis protoplasts were then transformed with pZR8. The mutAB genes from A. mediterranei were released from pYL28 (49) and cloned into the XhaI/HindIII sites of pSE34, resulting in pZC32.
Insertional inactivation of S. cinnamonensis meaA.
A shuttle vector pKC1139 containing the temperature-sensitive Streptomyces origin of replication from pSG5 was used to construct a meaA gene disruption plasmid (5, 25). A BamHI site suitable for the insertion of the erythromycin resistance gene (ermE) was created within the meaA ORF by PCR. Two fragments corresponding to the first and second halves of the meaA gene were PCR amplified from the positive cosmid clone pME291 by using the following primers: primer 1 (described above), primer 2 (described above), primer 3, 5′-ATCTGGATCCAAGATGTCCTCGTAC-3′, and primer 4, 5′-ATCTGAATTCCGACGGCTCGCACGTCGT-3′. Primer 3 contained a BamHI site, and primer 4 contained an EcoRI site (underlined). Primers 1 and 3 produced a 1.0-kb PCR fragment with XbaI and BamHI cohesive ends, and primers 2 and 4 produced a 1.0-kb PCR fragment with EcoRI and HindIII cohesive ends. The two fragments were cloned into the corresponding sites of pBluescript, resulting in the formation of pZR6. A 1.6-kb BglII ermE gene fragment was excised from pIJ4026 and cloned into the BamHI site of pZR6. The resulting pZR7 plasmid, with ermE transcribed in the same direction as meaA, was then isolated and confirmed by restriction analysis. A 3.6-kb XbaI/HindIII fragment containing the disrupted meaA gene and ermE insert was released from pZR7 and cloned into pKC1139 to yield pZR22, a meaA gene disruption plasmid. In order to increase transformation and homologous recombination efficiency, this plasmid was denatured by 1.0 N NaOH according to the method described by Oh et al. (29) before it was used to transform S. cinnamonensis protoplasts. The meaA mutant of S. cinnamonensis was then obtained following standard protocols (24) and confirmed by PCR.
Fatty acid analysis.
S. cinnamonensis cultures were grown in YEME medium at 30°C for 72 h, and the fatty acids were extracted and analyzed as described previously (45). When required, perdeuterated valine (200 mM) was added to the media at the time of inoculation.
Production and quantitation of monensins A and B.
Fermentations of S. cinnamonensis C730.1 cells and mutant derivatives were carried out in a two-stage fermentation process as described previously. The second-stage production medium was either a glucose-soybean meal (24) or an oil-based medium composed of soybean meal (1.5%), glucose (2.5%), soybean oil (1.5%), methyl oleate (2.0%), lard oil (1.0%), CaCO3 (0.3%), FeSO4 · 7H2O (0.03%), KCl (0.01%), and MnCl2 4H2O (0.003%). When required, valine (0.1 or 0.8 M) was added in three equal portions at 24, 60 and 72 h during fermentation. Monensins were isolated and quantitated by high-performance liquid chromatography (HPLC) analysis as described previously (34).
Isotope labeling experiments with ethyl [3,4-13C2] acetoacetate.
The conditions for the production of monensins A and B used in the labeling experiments were identical to those described above. A 30 mM 1:3 mixture of ethyl [3,4-13C2]acetoacetate and unlabeled ethyl acetoacetate was added batchwise to the liquid cultures in three equal portions after 24, 28, and 72 h of growth. Monensin A was purified from organic extracts of fermentations as described previously (34) and analyzed by 13C spectroscopy with proton decoupling.
Nucleotide sequence accession number.
The complete sequence of S. cinnamonensis meaA reported here has been deposited in the GenBank database under the accession no. AF303662.
RESULTS
Cloning and sequence analysis of S. cinnamonensis meaA.
The plasmid pHL1 (24), containing the ccr gene and 0.5 kb of meaA of S. cinnamonensis, was used as the DNA probe to screen a cosmid library. Eight positive cosmid clones with inserts of ∼35 kb were obtained. One of the positive cosmid clones, designated pME291, was chosen for further study. Restriction mapping of pME291 showed that meaA was contained within a 5.2-kb BglII/KpnI fragment. This fragment was cloned into pBluescript to generate pME291K. The complete nucleotide sequence of the meaA gene was determined. This ORF has a typical streptomycete codon bias (G+C content of 69.5%), extends from nucleotide 122 (ATG) to 2149 in the deposited sequence, and encodes a protein of 675 residues. The previously identified ccr gene is located 36 bp upstream of meaA (24), while the downstream sequence contained an incomplete downstream ORF, ORF1, encoding a protein with 48% sequence identity to malyl-CoA lyase of Methylobacterium extorquens (37) and 33% identity to citrate lyase of E. coli. The same gene order has previously been identified for S. collinus (16) and S. coelicolor (www.sanger.ac.uk). meaA possesses a stop codon overlapping the start codon of ORF1 at nucleotides 2147 to 2149 (ATGA), which is suggestive of translational coupling (17). The meaA and ORF1 ORFs are both preceded by a potential streptomycete ribosome binding sequence, GGAG, at nucleotides 107 to 110 and 2134 to 2137, respectively (4). No putative E.coli ς70-like promoter element was found upstream of either the meaA gene or ORF1 (40).
A phylogenetic tree of the coenzyme B12-dependent mutases, MCM, ICM, and MeaA, was made (data not shown). MeaA from S. cinnamonensis showed the highest amino acid sequence identity to corresponding proteins in S. coelicolor (89%), S. collinus (90%), and M. extorquens (60%). S. cinnamonensis MeaA is the third MCM-like coenzyme B12-dependent mutase from this organism to be studied and has amino acid sequence homology to the MCM large subunit (52% similarity, 41% identity), ICM large subunit (48% similarity, 33% identity), and ICM small subunit (62% similarity, 52% identity). Similar homologies were observed with the corresponding subunits of the S. coelicolor ICM enzyme. The motifs DXHXXG, SXL, and GG, which have been indicated as diagnostic characteristics for cobalamin binding from the E. coli methionine synthase (13) and the Propionibacterium freundenreichii subsp. shermanii MCM (26) crystal structures, are found perfectly conserved at the C-terminal end of MeaA. A crystal structure of MCM from P. freundenreichii subsp. shermanii with bound substrate has recently been revealed, in which amino acids in the active site form an interaction with methylmalonyl-CoA (26). An alignment of P. freundenreichii subsp. shermanii MCM with S. cinnamonensis MeaA revealed that many of these residues are conserved (data not shown).
