Abstract
Stem/progenitor cells, including cardiac-derived c-kit+ progenitor cells (CPCs), are under clinical evaluation for treatment of cardiac disease. Therapeutic efficacy of cardiac cell therapy can be attributed to paracrine signaling and the release of extracellular vesicles (EVs) carrying diverse cargo molecules. Despite some successes and demonstrated safety, large variation in cell populations and preclinical/clinical outcomes remains a problem. Here, we investigated this variability by sequencing coding and non-coding RNAs of CPCs and CPC-EVs from 30 congenital heart disease patients and used unsupervised learning methods to determine potential mechanistic insights. CPCs retained RNAs related to extracellular matrix organization and exported RNAs related to various signaling pathways to CPC-EVs. CPC-EVs are enriched in miRNA clusters related to cell proliferation and angiogenesis. With network analyses, we identified differences in non-coding RNAs which give insight into age-dependent functionality of CPCs. By taking a quantitative computational approach, we aimed to uncover sources of CPC cell therapy variability.
Keywords: Cardiac Cell Therapy, extracellular vesicle, exosome, RNA sequencing, miRNA, lncRNA, systems biology
Introduction
Heart disease remains the leading cause of morbidity and mortality in the US, and often results in irreversible damage to the myocardium 1. Initially treated by surgery, drugs, or transplant, cardiac cell therapy emerged in the early 2000s with the goal to regenerate healthy myocardium after injury or disease. Over the years, several stem or progenitor cell types have been investigated for the treatment of various ischaemic and congenital heart diseases 2,3. In particular, previous preclinical research has shown that cardiac-derived c-kit+ cells (CPCs) repair the myocardium after injury 4–6. Of note, our group is involved in a current phase I clinical trial investigating CPCs for treatment of hypoplastic left heart syndrome, a complex single ventricle congenital heart disease (NCT03406884).
Despite demonstrated safety and some efficacy in cardiac cell therapy preclinical and clinical trials, large variation in cell populations and patient outcomes remains a significant problem for further developing larger-scale, reliable therapies 2,7. Some of the variance in cell populations can be attributed to donor age. Additionally, once isolated, in vitro cell culture conditions or manipulations, like hypoxia and cell aggregation, also affect cell reparative effects 6,8. Considering the high variability of cell populations, it is thus important to identify specific mechanisms of action in order to enhance cell therapy efficacy for patients. To this extent, our group has demonstrated that CPCs can repair rat right ventricle failure in an age-dependent manner, with neonatal CPCs having the greatest reparative capacity 4,9,10. However, there is a dearth of quantitative studies investigating the underlying RNA cues driving CPC therapeutic efficacy. We have previously investigated the molecular basis for the differences between reparative and non-reparative CPCs with RNAseq experiments 4,10, however these studies were not comprehensive, with low sample sizes and consideration of only one or two types of RNA.
Originally, transplanted cells for cardiac disease treatment were thought to function via engraftment, proliferation, and differentiation. However, transplanted cellular retention is low and much of the therapeutic benefit is now attributed to paracrine signaling, including the release of extracellular vesicles (EVs) 2. EVs are lipid-bilayer vesicles released from cells via exocytosis or budding of the plasma membrane into the extracellular space. Once released, neighboring recipient cells may internalize EVs via endocytic processes, including direct membrane fusion, lipid-raft based uptake, and receptor-ligand interactions 11. Importantly, EVs carry and protect diverse molecules, including RNAs, proteins, and lipids. The crucial role of EVs in cell therapy has been highlighted in previous work demonstrating that the inhibition of EV release diminishes the reparative effect of stem and progenitor cells 12. In the context of cardiac cell therapy, this suggests CPCs themselves are not the only source of variable RNA signals contributing to repair. Studies have shown that the uptake of EVs by resident cardiac cells allows for the transfer of stem or progenitor cell EV cargo and stimulates repair in the injured tissue 2. Despite the well-established link between EV release/uptake and repair, our understanding of the signals or cargo molecules contributing to these effects is poor, especially given that EV cargo is highly heterogeneous 13. Importantly, CPCs and CPC-EV RNA content have been studied separately, but never together, comparatively.
Given the current limitations surrounding cardiac cell therapy variability, we aimed to investigate these discrepancies by performing bulk sequencing of primary CPCs from pediatric heart patients and their released EVs. First, we performed conventional differential expression analyses to identify key differences between cells and EVs and used unsupervised learning methods to reduce dataset dimensionality and summarize the data. Then, we systematically investigated the biological significance of our RNA-seq experiments with pathway and competitive endogenous RNA network analyses. We utilized publicly available datasets to provide greater context and determine the specificity of our CPC-EV results, in comparison to other cell types and EVs. Our results highlight the need for more quantitative investigations of cardiac cell therapy, and more personalized medicine approaches to cell therapy.
Results
Characterization of EVs from neonate, infant, and child CPCs
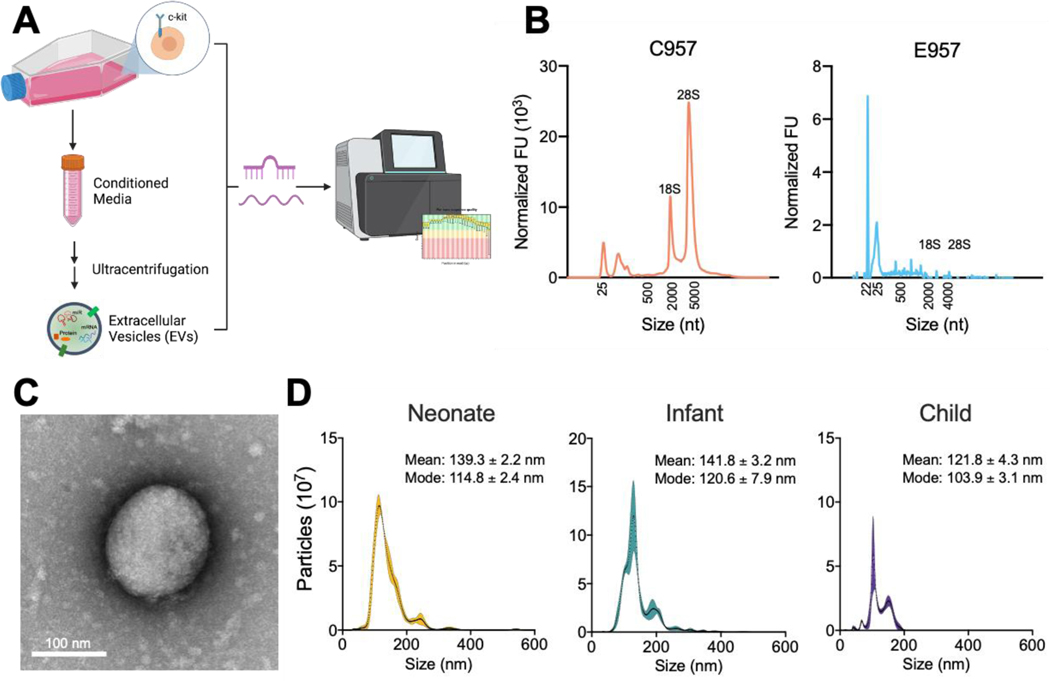
To characterize CPC-EVs, CPC populations were grown and expanded in 2D culture and EVs were isolated from the conditioned media via differential ultracentrifugation (Fig. 1a). CPCs were previously isolated from cardiac biopsies of neonate (<1 week, n=9), infant (1 week – 1 year, n=13), and child (>1 year, n=8) congenital heart disease patients (Supplementary Table 1). Bioanalyzer profiles of CPC and CPC-EV RNA revealed distinct 18S and 28S ribosomal peaks in the CPC RNA, but not in the CPC-EV RNA, confirming successful isolation of EVs without cellular contamination (Fig. 1b). Furthermore, total RNA from EVs was enriched in small RNAs with a peak ∼22nt, the size of miRNAs. CPC-EVs were imaged using transmission electron microscopy and analyzed for size and concentration with nanoparticle tracking analysis. Independent of patient cell source, all CPC-EVs were 100–140 nm, characteristic of exosomes or small EVs (Fig. 1c and 1d).
Figure 1. CPC-EV isolation and characterization.
a Schematic of study: CPCs and CPC EV miRNA and total RNA are sequenced and analyzed (created from Biorender.com).
b Bioanalyzer profile for patient matched CPC and CPC-EV RNA content.
c Transmission electron microscopy image of CPC EV. Scale bar, 100 nm.
d Vesicle size distribution histogram by nanoparticle tracking analysis in neonate, infant, and child CPC-derived EVs. Shaded region indicates standard error.