Overexpression of S. cinnamonensis meaA.
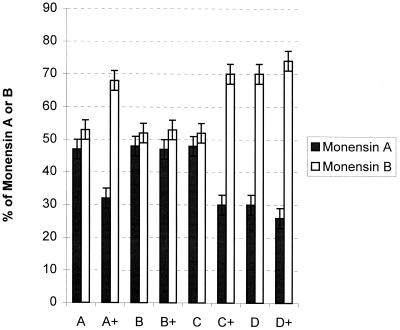
Earlier work demonstrated that the C730.1 strain produces monensins A and B in a ratio of about 1:1 under standard laboratory growth conditions (24,34). Neither icm inactivation nor mutAB inactivation has any effect on the monensin A-to-monensin B ratio or the total monensin titer (24, 44). A ccr mutant, however, causes a substantial decrease in this monensins A-to-B ratio from 1:1 to approximately to 1:4 (24). This change was attributed to a decrease in the pools of ethylmalonyl-CoA (formed by the action of CCR) relative to those of methylmalonyl-CoA (24). The S. cinnamonensis meaA gene was PCR amplified and used to generate pZR8, in which the meaA gene was expressed from the strong constitutive ermE∗ promoter (46). This plasmid was transformed into S. cinnamonensis C730.1 (wild type), as well as into icm (44), ccr (L1) (24), and mut (44) mutants. The monensin titers and monensin A-to-monensin B ratio for each of the resulting transformants was determined (Fig. 2). In all experiments, the overall monensin titers were not significantly altered. Plasmid-based overexpression of meaA, however, did significantly decrease the monensin A-to-monensin B ratio in the C730.1 strain and the mutAB mutant. These results are consistent with the expression of meaA leading to an increase in the pool of methylmalonyl-CoA relative to that of ethylmalonyl-CoA and suggestive of a role of MeaA in methylmalonyl-CoA formation. In the ccr mutant (L1), this ratio is already significantly reduced from that of the wild-type strain and only marginally increased by plasmid-based meaA overexpression. Surprisingly, meaA overexpression in the icm mutant did not change the monensin A-to-monensin B ratio.
FIG. 2.
Effects of plasmid-based meaA expression (pZR8) on the monensin A-to-monensin B ratio. (A) C730.1, wild-type strain; A+, C730.1/pZR8; B, icm mutant; B+, icm mutant/pZR8; C, mut mutant; C+, mut mutant/pZR8; D, ccr mutant (L1); D+, L1/pZR8.
Targeted disruption of S. cinnamonensis meaA and phenotype analysis.
An insertional inactivation strategy was used to disrupt the meaA gene in both S. cinnamonensis C730.1 and the icm mutant (44). The plasmid pZR22 used to create the meaA mutants was generated in E. coli ET12567 and denatured by alkali treatment (29) before it was used for the transformation of S. cinnamonensis cells. Colonies resistant to both apramycin and erythromycin (Ermr, Amr) were obtained. Mutants in which a single crossover between pZR22 and genomic DNA had occurred were selected by cultivating these transformants at 40°C in the presence of both antibiotics. Following several rounds of propagation of these single-crossover mutants in the absence of any antibiotics at 30°C, colonies resistant to erythromycin but sensitive to apramycin (Ermr, Ams) were obtained. The meaA::ermE disruption in C730.1 and the icm mutation of S. cinnamonensis were designated WM2 and WD2 strains, respectively. The double-crossover event in both these clones ware confirmed by PCR using primers 1 and 2 (see Materials and Methods).
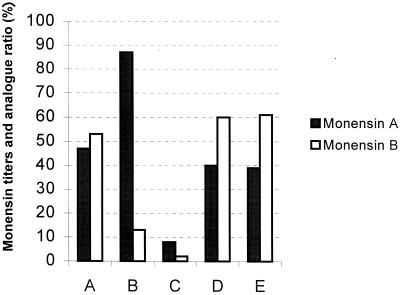
When grown in either a carbohydrate-based medium or an oil-based medium, the mutants WM2 (meaA mutation) and WD2 (icm and meaA mutations) and wild-type C730.1 strains grew equally well and displayed no morphological differences. The monensin titers and monensin A-to-monensin B ratio produced by each of these strains was analyzed (Fig. 3). The meaA mutant produced a significantly higher monensin A-to-monensin B ratio (4:1) compared to the C730.1 strain (1:1) in either production media. This ratio in the WM2 was more than restored with plasmid-based expression of the meaA gene. In fact, the S. cinnamonensis WM2/pZR8 strain produced slightly more monensin B than monensin A, presumably due to higher levels of meaA expression. These results are also consistent with a role of MeaA in production of methylmalonyl-CoA in S. cinnamonensis.
FIG. 3.
Monensin titers and analog ratios for various S. cinnamonensis mutants in carbohydrate-based fermentation medium. (A) C730.1, wild-type strain; B, meaA mutant (WM2); C, icm meaA mutant (WD2); D, WM2/pZR8 (plasmid-based expression of meaA); E, WD2/pZR8. Total monensin titers (monensins A and B) are expressed as a percentage of that obtained using the C730.1 strain. The same pattern of changes in ratios and titers was observed using the oil-based media.
The meaA mutant produced monensin titers comparable to those of the C730.1 strain, indicating that while the ratio of methylmalonyl-CoA to ethylmalonyl-CoA may have decreased, there was still sufficient methylmalonyl-CoA to support monensin production. The levels of methylmalonyl-CoA were limiting, however, in the icm meaA mutant (WD2), which produced only 10% of the monensin titers (predominantly as monensin A) seen with either the C730.1 meaA or icm mutants in either carbohydrate-based or oil-based media. Thus, in this mutant, meaA and icm together appear to be crucial for the monensin production in the tested media. As predicted, the monensin A-to-monensin B ratio and overall monensin titers were restored in the icm meaA mutant by plasmid-based expression of meaA (this experiment, like that with the WM2 strain, led to slightly higher levels of monensin B relative to those of monensin A).