CPCs retain ECM-related RNAs and export signaling pathway-related RNAs to EVs
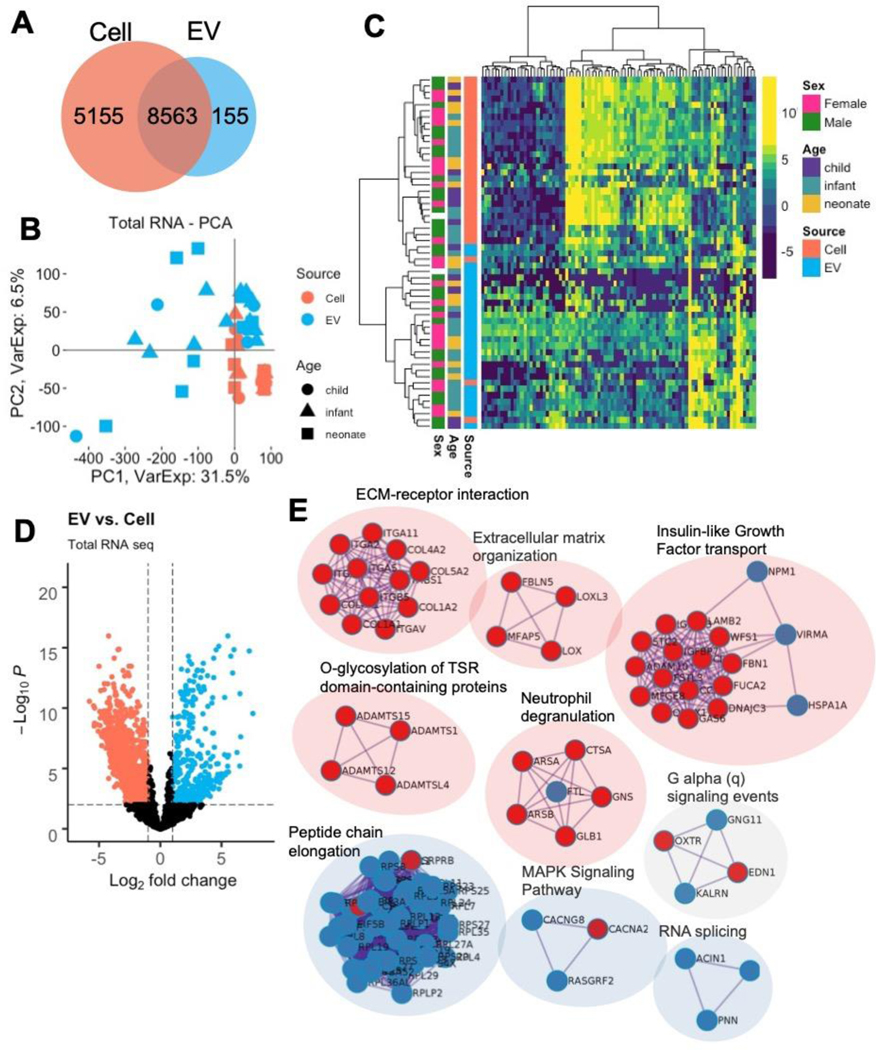
Total RNA sequencing results identified 13,718 and 8,718 expressed RNAs in CPCs and CPC-EVs, after filtering out lowly expressed RNAs (Fig. 2a) 14. Dimension reduction with principal component analysis (PCA) of the 8,563 commonly expressed RNAs revealed distinctive separation of CPC and CPC-EV samples across the first two components, cumulatively representing 38% of the total variance (Fig. 2b). Furthermore, an unbiased heatmap of the top 1% variable RNAs revealed hierarchical clustering by source: CPC vs. CPC-EV (Fig. 2c).
Figure 2. Differential expression of CPC and CPC-EV total RNAseq.
a RNAs expressed in CPCs and CPC EVs.
b PCA plot of CPC and CPC EVs show clustering by RNA source across PC1.
c Heatmap of top 1% variable RNAs show clustering of samples by source: cell and EV.
d Volcano plot of differentially expressed RNAs in CPCs and CPC EVs.
e Network of top enriched terms from differentially expressed RNAs upregulated in CPCs (red) vs. EVs (blue) using the Metascape tool.
Given that CPC and CPC-EV samples were matched, derived from each patient, we used the dream (differential expression for repeated measures) approach to determine differentially expressed RNAs between CPCs and CPC-EVs. In total, 4,898 RNAs, or 57% of the commonly expressed RNAs, were differentially expressed with adjusted p-values < 0.05 (Fig. 2d, Supplementary Table 2)15. The top differentially expressed RNAs in CPCs and CPC-EVs are displayed in Table 1. Notably, collagen type IV and VIII chains, integrin alpha V, dystroglycan 1, and growth arrest – specific 6 are upregulated in CPCs, and Ras-related protein Rab-13, dexamethasone-induced Ras-related protein 1, colony stimulating factor 1 receptor, and interleukins 33 and 16 are upregulated in CPC-EVs. To further determine the biological significance of these differentially expressed RNAs, we performed pathway analysis of the top 250 RNAs upregulated in CPCs and CPC-EVs (ranked by fold-change). Metascape pathway analysis revealed CPCs were enriched in RNAs associated with extracellular matrix (ECM) organization, ECM-receptor interaction, insulin-like growth factor transport and immune responses; whereas CPC-EVs were enriched in RNAs involved in peptide chain elongation, RNA splicing, and MAPK and G alpha (q) signaling (Fig. 2e, Supplementary Tables 3 and 4) 16.
Table 1.
RNAs preferentially released to EV and retained in cell
| RNA | Log2(EV/Cell) | padj | RNA | Log2(Cell/EV) | padj |
|---|---|---|---|---|---|
| CA11 | 7.56 | ** | CPA4 | 5.78 | *** |
| RAB13 | 7.26 | *** | OSMR | 5.44 | *** |
| RASD1 | 6.56 | ** | COL8A1 | 5.40 | ** |
| PLEKHA4 | 6.51 | *** | OXTR | 5.37 | ** |
| NEFM | 6.49 | ** | FSTL3 | 5.36 | *** |
| C1orf115 | 6.26 | ** | GDF6 | 5.15 | ** |
| ANP32B | 6.01 | *** | TNFRSF11B | 5.07 | ** |
| NET1 | 5.90 | *** | COL4A1 | 5.00 | *** |
| C22orf46 | 5.76 | *** | RECK | 4.94 | ** |
| CSF1R | 5.57 | ** | CHPF | 4.91 | ** |
| KIF21B | 5.57 | * | SLC16A3 | 4.89 | ** |
| TRAK2 | 5.52 | *** | P3H4 | 4.87 | *** |
| ERG | 5.50 | ** | EDIL3 | 4.82 | ** |
| CDC42BPG | 5.50 | ** | UGCG | 4.81 | ** |
| IL33 | 5.42 | * | TRAM2 | 4.79 | ** |
| IL16 | 5.31 | * | ANGPTL4 | 4.78 | ** |
| DNAH10 | 5.31 | * | MEGF6 | 4.75 | ** |
| CGNL1 | 5.26 | *** | GAS6 | 4.69 | ** |
| PITPNM3 | 5.25 | ** | IGFBP3 | 4.65 | ** |
| SPTBN4 | 5.06 | * | B4GALT1 | 4.64 | ** |
| ROCK1P1 | 5.00 | * | ARSJ | 4.63 | ** |
| GUCY1A2 | 4.88 | * | CD248 | 4.60 | ** |
| KIAA1211 | 4.87 | * | PKD1 | 4.59 | ** |
| CCDC88C | 4.79 | * | ITGA5 | 4.58 | ** |
| FLG | 4.79 | * | DAG1 | 4.58 | ** |
p < 0.01
p < 1e-5
p < 1e-10
CPC-EVs are enriched in miRNAs involved in cardiac development and cell signaling
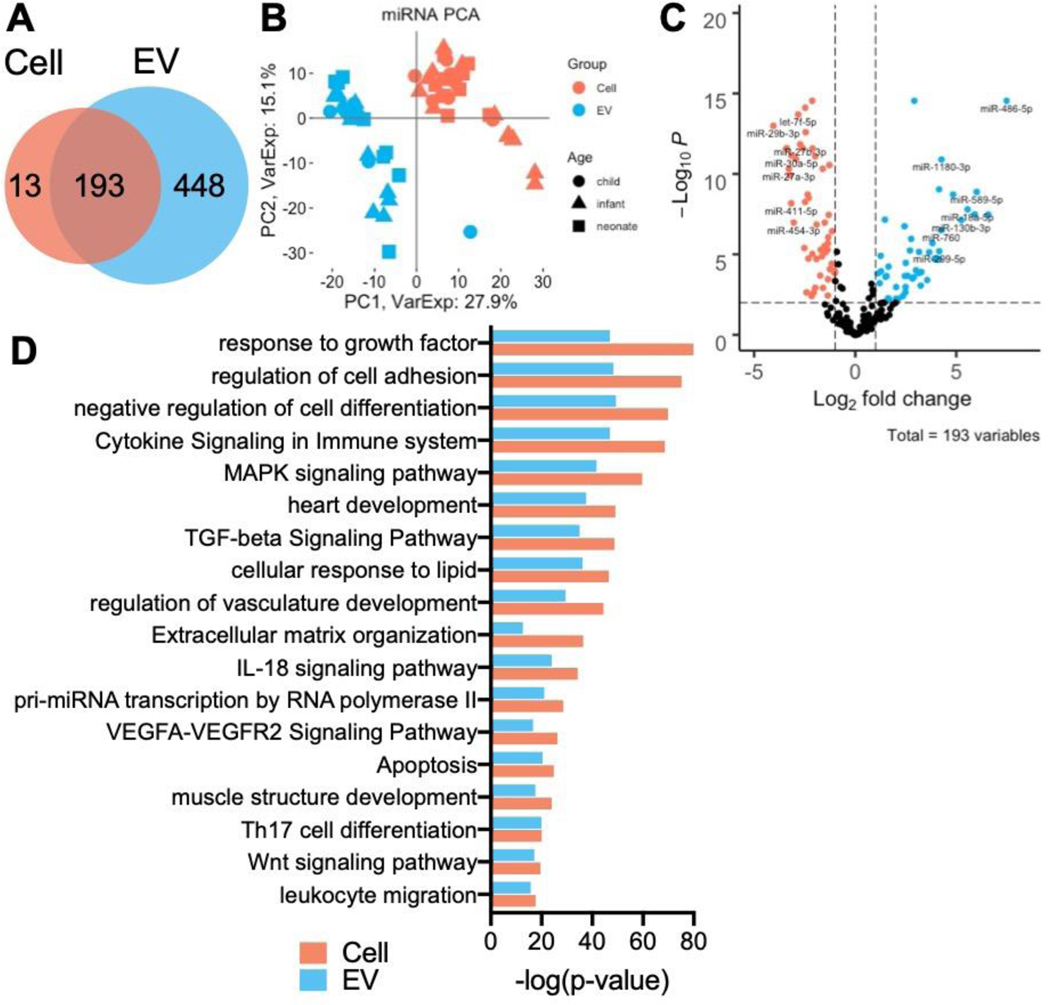
We then performed similar miRNAseq analyses: miRNAseq revealed 206 and 641 expressed miRNAs in CPCs and CPC-EVs, respectively, after filtering out lowly expressed miRNAs (Fig. 3a). PCA of the 193 commonly expressed miRNAs showed clear separation of CPC and CPC-EV samples across principal component 1, representing 27.9% of total variance (Fig. 3b). Linear regression using the dream approach was used to determine the differentially expressed miRNAs between CPCs and CPC-EVs. In total, 126 of 193, or 65% of miRNAs were differentially expressed with adjusted p-values <0.05 (Fig. 3c, Supplementary Table 5). The top differentially expressed miRNAs are displayed in Table 2. Next, we found gene targets for the differentially expressed miRNAs using miRTarBase with the criteria that targets be experimentally validated by at least 3 methods (Supplementary Table 6). Pathway analysis of the gene targets revealed enrichment of gene ontology (GO) pathways involved in vasculature and heart development, VEGFA-VEGFR2, and TGF-beta signaling, as well as cell adhesion, differentiation, and apoptosis (Fig. 3d, Supplementary Tables 7 and 8).
Figure 3. Differential expression of CPC and CPC EV miRNAseq.
a miRNAs expressed in CPCs and CPC EVs.
b PCA plot of CPC and CPC EVs show clustering by RNA source across PC1.
c Volcano plot of differentially expressed miRNAs in CPCs and CPC EVs.
d GO pathway enriched terms from differentially expressed miRNA gene targets, as determined by miRTarBase.
Table 2.
miRNAs preferentially released to EV and retained in cell
| miRNA | Log2(EV/Cell) | padj | RNA | Log2(Cell/EV) | padj |
|---|---|---|---|---|---|
| hsa-miR-486-5p | 7.49 | *** | hsa-miR-29b-3p | 4.05 | *** |
| hsa-miR-203a-3p | 6.58 | ** | hsa-miR-379-5p | 3.39 | *** |
| hsa-miR-589-5p | 6.01 | ** | hsa-miR-27a-3p | 3.28 | *** |
| hsa-miR-122-5p | 5.92 | ** | hsa-miR-31-5p | 3.24 | ** |
| hsa-miR-18a-5p | 5.56 | ** | hsa-miR-411-5p | 3.17 | ** |
| hsa-miR-130b-3p | 5.25 | ** | hsa-miR-30a-5p | 3.16 | *** |
| hsa-miR-320c | 4.84 | ** | hsa-miR-454-3p | 3.04 | ** |
| hsa-miR-1180-3p | 4.27 | *** | hsa-let-7e-5p | 2.93 | *** |
| hsa-miR-760 | 4.26 | ** | hsa-let-7f-5p | 2.82 | *** |
| hsa-miR-299-5p | 4.16 | ** | hsa-miR-27b-3p | 2.73 | *** |
| hsa-miR-92b-5p | 4.15 | ** | hsa-miR-29a-3p | 2.61 | *** |
| hsa-miR-339-3p | 4.15 | * | hsa-miR-143-5p | 2.52 | ** |
| hsa-miR-125a-3p | 3.93 | * | hsa-miR-30e-5p | 2.48 | ** |
| hsa-miR-494-3p | 3.82 | ** | hsa-miR-21-5p | 2.48 | *** |
| hsa-miR-501-3p | 3.66 | ** | hsa-miR-103a-3p | 2.45 | *** |
| hsa-miR-500a-3p | 3.56 | * | hsa-miR-27a-5p | 2.41 | * |
| hsa-miR-184 | 3.26 | * | hsa-let-7a-5p | 2.35 | ** |
| hsa-miR-127-5p | 3.23 | * | hsa-miR-134-5p | 2.32 | * |
| hsa-miR-4661-5p | 3.15 | * | hsa-miR-143-3p | 2.30 | ** |
| hsa-miR-197-3p | 3.15 | ** | hsa-miR-431-5p | 2.16 | * |
| hsa-miR-214-3p | 3.00 | * | hsa-miR-24-3p | 2.13 | *** |
| hsa-let-7d-3p | 2.91 | *** | hsa-miR-493-3p | 2.12 | ** |
| hsa-miR-1287-5p | 2.90 | * | hsa-miR-16-5p | 2.12 | *** |
| hsa-miR-320b | 2.82 | * | hsa-miR-450b-5p | 2.06 | * |
| hsa-miR-92b-3p | 2.76 | ** | hsa-miR-152-3p | 1.98 | *** |
p < 0.01
p < 1e-5
p < 1e-10
CPC-EVs contain vesicle biosynthesis and cell cycle-related miRNAs
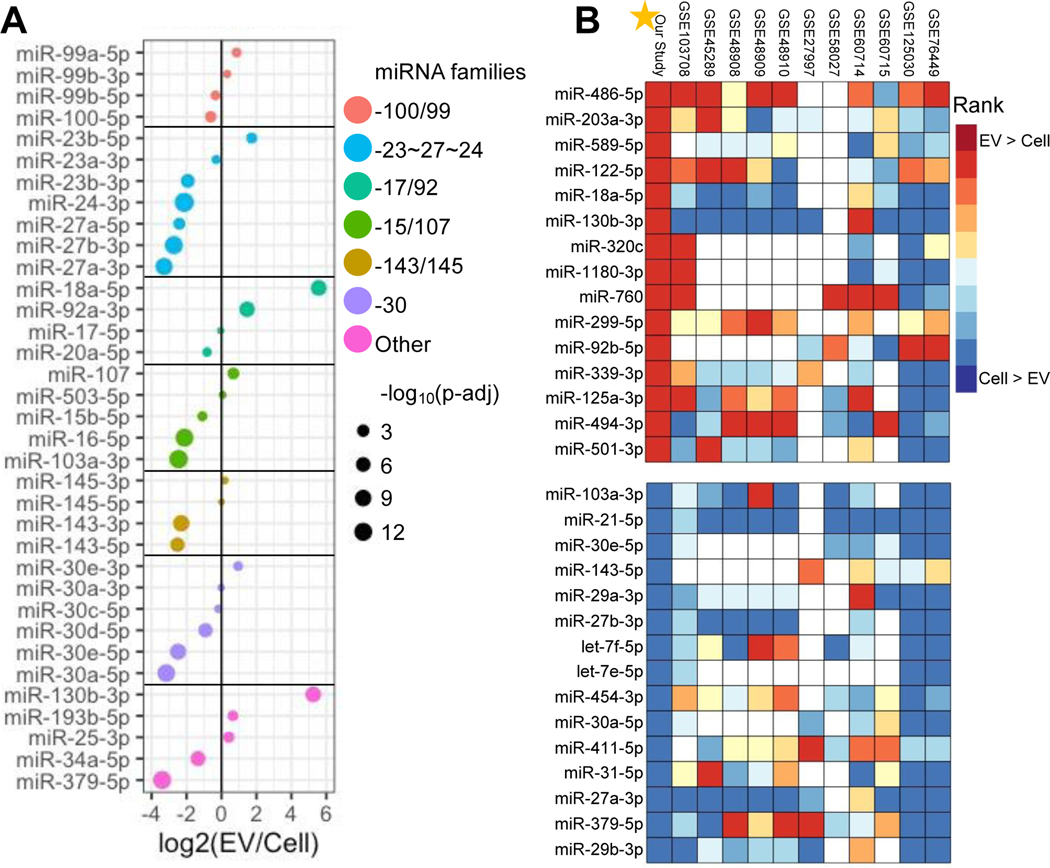
We investigated the enrichment of well-studied miRNA families and cardiac-related miRNAs, with known functions in CPC-EVs (Fig. 4a). Most notably, members of the miRNA 17/92 cluster (miR-18a-5p and miR-92a-3p) involved in development and cell proliferation are upregulated in CPC-EVs 17–19. Additionally, members of the miRNA 99/100 family (miR-99a-5p and miR-99b-3p) involved in hematopoietic stem cell renewal are also upregulated in CPC-EVs 20.