Plasmid-based overexpression of A. mediterrenei mutAB in S. cinnamonensis mutants.
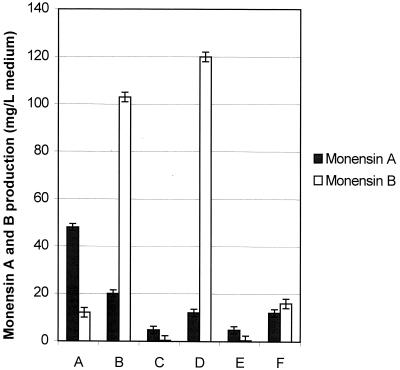
MCM is a well-studied enzyme and has long been thought to play an important role in providing methylmalony-CoA for polyketide formation in many organisms, including the erythromycin-producing S. erythraea (21), rifamycin-producing A. mediterranei (49), and monensin-producing S. cinnamonensis (6). Indeed, overexpression of MCM-encoding genes mutAB in S. cinnamonensis wild-type strains has recently been found to increase both the overall monensin titers and the amount of monensin B relative to that of monensin A (49). The same phenomenon was observed in this study with plasmid-based overexpression of the A. mediterranei mutAB genes in the meaA mutant (Fig. 4). In this case, the monensin titers were increased almost twofold (relative to those of either the C730.1 or meaA mutant) and the monensin A-to-monensin B ratio switched from 4:1 to 1:5. The overexpression of mutAB genes in the mutant WD2 (which produced only 10% of the monensin titers of the C730.1 or WM2 strain) led to a remarkable 20-fold increase in monensin titers. As in the case of the meaA mutant, plasmid-based expression of mutAB in the WD2 strain led to a significant decrease in the monensin A-to-monensin B ratio. Thus, in either the WM2 or WD2 strain, the shift toward the monensin A analog caused by insertional inactivation of meaA can be reversed by plasmid-based expression of either meaA or mutAB. Furthermore, expression of either of these genes can be used to restore monensin production in a meaA icm mutant (WD2) to at least wild-type levels. These observations are consistent with MeaA playing a major role in methylmalonyl-CoA formation.
FIG. 4.
Enhancement of monensin titers by either pZC32 plasmid-based expression of mutAB or addition of exogenous dl-valine. A, meaA mutant (WM2); B, WM2/pZC32; C, meaA icm mutant (WD2); D, WD2/pZC32; E, WD2 cells grown in the presence of 0.1 M dl-valine; F, WD2 cells grown with 0.8 M dl-valine.
Effect of valine feeding on monensin production in the WD2 strain.
Isotope labeling experiments with both polyether- and macrolide-producing streptomycetes have shown that valine can be catabolized to isobutyryl-CoA, and subsequently to methylmalonyl-CoA, for antibiotic biosynthesis (23, 30) (Fig. 1). Media supplemented with valine and isoleucine can sometimes stimulate macrolide antibiotic production, while increasing NH4+ concentrations can have negative effects on both macrolide production and valine dehydrogenase activity (31, 32), all suggesting that branched-chain amino acid catabolism may be an important source of building blocks for macrolide biosynthesis. Valine at 0.1 M was fed to fermentations of the meaA icm mutant in three separate aliquots (at 24, 48, and 72 h of fermentation in the production media). This experiment was also carried out using valine at a final concentration of 0.8 M. Feeding with 0.1 M valine had no significant effect on the monensin production by the WD2 mutant, while feeding with 0.8 M valine led to only moderate increases in the monensin titers (25% increase) (Fig. 4). In the latter case, the monensin titers were 40% of the level achieved by the wild-type C730.1 strain and the monensin A-to-monensin B ratios were essentially equivalent (1:1). Thus, the valine catabolic pathway in the meaA icm S. cinnamonensis mutant does not appear to be efficient process for producing methylmalonyl-CoA.
Incorporation of [3,4-13C2]acetoacetate into monensin A in the meaA mutant.
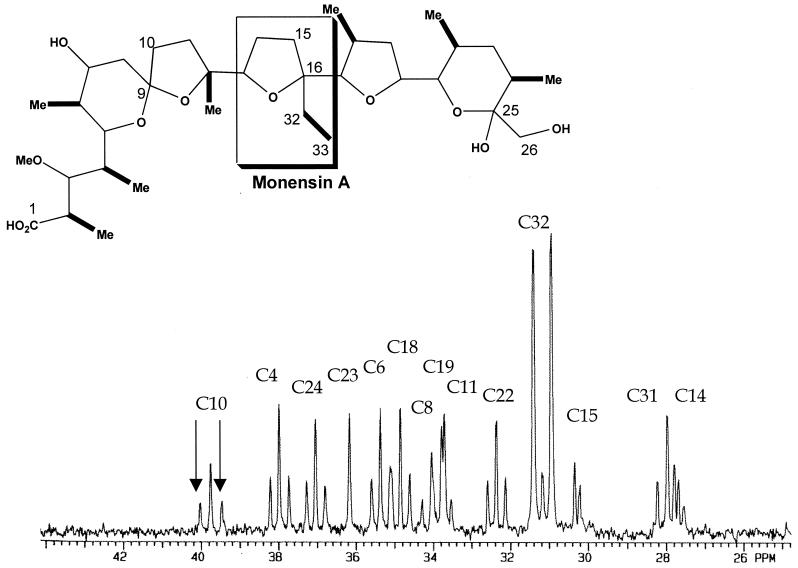
Biosynthetic studies in S. cinnamonensis have previously demonstrated that dual-labeled [1,3-13C2]acetoacetyl-CoA (generated by carrying out fermentations in the presence of the corresponding labeled ethyl acetoacetate) can be converted into [1,2-13C2]methylmalonyl-CoA in the absence of either ICM or MCM (44). The pathway by which this intact conversion, which requires a carbon skeleton rearrangement, occurs was undetermined. It was, however, suggested that MeaA might play a role in this pathway (44). To investigate this possibility, ethyl [3,4-13C2]acetoacetate was added to fermentations of C730.1 and the meaA mutant (WM2). Strong 13C doublets surrounding the natural abundance signal for both C-32 and C-33 of monensin A were observed (Fig. 5) for the WM2 mutant, which indicated the simultaneous presence of a 13C label at both carbons. The size of these doublets was substantially greater than the natural abundance signal and indicated an approximate 10-fold enrichment of 13C at these carbons due to the intact utilization of the 13C-labeled acetoacetate. The natural abundance 13C signals for all of the carbons of monensin A derived from C-2 and C-3 of methylmalonyl-CoA were similarly surrounded by enriched doublets. In this case, however, there was only about a twofold enrichment of 13C at these positions due to the intact utilization of the 13C-labeled acetoacetate. Indistinguishable labeling patterns were observed for monensin A isolated from the C730.1 strain grown under identical conditions. Thus, insertional inactivation of meaA affects the pool of methylmalonyl-CoA but does not seem to significantly affect the amount generated from acetoacetyl-CoA or related metabolites. These data combined with data from the previous study (44) indicate that acetoacetyl-CoA can be converted intact to methylmalonyl-CoA in S. cinnamonensis in the absence of either ICM, MCM, or MeaA. A similar study was also carried out with the icm meaA double mutant (WD2). In order to generate sufficient monensin A for nuclear magnetic resonance analysis, this experiment was carried out using the WD2/pZC32 strain (plasmid-based expression of mutAB). Similar low levels of intact labeling of the methymalonyl-CoA-derived positions of monensin from labeled ethyl acetoacetate were also seen in this experiment (data not shown), indicating that conversion of acetoacetyl-CoA to methylmalonyl-CoA can proceed in the absence of both ICM and MeaA.