Figure 4. Differential expression of well-studied CPC and CPC-EV miRNAs and miRNA clusters.
a Differential expression of well-studied miRNAs in CPCs and CPC-EVs.
b Our study’s top 15 miRNAs up (red) and down (blue) regulated in EV samples plotted with rank of top enriched miRNAs from 11 publicly available databases. GEO database numbers listed on top; white color indicates no available miRNA expression (NA).
To understand the specificity of our CPC vs. CPC-EV results, we searched the Gene Expression Omnibus (GEO) database for previously published datasets with miRNAseq from EVs and their parent cells. We found eleven datasets comprised of many cell types, including, various cancer cells, immune cells, and bone marrow mesenchymal stem cells 21–28. We calculated fold-change values (EV/Cell) for each data set and ranked the miRNAs in order of decreasing fold-change value (Fig. 4b). We found our data set was congruent with other data sets: our CPC-EVs were enriched in some miRNAs upregulated in other EV types, and our CPCs retained some similar miRNAs to other cell types. Specifically, well-studied miR-122-5p, which has been implicated in miRNA EV cargo sorting, was top ranked in multiple data sets 29. Nevertheless, there were a handful of unique miRNAs, enriched in CPC-EVs, but not in EVs from other cell types. These miRNAs included miR-18a-5p and miR-130b-3p. Evidence suggests that miR-18a-5p is anti-apoptotic 30,31, while miR-130b-3p is pro-apoptotic, targets insulin-like growth factor 1, and may be cardiac harmful 32,33.
Construction of the ceRNA network
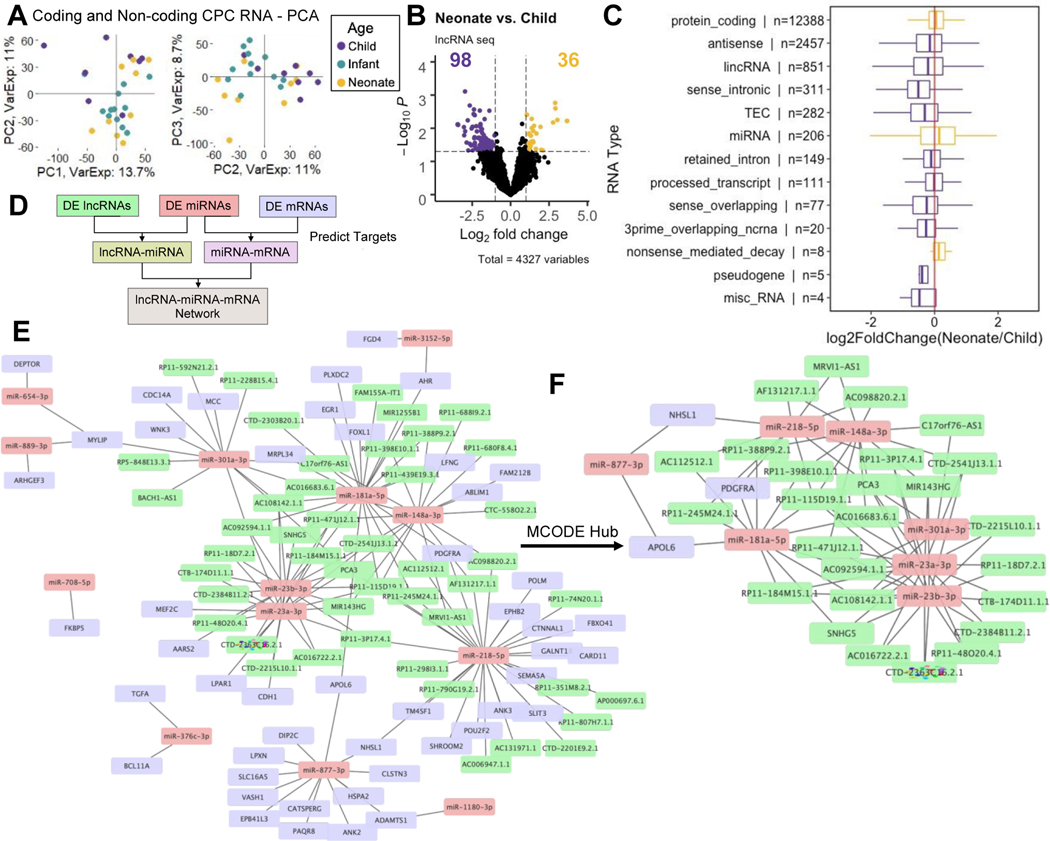
Evidence suggests that lncRNAs act as competing endogenous RNAs (ceRNAs) and play a key role in regulating RNA expression 34–36. Furthermore, previous work has demonstrated that CPCs function in an age-dependent manner: CPCs from neonate patients are more reparative than CPCs from older patients 4,37. To gain a comprehensive level of understanding of age-dependent CPC ceRNA interactions, we investigated the differentially expressed mRNAs, lncRNAs, and miRNAs between neonate and child CPCs. Overall, child and neonate CPC RNA profiles were the most dissimilar (Supplementary Fig. 1). PCA of coding and non-coding RNAs showed separation of neonate and child CPCs across the first three components (Fig. 5a).
Figure 5. ceRNA network of neonate and child CPCs.
a PCA plot of CPC coding and non-coding CPC RNAs (PC1, PC2, PC3) show clustering by patient age group.
b Volcano plot of differentially expressed non-coding RNAs between neonate and child CPCs. Thirty-six and 98 RNAs are upregulated in neonate and child cells, respectively.
c Fold-change values for each RNA in various categories. RNAs upregulated in child CPCs (purple), compared to neonate CPCs (yellow). N represents number of measured RNAs in each category.
d Schematic for ceRNA network construction: differentially expressed RNAs between neonate and child CPCs were matched in miRcode and miRTarBase by putative target sites.
e Full ceRNA network of differentially expressed RNAs (neonate vs. child CPCs) with 107 nodes and 144 edges.
f Reduced MCODE network of the most highly connected RNA nodes.