FIG. 5.
A portion of the proton decoupled-13C nuclear magnetic resonance spectrum of monensin A isolated after feeding ethyl [3,4-13C2]acetoacetate to a meaA mutant of S. cinnamonensis WM2. Enriched doublets surrounding the natural abundance signals for positions C-2, C-4, C-6, C-12, C-18, C-22, and C-24 and the corresponding methyl substituents were observed, consistent with intact incorporation of [2,3-13C2]methylmalonyl-CoA. Substantially larger enriched doublets were observed at C-32 and C-33 of monensin A, consistent with the intact incorporation of [3,4-13C2]ethylmalonyl-CoA. Arrows indicate the enriched doublet for the malonyl-CoA-derived C-10 position. No significant doulets were observed for monensin A carbons derived from C-1 of methylmalonyl-CoA. Carbon-carbon bonds in monensin A labeled intact from C-3 and C-4 of methylmalonyl-CoA are shown in bold. A similar pattern of labeling was observed for the wild-type C730.1 strain and the WD2/pZC32 strain.
Fatty acid analyses.
It has previously been shown that alterations in the levels of butyryl-CoA or isobutyryl-CoA pool can directly alter the types and amounts of branched-chain and straight-chain fatty acids in streptomycetes (45). This present study clearly demonstrated that disruption and overexpression of meaA affected the levels of the related metabolites and the methylmalonyl-CoA and ethylmalonyl-CoA pool and raised the possibility that changes in the fatty acid profiles might also be observed. Fatty acid profiles for the S. cinnamonensis wild-type C730.1 strain and meaA and icm meaA mutants were determined and shown to be essentially indistinguishable.
Fatty acid analyses were also carried out for these strains and the icm mutant grown in the presence of perdeuterated valine. It has previously been shown that approximately 60% of the isopalmitate in streptomycetes grown in the presence of perdeuterated valine is generated from use of the corresponding perdeuterated isobutyryl-CoA catabolite. In these experiments, the action of ICM on the labeled isobutyryl-CoA provides perdeuterated butyryl-CoA which can label intact as much as 10% of the palmitate pool. In the present experiments, only labeled palmitate was observed for the C730.1 and MeaA mutants grown in the presence of 200 mM perdeuterated valine. No labeled palmitate was observed for either the icm or meaA icm mutant, while labeled isopalmitate was observed in all four strains (data not shown). These observations are strongly suggestive of the absence of any significant in vivo ICM activity in mutants with an insertional inactivation of the icm gene under our test conditions.
DISCUSSION
Role of methylmalonyl-CoA in polyketide biosynthesis.
The identification of numerous gene clusters involved in polyketide antibiotics has led to a better understanding of the genetic and biochemical elements governing the biosynthesis of these compounds (2, 20). By contrast, less information is available on the enzymes and pathways that provide the required biosynthetic precursors for these processes. Perhaps the best example is methylmalonyl-CoA, one of the most common precursors used in polyketide biosynthesis. Several different pathways have been proposed to be involved in the generation of this precursor, yet the relative importance of each of these for methylmalonyl-CoA production within different polyketide-producing organisms is unknown. Furthermore, it is not yet clear if there are additional pathways for producing methylmalonyl-CoA beyond those known to date. Substrate availability can clearly affect the type of polyketide analogs made in a fermentation (24, 38) and in some cases can be a limiting factor controlling overall titers. Knowledge of the enzymes and pathways involved in the precursor supply thus offers an opportunity for rational approaches for increasing fermentation titers. In this study, we reported the cloning and characterization of meaA, which encodes a novel coenzyme B12-dependent mutase in monensin A-producing S. cinnamonensis. This gene has been shown to be necessary in the methanol and ethanol assimilation in M. extorquens AM1 and to be important in growth of S. collinus on acetate (9, 15, 37). The present study provides evidence that MeaA has a significant role in providing methylmalonyl-CoA for the biosynthesis of the monensin polyketide.
MeaA, a putative coenzyme B12-dependent mutase.
Sequence analysis predicts that S. cinnamonensis meaA encodes a 74-kDa protein with a perfectly conserved coenzyme B12 binding motif at the C terminus and 36 to 40% amino acid sequence identity to the two known coenzyme B12-dependent mutases (MCM and ICM) (6, 27, 47). Isobutyryl-CoA and methylmalonyl-CoA (substrates for ICM and MCM, respectively) differ only in the oxidation state of one carbon. The two enzymes show significant amino acid sequence homology, particularly in the region shown from the crystal structure of the P. freundenreichii subsp. shermanii MCM shown to be involved in substrate binding (26). Most of these amino acids are also conserved in MeaA (data not shown). In MCM, the TyrA89 conserved in all MCM sequences and ArgA208 are located near the bottom of the substrate binding hole and appear to form hydrogen bonds with the carboxyl group of the methylmalonyl-CoA substrate. These residues are not conserved in ICM (the substrate in this case contains the reduced methyl carbon substituent) but are conserved in the predicted amino acid sequences of all known MeaA proteins. Thus, the MeaA substrate might also contain two oxidized and possibly carboxylated substituents, with one activated as a CoA thioester.
Role of MeaA in providing methylmalonyl-CoA.
The role of meaA in providing methylmalonyl-CoA for monensin biosynthesis was studied by two approaches. Firstly, meaA was overexpressed from a multicopy plasmid using the strong constitutive ermE promoter. This system has been used successfully in the past for overexpression of the ccr gene in S. cinnamonensis (24). Overexpression of the meaA gene resulted in a significant increase of methylmalonyl-CoA-derived monensin B compared to the ethylmalonyl-CoA-derived monensin A in the C730.1 type strain and mutAB-blocked mutants. These observations are consistent with an increased pool of methylmalonyl-CoA relative to that of ethylmalonyl-CoA and suggest that MeaA is able to generate methylmalonyl-CoA. The same conclusion was reached by generating a meaA mutant of S. cinnamonensis and observing that it made significantly higher ratios of monensin A compared to monensin B. No significant change in total monensin titers was observed in either of these experiments, indicating that under these conditions, another process provides the methylmalonyl-CoA for monensin biosynthesis. This observation was not made for the meaA icm mutant that produced less than 10% of the monensin titers than the corresponding control strains. Evidence that this decrease was due to a limitation in methylmalonyl-CoA was provided by the observation that plasmid-based expression of either mutAB or meaA restored monensin production at least to the same levels as those seen in the control. The reason that inactivation of insertional inactivation of meaA has a more pronounced effect on the icm mutant than on the C730.1 strain is unclear at present. It is interesting to note that only in the icm mutant did plasmid-based overexpression of meaA not lead to an increase in the monensin A-to-monensin B ratio. Regardless, these experiments have clearly indicated that alterations in the levels of MeaA in S. cinnamonensis can lead to significant changes in the amounts of methylmalonyl-CoA available for monensin biosynthesis and that its role can be efficiently replaced by MCM. Previous studies of MeaA in M. extorquens AM1 (9, 37) and S. collinus (15) have indicated that MeaA is neither an MCM nor an ICM. Thus, it seems likely that under the tested conditions, this methylmalonyl-CoA is being produced by a pathway involving a different type of coenzyme B12-dependent mutase.
Role of valine degradation in providing methylmalonyl-CoA.
Valine catabolism as a source for methylmalonyl-CoA has been well studied for some streptomycetes strains, including S. coelicolor (41), Streptomyces ambofaciens, and Streptomyces fradiae (42). Labeling studies, including those carried out with S. cinnamonensis cells, all indicate that the most likely pathway in a complex fermentation medium involves direct oxidation of one of the methyl groups of valine catabolite isobutyryl-CoA to yield methylmalonyl-CoA (Fig. 1) (34, 44). The decreased monensin titers in the meaA icm mutant clearly demonstrate that valine catabolism involving direct oxidation of isobutyryl-CoA is not a major contributor to methylmalonyl-CoA production (this process does not involve a carbon skeleton rearrangement and thus is not predicted to require a coenzyme B12-dependent mutase). A partial restoration of monensin titers with high levels of exogenously supplied valine indicates that such a B12-independent process can provide at least some of the methylmalonyl-CoA. Nonetheless, significantly higher monensin titers could be achieved in the icm meaA mutant strain by introduction of either MeaA or MCM, suggesting that processes using these enzymes are more efficient at the generation of methylmalonyl-CoA.
A multitude of labeling studies carried out with isobutyrate, butyrate, and ethyl acetoacetate, in this and previous studies of S. cinnamonensis, have consistently shown significantly greater labeling of the butyryl-CoA (ethylmalonyl-CoA)-derived position of monensin A than the methylmalonyl-CoA-derived positions. These observations would also be consistent with the valine catabolite isobutyryl-CoA not being an intermediate in the major pathway for methylmalonyl-CoA production. A similar conclusion can be drawn concerning the role of valine catabolism in Streptomyces avermitilis. A bkd mutation of this strain is blocked completely in valine degradation, resulting in a complete lack of the isobutyryl-CoA starter unit required for biosynthesis of the polyketide avermectin (10). When alternative starter units, such as cyclohexanecarboxylic acid, are provided to this strain, novel avermectins requiring methylmalonyl-CoA precursors as extender units can be made, despite the lack of any isobutyryl-CoA (10). More recently, we have described that addition of labeled isobutyric acid to this strain results in production of avermectin B1b labeled 16-fold more efficiently at the isobutyryl-CoA-derived position than the methylmalonyl-CoA-derived positions (50). Thus, it is quite clear that under the tested conditions, isobutyryl-CoA derived from valine catabolism does not play a significant role in providing methylmalonyl-CoA for monensin or avermectin biosynthesis. These observations do not preclude the possibility that under certain conditions or with different organisms, valine catabolism may be playing a more significant role in providing methylmalonyl-CoA.
Multiple pathways linking acetoacetyl-CoA or butyryl-CoA and methylmalonyl-CoA.