We used edgeR/limma to find the top differentially expressed miRNAs (18), lncRNAs (134), and mRNAs (505) (|fold-change|>2 and p<0.05) between neonate and child CPCs (Supplementary Table 9). Overall, child CPCs were enriched in various non-coding RNAs, as compared to neonate CPCs (Fig. 5b and 5c). We then matched differentially expressed miRNAs to differentially expressed lncRNAs and mRNAs by putative target sites using miRcode and miRTarBase as shown in Fig. 5d 38,39. The resulting ceRNA network consisted of 107 nodes and 144 edges (Fig. 5e). The most highly connected nodes included miRNAs: miR-218-5p and −8773p upregulated in child CPCs, and −23a-3p, −23b-3p, and −301a-3p, upregulated in neonate CPCs. (Table 3, Supplementary Fig. 2). Metascape enrichment analysis revealed these nodes are enriched in pathways including, blood vessel development, positive regulation of cell cycle, and regulation of Wnt signaling pathway (Supplementary Table 10). We analyzed the hub genes in the full ceRNA (Fig. 5e) network using the Cytoscape MCODE plug-in. The full network was reduced to the most highly connected 36 hub nodes, including miRNAs: miR-23a-3p, −23b-3p, 148a-3p, −181a-5p, −218-5p, −301a-3p, and −877-3p (Fig. 5f).
Table 3.
ceRNA network hub connectivity
| RNA Type | Name | Node Degree |
|---|---|---|
| miRNA | miR-218-5p | 31 |
| miRNA | miR-181a-5p | 22 |
| miRNA | miR-23a-3p | 20 |
| miRNA | miR-23b-3p | 18 |
| miRNA | miR-148a-3p | 17 |
| miRNA | miR-877-3p | 13 |
| miRNA | miR-301a-3p | 13 |
| lncRNA | RP11-115D19.1.1 | 5 |
| lncRNA | PCA3 | 5 |
| lncRNA | AC092594.1.1 | 4 |
| lncRNA | AC108142.1.1 | 4 |
| mRNA | MYLIP | 3 |
| lncRNA | CTD-2541J13.1.1 | 3 |
| lncRNA | MIR143HG | 3 |
| lncRNA | RP11-184M15.1.1 | 3 |
| lncRNA | RP11-3P17.4.1 | 3 |
| lncRNA | RP11-471J12.1.1 | 3 |
| lncRNA | SNHG5 | 3 |
| lncRNA | AC016683.6.1 | 3 |
Discussion
Several cell types are being clinically evaluated for use in cardiac cell therapy, including CPCs (NCT02501811, NCT03351400, NCT03406884). Despite promising preclinical results, there remain concerns over variability from different cell populations and cell therapy outcomes 40. Specifically, several groups have shown that CPC therapeutic efficacy is dependent on patient age and disease, as well as cell culture and expansion conditions 4,6,41,42. Furthermore, research has shown that cardiac cell therapy efficacy can be attributed to paracrine signaling and the release of EVs, rather than cellular engraftment, proliferation, and differentiation 2. Therefore, to investigate sources of CPC-therapy variability, we used next generation sequencing methods to determine differences among pediatric primary CPCs (n=30 patients) and their respective EVs released in vitro. We analyzed CPC and CPC-EV coding and non-coding RNA content and investigated the differences (1) between patient matched CPCs and CPC-EVs, and (2) across CPC patient age groups. Previous research has investigated differences between stem cells and their EVs, but these studies were limited in sample size and focused on miRNA only 43,44. By taking a comprehensive computational approach, we gained insight into potential mechanisms of action and sources of variability of CPC cell therapy.
First, we determined the differences between CPC and CPC-EV RNA content. For this study, we profiled coding and long non-coding RNA using total RNAseq and miRNA with small RNAseq. As expected, we found that the greatest contributor to variance across the entire dataset was source (cell vs. EV), rather than patient age or sex (Fig. 2b, 2c, 3b, Supplementary Fig. 3c and 3d). Using differential expression analysis, we discovered that a large portion of RNAs and miRNAs were expressed in only cells or EVs. Further, of the intersecting set of RNAs expressed in both cells and EVs, many were differentially expressed (adjusted p-values < 0.05, Fig. 2a, 2d, 3a, and 3c). In total, CPCs retained RNAs involved in extracellular matrix organization and exported RNAs to EVs involved in various signaling pathways (Fig. 2e and 3d).
Importantly, cell type and cell environment affect EV cargo 9,45,46. In the context of cardiac cell therapy, stem and progenitor cell-EVs have been shown to carry RNAs with beneficial pleiotropic effects—immunomodulatory, anti-fibrotic, anti-apoptotic, pro-angiogenic, pro-migratory, and pro-proliferative—as compared to their non-progenitor cardiac cell type counterparts 2,12,47. Additionally, manipulations affecting parent stem and progenitor cell environment, like hypoxia and cell aggregation, affect EV RNA cargo and resulting EV efficacy 9,45. Special attention has been paid to EV miRNA cargo, as it plays an important role in cardiac repair: signaling between stem or progenitor cells and resident cardiac cells 48. Circulating EVs protect internal cargo from degradation and are thus a rich source of circulating miRNAs. Our results indicate that CPC-EVs are upregulated in several important and well-studied miRNA clusters (Fig. 4a). In particular, members of miRNA cluster 17/92 are upregulated in CPC-EVs. The miRNA 17/92 cluster was initially discovered as an oncogene and has been shown to promote cardiomyocyte proliferation 17,19. Furthermore, miR-92a is highly expressed in endothelial cells and is upregulated in CPC-EVs (log2fold-change = 1.46, as compared to CPC). Previous studies have shown that increasing cellular expression of miR-92a-3p specifically via EV delivery is pro-angiogenic—promoting cell cycle progression and endothelial-to-mesenchymal transition in endothelial cells—whereas direct cell overexpression of miR-92a-3p may be anti-angiogenic 49–51. Additionally, members of the miR-23/−24/−27 cluster are implicated in both positive and negative regulation of neovascularization and are differentially expressed in CPC and CPC-EVs 52,53. Overall, identifying well-studied miRNA clusters with known biological roles in our CPC-EVs provides greater insight into their potential mechanisms of action in vitro or in vivo.
Furthermore, we aimed to differentiate non-specific, EV biogenesis-related RNA cargo from CPC-specific, potentially pro-reparative cargo. To do so, we compared the top 15 miRNAs enriched in CPC-EVs and CPCs to publicly available data sets from various cell types—immune cells, cancer cells, and mesenchymal stem cells—and we found both similarities and differences in the top ranked miRNAs. The 4th most upregulated miRNA in CPC-EVs, miR-122-5p (log2fold-change = 5.92, Table 2), is upregulated in EVs from other cell types (Fig. 4b) and is loaded into EVs via Lupus La protein binding 29. Several other miRNA EV sorting mechanisms have been identified, including other RNA-binding protein mechanisms and membrane proteins involved in EV biogenesis. For example, three of the miRNAs upregulated in CPC-EVs, miR320a, −193a, and −92a (log2fold-change = 1.25, 0.80, 1.46) have been shown to be actively loaded into EVs via Argonaute 2, major vault protein, and vacuolar protein sorting-associated protein 4 binding, respectively 54–56.
On the other hand, in our comparison to other datasets, we also identified miRNAs enriched in CPC-EVs that are not enriched in EVs from other cell types. Most notably, miR-18a-5p is upregulated in CPC-EVs (log2fold-change = 5.56) and is a member of the pro-proliferative and anti-apoptotic 17/92 miRNA cluster. In contrast to some other cell type EVs, miR-501-3p was also identified as a top upregulated miRNA in CPC-EVs. A previous report determined that macrophage-derived exosome miR-501-3p promoted pancreatic ductal adenocarcinoma cell migration and proliferation 57. Considering treatment strategies for cardiac repair strive to promote cell proliferation, miR-18a-5p and miR-501-3p may be potent, progenitor cell-specific EV signals driving therapeutic response. Further investigation of these miRNAs is warranted. Overall, when understanding the function of released EVs in cell therapy, it will be important to discriminate between non-specific, machinery-related RNA cargo vs. CPC-specific, potentially beneficial RNA. Elucidating CPC-specific EV cargo molecules will help us understand which RNA molecules are driving cardiac cell therapy efficacy.