The intact conversion of [1,3-13C2]acetoacetyl-CoA into the methylmalonyl-CoA-derived positions of monensin has long been considered to be indicative of a pathway (Fig. 1) involving reduction to butyryl-CoA, isomerization to isobutyryl-CoA, and subsequent oxidation (38, 44). The retention of all three deuteriums of [13CD3] acetate into the methyl group of methylmalonyl-CoA in S. cinnamonensis is also consistent with such a process and inconsistent with other pathways involving MCM or PCC and MMT (Fig. 1) The recent observation that intact conversion of labeled acetoacetate into methylmalonyl-CoA was not affected by insertional in activation of the icm gene was surprising and suggested that another enzyme-catalyzed process must account for this conversion. In the present study, the same low levels of intact conversion of acetoacetyl-CoA into methylmalonyl-CoA were observed for meaA and meaA icm mutants and a wild-type strain, suggesting that meaA is not involved in converting acetoacetyl-CoA or related metabolites (butyryl-CoA) to methylmalonyl-CoA. Furthermore, it is evident from these data and a previous analysis (44) that a process not involving the cloned icm, mcm, or meaA genes is involved in this conversion (Fig. 1). One possible explanation might be the presence of a second copy of icm expressed under certain conditions. Recently, a putative second icm gene was discovered in the genome of S. coelicolor (accession no. CAB71920 and CAB71846 for the large and small subunits in the NCBI GenBank database, respectively). This gene contains two ORFs (designated the mutB and icmA genes, respectively) encoding 531 and 159 residues, respectively, which are similar in size to IcmA and IcmB of both S. cinnamonensis and S. coelicolor (33, 48). mutB encodes a protein with 51% identity to IcmB, while icmA encodes a protein with 68% identity to IcmA of S. coelicolor. Unlike the first icmAB genes characterized (33, 48), these two ORFs are separated by only 40 bp, suggesting that they may be translationally coupled. An alignment of this putative ICM with the MCM of P. freundenreichii subsp. shermanii (data not shown) revealed that many of the same amino acids lining the active site of MCM are conserved. The TyrA89 and Arg207 present in MCM and in MeaA are absent in both this putative S. coelicolor ICM as well as the S. cinnamonensis ICM (data not shown). Thus, sequence data suggest, but do not prove, that there may be two copies of the genes encoding ICM in S. coelicolor. Such a possibility cannot be completely ruled out for S. cinnamonensis. However, it has been demonstrated that there is no detectable ICM activity in the S. cinnamonensis icm mutant (48). Furthermore, the fatty acid analyses carried out in the presence of valine in the present study showed that there is no detectable in vivo conversion of isobutyryl-CoA to butyryl-CoA in either the icm or icm meaA mutants using a minimal medium. We were unable to make this determination in a complex fermentation medium because only trace amounts of the straight-chain fatty acids required for this analysis were produced. The manner in which butyryl-CoA and acetoacetyl-CoA are converted into methylmalonyl-CoA in fermentations of these mutants thus remains uncertain.
Conclusion.
In conclusion, MeaA plays an important role via an unknown pathway in providing methylmalonyl-CoA precursors for monensin biosynthesis in S. cinnamonensis. Increased methylmalonyl-CoA pools can be accomplished by plasmid-based expression of meaA. Conversely, these pools can be decreased by insertional inactivation of meaA. In the case of an icm meaA mutant, these decreased pools significantly reduce monensin titers. Restoration of monensin titers can be accomplished by plasmid-based expression of genes encoding either MeaA or MCM. By comparison, isobutyryl-CoA obtained either from valine catabolism or by ICM-catalyzed isomerization of butyryl-CoA plays a more minor role in providing methylmalonyl-CoA for monensin biosynthesis. Finally, an enzyme other than those encoded by the cloned icm meaA and mcm genes of S. cinnamonensis can catalyze the intact conversion of acetoacetyl-CoA to methylmalonyl-CoA.
ACKNOWLEDGMENTS
We thank John Robinson (Institute of Organic Chemistry, University of Zurich-IRCHEL) for helpful discussions and contributing to the construction of the S. cinnamonensis DNA library. We are grateful to Insmed Pharmaceuticals for the use of a refractive index detector for HPLC analysis of monensin production, W. H. Jiang (Shanghai Institute of Plant Physiology, CAS) for providing the A. mediterranei mutAB mutant containing the pYL28 plasmid, and Eli Lilly & Company for providing S. cinnamonensis C730.1.
Financial support was provided to K.A.R. by the National Institutes of Health (Grant GM50541) and Eli Lilly & Company.
REFERENCES
- 1.Alting-Mess A A, Short J M. pBluescript II: gene mapping vectors. Nucleic Acids Res. 1989;17:9494. doi: 10.1093/nar/17.22.9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.August P R, Tang L, Yoon Y J, Ning S, Muller R, Yu T W, Taylor M, Hoffmann D, Kim C G, Zhang X, Hutchinson C R, Floss H G. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterraneiS699. Chem Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 3.Beran M, Zima J. Determination of monensin A and B in the fermentation broth of Streptomyces cinnamonensisby high performance liquid chromatograph. Chromatographia. 1993;35:206–208. [Google Scholar]
- 4.Bibb M J, Cohen S N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividansMol. Gen Genet. 1982;187:265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- 5.Bierman M, Logan R, Brien K O, Seno E T, Rao R N, Schoner B E. Plasmid cloning vectors for the conjugal transfer of DNA from Escherichia coli to Streptomyces spp. Gene. 1992;116:43–49. doi: 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- 6.Birch A, Leiser A, Robinson J A. Cloning, sequencing, and expression of the gene encoding methylmalonyl-coenzyme A mutase from Streptomyces cinnamonensis. J Bacteriol. 1993;175:3511–3519. doi: 10.1128/jb.175.11.3511-3519.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bramwell H, Hunter I S, Coggins J R, Nimmo H G. Propionyl-CoA carboxylase from Streptomyces coelicolorA3(2): cloning of the gene encoding the biotin-containing subunit. Microbiology. 1999;142:649–655. doi: 10.1099/13500872-142-3-649. [DOI] [PubMed] [Google Scholar]
- 8.Bullock W O, Fernandez J M, Short J M. XL1-Blue: a high efficiency plasmid transforming recA Escherichia colistrain with beta-galactosidase selection. BioTechniques. 1987;5:367–379. [Google Scholar]
- 9.Chistoserdova L V, Lidstorm M E. Molecular characterization of a chromosomal region involved in the oxidation of acetyl-CoA to glyoxylate in the isocitrate-lyase-negative methylotroph Methylobacterium extorquensAM1. Microbiology. 1996;142:1459–1468. doi: 10.1099/13500872-142-6-1459. [DOI] [PubMed] [Google Scholar]
- 10.Cropp T A, Smogowicz A A, Hafner E W, Denoya C D, McArthur H A I, Reynolds K A. Fatty-acid biosynthesis in a branched-chain alpha-keto acid dehydrogenase mutant of Streptomyces avermitilis. Can J Microbiol. 2000;46:506–514. [PubMed] [Google Scholar]
- 11.Donadio S, Staver M J, McAlpine J B, Swanson S J, Katz L. Modular organization of genes required for complex polyketide biosynthesis. Science. 1991;252:675–679. doi: 10.1126/science.2024119. [DOI] [PubMed] [Google Scholar]
- 12.Donadio S, Staver M J, Katz L. Erythromycin production in Saccharopolyspora erythraeadoes not require a functional propionyl-CoA carboxylase. Mol Microbiol. 1996;19:977–984. doi: 10.1046/j.1365-2958.1996.439969.x. [DOI] [PubMed] [Google Scholar]
- 13.Drennan C L, Huang S, Drummond J T, Matthews R G, Lidwig M L. How a protein binds B12: a 3.0 A X-ray structure of B12-binding domains of methionine synthase. Science. 1994;266:1669–1674. doi: 10.1126/science.7992050. [DOI] [PubMed] [Google Scholar]
- 14.Gorman M, Chamberlin J W, Hamill R L. Monensin, a new biologically active compound. V. Compounds related to monensin. Antimicrob Agents Chemother. 1967;7:363–368. [PubMed] [Google Scholar]
- 15.Han L, Reynolds K A. A novel alternate anaplerotic pathway to the glyoxylate cycle in Streptomyces. J Bacteriol. 1997;179:5157–5164. doi: 10.1128/jb.179.16.5157-5164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Han L, Yang K, Kulowski K, Wendt-Pienkowski E, Hutchinson C R, Vining L C. An acyl-coenzyme A carboxylase encoding gene associated with jadomycin biosynthesis in Streptomyces venezuelaeISP5230. Microbiology. 2000;146:903–910. doi: 10.1099/00221287-146-4-903. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan D. Studies on transformation of Escherichia coliwith plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 18.Hector M L, Fall R R. Multiple acyl-coenzyme A carboxylases in Pseudomonas citronellolis. Biochemistry. 1976;15:3465–3471. doi: 10.1021/bi00661a011. [DOI] [PubMed] [Google Scholar]
- 19.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, England: John Innes Foundation; 1985. [Google Scholar]
- 20.Hopwood D A. Genetic contributions to understanding polyketide synthases. Chem Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 21.Hunaiti A R, Kolattukudy P E. Source of methylmalonyl-coenzyme A for erythromycin synthesis: methylmalonyl-coenzyme A mutase from Streptomyces erythreus. Antimicrob Agents Chemother. 1984;25:173–178. doi: 10.1128/aac.25.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunaiti A R, Kolattukudy P E. Isolation and characterization of an acyl-coenzyme A carboxylase from an erythromycin-producing Streptomyces erythreus. Arch Biochem Biophys. 1982;216:362–371. doi: 10.1016/0003-9861(82)90222-3. [DOI] [PubMed] [Google Scholar]
- 23.Leiser A, Birch A, Robinson J A. Cloning, sequencing, over-expression in Escherichia coli, and inactivation of the valine dehydrogenase gene in the polyether antibiotic producer Streptomyces cinnamonensis. Gene. 1996;177:217–222. doi: 10.1016/0378-1119(96)00305-8. [DOI] [PubMed] [Google Scholar]
- 24.Liu H, Reynolds K A. Role of crotonyl coenzyme A reductase in determining the ratio of polyketides monensin A and monensin B produced by Streptomyces cinnamonensis. J Bacteriol. 1999;181:6806–6813. doi: 10.1128/jb.181.21.6806-6813.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacNeil D J, Gewain K M, Ruby C L, Dezeny G, Gibbons P H, MacNeil T. Analysis of Streptomyces avermitilisgenes required for avermectin biosynthesis utilizing a novel intergration vector. Gene. 1992;111:61–68. doi: 10.1016/0378-1119(92)90603-m. [DOI] [PubMed] [Google Scholar]
- 26.Mancia F, Keep N H, Nakagawa A, Leadlay P F, McSweeney S, Rasmussen B, Bosecke P, Dia O, Evans P R. How coenzyme B12 radicals are generated: the crystal structure of methylmalonyl-coenzyme A mutase at 2 Åresolution. Structure. 1996;4:339–350. doi: 10.1016/s0969-2126(96)00037-8. [DOI] [PubMed] [Google Scholar]
- 27.Marsh E N, McKie N, Davis N K, Leadlay P F. Cloning and structural characterization of the genes coding for adenosylcobalamin-dependent methylmalonyl-CoA mutase from Propionibacterium shermanii. Biochem J. 1989;260:345–352. doi: 10.1042/bj2600345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marsh E N. Coenzyme B12(cobalamin)-dependent enzymes. Essays Biochem. 1999;34:139–154. doi: 10.1042/bse0340139. [DOI] [PubMed] [Google Scholar]
- 29.Oh S, Chater K F. Denaturation of circular or linear DNA facilitates targeted intergrative transformation of Streptomyces coelicolorA3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Omura S, Tsuzuki K, Tanaka Y, Sakakibara H, Aizawa M, Lukacs G. Valine as a precursor of n-butyrate unit in the biosynthesis of macrolide aglycone. J Antibiot (Tokyo) 1983;36:614–616. doi: 10.7164/antibiotics.36.614. [DOI] [PubMed] [Google Scholar]