We have previously examined and reported differential CPC mRNA expression across neonate, infant, and child age groups in a smaller sample size 10. We have not, however, investigated the full RNA landscape and potential interactions of coding and non-coding RNAs in CPCs. Importantly, in 2011, Salmena et al. introduced the competing endogenous RNA (ceRNA) hypothesis, suggesting that RNA transcripts “talk” to each other via miRNA response elements, forming a large-scale transcriptome regulatory network 36. A growing body of evidence suggests that lncRNAs play a key role in protein-coding gene regulation by acting as miRNA sponges 34–36. Furthermore, non-coding RNAs play an important role in cardiac development 58, and CPCs lose their therapeutic functionality as they age 4,37. Thus, to gain a comprehensive level of understanding of age-dependent CPC ceRNA interactions, we investigated the differentially expressed miRNAs, lncRNAs, and mRNAs between neonate and child CPCs. We determined that neonate and child CPCs had the greatest differences in RNA content. Infant CPCs had an “intermediate” RNA profile and were not included in the pairwise ceRNA network analysis (Fig. 5a and Supplementary Fig. 1) Largely, child CPCs had higher expression of non-coding RNAs, compared to neonate CPCs (Fig. 5c). Using the miRcode and miRTarBase target prediction databases, we mapped a lncRNA-miRNA-mRNA network of differentially expressed RNAs between neonate and child CPCs (Fig. 5d, 5e, and 5f). The resulting network highlighted the importance of highly connected miR-218-5p, −181a-5p, −23a-3p, and −23b-3p (Table 3).
Our findings presented in this report are constrained by CPC in vitro culture conditions. Expansion of CPCs for the collection of EVs demands tens of millions of cells, requiring in vitro passaging of cells and potentially introducing transcriptome drift. To limit this issue, we kept cultures less than or equal to passage nine. The interpretation of these results is also limited due to the CPC two-dimensional culture experimental design, which is not ideal to recapitulate the in vivo cellular environment. Furthermore, CPC-EVs released from in vitro cell cultures may vary from CPC-EVs released from transplanted cells in in vivo and clinical models. EV cargo is highly heterogenous and affected by parent cell conditions and environment. Previous research in allogenic cardiac cell therapy has addressed this issue by collecting exosomes released from human CPCs in rat plasma, after cell transplantation, via major histocompatibility complex class I 5. Future efforts to identify CPC-EV markers for autologous and allogenic transplant models will assist our efforts to understand CPC-EV cardiac repair mechanisms of action.
In conclusion, we sequenced patient-derived CPCs and CPC-EVs and examined the RNA profiles with unsupervised learning methods to explore differences in coding and non-coding RNAs. We determined that CPC and CPC-EV RNA profiles differ and that there are age-dependent differences in non-coding RNAs of CPCs. More specifically, CPCs retain cell adhesion-related RNAs and export both generic EV transport-related RNAs and potential progenitor cell-specific and pro-reparative RNAs involved in cell proliferation and neovascularization. Further, child CPCs contain elevated levels of non-coding RNAs, compared to neonate CPCs. With this study, we hope to highlight the value of using unbiased methods as “precursors” to quickly hone in on potentially important CPC and CPC-EV RNAs so that more targeted experimental tests may be performed. Cell therapy for children is currently in early clinical trials, and data from our laboratory and others show that cells and EVs may independently, or in concert, repair the damaged myocardium 5,12. Using bulk sequencing to develop tools to computationally assess mechanisms and biomarkers in an unbiased manner could improve the outcomes of this promising approach. Our work provides further perspective for understanding the mechanism of action of CPCs, which is valuable for addressing clinical trial variability.
Materials and Methods
Isolation and Culture of c-kit+ Progenitor Cells (CPCs)
This study was approved by the Institutional Review Board at Children’s Healthcare of Atlanta and Emory University. CPCs were isolated by c-kit magnetic bead sorting from cardiac biopsies of congenital heart disease patients (Supplementary Table 1) as previously described4,59. CPCs were cultured using Hams F-12 medium with 10% fetal bovine serum, 1% Penicillin-Streptonmycin, 1% L-glutamine, and 0.04% fibroblast growth factor 2.
Extracellular Vesicle (EV) Collection and Characterization
EVs were successfully isolated from 27 of the 30 CPC populations. Briefly, CPCs were grown to 90% confluency, washed with PBS, and quiesced with serum free medium for 24 hours. Conditioned media was collected and subjected to sequential centrifugation: 3000 g for 10 min to remove cells, 28,000 g for 30 min to remove cell debris, and 118,000 g for 1 hr 54 min to pellet EVs (Optima XPN-100 ultracentrifuge; Beckman Coulter SW 41 Ti rotor). EV size and concentration was determined by NanoSight NS300. Samples were diluted 1:10 in PBS, and three, 60-second video images were captured per sample and analyzed by NanoSight NTA 3.4 software.
Next Generation Sequencing
RNA from CPCs and CPC-EVs were isolated with the miRNeasy Mini Kit (Qiagen), according to manufacturer’s instructions. Purified RNA was analyzed (2100 Bioanalyzer and TapeStation Controller, Agilent Genomics) for miRNA and RNA size, quality, and quantity. RNA library preparation and sequencing was conducted by Novogene Co., Ltd (Illumina NovaSeq 6000 with PE150 platform) or the Emory Yerkes Nonhuman Primate Genomics core (Illumina HiSeq 3000). Sequencing source information for each CPC patient is displayed in Supplementary Table 1.
Small RNA sequencing performed at Novogene were trimmed and filtered in the FASTQ Toolkit Illumina Basespace app. Truseq adapters (AGATCGGAAGAGC) were trimmed with an adapter trim stringency set to 0.90. Reads were filtered to 18–51 length with reads passing the FASTQ Toolkit filter. Then, reads were mapped with Bowtie aligner and hg19 and miRBase v21 references, and mature miRNA hits were determined using the small RNA Illumina Basespace app. Small RNA sequencing performed at Emory Yerkes Nonhuman Primate Genomics core were aligned and hits were determined using the Qiagen GeneGlobe console with QIAseq miRNA Quantification tool. Default parameters were used: 3’ adapters were trimmed using cutadapt, reads with less than 16 base pair insert sequences or less than 10 base pair unique molecular index sequences were removed, reads were aligned with Bowtie aligner, GRCh38 and miRBase v21 references.
Total RNAseq files from Novogene and Emory Yerkes Nonhuman Primate Genomics core were aligned, and gene counts were determined with the STAR aligner in the Illumina BaseSpace app, RNA-Seq Alignment. Reads were aligned with hg19 reference genome. Biotypes were matched to alignment results using the Ensembl based annotation package (EnsDb.Hsapiens.v79). miRNAs were considered only from small RNA sequencing and were thus removed from the total RNA sequencing set. All of CPC lncRNAseq was performed with Novogene. lncRNAs were aligned using STAR with the quantMode GeneCounts option and GRCh38 reference 60. Total RNA sequencing counts after filtering are presented in Supplementary Tables 11 and 12; miRNA sequencing counts after filtering are presented in Supplementary Tables 13 and 14. RNA sequencing and alignment metrics are presented in Supplementary Tables 15 and 16.
RNA Sequencing Data Analysis
Data analysis was completed in R. First, raw aligned RNA counts for CPC and CPC-EVs were filtered: we removed RNA with zero count entries in ten or more samples and used edgeR package’s ‘filterByExpr’ function using the default parameter settings. RNA counts were normalized using the edgeR weighted trimmed mean of M-values method (default parameters), and transformed into log2 counts per million (logCPM). Principal component analyses (PCA) were performed using the ‘prcomp’ built in function. Heatmaps were generated using the pheatmap package with Manhattan distance calculations and ward.D2 clustering method.
Batch correction of sequencing data (in logCPM) was implemented using the sva package’s ‘ComBat’ function to correct for sequencing performed at separate sites: Novogene and Emory Yerkes Nonhuman Primate Genomics Core (Supplementary Table 1)61. PCA plots before and after batch correction are displayed in Supplementary Figs. 3a and 3b. Variance explained by each covariate (patient, sex, age (in log10months), type (cell or EV), and sequencing site) was assessed before and after batch correction with variancePartition package (Supplementary Figs. 3c and 3d) 62. Age was treated as a fixed effect and all other categorical covariates were treated as random effects.
For differential expression analysis of CPC and CPC-EV RNA content, we used the dream (differential expression for repeated measures) linear mixed modeling approach from the variancePartition package to account for patient matched cell and EV data 15. Patient was treated as a random effect and the following co-variates were considered: patient, sex, age (in log10months), type (cell or EV), batch (sequencing site). Weights were estimated with the voomWithDreamWeights function and the cell vs. EV hypothesis test was conducted with the dream function (Satterthwaite approximation method). Cell vs. EV differential expression analyses were conducted for n=26 patients: Patient 894 was removed because sex was not known, and cell sequencing from patients 938, 1048, and 1092, were not included as EVs were not sequenced from these patients.