- 31.Omura S, Taki A, Matsuda K, Tanaka Y. Ammonium ions suppress the amino acid metabolism involved in the biosynthesis of protylonolide in a mutant of Streptomyces fradiae. J Antibiot (Tokyo) 1984;37:1362–1369. doi: 10.7164/antibiotics.37.1362. [DOI] [PubMed] [Google Scholar]
- 32.Omura S, Tanaka Y, Mamada H, Masuma R. Effect of ammonium ion, inorganic phosphate and amino acids on the biosynthesis of protylonolide, a precursor of tylosin aglycone. J Antibiot (Tokyo) 1984;37:494–502. doi: 10.7164/antibiotics.37.494. [DOI] [PubMed] [Google Scholar]
- 33.Ratnatilleke A, Vrijbloed J W, Robinson J A. Cloning and sequencing of the coenzyme B12-binding domain of isobutyryl-CoA mutase from Streptomyces cinnamonensis, reconstitution of mutase activity, and characterization of the recombinant enzyme produced in Escherichia coli. J Biol Chem. 1999;274:31679–31685. doi: 10.1074/jbc.274.44.31679. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds K A, O'Hagan D, Gani D, Robinson J A. Butyrate metabolism in streptomycetes. Characterization of an intramolecular vicinal interchange rearrangement linking isobutyrate and n-butyrate in Streptomyces cinnamonensis. J Chem Soc Perkin Trans I. 1988;1988:3197–3207. [Google Scholar]
- 35.Rodriguez E, Gramajo H. Genetic and biochemical characterization of the alpha and beta components of a propionyl-CoA carboxylase complex of Streptomyces coelicolorA3(2) Microbiology. 1999;145:3109–3119. doi: 10.1099/00221287-145-11-3109. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Smith L M, Meijer W G, Dijkhuizen L, Goodwin P M. A protein having similarity with methylmalonyl-CoA mutase is required for the assimilation of methanol and ethanol by Methylobacterium extorquensAMI. Microbiology. 1996;142:675–684. doi: 10.1099/13500872-142-3-675. [DOI] [PubMed] [Google Scholar]
- 38.Sohling B, Gottschalk G. Molecular analysis of the anaerobic succinate degradation pathway in Clostridium kluyveri. J Bacteriol. 1996;178:871–880. doi: 10.1128/jb.178.3.871-880.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stassi D L, Kakavas S J, Reynolds K A, Gunawardana G, Swanson S, Zeidner D, Jackson M, Liu H, Buko A, Katz L. Ethyl-substituted erythromycin derivatives produced by directed metabolic engineering. Proc Natl Acad Sci USA. 1998;95:7305–7309. doi: 10.1073/pnas.95.13.7305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strohl W R. Compilation and analysis of DNA sequences associated with apparent streptomycete promoters. Nucleic Acids Res. 1992;20:961–974. doi: 10.1093/nar/20.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang L, Hutchinson C R. Sequence, transcriptional, and functional analyses of the valine (branched-chain amino acid) dehydrogenase gene of Streptomyces coelicolor. J Bacteriol. 1993;175:4176–4185. doi: 10.1128/jb.175.13.4176-4185.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang L, Zhang Y X, Hutchinson C R. Amino acid catabolism and antibiotic synthesis: valine is a source of precursors for macrolide biosynthesis in Streptomyces ambofaciens and Streptomyces fradiae. J Bacteriol. 1994;176:6107–6119. doi: 10.1128/jb.176.19.6107-6119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thoma N H, Leadlay P F. Mechanistic and structural studies on methylmalonyl-CoA mutase. Biochem Soc Trans. 1998;26:293–298. doi: 10.1042/bst0260293. [DOI] [PubMed] [Google Scholar]
- 44.Vrijbloed J W, Zerbe-Burkhardt K, Ratnatilleke A, Grubelink-Leiser A, Robinson J A. Insertional inactivation of methylmalonyl coenzyme A (CoA) mutase and isobutyryl-CoA mutase genes in Streptomyces cinnamonensis: influence on polyketide antibiotic biosynthesis. J Bacteriol. 1999;181:5600–5605. doi: 10.1128/jb.181.18.5600-5605.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wallace K K, Zhao B, McArther H A I, Reynolds K A. In vivo analysis of straight-chain and branched-chain fatty acid biosynthesis in three actinomycetes. FEMS Microbiol Lett. 1995;131:227–234. doi: 10.1111/j.1574-6968.1995.tb07781.x. [DOI] [PubMed] [Google Scholar]
- 46.Weber J M, Schoner B, Losick R. Identification of a gene required for the terminal step in erythromycin A biosynthesis in Saccharopolyspora erythraea (Streptomyces erythreus) Gene. 1989;75:235–241. doi: 10.1016/0378-1119(89)90269-2. [DOI] [PubMed] [Google Scholar]
- 47.Wilkemeyer M F, Crane A M, Ledley F D. Primary structure and activity of mouse methylmalonyl-CoA mutase. Biochem J. 1990;271:449–455. doi: 10.1042/bj2710449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zerbe-Burkhardt K, Ratnatilleke A, Philippon N, Birch A, Leiser A, Vrijbloed J W, Hess D, Hunziker P, Robinson J A. Cloning, sequencing, expression and insertional inactivation of the gene for the large subunit of the coenzyme B12-dependent isobutyryl-CoA mutase from Streptomyces cinnamonensis. J Biol Chem. 1998;273:6508–6517. doi: 10.1074/jbc.273.11.6508. [DOI] [PubMed] [Google Scholar]
- 49.Zhang W, Yang L, Jiang W, Zhao G, Yang Y, Chiao J S. Molecular analysis and heterologous expression of the gene encoding methylmalonyl-CoA mutase from a rifamycin SV-producing Amycolatopsis mediterraneiU32. Appl Biochem Biotechnol. 1999;82:209–225. doi: 10.1385/abab:82:3:209. [DOI] [PubMed] [Google Scholar]
- 50.Zhang Y X, Denoya C D, Skinner D D, Fedechko R W, McArthur H A I, Morgenstern M R, Davies R A, Lobo S, Reynolds K A, Hutchinson C R. Cloning and characterization of genes encoding acyl-CoA dehydrogenase (AcdH) homologs from Streptomyces coelicolor and Streptomyces avermitilis. Microbiology. 1999;145:2323–2334. doi: 10.1099/00221287-145-9-2323. [DOI] [PubMed] [Google Scholar]
- 51.Zhou C, Yang Y, Jong A Y. Miniprep in 10 minutes. BioTechniques. 1990;8:172–173. [PubMed] [Google Scholar]