Volcano plots were constructed from linear mixed model results using the EnhancedVolcano package. Correspondence between PCA variable loadings and differential expression analysis results, presented in volcano plots, is displayed in Supplementary Fig. 4. miRTarBase was used to identify miRNAs with known targets (validated by at least three assays) 39. Biological pathway enrichment analyses were performed using Metascape 16.
Data Mining
We probed previously published datasets containing miRNA data from parent cells and their EVs 21–28. GEO2R was used to determine the differential expression of EV vs. cell miRNAs (Benjamini Hochberg p-value adjustment)63. miRNAs in each dataset were ranked from largest to smallest log2fold-change (EV/Cell). To account for different sized datasets, ranks were scaled within their respective study using the ‘smoothPalette’ function in the tagcloud package, before plotting with the scales package in R. Data mining results are presented in Supplementary Table 17.
ceRNA Network Construction
Differentially expressed RNAs (neonate vs. child CPCs) were determined using edgeR and limma/voom method. Reads from total RNA, lncRNA, and miRNA CPC sequencing were filtered: we removed RNA with zero count entries in ten or more samples and used edgeR package’s ‘filterByExpr’ function with the default parameter settings. Total RNA, miRNA, and lncRNA models were built using all CPC data (neonate, infant, and child) with the following covariates: age group, sex, and batch (sequencing site). Counts were transformed to logCPM values and weights for linear modeling were computed using the limma voom function. Linear models were fit. Then, contrast matrices for age groups were created (neonate – child, neonate – infant, infant – child), coefficients were estimated, and moderated statistics were computed with empirical Bayes moderation. Differentially expressed RNAs between neonate and child groups were identified using the topTable function. Differential expression results for lncRNA sequencing are presented in Supplementary Table 18. The differentially expressed miRNAs (18) were matched to differentially expressed lncRNAs (134) by miRcode and differentially expressed mRNAs (505) by miRTarBase (|fold-change|>2 and p<0.05). The resulting RNAs that were matched within the set were displayed in a network with Cytoscape 64. The full network (107 nodes, 144 edges) was reduced to the most highly connected hubs (36 nodes, 75 edges) using the MCODE plug in.
Furthermore, we investigated the age-related differences in CPC RNAs in a quantitative approach by transforming patient age to log10months. We constructed linear models with age (continuous), sex, and batch (if applicable) covariates, as before. We compared the differentially expressed RNAs (p<0.05) determined by this quantitative method with the differentially expressed RNAs determined by the categorical method above (age as neonate, infant, or child). The results are displayed in Supplementary Fig. 5.
Supplementary Material
Highlights.
C-kit+ progenitor cells retain and release specific RNAs to EVs
CPC-EVs contain miRNA related to cell proliferation, not found in other cell type EVs
CPCs derived from older patients are enriched in non-coding RNA
Acknowledgements
We thank Dr. Rabindra Tirouvanziam and Brian Dobosh for assistance with NanoSight nanoparticle tracking analysis. We also gratefully acknowledge Dr. Kathryn Pellegrini and the Emory Yerkes Nonhuman Primate Genomics Core for their assistance with sequencing, as well as Dr. Ricardo Guerrero-Ferreira and the Emory Integrated Electron Microscopy Core for CPC-EV imaging.
Funding
National Institutes of Health grant R01HL145644 (MED)
National Institutes of Health grant F31HL154725 (JRH)
National Institutes of Health grant T32GM008602 (JRH)
Footnotes
Declaration of interests
Authors declare that they have no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Data Availability
Relevant data are available in the main text or the supplementary materials.
Sequencing data have been deposited:
GSE196556 (CPC lncRNAseq)
GSE (CPC and CPC-EV miRNAseq) – submitted to GEO, awaiting accession number
GSE (CPC and CPC-EV total RNAseq) – submitted to GEO, awaiting accession number
References
- 1.Kochanek KD, Xu J & Arias E Mortality in the United States, 2019. NCHS Data Brief, no 395. Hyattsville, MD. National Center for Health Statistics; (2020). [PubMed] [Google Scholar]
- 2.Marban E A mechanistic roadmap for the clinical application of cardiac cell therapies. Nat Biomed Eng 2, 353–361, doi: 10.1038/s41551-018-0216-z (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bittle GJ et al. Stem Cell Therapy for Hypoplastic Left Heart Syndrome: Mechanism, Clinical Application, and Future Directions. Circ Res 123, 288–300, doi: 10.1161/CIRCRESAHA.117.311206 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal U et al. Age-Dependent Effect of Pediatric Cardiac Progenitor Cells After Juvenile Heart Failure. Stem Cells Transl Med 5, 883–892, doi: 10.5966/sctm.2015-0241 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saha P et al. Circulating exosomes derived from transplanted progenitor cells aid the functional recovery of ischemic myocardium. Sci Transl Med 11, doi: 10.1126/scitranslmed.aau1168 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Trac D, Maxwell JT, Brown ME, Xu C & Davis ME Aggregation of Child Cardiac Progenitor Cells Into Spheres Activates Notch Signaling and Improves Treatment of Right Ventricular Heart Failure. Circ Res 124, 526–538, doi: 10.1161/CIRCRESAHA.118.313845 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen PK, Neofytou E, Rhee JW & Wu JC Potential Strategies to Address the Major Clinical Barriers Facing Stem Cell Regenerative Therapy for Cardiovascular Disease: A Review. JAMA Cardiol 1, 953–962, doi: 10.1001/jamacardio.2016.2750 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray WD et al. Identification of therapeutic covariant microRNA clusters in hypoxiatreated cardiac progenitor cell exosomes using systems biology. Circ Res 116, 255–263, doi: 10.1161/CIRCRESAHA.116.304360 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Agarwal U et al. Experimental, Systems, and Computational Approaches to Understanding the MicroRNA-Mediated Reparative Potential of Cardiac Progenitor Cell-Derived Exosomes From Pediatric Patients. Circ Res 120, 701–712, doi: 10.1161/CIRCRESAHA.116.309935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shoja-Taheri F et al. Using Statistical Modeling to Understand and Predict Pediatric Stem Cell Function. Circ Genom Precis Med 12, e002403, doi: 10.1161/CIRCGEN.118.002403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bheri S, Hoffman JR, Park HJ & Davis ME Biomimetic nanovesicle design for cardiac tissue repair. Nanomedicine (Lond) 15, 1873–1896, doi: 10.2217/nnm-2020-0097 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ibrahim AG, Cheng K & Marban E Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Reports 2, 606–619, doi: 10.1016/j.stemcr.2014.04.006 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Willms E et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep 6, 22519, doi: 10.1038/srep22519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson MD, McCarthy DJ & Smyth GK edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140, doi: 10.1093/bioinformatics/btp616 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman GE & Roussos P Dream: powerful differential expression analysis for repeated measures designs. Bioinformatics 37, 192–201, doi: 10.1093/bioinformatics/btaa687 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou Y et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 10, 1523, doi: 10.1038/s41467-019-09234-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He L et al. A microRNA polycistron as a potential human oncogene. Nature 435, 828–833, doi: 10.1038/nature03552 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lu Y, Thomson JM, Wong HY, Hammond SM & Hogan BL Transgenic overexpression of the microRNA miR-17–92 cluster promotes proliferation and inhibits differentiation of lung epithelial progenitor cells. Dev Biol 310, 442–453, doi: 10.1016/j.ydbio.2007.08.007 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen J et al. mir-17–92 cluster is required for and sufficient to induce cardiomyocyte proliferation in postnatal and adult hearts. Circ Res 112, 1557–1566, doi: 10.1161/CIRCRESAHA.112.300658 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emmrich S et al. miR-99a/100∼125b tricistrons regulate hematopoietic stem and progenitor cell homeostasis by shifting the balance between TGFbeta and Wnt signaling. Genes Dev 28, 858–874, doi: 10.1101/gad.233791.113 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanlikilicer P et al. Ubiquitous Release of Exosomal Tumor Suppressor miR-6126 from Ovarian Cancer Cells. Cancer Res 76, 7194–7207, doi: 10.1158/0008-5472.CAN-16-0714 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Melo SA et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell 26, 707–721, doi: 10.1016/j.ccell.2014.09.005 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mittelbrunn M et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun 2, 282, doi: 10.1038/ncomms1285 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ono M et al. Exosomes from bone marrow mesenchymal stem cells contain a microRNA that promotes dormancy in metastatic breast cancer cells. Sci Signal 7, ra63, doi: 10.1126/scisignal.2005231 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Tadokoro H, Umezu T, Ohyashiki K, Hirano T & Ohyashiki JH Exosomes derived from hypoxic leukemia cells enhance tube formation in endothelial cells. J Biol Chem 288, 34343–34351, doi: 10.1074/jbc.M113.480822 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Umezu T et al. Exosomal miR-135b shed from hypoxic multiple myeloma cells enhances angiogenesis by targeting factor-inhibiting HIF-1. Blood 124, 3748–3757, doi: 10.1182/blood-2014-05-576116 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vignard V et al. MicroRNAs in Tumor Exosomes Drive Immune Escape in Melanoma. Cancer Immunol Res 8, 255–267, doi: 10.1158/2326-6066.CIR-19-0522 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Yokoi A et al. Integrated extracellular microRNA profiling for ovarian cancer screening. Nat Commun 9, 4319, doi: 10.1038/s41467-018-06434-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Temoche-Diaz MM et al. Distinct mechanisms of microRNA sorting into cancer cell-derived extracellular vesicle subtypes. Elife 8, doi: 10.7554/eLife.47544 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mogilyansky E & Rigoutsos I The miR-17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ 20, 1603–1614, doi: 10.1038/cdd.2013.125 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T, Ding ZL, Zheng YL & Wang W MiR-484 promotes non-small-cell lung cancer (NSCLC) progression through inhibiting Apaf-1 associated with the suppression of apoptosis. Biomed Pharmacother 96, 153–164, doi: 10.1016/j.biopha.2017.09.102 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Li S et al. miR-130b-3p Modulates Epithelial-Mesenchymal Crosstalk in Lung Fibrosis by Targeting IGF-1. PLoS One 11, e0150418, doi: 10.1371/journal.pone.0150418 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gan L et al. Small Extracellular Microvesicles Mediated Pathological Communications Between Dysfunctional Adipocytes and Cardiomyocytes as a Novel Mechanism Exacerbating Ischemia/Reperfusion Injury in Diabetic Mice. Circulation 141, 968–983, doi: 10.1161/CIRCULATIONAHA.119.042640 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma Z et al. A lncRNA-miRNA-mRNA network for human primed, naive and extended pluripotent stem cells. PLoS One 15, e0234628, doi: 10.1371/journal.pone.0234628 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qi X et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet 52, 710–718, doi: 10.1136/jmedgenet-2015-103334 (2015). [DOI] [PubMed] [Google Scholar]
- 36.Salmena L, Poliseno L, Tay Y, Kats L & Pandolfi PP A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell 146, 353–358, doi: 10.1016/j.cell.2011.07.014 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S et al. A Deep Proteome Analysis Identifies the Complete Secretome as the Functional Unit of Human Cardiac Progenitor Cells. Circ Res 120, 816–834, doi: 10.1161/CIRCRESAHA.116.309782 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeggari A, Marks DS & Larsson E miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 28, 2062–2063, doi: 10.1093/bioinformatics/bts344 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu SD et al. miRTarBase: a database curates experimentally validated microRNAtarget interactions. Nucleic Acids Res 39, D163–169, doi: 10.1093/nar/gkq1107 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosen MR, Myerburg RJ, Francis DP, Cole GD & Marban E Translating stem cell research to cardiac disease therapies: pitfalls and prospects for improvement. J Am Coll Cardiol 64, 922–937, doi: 10.1016/j.jacc.2014.06.1175 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mishra R et al. Characterization and functionality of cardiac progenitor cells in congenital heart patients. Circulation 123, 364–373, doi: 10.1161/CIRCULATIONAHA.110.971622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yan F et al. Hypoxic preconditioning improves survival of cardiac progenitor cells: role of stromal cell derived factor-1alpha-CXCR4 axis. PLoS One 7, e37948, doi: 10.1371/journal.pone.0037948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adamiak M et al. Induced Pluripotent Stem Cell (iPSC)-Derived Extracellular Vesicles Are Safer and More Effective for Cardiac Repair Than iPSCs. Circ Res 122, 296–309, doi: 10.1161/CIRCRESAHA.117.311769 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xin L et al. A scaffold laden with mesenchymal stem cell-derived exosomes for promoting endometrium regeneration and fertility restoration through macrophage immunomodulation. Acta Biomater 113, 252–266, doi: 10.1016/j.actbio.2020.06.029 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Trac D et al. Predicting Functional Responses of Progenitor Cell Exosome Potential with Computational Modeling. Stem Cells Transl Med 8, 1212–1221, doi: 10.1002/sctm.19-0059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sahoo S & Losordo DW Exosomes and cardiac repair after myocardial infarction. Circ Res 114, 333–344, doi: 10.1161/CIRCRESAHA.114.300639 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Barile L, Moccetti T, Marban E & Vassalli G Roles of exosomes in cardioprotection. Eur Heart J 38, 1372–1379, doi: 10.1093/eurheartj/ehw304 (2017). [DOI] [PubMed] [Google Scholar]
- 48.Emanueli C, Shearn AI, Angelini GD & Sahoo S Exosomes and exosomal miRNAs in cardiovascular protection and repair. Vascul Pharmacol 71, 24–30, doi: 10.1016/j.vph.2015.02.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamada NO, Heishima K, Akao Y & Senda T Extracellular Vesicles Containing MicroRNA-92a-3p Facilitate Partial Endothelial-Mesenchymal Transition and Angiogenesis in Endothelial Cells. Int J Mol Sci 20, doi: 10.3390/ijms20184406 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Umezu T, Ohyashiki K, Kuroda M & Ohyashiki JH Leukemia cell to endothelial cell communication via exosomal miRNAs. Oncogene 32, 2747–2755, doi: 10.1038/onc.2012.295 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Liu Y et al. Atherosclerotic Conditions Promote the Packaging of Functional MicroRNA-92a-3p Into Endothelial Microvesicles. Circ Res 124, 575–587, doi: 10.1161/CIRCRESAHA.118.314010 (2019). [DOI] [PubMed] [Google Scholar]
- 52.Kesidou D et al. Extracellular Vesicle miRNAs in the Promotion of Cardiac Neovascularisation. Front Physiol 11, 579892, doi: 10.3389/fphys.2020.579892 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J et al. The Poly-cistronic miR-23–27-24 Complexes Target Endothelial Cell Junctions: Differential Functional and Molecular Effects of miR-23a and miR-23b. Mol Ther Nucleic Acids 5, e354, doi: 10.1038/mtna.2016.62 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jackson CE, Scruggs BS, Schaffer JE & Hanson PI Effects of Inhibiting VPS4 Support a General Role for ESCRTs in Extracellular Vesicle Biogenesis. Biophys J 113, 1342–1352, doi: 10.1016/j.bpj.2017.05.032 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McKenzie AJ et al. KRAS-MEK Signaling Controls Ago2 Sorting into Exosomes. Cell Rep 15, 978–987, doi: 10.1016/j.celrep.2016.03.085 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Teng Y et al. MVP-mediated exosomal sorting of miR-193a promotes colon cancer progression. Nat Commun 8, 14448, doi: 10.1038/ncomms14448 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yin Z et al. Macrophage-derived exosomal microRNA-501-3p promotes progression of pancreatic ductal adenocarcinoma through the TGFBR3-mediated TGF-beta signaling pathway. J Exp Clin Cancer Res 38, 310, doi: 10.1186/s13046-019-1313-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Frank S, Aguirre A, Hescheler J & Kurian L A lncRNA Perspective into (Re)Building the Heart. Front Cell Dev Biol 4, 128, doi: 10.3389/fcell.2016.00128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.French KM & Davis ME Isolation and expansion of c-kit-positive cardiac progenitor cells by magnetic cell sorting. Methods Mol Biol 1181, 39–50, doi: 10.1007/978-1-49391047-2_4 (2014). [DOI] [PubMed] [Google Scholar]
- 60.Dobin A et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leek JT, Johnson WE, Parker HS, Jaffe AE & Storey JD The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics 28, 882–883, doi: 10.1093/bioinformatics/bts034 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hoffman GE & Schadt EE variancePartition: interpreting drivers of variation in complex gene expression studies. BMC Bioinformatics 17, 483, doi: 10.1186/s12859-0161323-z (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barrett T et al. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res 41, D991–995, doi: 10.1093/nar/gks1193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shannon P et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13, 2498–2504, doi: 10.1101/gr.1239303 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Relevant data are available in the main text or the supplementary materials.
Sequencing data have been deposited:
GSE196556 (CPC lncRNAseq)
GSE (CPC and CPC-EV miRNAseq) – submitted to GEO, awaiting accession number
GSE (CPC and CPC-EV total RNAseq) – submitted to GEO, awaiting accession number